Cell-Free DNA Methylation: The New Frontiers of Pancreatic Cancer Biomarkers’ Discovery
Abstract
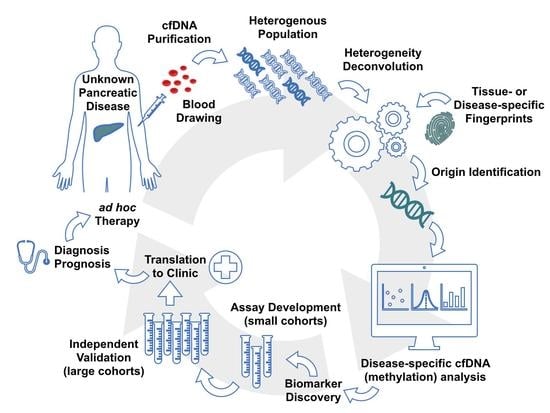
1. Introduction
2. Genetic and Epigenetic Implications of cfDNA in Cancer Diagnosis
2.1. Pitfalls in cfDNA Data Interpretation
2.2. Exploiting DNA Methylation Signatures for the Identification of Circulating cfDNA
3. cfDNA Methylation Biomarkers in Pancreatic Cancer or Pancreatic Pre-Neoplastic Conditions
3.1. cfDNA Hypomethylated or Non-Methylated Targets
3.2. cfDNA Hypermethylated Targets
| Genes Useful to Identify Circulating Exocrine Pancreatic cfDNA | ||||||
|---|---|---|---|---|---|---|
| CUX2 | Ductal cell marker | Increased signal in PDAC | ||||
| REG1A | Ductal and acing cell marker | Increasing signal in CP | ||||
| Frequency of cfDNA methylation biomarkers (%) | ||||||
| Gene | healthy | CP | PIN | PDAC I | PDAC II | PDAC III/IV |
| ADAMTS1 | <3 | - | 25 | 25–88 | 78–90 | 55 |
| BNC1 | 3–7 | 5 | 70 | 63–95 | 56–95 | 100 |
| CCND2 | 17 | 82 | - | - | - | 24 |
| CDKN1C | 60 | 90 | - | - | - | 27 |
| MLH1 | 22 | 78 | - | - | - | 27 |
| PGR (prox) | 14 | 76 | - | - | - | 37 |
| SYK | 57 | 89 | - | - | - | 57 |
4. Discussion and Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AP | Acute pancreatitis |
| ASA | American Society of Anesthesiologists |
| cfDNA | Cell-free DNA |
| CP | Chronic pancreatitis |
| CTCs | Circulating tumor cells |
| DNMTs | DNA methyl transferases |
| HR | Hazard ratio |
| IMPN | Intramucinous pancreatic neoplasia |
| MHBs | Methylation haplotype blocks |
| MI | Methylation index |
| MSP | Methylation-specific PCR |
| NGS | Next-generation sequencing |
| PDAC | Pancreatic ductal adenocarcinoma |
| PIN | Pancreatic intra-epithelial neoplasia |
| RRBS | Reduced representation bisulfite sequencing |
References
- Raimondi, S.; Lowenfels, A.B.; Morselli-Labate, A.M.; Maisonneuve, P.; Pezzilli, R. Pancreatic cancer in chronic pancreatitis; aetiology, incidence, and early detection. Best Pract. Res. Clin. Gastroenterol. 2010, 24, 349–358. [Google Scholar] [CrossRef] [PubMed]
- NIH. Cancer Stat Facts: Pancreatic Cancer, 2019. Available online: https://seer.cancer.gov/statfacts/html/pancreas.html (accessed on 20 November 2019).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Karmazanovsky, G.; Fedorov, V.; Kubyshkin, V.; Kotchatkov, A. Pancreatic head cancer: Accuracy of CT in determination of resectability. Abdom. Imaging 2005, 30, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Ebner, D.K.; Tinganelli, W.; Helm, A.; Bisio, A.; Simoniello, P.; Natale, F.; Yamada, S.; Kamada, T.; Shimokawa, T.; Durante, M. Generating and grading the abscopal effect: Proposal for comprehensive evaluation of combination immunoradiotherapy in mouse models. Transl. Cancer Res. 2017, 6, S892–S899. [Google Scholar] [CrossRef]
- Garrido-Laguna, I.; Hidalgo, M. Pancreatic cancer: From state-of-the-art treatments to promising novel therapies. Nat. Rev. Clin. Oncol. 2015, 12, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Yachida, S.; Jones, S.; Bozic, I.; Antal, T.; Leary, R.; Fu, B.; Kamiyama, M.; Hruban, R.H.; Eshleman, J.R.; Nowak, M.A.; et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010, 467, 1114–1117. [Google Scholar] [CrossRef]
