Transcriptome Analysis of Amyloodinium ocellatum Tomonts Revealed Basic Information on the Major Potential Virulence Factors
Abstract
:1. Introduction
2. Materials and Methods
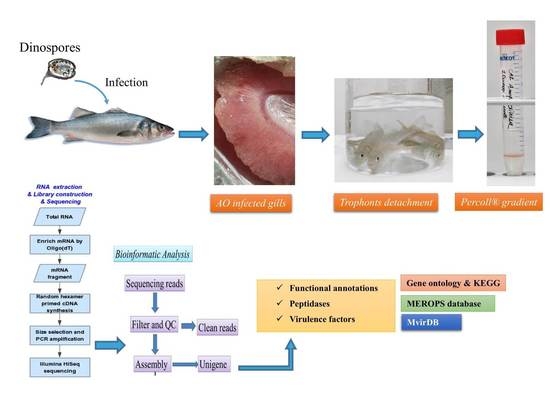
2.1. Parasite and Total RNA Preparation for Sequencing
2.2. Sequence Assembly
2.3. Assigning Functional Annotations to Transcripts Belonging to Alveolata
2.4. Assigning Taxonomy to Transcripts
2.5. Identification of Peptidases and Virulence Factors
2.6. In Silico Analysis of AO Heat Shock Protein 70 and Casein Kinase II Alpha
3. Results
3.1. De Novo Sequence Assembly of AO Transcriptome
3.2. Functional Annotation of AO Contigs
3.3. Peptidases Identified from AO Transcriptome Data
3.4. Potential Virulent Factors from AO Transcriptome
3.5. Sequence Analysis of AO Heat Shock Protein 70 and Casein Kinase II Alpha
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lom, J.; Lawler, A.R. An ultrastructural study on the mode of attachment in dinoflagellates invading gills of Cyprinodontidae. Protistologica 1973, 2, 293–309. [Google Scholar]
- Noga, E.J.; Levy, M.G. Phylum Dinoflagellata. In Fish Diseases and Disorders, Volume 1: Protozoan and Metazoan Infections, 2nd ed.; Woo, P.T.K., Ed.; CABI International: King’s Lynn, UK, 2006; p. 791. [Google Scholar]
- Paperna, I. Reproduction Cycle and Tolerance to Temperature and Salinity of Amyloodinium ocellatum (Brown, 1931) (Dinoflagellida). Ann. Parasitol. Hum. Comparée 1984, 59, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noga, E.J. Propagation in Cell Culture of the Dinoflagellate Amyloodinium, an Ectoparasite of Marine Fishes. Science 1987, 236, 1302–1304. [Google Scholar] [CrossRef] [PubMed]
- Byadgi, O.; Beraldo, P.; Volpatti, D.; Massimo, M.; Bulfon, C.; Galeotti, M. Expression of infection-related immune response in European sea bass (Dicentrarchus labrax) during a natural outbreak from a unique dinoflagellate Amyloodinium ocellatum. Fish Shellfish Immunol. 2019, 84, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Mo, Z.Q.; Li, Y.W.; Wang, H.Q.; Wang, J.L.; Ni, L.Y.; Yang, M.; Lao, G.F.; Luo, X.C.; Li, A.X.; Dan, X.M. Comparative transcriptional profile of the fish parasite Cryptocaryon irritans. Parasites Vectors 2016, 9, 630. [Google Scholar] [CrossRef] [Green Version]
- Cassidy-Hanley, D.M.; Cordonnier-Pratt, M.M.; Pratt, L.H.; Devine, C.; Mozammal, M.; Dickerson, H.W.; Clark, T.G. Transcriptional profiling of stage specific gene expression in the parasitic ciliate Ichthyophthirius multifiliis. Mol. Biochem. Parasitol. 2011, 178, 29–39. [Google Scholar] [CrossRef]
- Yin, F.; Sun, P.; Wang, J.-T.; Gao, Q. Transcriptome analysis of dormant tomonts of the marine fish ectoparasitic ciliate Cryptocaryon irritans under low temperature. Parasites Vectors 2016, 9, 280. [Google Scholar] [CrossRef] [Green Version]
- Beraldo, P.; Massimo, M.; Galeotti, M. SOP for Amyloodinium ocellatum. In Fish Parasites: A Handbook of Protocols for Their Isolation, Culture and Transmission; Sitjà-Bobadilla, A., Bron, J.E., Wiegertjes, G.F., Piazzon, M.C., Eds.; 5M Publishing: Sheffield, UK, 2020; in press. [Google Scholar]
- The UniProt Consortium. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tenenbaum, D. KEGGREST: Client-Side REST Access to KEGG 2020, R package version 1.29.0.; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2015, 44, D286–D293. [Google Scholar] [CrossRef] [Green Version]
