Developmental scRNAseq Trajectories in Gene- and Cell-State Space—The Flatworm Example
Abstract
1. Introduction
2. Materials and Methods
2.1. scRNA Seq Data of Planarian Development
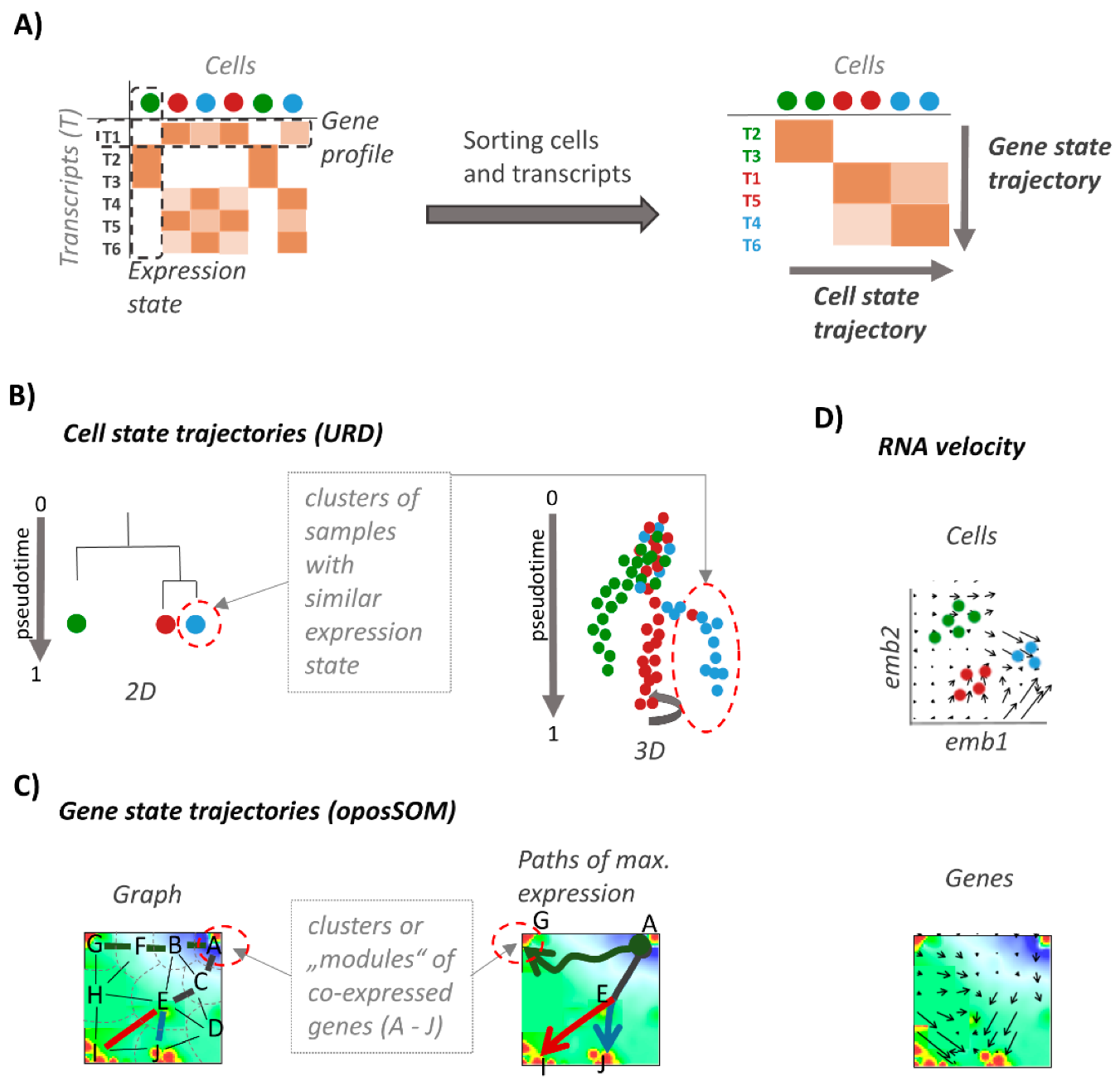
2.2. Single Cell SOM Portrayal
2.3. Cell Clustering, Cell State Trajectories, and Pseudotime
2.4. Gene State Trajectories in SOM Space
2.5. RNA-Velocity Dynamics
2.6. Function Analysis
2.7. Methods Availability
3. Results
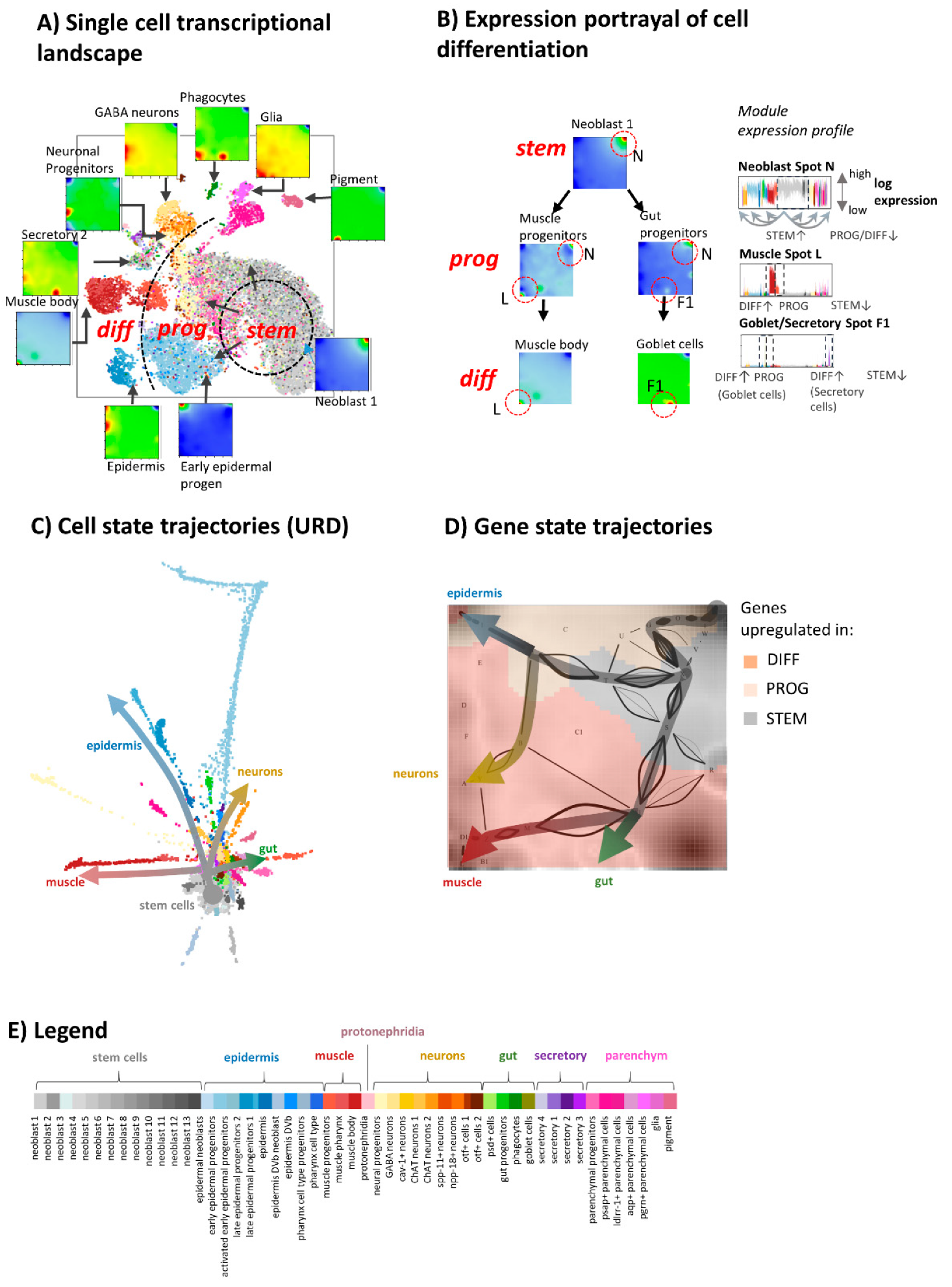
3.1. Portrayal of Developing Single Cell Transcriptomes
3.2. The Transcriptome Landscape of Planarian
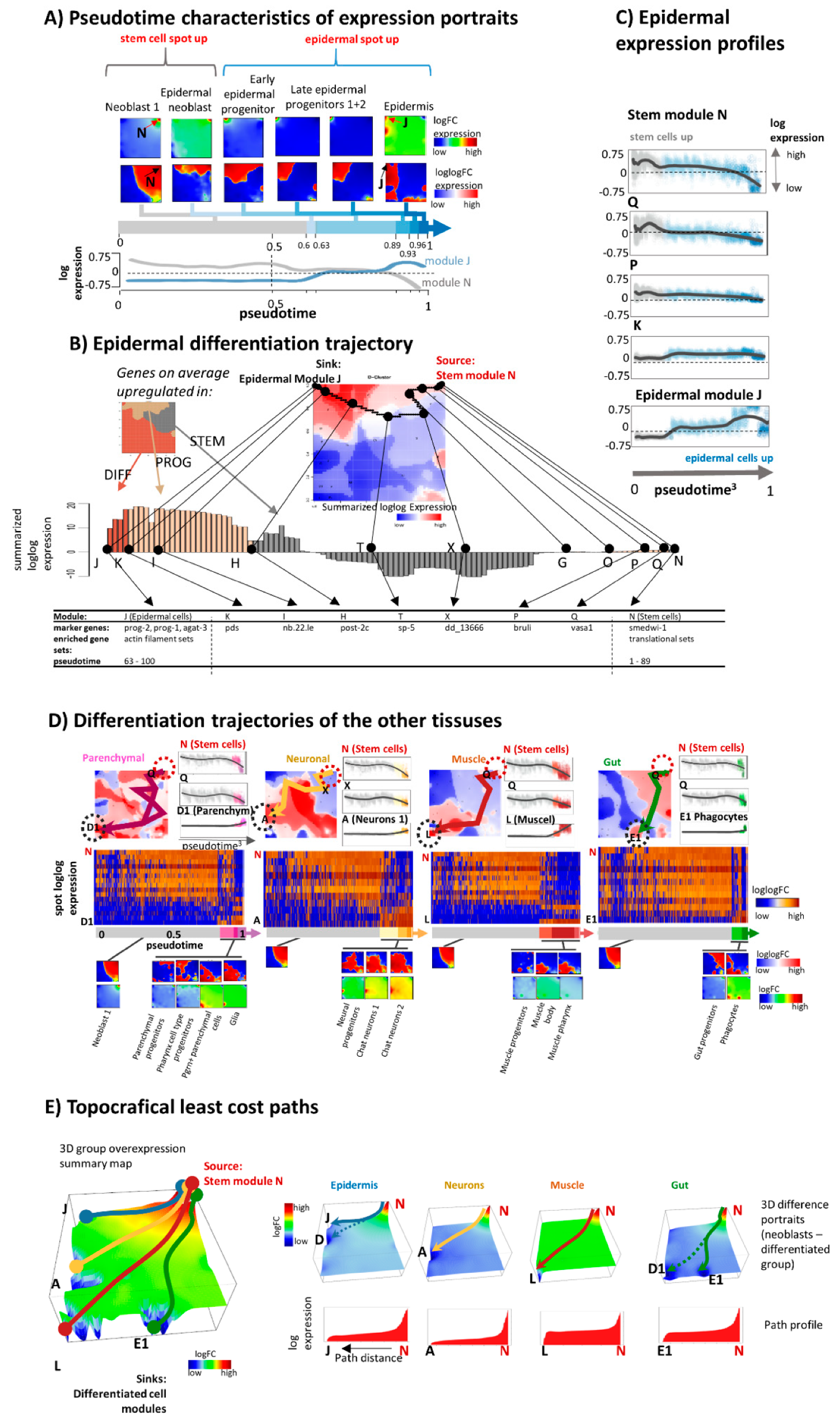
3.3. Portrayal of Developmental Paths
3.4. Development along Branched Trees
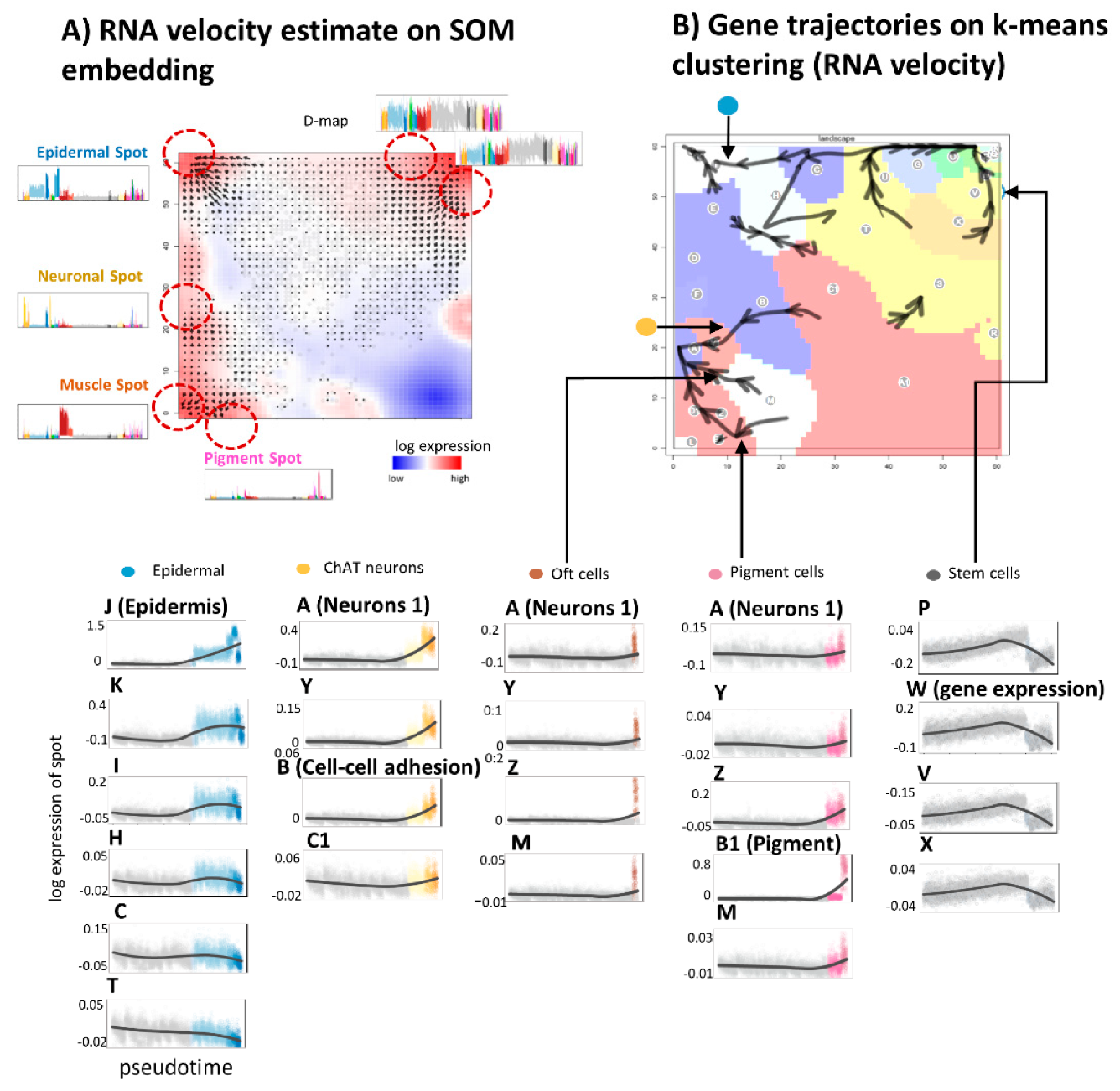
3.5. RNA Velocity Trajectories
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kalisky, T.; Quake, S.R. Single-cell genomics. Nat. Methods 2011, 8, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, I.C.; Voet, T. Single Cell Genomics: Advances and Future Perspectives. PLoS Genet. 2014, 10, e1004126. [Google Scholar] [CrossRef] [PubMed]
- Bendall, S.C.; Davis, K.L.; Amir, E.A.D.; Tadmor, M.D.; Simonds, E.F.; Chen, T.J.; Shenfeld, D.K.; Nolan, G.P.; Pe’Er, D. Single-cell trajectory detection uncovers progression and regulatory coordination in human b cell development. Cell 2014, 157, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Haghverdi, L.; Büttner, M.; Wolf, F.A.; Buettner, F.; Theis, F.J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 2016, 13, 845–848. [Google Scholar] [CrossRef]
- Hanchate, N.K.; Kondoh, K.; Lu, Z.; Kuang, D.; Ye, X.; Qiu, X.; Pachter, L.; Trapnell, C.; Buck, L.B. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science 2015, 350, 1251–1255. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Campbell, K.R.; Yau, C. A descriptive marker gene approach to single-cell pseudotime inference. Bioinformatics 2019, 35, 28–35. [Google Scholar] [CrossRef]
