Features of DNA Repair in the Early Stages of Mammalian Embryonic Development
Abstract
1. Introduction
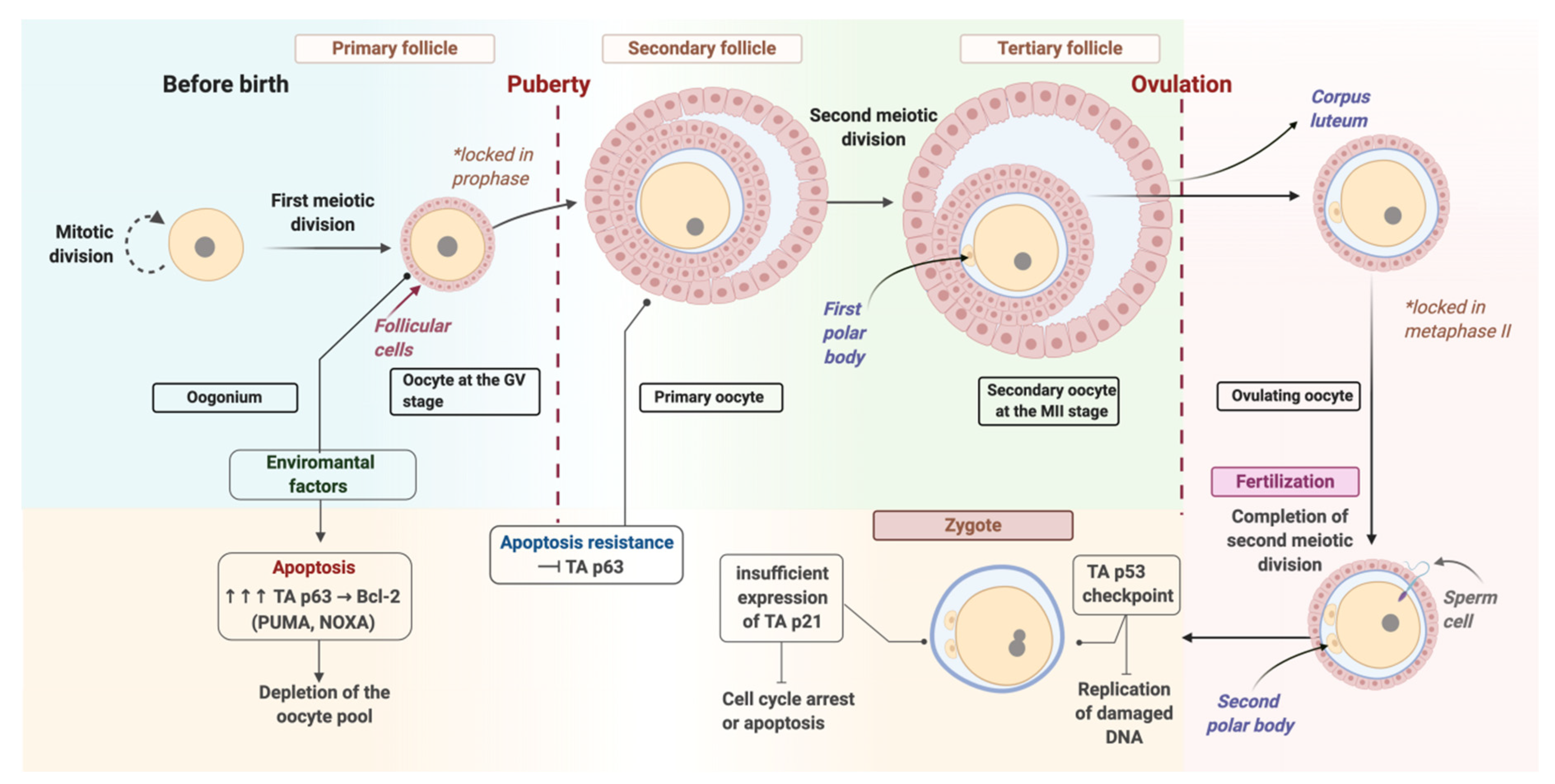
2. Oocyte Repair
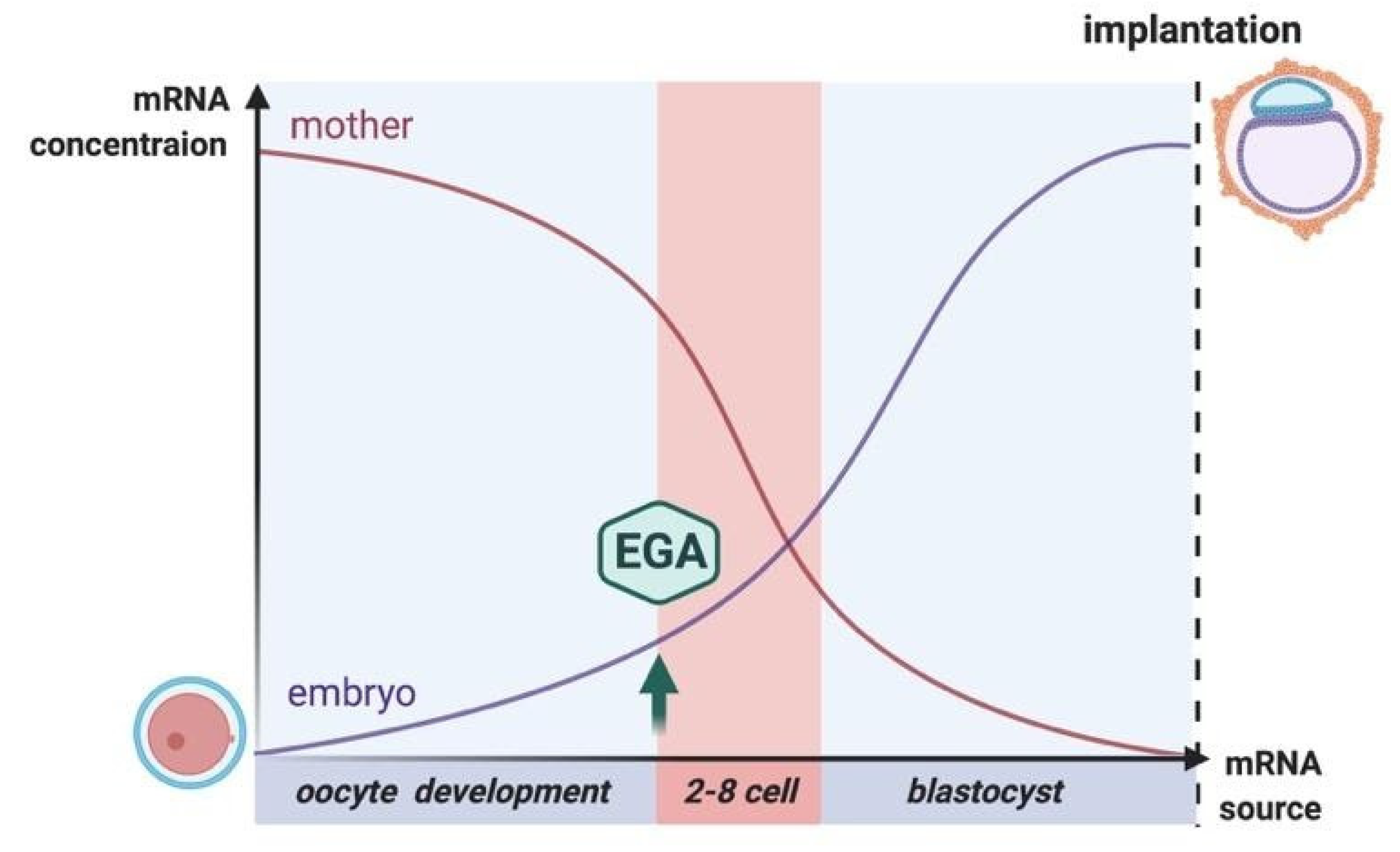
3. Repair at the Zygote Stage
4. Repair at the Cleavage and Blastocyst Stages
5. Embryonic Stem Cells
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Juan, H.-C.; Lin, Y.; Chen, H.-R.; Fann, M.-J. Cdk12 is essential for embryonic development and the maintenance of genomic stability. Cell Death Differ. 2016, 23, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Hamdoun, A.; Epel, D. Embryo stability and vulnerability in an always changing world. Proc. Natl. Acad. Sci. USA 2007, 104, 1745–1750. [Google Scholar] [CrossRef]
- Smith, M.L.; Fornace, A.J. Mammalian DNA damage-inducible genes associated with growth arrest and apoptosis. Mutat. Res. Rev. Genet. Toxicol. 1996, 340, 109–124. [Google Scholar] [CrossRef]
- Harrison, R.H.; Kuo, H.-C.; Scriven, P.N.; Handyside, A.H.; Ogilvie, C.M. Lack of cell cycle checkpoints in human cleavage stage embryos revealed by a clonal pattern of chromosomal mosaicism analysed by sequential multicolour FISH. Zygote 2000, 8, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Rimon-Dahari, N.; Yerushalmi-Heinemann, L.; Alyagor, L.; Dekel, N. Ovarian Folliculogenesis: Molecular Mechanisms of Cell Differentiation in Gonad Development. In Results and Problems in Cell Differentiation; Piprek, R.P., Ed.; Springer International Publishing: Cham, Switzerland, 2016; Volume 58, pp. 167–190. ISBN 978-3-319-31971-1. [Google Scholar]
- Martin, J.H.; Bromfield, E.G.; Aitken, R.J.; Nixon, B. Biochemical alterations in the oocyte in support of early embryonic development. Cell. Mol. Life Sci. 2017, 74, 469–485. [Google Scholar] [CrossRef]
- Kerr, J.B.; Brogan, L.; Myers, M.; Hutt, K.J.; Mladenovska, T.; Ricardo, S.; Hamza, K.; Scott, C.L.; Strasser, A.; Findlay, J.K. The primordial follicle reserve is not renewed after chemical or γ-irradiation mediated depletion. Reproduction 2012, 143, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Kujjo, L.L.; Laine, T.; Pereira, R.J.G.; Kagawa, W.; Kurumizaka, H.; Yokoyama, S.; Perez, G.I. Enhancing Survival of Mouse Oocytes Following Chemotherapy or Aging by Targeting Bax and Rad51. PLoS ONE 2010, 5, e9204. [Google Scholar] [CrossRef]
- Hunt, P.A.; Lawson, C.; Gieske, M.; Murdoch, B.; Smith, H.; Marre, A.; Hassold, T.; VandeVoort, C.A. Bisphenol A alters early oogenesis and follicle formation in the fetal ovary of the rhesus monkey. Proc. Natl. Acad. Sci. USA 2012, 109, 17525–17530. [Google Scholar] [CrossRef]