- Natale, F.; Vivo, M.; Falco, G.; Angrisano, T. Deciphering DNA methylation signatures of pancreatic cancer and pancreatitis. Clin. Epigenet. 2019, 11, 132. [Google Scholar] [CrossRef]
- Goonetilleke, K.; Siriwardena, A. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur. J. Surg. Oncol. (EJSO) 2007, 33, 266–270. [Google Scholar] [CrossRef]
- Trikudanathan, G.; Vega-Peralta, J.; Malli, A.; Munigala, S.; Han, Y.; Bellin, M.; Barlass, U.; Chinnakotla, S.; Dunn, T.; Pruett, T.; et al. Diagnostic Performance of Endoscopic Ultrasound (EUS) for Non-Calcific Chronic Pancreatitis (NCCP) Based on Histopathology. Am. J. Gastroenterol. 2016, 111, 568–574. [Google Scholar] [CrossRef]
- Kisiel, J.; Raimondo, M.; Taylor, W.; Yab, T.; Mahoney, D.; Sun, Z.; Middha, S.; Baheti, S.; Zou, H.; Smyrk, T.; et al. New DNA Methylation Markers for Pancreatic Cancer: Discovery, Tissue Validation, and Pilot Testing in Pancreatic Juice. Clin. Cancer Res. 2015, 21, 4473–4481. [Google Scholar] [CrossRef]
- Matsubayashi, H.; Canto, M.; Sato, N.; Klein, A.; Abe, T.; Yamashita, K.; Yeo, C.; Kalloo, A.; Hruban, R.; Goggins, M. DNA Methylation Alterations in the Pancreatic Juice of Patients with Suspected Pancreatic Disease. Cancer Res. 2006, 66, 1208–1217. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; D’Alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B.; Jones, P.A. Epigenetic Determinants of Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019505. [Google Scholar] [CrossRef]
- Chung, W.; Bondaruk, J.; Jelinek, J.; Lotan, Y.; Liang, S.; Czerniak, B.; Issa, J.P. Detection of Bladder Cancer Using Novel DNA Methylation Biomarkers in Urine Sediments. Cancer Epidemiol. Prev. Biomark. 2011, 20, 1483–1491. [Google Scholar] [CrossRef]
- Jeschke, J.; Van Neste, L.; Glöckner, S.C.; Dhir, M.; Calmon, M.F.; Deregowski, V.; Van Criekinge, W.; Vlassenbroeck, I.; Koch, A.; Chan, T.A.; et al. Biomarkers for detection and prognosis of breast cancer identified by a functional hypermethylome screen. Epigenetics 2012, 7, 701–709. [Google Scholar] [CrossRef]
- Schuebel, K.E.; Chen, W.; Cope, L.; Glöckner, S.C.; Suzuki, H.; Yi, J.M.; Chan, T.A.; Neste, L.V.; Criekinge, W.V.; Bosch, S.V.D.; et al. Comparing the DNA Hypermethylome with Gene Mutations in Human Colorectal Cancer. PLoS Genet. 2007, 3, e157. [Google Scholar] [CrossRef]
- Su, Y.; Fang, H.B.; Jiang, F. An epigenetic classifier for early stage lung cancer. Clin. Epigenet. 2018, 10, 68. [Google Scholar] [CrossRef]
- Zochbauer-Muller, S. Aberrant DNA Methylation in Lung Cancer: Biological and Clinical Implications. Oncologist 2002, 7, 451–457. [Google Scholar] [CrossRef]
- Yi, J.; Guzzetta, A.; Bailey, V.; Downing, S.; Van+Neste, L.; Chiappinelli, K.; Keeley, B.; Stark, A.; Herrera, A.; Wolfgang, C.; et al. Novel Methylation Biomarker Panel for the Early Detection of Pancreatic Cancer. Clin. Cancer Res. 2013, 19, 6544–6555. [Google Scholar] [CrossRef]