- Rawlings, N.D.; Waller, M.; Barrett, A.J.; Bateman, A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014, 42, D503–D509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, C.; Smith, J.; Lam, M.; Zemla, A.; Dyer, M.D.; Slezak, T. MvirDB—A microbial database of protein toxins, virulence factors and antibiotic resistance genes for bio-defence applications. Nucleic Acids Res. 2006, 35, D391–D394. [Google Scholar] [CrossRef] [PubMed]
- Ramana, J.; Gupta, D. ProtVirDB: A database of protozoan virulent proteins. Bioinformatics 2009, 25, 1568–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.A.; Sternberg, M.J.E. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009, 4, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Nozzi, V.; Strofaldi, S.; Forner Piquer, I.; Di Crescenzo, D.; Olivotto, I.; Carnevali, O. Amyloodinum ocellatum in Dicentrarchus labrax: Study of infection in salt water and freshwater aquaponics. Fish Shellfish Immunol. 2016, 57, 179–185. [Google Scholar] [CrossRef]
- Aranda, M.; Li, Y.; Liew, Y.J.; Baumgarten, S.; Simakov, O.; Wilson, M.C.; Piel, J.; Ashoor, H.; Bougouffa, S.; Bajic, V.B.; et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016, 6, 39734. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Jagus, R.; Morse, D. Translation and Translational Control in Dinoflagellates. Microorganisms 2018, 6, 30. [Google Scholar] [CrossRef] [Green Version]
- Rhee, S.Y.; Wood, V.; Dolinski, K.; Draghici, S. Use and misuse of the gene ontology annotations. Nat. Rev. Genet. 2008, 9, 509–515. [Google Scholar] [CrossRef]
- Lokanathan, Y.; Mohd-Adnan, A.; Wan, K.-L.; Nathan, S. Transcriptome analysis of the Cryptocaryon irritans tomont stage identifies potential genes for the detection and control of cryptocaryonosis. BMC Genom. 2010, 11, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Tan, Y.; Cui, G.; Feng, Y.; Cui, Q.; Song, X. Transcriptome and gene expression analysis of DHA producer Aurantiochytrium under low temperature conditions. Sci. Rep. 2015, 5, 14446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, J.S.; Jeon, E.J.; Jung, S.H.; Park, M.A.; Kim, J.W.; Kim, K.H.; Woo, S.H.; Lee, E.H. Molecular cloning and expression analysis of peptidase genes in the fish-pathogenic scuticociliate Miamiensis avidus. BMC Vet. Res. 2013, 9, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado-Aguayo, W.; Gallardo-Escárate, C. Increasing transcriptome response of serpins during the ontogenetic stages in the salmon louse Caligus rogercresseyi (Copepoda: Caligidae). Mar. Genom. 2014, 15, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Hirt, R.; de Miguel, N.; Nakjang, S.; Dessi, D.; Liu, Y.C.; Diaz, N.; Rappelli, P.; Acosta-Serano, A.; Fiori, P.L.; Mottram, J.C. Trichomonas vaginalis pathobiology: New insights from the genome sequence. Adv. Parasitol. 2011, 77, 87–140. [Google Scholar]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Hartl, F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef]
- Davis-Hayman, S.R.; Shah, P.H.; Finley, R.W.; Lushbaugh, W.B.; Meade, J.C. Trichomonas vaginalis: Analysis of a heat-inducible member of the cytosolic heat-shock protein 70 multigene family. Parasitol. Res. 2000, 86, 608–612. [Google Scholar] [CrossRef]
- Gould, S.B.; Woehle, C.; Kusdian, G.; Landan, G.; TachezyJ, Z.V.; Martin, W.F. Deep sequencing of Trichomonas vaginalis during the early infection of vaginal epithelial cells and amoeboid transition. Int. J. Parasitol. 2013, 43, 707–719. [Google Scholar] [CrossRef]