- Winship, A.L.; Stringer, J.M.; Liew, S.H.; Hutt, K.J. The importance of DNA repair for maintaining oocyte quality in response to anti-cancer treatments, environmental toxins and maternal ageing. Hum. Reprod. Update 2018, 24, 119–134. [Google Scholar] [CrossRef]
- Stringer, J.M.; Winship, A.; Liew, S.H.; Hutt, K. The capacity of oocytes for DNA repair. Cell. Mol. Life Sci. 2018, 75, 2777–2792. [Google Scholar] [CrossRef]
- Hu, W. The Role of p53 Gene Family in Reproduction. Cold Spring Harb. Perspect. Biol. 2009, 1, a001073. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.B.; Hutt, K.J.; Michalak, E.M.; Cook, M.; Vandenberg, C.J.; Liew, S.H.; Bouillet, P.; Mills, A.; Scott, C.L.; Findlay, J.K.; et al. DNA Damage-Induced Primordial Follicle Oocyte Apoptosis and Loss of Fertility Require TAp63-Mediated Induction of Puma and Noxa. Mol. Cell 2012, 48, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Suh, E.-K.; Yang, A.; Kettenbach, A.; Bamberger, C.; Michaelis, A.H.; Zhu, Z.; Elvin, J.A.; Bronson, R.T.; Crum, C.P.; McKeon, F. p63 protects the female germ line during meiotic arrest. Nature 2006, 444, 624–628. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kou, Z.; Jing, Z.; Zhang, Y.; Guo, X.; Dong, M.; Wilmut, I.; Gao, S. Proteome of mouse oocytes at different developmental stages. Proc. Natl. Acad. Sci. USA 2010, 107, 17639–17644. [Google Scholar] [CrossRef]
- Zheng, P.; Schramm, R.D.; Latham, K.E. Developmental Regulation and In Vitro Culture Effects on Expression of DNA Repair and Cell Cycle Checkpoint Control Genes in Rhesus Monkey Oocytes and Embryos. Biol. Reprod. 2005, 72, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Jaroudi, S.; Kakourou, G.; Cawood, S.; Doshi, A.; Ranieri, D.M.; Serhal, P.; Harper, J.C.; SenGupta, S.B. Expression profiling of DNA repair genes in human oocytes and blastocysts using microarrays. Hum. Reprod. 2009, 24, 2649–2655. [Google Scholar] [CrossRef]
- Tulay, P.; Naja, R.P.; Cascales-Roman, O.; Doshi, A.; Serhal, P.; SenGupta, S.B. Investigation of microRNA expression and DNA repair gene transcripts in human oocytes and blastocysts. J. Assist. Reprod. Genet. 2015, 32, 1757–1764. [Google Scholar] [CrossRef]
- Menezo, Y.J.; Russo, G.; Tosti, E.; Mouatassim, S.E.; Benkhalifa, M. Expression profile of genes coding for DNA repair in human oocytes using pangenomic microarrays, with a special focus on ROS linked decays. J. Assist. Reprod. Genet. 2007, 24, 513–520. [Google Scholar] [CrossRef]