- Hong, S.M.; Omura, N.; Vincent, A.; Li, A.; Knight, S.; Yu, J.; Hruban, R.; Goggins, M. Genome-Wide CpG Island Profiling of Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin. Cancer Res. 2012, 18, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.M.; Kelly, D.; Griffith, M.; Omura, N.; Li, A.; Li, C.P.; Hruban, R.H.; Goggins, M. Multiple genes are hypermethylated in intraductal papillary mucinous neoplasms of the pancreas. Modern Pathol. 2008, 21, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Fukushima, N.; Hruban, R.H.; Goggins, M. CpG island methylation profile of pancreatic intraepithelial neoplasia. Modern Pathol. 2007, 21, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Lehmann-Werman, R.; Neiman, D.; Zemmour, H.; Moss, J.; Magenheim, J.; Vaknin-Dembinsky, A.; Rubertsson, S.; Nellgård, B.; Blennow, K.; Zetterberg, H.; et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc. Natl. Acad. Sci. USA 2016, 113, E1826–E1834. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Jiang, P.; Chan, K.; Wong, J.; Cheng, Y.; Liang, R.; Chan, W.K.; Ma, E.; Chan, S.; Cheng, S.; et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc. Natl. Acad. Sci. USA 2015, 112, E5503–E5512. [Google Scholar] [CrossRef]
- Imperiale, T.F.; Ransohoff, D.F.; Itzkowitz, S.H.; Levin, T.R.; Lavin, P.; Lidgard, G.P.; Ahlquist, D.A.; Berger, B.M. Multitarget Stool DNA Testing for Colorectal-Cancer Screening. N. Engl. J. Med. 2014, 370, 1287–1297. [Google Scholar] [CrossRef]
- Gahan, P.B.; Swaminathan, R. Circulating Nucleic Acids in Plasma and Serum. Ann. N. Y. Acad. Sci. 2008, 1137, 1–6. [Google Scholar] [CrossRef]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.; Hesch, R.D.; Knippers, R. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin from Apoptotic and Necrotic Cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Lo, Y.; Rainer, T.; Chan, L.; Hjelm, N.; Cocks, R. Plasma DNA as a Prognostic Marker in Trauma Patients. Clin. Chem. 2000, 46, 319–323. [Google Scholar]
- Rainer, T.; Wong, L.; Lam, W.; Yuen, E.; Lam, N.; Metreweli, C.; Lo, Y. Prognostic Use of Circulating Plasma Nucleic Acid Concentrations in Patients with Acute Stroke. Clin. Chem. 2003, 49, 562–569. [Google Scholar] [CrossRef]
- Atamaniuk, J.; Vidotto, C.; Kinzlbauer, M.; Bachl, N.; Tiran, B.; Tschan, H. Cell-free plasma DNA and purine nucleotide degradation markers following weightlifting exercise. Eur. J. Appl. Physiol. 2010, 110, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Alborelli, I.; Generali, D.; Jermann, P.; Cappelletti, M.R.; Ferrero, G.; Scaggiante, B.; Bortul, M.; Zanconati, F.; Nicolet, S.; Haegele, J.; et al. Cell-free DNA analysis in healthy individuals by next-generation sequencing: A proof of concept and technical validation study. Cell Death Dis. 2019, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1324. [Google Scholar] [CrossRef] [PubMed]
- Karampini, E.; McCaughan, F. Circulating DNA in solid organ cancers-analysis and clinical application. QJM Int. J. Med. 2015, 109, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Fleischhacker, M.; Rabien, A. Cell-free DNA in the blood as a solid tumor biomarker—A critical appraisal of the literature. Clin. Chim. Acta 2010, 411, 1611–1624. [Google Scholar] [CrossRef]
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim. Biophys. Acta (BBA) Rev. Cancer 2007, 1775, 181–232. [Google Scholar] [CrossRef]