- Reid, J.E.; Wernisch, L. Pseudotime estimation: Deconfounding single cell time series. Bioinformatics 2016, 32, 2973–2980. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.R.; Yau, C. Order Under Uncertainty: Robust Differential Expression Analysis Using Probabilistic Models for Pseudotime Inference. PLoS Comput. Biol. 2016, 12, e1005212. [Google Scholar] [CrossRef]
- Saelens, W.; Cannoodt, R.; Todorov, H.; Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019, 37, 547–554. [Google Scholar] [CrossRef]
- Shin, J.; Berg, D.A.; Zhu, Y.; Shin, J.Y.; Song, J.; Bonaguidi, M.A.; Enikolopov, G.; Nauen, D.W.; Christian, K.M.; Ming, G.L.; et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 2015, 17, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Street, K.; Risso, D.; Fletcher, R.B.; Das, D.; Ngai, J.; Yosef, N.; Purdom, E.; Dudoit, S. Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018, 19, 477. [Google Scholar] [CrossRef] [PubMed]
- Setty, M.; Tadmor, M.D.; Reich-Zeliger, S.; Angel, O.; Salame, T.M.; Kathail, P.; Choi, K.; Bendall, S.; Friedman, N.; Pe’Er, D. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 2016, 34, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Wolf, F.A.; Hamey, F.K.; Plass, M.; Solana, J.; Dahlin, J.S.; Göttgens, B.; Rajewsky, N.; Simon, L.; Theis, F.J. PAGA: Graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 2019, 20, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.E.; Klein, A.M. Lineage tracing meets single-cell omics: Opportunities and challenges. Nat. Rev. Genet. 2020, 21, 410–427. [Google Scholar] [CrossRef] [PubMed]
- Orman, M.A.; Berthiaume, F.; Androulakis, I.P.; Ierapetritou, M.G. Advanced stoichiometric analysis of metabolic networks of mammalian systems. Crit. Rev. Biomed. Eng. 2011, 39, 511–534. [Google Scholar] [CrossRef]
- Tritschler, S.; Büttner, M.; Fischer, D.S.; Lange, M.; Bergen, V.; Lickert, H.; Theis, F.J. Concepts and limitations for learning developmental trajectories from single cell genomics. Development 2019, 146, dev170506. [Google Scholar] [CrossRef]