- Zeng, F.; Baldwin, D.A.; Schultz, R.M. Transcript profiling during preimplantation mouse development. Dev. Biol. 2004, 272, 483–496. [Google Scholar] [CrossRef]
- Men, N.T.; Kikuchi, K.; Furusawa, T.; Dang-Nguyen, T.Q.; Nakai, M.; Fukuda, A.; Noguchi, J.; Kaneko, H.; Viet Linh, N.; Xuan Nguyen, B.; et al. Expression of DNA repair genes in porcine oocytes before and after fertilization by ICSI using freeze-dried sperm: DNA Repair Genes in Porcine Oocytes. Anim. Sci. J. 2016, 87, 1325–1333. [Google Scholar] [CrossRef]
- Gunes, S.; Sertyel, S. Sperm DNA Damage and Oocyte Repair Capability. In A Clinician’s Guide to Sperm DNA and Chromatin Damage; Zini, A., Agarwal, A., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 321–346. ISBN 978-3-319-71814-9. [Google Scholar]
- Virant-Klun, I.; Knez, K.; Tomazevic, T.; Skutella, T. Gene Expression Profiling of Human Oocytes Developed and Matured In Vivo or In Vitro. Biomed Res. Int. 2013, 2013, 1–20. [Google Scholar] [CrossRef]
- Gasca, S.; Pellestor, F.; Assou, S.; Loup, V.; Anahory, T.; Dechaud, H.; De Vos, J.; Hamamah, S. Identifying new human oocyte marker genes: A microarray approach. Reprod. Biomed. Online 2007, 14, 175–183. [Google Scholar] [CrossRef]
- Lee, N.S.; Kim, S.; Jung, Y.W.; Kim, H. Eukaryotic DNA damage responses: Homologous recombination factors and ubiquitin modification. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2018, 809, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Giritharan, G.; Talbi, S.; Donjacour, A.; Di Sebastiano, F.; Dobson, A.T.; Rinaudo, P.F. Effect of in vitro fertilization on gene expression and development of mouse preimplantation embryos. Reproduction 2007, 134, 63–72. [Google Scholar] [CrossRef]
- Zhao, H.; Li, T.; Zhao, Y.; Tan, T.; Liu, C.; Liu, Y.; Chang, L.; Huang, N.; Li, C.; Fan, Y.; et al. Single-Cell Transcriptomics of Human Oocytes: Environment-Driven Metabolic Competition and Compensatory Mechanisms During Oocyte Maturation. Antioxid. Redox Signal. 2019, 30, 542–559. [Google Scholar] [CrossRef]
- Jaroudi, S.; SenGupta, S. DNA repair in mammalian embryos. Mutat. Res. Rev. Mutat. Res. 2007, 635, 53–77. [Google Scholar] [CrossRef]
- Ménézo, Y.; Dale, B.; Cohen, M. DNA damage and repair in human oocytes and embryos: A review. Zygote 2010, 18, 357–365. [Google Scholar] [CrossRef]
- Stitzel, M.L.; Seydoux, G. Regulation of the Oocyte-to-Zygote Transition. Science 2007, 316, 407–408. [Google Scholar] [CrossRef]