- Josepriya, T.A.; Chien, K.H.; Lin, H.Y.; Huang, H.N.; Wu, C.J.; Song, Y.L. Immobilization antigen vaccine adjuvanted by parasitic heat shock protein 70C confers high protection in fish against cryptocaryonosis. Fish Shellfish Immunol. 2015, 45, 517–527. [Google Scholar] [CrossRef]



| Nber | N50 (bp) | Max Length (kb) | Mean Length (bp) | Total Assembly Length (Mb) | GC% | |
|---|---|---|---|---|---|---|
| Contigs | 175,030 | 624 | 5.435 | 517.77 | 90.6 | 56 |
| Contigs mapping against nr | 48,188 | NA | NA | NA | NA | NA |
| Contigs assigned to Alveolata | 29,691 | 1191 | 5.435 | 875.9 | 26.6 | 62 |
| Assigned | Total No of Contigs | Similarity (%) | |
|---|---|---|---|
| Superkingdom | Eukaryota | 45,008 | 93.4 |
| Class | Dinophycaae | 25,258 | 56.12 |
| Species | Symbiodinium microadriaticum | 23,897 | 94.61 |
| AO Contig Number | Major Peptidases Family | Content of Family | Functions |
|---|---|---|---|
| Contig_10007 | MER1360781-family A31 non-peptidase homologues | Endopeptidases | Cleavage of active enzymes |
| Contig_22247 | MER1200850-family M8 unassigned peptidases | Zinc metallopeptidase | Role in cell migration and invasion |
| Contig_114405 | MER1171318 - family C12 unassigned peptidases | Ubiquitinyl hydrolases | Removal of ubiquitin (Ub) from protein substrates, involvement in numerous biological processes |
| Contig_32531 | MER0253524 - family S82 unassigned homologues | Autocrine proliferation repressor protein A | Regulation of cell proliferation/number |
| Virulence Group | AO Potential Virulence Factors | Contig Number |
|---|---|---|
| Adhesin | AP120 | Contig_9557 |
| Invasion | Gp63 (surface metalloprotease) | Contig_142760 |
| P0 (Ribosomal phosphoprotein) | Contig_3026 | |
| ROM1 (Rhomboid-like protease) | Contig_40819 | |
| RabA | Contig_83170 | |
| Heat shock protein | Hsp70 | Contig_50520 |
| Establishment | Casein kinase II alpha | Contig_10331 |
| Rab7A | Contig_103654 | |
| Vps35 | Contig_1094 | |
| Rab11B | Contig_112882 | |
| Vps29 | Contig_116994 | |
| Rab5 | Contig_129899 | |
| CPSII | Contig_60947 | |
| Proteases | Brucipain | Contig_105984 |
| Plasmepsin II | Contig_10808 | |
| Plasmepsin I | Contig_12020 | |
| Plasmepsin IV | Contig_19630 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Byadgi, O.; Marroni, F.; Dirks, R.; Massimo, M.; Volpatti, D.; Galeotti, M.; Beraldo, P. Transcriptome Analysis of Amyloodinium ocellatum Tomonts Revealed Basic Information on the Major Potential Virulence Factors. Genes 2020, 11, 1252. https://doi.org/10.3390/genes11111252
Byadgi O, Marroni F, Dirks R, Massimo M, Volpatti D, Galeotti M, Beraldo P. Transcriptome Analysis of Amyloodinium ocellatum Tomonts Revealed Basic Information on the Major Potential Virulence Factors. Genes. 2020; 11(11):1252. https://doi.org/10.3390/genes11111252
Chicago/Turabian StyleByadgi, Omkar, Fabio Marroni, Ron Dirks, Michela Massimo, Donatella Volpatti, Marco Galeotti, and Paola Beraldo. 2020. "Transcriptome Analysis of Amyloodinium ocellatum Tomonts Revealed Basic Information on the Major Potential Virulence Factors" Genes 11, no. 11: 1252. https://doi.org/10.3390/genes11111252
APA StyleByadgi, O., Marroni, F., Dirks, R., Massimo, M., Volpatti, D., Galeotti, M., & Beraldo, P. (2020). Transcriptome Analysis of Amyloodinium ocellatum Tomonts Revealed Basic Information on the Major Potential Virulence Factors. Genes, 11(11), 1252. https://doi.org/10.3390/genes11111252