- Loeffler-Wirth, H.; Binder, H.; Willscher, E.; Gerber, T.; Kunz, M. Pseudotime dynamics in melanoma single-cell transcriptomes reveals different mechanisms of tumor progression. Biology 2018, 7, 23. [Google Scholar] [CrossRef]
- Fan, J.; Slowikowski, K.; Zhang, F. Single-cell transcriptomics in cancer: Computational challenges and opportunities. Exp. Mol. Med. 2020, 52, 1452–1465. [Google Scholar] [CrossRef]
- Valdes-Mora, F.; Handler, K.; Law, A.M.K.; Salomon, R.; Oakes, S.R.; Ormandy, C.J.; Gallego-Ortega, D. Single-cell transcriptomics in cancer immunobiology: The future of precision oncology. Front. Immunol. 2018, 9, 2582. [Google Scholar] [CrossRef] [PubMed]
- Wirth, H.; Löffler, M.; Von Bergen, M.; Binder, H. Expression cartography of human tissues using self organizing maps. BMC Bioinform. 2011, 12, 306. [Google Scholar] [CrossRef] [PubMed]
- Wirth, H.; von Bergen, M.; Binder, H. Mining SOM expression portraits: Feature selection and integrating concepts of molecular function. BioData Min. 2012, 5, 18. [Google Scholar] [CrossRef]
- Kunz, M.; Löffler-Wirth, H.; Dannemann, M.; Willscher, E.; Doose, G.; Kelso, J.; Kottek, T.; Nickel, B.; Hopp, L.; Landsberg, J.; et al. RNA-seq analysis identifies different transcriptomic types and developmental trajectories of primary melanomas. Oncogene 2018, 37, 6136–6151. [Google Scholar] [CrossRef]
- Gerber, T.; Willscher, E.; Loeffler-wirth, H.; Hopp, L.; Schartl, M.; Anderegg, U.; Camp, G.; Treutlein, B. Mapping heterogeneity in patient-derived melanoma cultures by. Oncotarget 2017, 8, 846–862. [Google Scholar] [CrossRef]
- Camp, J.G.; Sekine, K.; Gerber, T.; Loeffler-Wirth, H.; Binder, H.; Gac, M.; Kanton, S.; Kageyama, J.; Damm, G.; Seehofer, D.; et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017, 546, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Binder, H.; Löffler-Wirth, H. Analysis of large-scale OMIC data using Self Organizing Maps. Encycl. Inf. Sci. Technol. Third Ed. 2014, 1642–1654. [Google Scholar] [CrossRef]