- Dai, X.-X.; Jiang, J.-C.; Sha, Q.-Q.; Jiang, Y.; Ou, X.-H.; Fan, H.-Y. A combinatorial code for mRNA 3′-UTR-mediated translational control in the mouse oocyte. Nucleic Acids Res. 2019, 47, 328–340. [Google Scholar] [CrossRef]
- Stebbins-Boaz, B.; Cao, Q.; de Moor, C.H.; Mendez, R.; Richter, J.D. Maskin Is a CPEB-Associated Factor That Transiently Interacts with eIF-4E. Mol. Cell 1999, 4, 1017–1027. [Google Scholar] [CrossRef]
- Bao, J.; Bedford, M.T. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction 2016, 151, R55–R70. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Li, X.; Liang, D.; Li, T.; Zhu, P.; Guo, H.; Wu, X.; Wen, L.; Gu, T.-P.; Hu, B.; et al. Active and Passive Demethylation of Male and Female Pronuclear DNA in the Mammalian Zygote. Cell Stem Cell 2014, 15, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Active DNA Demethylation Mediated by DNA Glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef] [PubMed]
- Wossidlo, M.; Arand, J.; Sebastiano, V.; Lepikhov, K.; Boiani, M.; Reinhardt, R.; Schöler, H.; Walter, J. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010, 29, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Gehring, M.; Reik, W.; Henikoff, S. DNA demethylation by DNA repair. Trends Genet. 2009, 25, 82–90. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, Y. Reversing DNA Methylation: Mechanisms, Genomics, and Biological Functions. Cell 2014, 156, 45–68. [Google Scholar] [CrossRef]
- Ladstätter, S.; Tachibana-Konwalski, K. A Surveillance Mechanism Ensures Repair of DNA Lesions during Zygotic Reprogramming. Cell 2016, 167, 1774–1787.e13. [Google Scholar] [CrossRef]
- García-Rodríguez, A.; Gosálvez, J.; Agarwal, A.; Roy, R.; Johnston, S. DNA Damage and Repair in Human Reproductive Cells. Int. J. Mol. Sci. 2018, 20, 31. [Google Scholar] [CrossRef]
- Smith, T.B.; Dun, M.D.; Smith, N.D.; Curry, B.J.; Connaughton, H.S.; Aitken, R.J. The presence of a truncated base excision repair pathway in human spermatozoa that is mediated by OGG1. J. Cell Sci. 2013, 126, 1488–1497. [Google Scholar] [CrossRef]
- Borini, A.; Tarozzi, N.; Bizzaro, D.; Bonu, M.A.; Fava, L.; Flamigni, C.; Coticchio, G. Sperm DNA fragmentation: Paternal effect on early post-implantation embryo development in ART. Hum. Reprod. 2006, 21, 2876–2881. [Google Scholar] [CrossRef]
- Rothkamm, K.; Krüger, I.; Thompson, L.H.; Löbrich, M. Pathways of DNA Double-Strand Break Repair during the Mammalian Cell Cycle. MCB 2003, 23, 5706–5715. [Google Scholar] [CrossRef] [PubMed]