- Giacona, M.B. Cell-Free DNA in Human Blood Plasma: Length Measurements in Patients with Pancreatic Cancer and Healthy Controls. Pancreas 1998, 17, 89–97. [Google Scholar] [CrossRef]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.; Goodman, S.; David, K.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef]
- Tada, M. Detection or ras Gene Mutations in Pancreatic Juice and Peripheral Blood of Patients with Pancreatic Adenocarcinoma. Cancer Res. 1993, 53, 2472–2474. [Google Scholar]
- Adamo, P.; Cowley, C.M.; Neal, C.P.; Mistry, V.; Page, K.; Dennison, A.R.; Isherwood, J.; Hastings, R.; Luo, J.; Moore, D.A.; et al. Profiling tumor heterogeneity through circulating tumor DNA in patients with pancreatic cancer. Oncotarget 2017, 8, 87221. [Google Scholar] [CrossRef] [PubMed]
- Le Calvez-Kelm, F.; Foll, M.; Wozniak, M.B.; Delhomme, T.M.; Durand, G.; Chopard, P.; Pertesi, M.; Fabianova, E.; Adamcakova, Z.; Holcatova, I.; et al. KRAS mutations in blood circulating cell-free DNA: A pancreatic cancer case-control. Oncotarget 2016, 7, 78827. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.; Wang, H.; Luber, B.; Alani, R.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed]
- Gall, T.M.; Belete, S.; Khanderia, E.; Frampton, A.E.; Jiao, L.R. Circulating Tumor Cells and Cell-Free DNA in Pancreatic Ductal Adenocarcinoma. Am. J. Pathol. 2019, 189, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2017, 8, a031435. [Google Scholar] [CrossRef]
- Bailey, V.; Zhang, Y.; Keeley, B.; Yin, C.; Pelosky, K.; Brock, M.; Baylin, S.; Herman, J.; Wang, T.H. Single-Tube Analysis of DNA Methylation with Silica Superparamagnetic Beads. Clin. Chem. 2010, 56, 1022–1025. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Schwarzenbach, H.; Pantel, K. Circulating Tumor Cells and Circulating Tumor DNA. Annu. Rev. Med. 2012, 63, 199–215. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef]
- Jamal-Hanjani, M.; Wilson, G.A.; Horswell, S.; Mitter, R.; Sakarya, O.; Constantin, T.; Salari, R.; Kirkizlar, E.; Sigurjonsson, S.; Pelham, R.; et al. Detection of ubiquitous and heterogeneous mutations in cell-free DNA from patients with early-stage non-small-cell lung cancer. Ann. Oncol. 2016, 27, 862–867. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.; Zhou, S.; Diaz, L.; Kinzler, K. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef]
- Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; Ziller, M.J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, X.; Lu, X.; You, L.; Song, Y.; Luo, Z.; Zhang, J.; Nie, J.; Zheng, W.; Xu, D.; et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017, 27, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.D.; Wu, H.; Li, Y.; Liu, X.; Zhang, Z.; Yu, L.; Qin, Z.; Su, Z.; Liu, R.; He, Q.; et al. 686P Early detection of pancreatic ductal adenocarcinoma using methylation signatures in circulating tumor DNA. Ann. Oncol. 2019, 30, mdz247.013. [Google Scholar] [CrossRef]