- Binder, H.; Wirth, H.; Arakelyan, A.; Lembcke, K.; Tiys, E.S.; Ivanisenko, V.A.; Kolchanov, N.A.; Kononikhin, A.; Popov, I.; Nikolaev, E.N.; et al. Time-course human urine proteomics in space-flight simulation experiments. BMC Genom. 2014, 15, 1–19. [Google Scholar] [CrossRef]
- Plass, M.; Solana, J.; Wolf, F.A.; Ayoub, S.; Misios, A.; Glažar, P.; Obermayer, B.; Theis, F.J.; Kocks, C.; Rajewsky, N. Supplementary Materials for Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 2018, 360, eaaq1723. [Google Scholar] [CrossRef]
- Huang, S.; Eichler, G.; Bar-Yam, Y.; Ingber, D.E. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys. Rev. Lett. 2005, 94, 128701. [Google Scholar] [CrossRef]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Bergen, V.; Lange, M.; Peidli, S.; Wolf, F.A.; Theis, F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Asami, M.; Higuchi, S.; Shibata, N.; Agata, K. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev. Growth Differ. 2006, 48, 371–380. [Google Scholar] [CrossRef]
- Önal, P.; Grün, D.; Adamidi, C.; Rybak, A.; Solana, J.; Mastrobuoni, G.; Wang, Y.; Rahn, H.P.; Chen, W.; Kempa, S.; et al. Gene expression of pluripotency determinants is conserved between mammalian and planarian stem cells. EMBO J. 2012, 31, 2755–2769. [Google Scholar] [CrossRef] [PubMed]
- Loeffler-Wirth, H.; Kreuz, M.; Hopp, L.; Arakelyan, A.; Haake, A.; Cogliatti, S.B.; Feller, A.C.; Hansmann, M.L.; Lenze, D.; Möller, P.; et al. A modular transcriptome map of mature B cell lymphomas. Genome Med. 2019, 11, 27. [Google Scholar] [CrossRef]
- Loeffler-Wirth, H.; Kalcher, M.; Binder, H. OposSOM: R-package for high-dimensional portraying of genome-wide expression landscapes on bioconductor. Bioinformatics 2015, 31, 3225–3227. [Google Scholar] [CrossRef]