- Derijck, A.; van der Heijden, G.; Giele, M.; Philippens, M.; de Boer, P. DNA double-strand break repair in parental chromatin of mouse zygotes, the first cell cycle as an origin of de novo mutation. Hum. Mol. Genet. 2008, 17, 1922–1937. [Google Scholar] [CrossRef] [PubMed]
- Osada, T.; Ogino, H.; Hino, T.; Ichinose, S.; Nakamura, K.; Omori, A.; Noce, T.; Masutani, M. PolyADP-Ribosylation Is Required for Pronuclear Fusion during Postfertilization in Mice. PLoS ONE 2010, 5, e12526. [Google Scholar] [CrossRef] [PubMed]
- Izumi, T.; Mellon, I. Base Excision Repair and Nucleotide Excision Repair. In Genome Stability; Elsevier: Amsterdam, The Netherlands, 2016; pp. 275–302. ISBN 978-0-12-803309-8. [Google Scholar]
- Tong, W.-M.; Cortes, U.; Wang, Z.-Q. Poly (ADP-ribose) polymerase: A guardian angel protecting the genome and suppressing tumorigenesis. Biochim. Biophys. Acta BBA Rev. Cancer 2001, 1552, 27–37. [Google Scholar] [CrossRef]
- Gawecka, J.E.; Marh, J.; Ortega, M.; Yamauchi, Y.; Ward, M.A.; Ward, W.S. Mouse Zygotes Respond to Severe Sperm DNA Damage by Delaying Paternal DNA Replication and Embryonic Development. PLoS ONE 2013, 8, e56385. [Google Scholar] [CrossRef] [PubMed]
- Adiga, S.K.; Toyoshima, M.; Shiraishi, K.; Shimura, T.; Takeda, J.; Taga, M.; Nagai, H.; Kumar, P.; Niwa, O. p21 provides stage specific DNA damage control to preimplantation embryos. Oncogene 2007, 26, 6141–6149. [Google Scholar] [CrossRef]
- Bielanska, M.; Jin, S.; Bernier, M.; Tan, S.L.; Ao, A. Diploid-aneuploid mosaicism in human embryos cultured to the blastocyst stage. Fertil. Steril. 2005, 84, 336–342. [Google Scholar] [CrossRef]
- Pfeiffer, P. Mechanisms of DNA double-strand break repair and their potential to induce chromosomal aberrations. Mutagenesis 2000, 15, 289–302. [Google Scholar] [CrossRef]
- Petropoulos, S.; Panula, S.P.; Schell, J.P.; Lanner, F. Single-cell RNA sequencing: Revealing human pre-implantation development, pluripotency and germline development. J. Intern. Med. 2016, 280, 252–264. [Google Scholar] [CrossRef]
- Lavagi, I.; Krebs, S.; Simmet, K.; Beck, A.; Zakhartchenko, V.; Wolf, E.; Blum, H. Single-cell RNA sequencing reveals developmental heterogeneity of blastomeres during major genome activation in bovine embryos. Sci. Rep. 2018, 8, 4071. [Google Scholar] [CrossRef]
- Groff, A.F.; Resetkova, N.; DiDomenico, F.; Sakkas, D.; Penzias, A.; Rinn, J.L.; Eggan, K. RNA-seq as a tool for evaluating human embryo competence. Genome Res. 2019, 29, 1705–1718. [Google Scholar] [CrossRef] [PubMed]