- Liggett, T.; Melnikov, A.; Yi, Q.L.; Replogle, C.; Brand, R.; Kaul, K.; Talamonti, M.; Abrams, R.A.; Levenson, V. Differential methylation of cell-free circulating DNA among patients with pancreatic cancer versus chronic pancreatitis. Cancer 2010, 116, 1674–1680. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, A.A.; Scholtens, D.; Talamonti, M.S.; Bentrem, D.J.; Levenson, V.V. Methylation profile of circulating plasma DNA in patients with pancreatic cancer. J. Surg. Oncol. 2009, 99, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Melnikov, A.A.; Scholtens, D.M.; Wiley, E.L.; Khan, S.A.; Levenson, V.V. Array-Based Multiplex Analysis of DNA Methylation in Breast Cancer Tissues. J. Mol. Diagnost. 2008, 10, 93–101. [Google Scholar] [CrossRef]
- Park, J.W.; Baek, I.H.; Kim, Y.T. Preliminary Study Analyzing the Methylated Genes in the Plasma of Patients with Pancreatic Cancer. Scand. J. Surg. 2012, 101, 38–44. [Google Scholar] [CrossRef]
- Dammann, R.; Schagdarsurengin, U.; Liu, L.; Otto, N.; Gimm, O.; Dralle, H.; Boehm, B.O.; Pfeifer, G.P.; Hoang-Vu, C. Frequent RASSF1A promoter hypermethylation and K-ras mutations in pancreatic carcinoma. Oncogene 2003, 22, 3806–3812. [Google Scholar] [CrossRef]
- Park, J.K.; Ryu, J.K.; Yoon, W.J.; Lee, S.H.; Lee, G.Y.; Jeong, K.S.; Kim, Y.T.; Yoon, Y.B. The Role of Quantitative NPTX2 Hypermethylation as a Novel Serum Diagnostic Marker in Pancreatic Cancer. Pancreas 2012, 41, 95–101. [Google Scholar] [CrossRef]
- Singh, N.; Gupta, S.; Rashid, S.S.; Rashid, S.; Dash, N.R.; Saraya, A. Tu2033 – Quantitation of Methylation Load of Tumor Suppressor Gene Promoter Methylation in Pancreatic Cancer. Gastroenterology 2019, 156, S-1176. [Google Scholar] [CrossRef]
- Eissa, M.A.L.; Lerner, L.; Abdelfatah, E.; Shankar, N.; Canner, J.K.; Hasan, N.M.; Yaghoobi, V.; Huang, B.; Kerner, Z.; Takaesu, F.; et al. Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin. Epigenet. 2019, 11. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, S.D.; Madsen, P.H.; Larsen, A.C.; Johansen, M.B.; Drewes, A.M.; Pedersen, I.S.; Krarup, H.; Thorlacius-Ussing, O. Cell-free DNA promoter hypermethylation in plasma as a diagnostic marker for pancreatic adenocarcinoma. Clin. Epigenet. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Kinugawa, Y.; Uehara, T.; Sano, K.; Matsuda, K.; Maruyama, Y.; Kobayashi, Y.; Nakajima, T.; Hamano, H.; Kawa, S.; Higuchi, K.; et al. Methylation of Tumor Suppressor Genes in Autoimmune Pancreatitis. Pancreas 2017, 46, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Dam Henriksen, S.; Henning Madsen, P.; Christian Larsen, A.; Berg Johansen, M.; Søkilde Pedersen, I.; Krarup, H.; Thorlacius-Ussing, O. Cell-free DNA promoter hypermethylation in plasma as a predictive marker for survival of patients with pancreatic adenocarcinoma. Oncotarget 2017, 8. [Google Scholar] [CrossRef]
- Guo, S.; Diep, D.; Plongthongkum, N.; Fung, H.L.; Zhang, K.; Zhang, K. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 2017, 49, 635–642. [Google Scholar] [CrossRef]
- Fernández-Carballo, B.; Broger, T.; Wyss, R.; Banaei, N.; Denkinger, C. Toward the Development of a Circulating Free DNA-Based In Vitro Diagnostic Test for Infectious Diseases: A Review of Evidence for Tuberculosis. J. Clin. Microbiol. 2019, 57. [Google Scholar] [CrossRef]
- Hyun, K.A.; Gwak, H.; Lee, J.; Kwak, B.; Jung, H.I. Salivary Exosome and Cell-Free DNA for Cancer Detection. Micromachines 2018, 9, 340. [Google Scholar] [CrossRef]