- Hopp, L.; Loeffler-Wirth, H.; Nersisyan, L.; Arakelyan, A.; Binder, H. Footprints of Sepsis Framed Within Community Acquired Pneumonia in the Blood Transcriptome. Front. Immunol. 2018, 9, 1620. [Google Scholar] [CrossRef]
- Van Der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn Res. 2008, 9, 2579–2605. [Google Scholar]
- Farrell, J.A.; Wang, Y.; Riesenfeld, S.J.; Shekhar, K.; Regev, A.; Schier, A.F. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 2018, 360, eaar3131. [Google Scholar] [CrossRef]
- Goldberg, A.V.; Tarjan, R.E. A New Approach to the Maximum-Flow Problem. J. ACM 1988, 35, 921–940. [Google Scholar] [CrossRef]
- Wang, I.J. Topographic path analysis for modelling dispersal and functional connectivity: Calculating topographic distances using the topoDistance r package. Methods Ecol. Evol. 2020, 11, 265–272. [Google Scholar] [CrossRef]
- Brandl, H.; Moon, H.K.; Vila-Farré, M.; Liu, S.Y.; Henry, I.; Rink, J.C. PlanMine—A mineable resource of planarian biology and biodiversity. Nucleic Acids Res. 2016, 44, D764–D773. [Google Scholar] [CrossRef] [PubMed]
- Fincher, C.T.; Wurtzel, O.; de Hoog, T.; Kravarik, K.M.; Reddien, P.W. Cell type transcriptome atlas for the planarian Schmidtea mediterranea. Science 2018, 360, eaaq1736. [Google Scholar] [CrossRef]
- Waddington, C.H. The Strategy of the Genes; George Allen & Unwin, Ltd: London, UK, 1957; ISBN 978-1-315-77932-4. [Google Scholar]
- Lareau, C.A.; Duarte, F.M.; Chew, J.G.; Kartha, V.K.; Burkett, Z.D.; Kohlway, A.S.; Pokholok, D.; Aryee, M.J.; Steemers, F.J.; Lebofsky, R.; et al. Droplet-based combinatorial indexing for massive-scale single-cell chromatin accessibility. Nat. Biotechnol. 2019, 37, 916–924. [Google Scholar] [CrossRef]
- Budnik, B.; Levy, E.; Harmange, G.; Slavov, N. SCoPE-MS: Mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol. 2018, 19, 161. [Google Scholar] [CrossRef]