- Tichy, E.D.; Stambrook, P.J. DNA repair in murine embryonic stem cells and differentiated cells. Exp. Cell Res. 2008, 314, 1929–1936. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.-H.; Yoon, S.; Park, K.-S.; Kim, K.P. The Homologous Recombination Machinery Orchestrates Post-replication DNA Repair During Self-renewal of Mouse Embryonic Stem Cells. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, A.K.; Jodkowska, K.; Teloni, F.; Bizard, A.H.; Zellweger, R.; Herrador, R.; Ortega, S.; Hickson, I.D.; Altmeyer, M.; Mendez, J.; et al. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 2016, 7, 10660. [Google Scholar] [CrossRef]
- Van Sloun, P.P.H.; Jansen, J.G.; Weeda, G.; Mullenders, L.H.F.; van Zeeland, A.A.; Lohman, P.H.M.; Vrieling, H. The role of nucleotide excision repair in protecting embryonic stem cells from genotoxic effects of UV-induced DNA damage. Nucleic Acids Res. 1999, 27, 3276–3282. [Google Scholar] [CrossRef] [PubMed]
- Kermi, C.; Aze, A.; Maiorano, D. Preserving Genome Integrity during the Early Embryonic DNA Replication Cycles. Genes 2019, 10, 398. [Google Scholar] [CrossRef]
- Neganova, I.; Lako, M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J. Anat. 2008, 213, 30–44. [Google Scholar] [CrossRef]
- Van der Laan, S.; Tsanov, N.; Crozet, C.; Maiorano, D. High Dub3 Expression in Mouse ESCs Couples the G1/S Checkpoint to Pluripotency. Mol. Cell 2013, 52, 366–379. [Google Scholar] [CrossRef]
- Liu, L.; Michowski, W.; Kolodziejczyk, A.; Sicinski, P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell Biol. 2019, 21, 1060–1067. [Google Scholar] [CrossRef]
- Fu, X.; Cui, K.; Yi, Q.; Yu, L.; Xu, Y. DNA repair mechanisms in embryonic stem cells. Cell. Mol. Life Sci. 2017, 74, 487–493. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khokhlova, E.V.; Fesenko, Z.S.; Sopova, J.V.; Leonova, E.I. Features of DNA Repair in the Early Stages of Mammalian Embryonic Development. Genes 2020, 11, 1138. https://doi.org/10.3390/genes11101138
Khokhlova EV, Fesenko ZS, Sopova JV, Leonova EI. Features of DNA Repair in the Early Stages of Mammalian Embryonic Development. Genes. 2020; 11(10):1138. https://doi.org/10.3390/genes11101138
Chicago/Turabian StyleKhokhlova, Evgenia V., Zoia S. Fesenko, Julia V. Sopova, and Elena I. Leonova. 2020. "Features of DNA Repair in the Early Stages of Mammalian Embryonic Development" Genes 11, no. 10: 1138. https://doi.org/10.3390/genes11101138
APA StyleKhokhlova, E. V., Fesenko, Z. S., Sopova, J. V., & Leonova, E. I. (2020). Features of DNA Repair in the Early Stages of Mammalian Embryonic Development. Genes, 11(10), 1138. https://doi.org/10.3390/genes11101138