- Humeau, M.; Vignolle-Vidoni, A.; Sicard, F.; Martins, F.; Bournet, B.; Buscail, L.; Torrisani, J.; Cordelier, P. Salivary MicroRNA in Pancreatic Cancer Patients. PLoS ONE 2015, 10, e0130996. [Google Scholar] [CrossRef]
- Su, Q.; Zhu, E.C.; Qu, Y.L.; Wang, D.Y.; Qu, W.W.; Zhang, C.G.; Wu, T.; Gao, Z.H. Serum level of co-expressed hub miRNAs as diagnostic and prognostic biomarkers for pancreatic ductal adenocarcinoma. J. Cancer 2018, 9, 3991–3999. [Google Scholar] [CrossRef]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenet. 2018, 10. [Google Scholar] [CrossRef]
- Pero, R.; Angrisano, T.; Brancaccio, M.; Falanga, A.; Lombardi, L.; Natale, F.; Laneri, S.; Lombardo, B.; Galdiero, S.; Scudiero, O. Beta-defensins and analogs in Helicobacter pylori infections: mRNA expression levels, DNA methylation, and antibacterial activity. PLoS ONE 2019, 14, e0222295. [Google Scholar] [CrossRef] [PubMed]
- Angrisano, T.; Pero, R.; Brancaccio, M.; Coretti, L.; Florio, E.; Pezone, A.; Calabrò, V.; Falco, G.; Keller, S.; Lembo, F.; et al. Cyclical DNA Methylation and Histone Changes Are Induced by LPS to Activate COX-2 in Human Intestinal Epithelial Cells. PLoS ONE 2016, 11, e0156671. [Google Scholar] [CrossRef] [PubMed]

| Method | |||||
|---|---|---|---|---|---|
| Features | MethDet-M3 | MSP | QMSP | Targeted Amplicon Sequencing | RRBS |
| DNA methylation discrimination | Methylation-sensitive endonuclease | Bisulfite Treatment | Bisulfite Treatment | Bisulfite Treatment | Bisulfite Treatment |
| Coverage | Medium/Low | Targeted | Targeted | Targeted | Medium |
| Throughput | Medium | Low | Medium | Medium/High | High |
| Analytical Sensitivity | Medium | Low | Medium | Low | Low |
| Analytical Specificity | High | Low/Medium | Medium | Medium | Medium |
| Advantages | Scalable (e.g., microarray) | Fast results Cost-effective | Quantitative | Scalable (e.g., NGS) Single CpG methylation status | Scalable (e.g., NGS) Single CpG methylation status |
| Disadvantages | Risk of false positive/negative | Limited coverage | Limited coverage | May require extensive data analysis | Extensive data analysis |
| Popularity | Unpopular | Very Popular | Popular | Popular | Popular |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brancaccio, M.; Natale, F.; Falco, G.; Angrisano, T. Cell-Free DNA Methylation: The New Frontiers of Pancreatic Cancer Biomarkers’ Discovery. Genes 2020, 11, 14. https://doi.org/10.3390/genes11010014
Brancaccio M, Natale F, Falco G, Angrisano T. Cell-Free DNA Methylation: The New Frontiers of Pancreatic Cancer Biomarkers’ Discovery. Genes. 2020; 11(1):14. https://doi.org/10.3390/genes11010014
Chicago/Turabian StyleBrancaccio, Mariarita, Francesco Natale, Geppino Falco, and Tiziana Angrisano. 2020. "Cell-Free DNA Methylation: The New Frontiers of Pancreatic Cancer Biomarkers’ Discovery" Genes 11, no. 1: 14. https://doi.org/10.3390/genes11010014
APA StyleBrancaccio, M., Natale, F., Falco, G., & Angrisano, T. (2020). Cell-Free DNA Methylation: The New Frontiers of Pancreatic Cancer Biomarkers’ Discovery. Genes, 11(1), 14. https://doi.org/10.3390/genes11010014