- Duncan, K.D.; Fyrestam, J.; Lanekoff, I. Advances in mass spectrometry based single-cell metabolomics. Analyst 2019, 144, 782–793. [Google Scholar] [CrossRef]
- Dey, S.S.; Kester, L.; Spanjaard, B.; Bienko, M.; Van Oudenaarden, A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 2015, 33, 285–289. [Google Scholar] [CrossRef]
- Lubeck, E.; Coskun, A.F.; Zhiyentayev, T.; Ahmad, M.; Cai, L. Single-cell in situ RNA profiling by sequential hybridization. Nat. Methods 2014, 11, 360–361. [Google Scholar] [CrossRef]
- Sebastian, A.; Hum, N.R.; Martin, K.A.; Gilmore, S.F.; Peran, I.; Byers, S.W.; Wheeler, E.K.; Coleman, M.A.; Loots, G.G. Single-Cell Transcriptomic Analysis of Tumor-Derived Fibroblasts and Normal Tissue-Resident Fibroblasts Reveals Fibroblast Heterogeneity in Breast Cancer. Cancers 2020, 12, 1307. [Google Scholar] [CrossRef]
- Qian, J.; Olbrecht, S.; Boeckx, B.; Vos, H.; Laoui, D.; Etlioglu, E.; Wauters, E.; Pomella, V.; Verbandt, S.; Busschaert, P.; et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020, 30, 745–762. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Hu, Y.; Wu, F.; Guo, Q.; Qian, Z.; Hu, W.; Chen, J.; Wang, K.; Fan, X.; Wu, X.; et al. Surveying brain tumor heterogeneity by single-cell RNA-sequencing of multi-sector biopsies. Natl. Sci. Rev. 2020, 7, 1306–1318. [Google Scholar] [CrossRef]
- Tirosh, I.; Suvà, M.L. Deciphering Human Tumor Biology by Single-Cell Expression Profiling. Annu. Rev. Cancer Biol. 2019, 3, 151–166. [Google Scholar] [CrossRef]
- Mar, J.C.; Quackenbush, J. Decomposition of Gene Expression State Space Trajectories. PLoS Comput. Biol. 2009, 5, e1000626. [Google Scholar] [CrossRef] [PubMed]
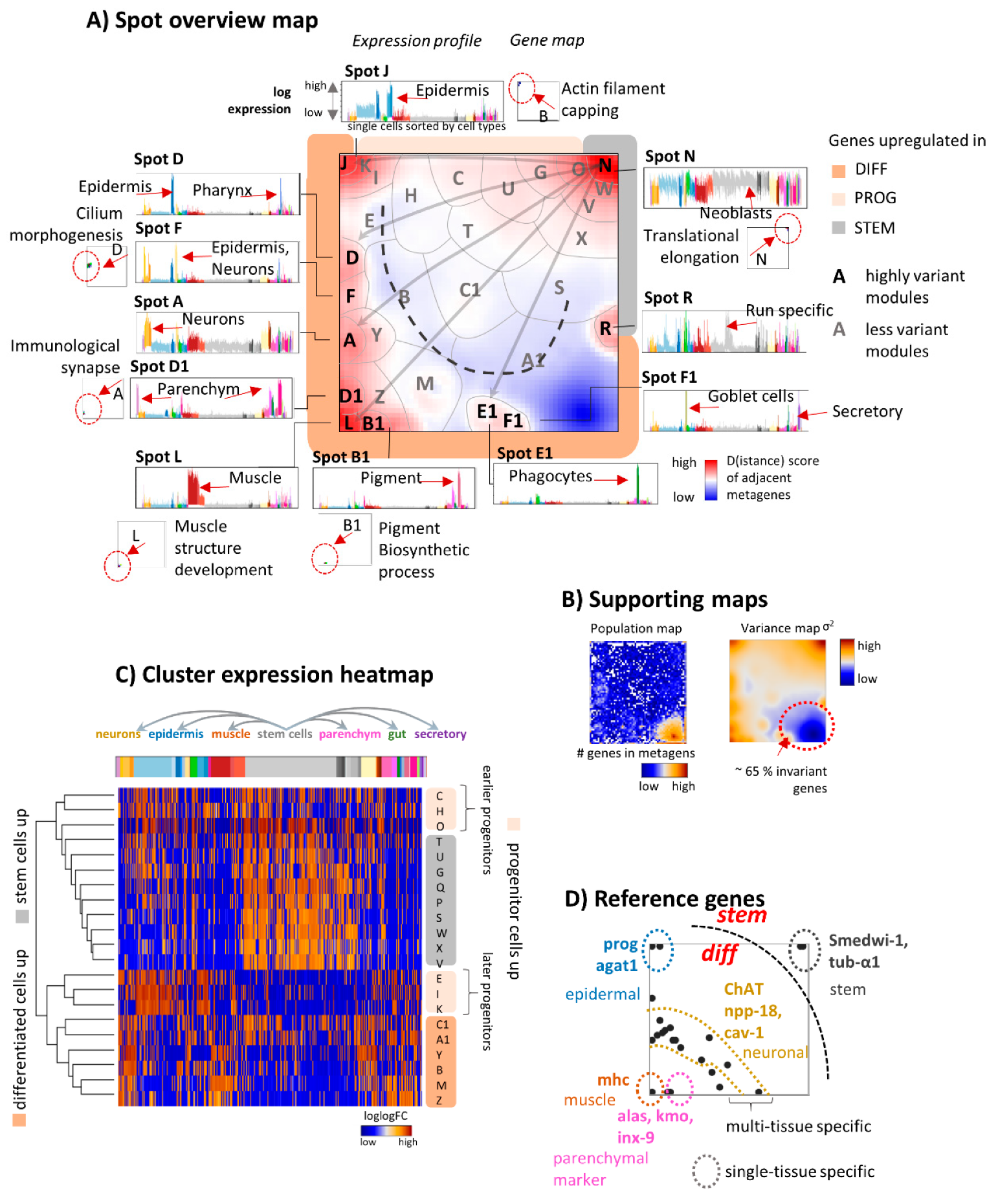

| Cluster | Genes | Name | Enriched BP Gene Sets (log p Enrichment) | Marker Genes | Tissue |
|---|---|---|---|---|---|
| A | 235 | Neurons 1 | Neurotransmitter secretion (−6) Synaptic vesicle localization (−5) Neurotransmitter transport (−5) Regulation of neurotransmitter levels (−5) Synaptic vesicle transport (−4) | pc2, ChAT, gpas | cav-1+ neurons, ChAT neurons 1, ChAT neurons 2 |
| B | 1314 | Phosphorus metabolic process (−4) Phosphate-containing compound metabolic process (−4) Homopholic cell adhesion via plasma membrane adhesion molecules (−4) Cell-cell adhesion via plasma-membrane adhesion molecules (−4) Cell communication (−3) | GABRB3 (dd21541), CAVII-like, th | ||
| C | 206 | ||||
| D | 265 | Pharynx | vim-1,VIT (dd1071), NPEPL1 (dd181) | Epidermis, Pharynx | |
| E | 383 | 1.G9.2, ifn | |||
| F | 370 | Neurons 2 | Microtubule-based movement (−18) Microtubule-based process (−15) cilium morphogenesis/organization/assembly (≥−14) movement of cell/subcellular component (−9) | pkd2l-2, rootletin (dd6573), cav-1 | cav-1+ neurons, GABA neurons |
| G | 218 | ||||
| H | 330 | Post-2c | |||
| I | 228 | nb.22.le | |||
| J | 111 | Epidermis | actin filament capping (−2) negative regulation of actin filament depolymerization (−2) | prog-2, prog-1, agat-3 | Epidermis, late epidermal progenitors 1/2 |
| K | 100 | pds | |||
| L | 150 | Muscle | muscle structure development (−9) tissue development (−8) actin filament-based process (−7) anatomical structure development (−6) muscle organ development (−6) | COL4A6A (dd2337), collagen, COL21A1 (dd9565) | Muscle body |
| M | 1032 | cali, if-1, HSPG2 (dd8356) | |||
| N | 121 | Stem cells | translational elongation and termination (≥−79) cotranslational protein targeting to membrane (≥−73) protein targeting to ER (−72) Peptide and amid biosynthetic process (≥−71) | smedwi-1, dd_6998 | Stem cells |
| O | 143 | ||||
| P | 114 | bruli | |||
| Q | 44 | vasa-1 | |||
| R | 119 | Run specific | Unknown origin, presumably due to batch effects | Run specific cells | |
| S | 974 | dd_5560, SAMD15 (dd19710), wntP-3 | |||
| T | 404 | sp-5 | |||
| U | 235 | ||||
| V | 265 | TYMS, gH4 | |||
| W | 146 | Gene expression (−8) Nucleic acid metabolic process (−7) Cellular nitrogen compound metabolic (−6) Cellular macromolecule metabolic process (−5) Nucleosome organization (−5) | |||
| X | 398 | dd_13666 | |||
| Y | 445 | GLIPR1 (dd210), npp-18 | |||
| Z | 575 | TMPRSS9 (dd7966), CTSL2 (dd582), gata 4/5/6 | |||
| A1 | 18239 | G−protein coupled receptor signaling pathway (≥−47) cell communication (−12) Signaling (−11) single organism signaling (−11) | glipr-1, PI16 (dd940), ASCL4 (dd1854) | ||
| B1 | 181 | Pigment | organonitrogen compound catabolic process (−6) tyrosine catabolic process (−5) small molecule metabolic process (−4) response to transition metal nanoparticle (−4) L-phenylalanine catabolic process (−4) erythrose 4− | pgbd-1, PSAPL1 (dd1706), KMO (dd7884) | Pigment cells |
| C1 | 458 | ||||
| D1 | 262 | Parenchym | regulation of intracellular signal transduction (−3) negative regulation of antigen receptor-mediated signaling pathway (−3) | ctsl2, CTP, PCN, dd_Smed_v6_3_0, PSAP | aqp+ parenchymal cells, pgrn+ parenchymal cells, psap+ parenchymal cells |
| E1 | 226 | Phagocytes | mat | Phagocytes | |
| F1 | 725 | Goblet, secretory cells | Goblet and secretory cells |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmidt, M.; Loeffler-Wirth, H.; Binder, H. Developmental scRNAseq Trajectories in Gene- and Cell-State Space—The Flatworm Example. Genes 2020, 11, 1214. https://doi.org/10.3390/genes11101214
Schmidt M, Loeffler-Wirth H, Binder H. Developmental scRNAseq Trajectories in Gene- and Cell-State Space—The Flatworm Example. Genes. 2020; 11(10):1214. https://doi.org/10.3390/genes11101214
Chicago/Turabian StyleSchmidt, Maria, Henry Loeffler-Wirth, and Hans Binder. 2020. "Developmental scRNAseq Trajectories in Gene- and Cell-State Space—The Flatworm Example" Genes 11, no. 10: 1214. https://doi.org/10.3390/genes11101214
APA StyleSchmidt, M., Loeffler-Wirth, H., & Binder, H. (2020). Developmental scRNAseq Trajectories in Gene- and Cell-State Space—The Flatworm Example. Genes, 11(10), 1214. https://doi.org/10.3390/genes11101214






