Advances of Molecular Markers and Their Application for Body Variables and Carcass Traits in Qinchuan Cattle
Abstract
1. Introduction
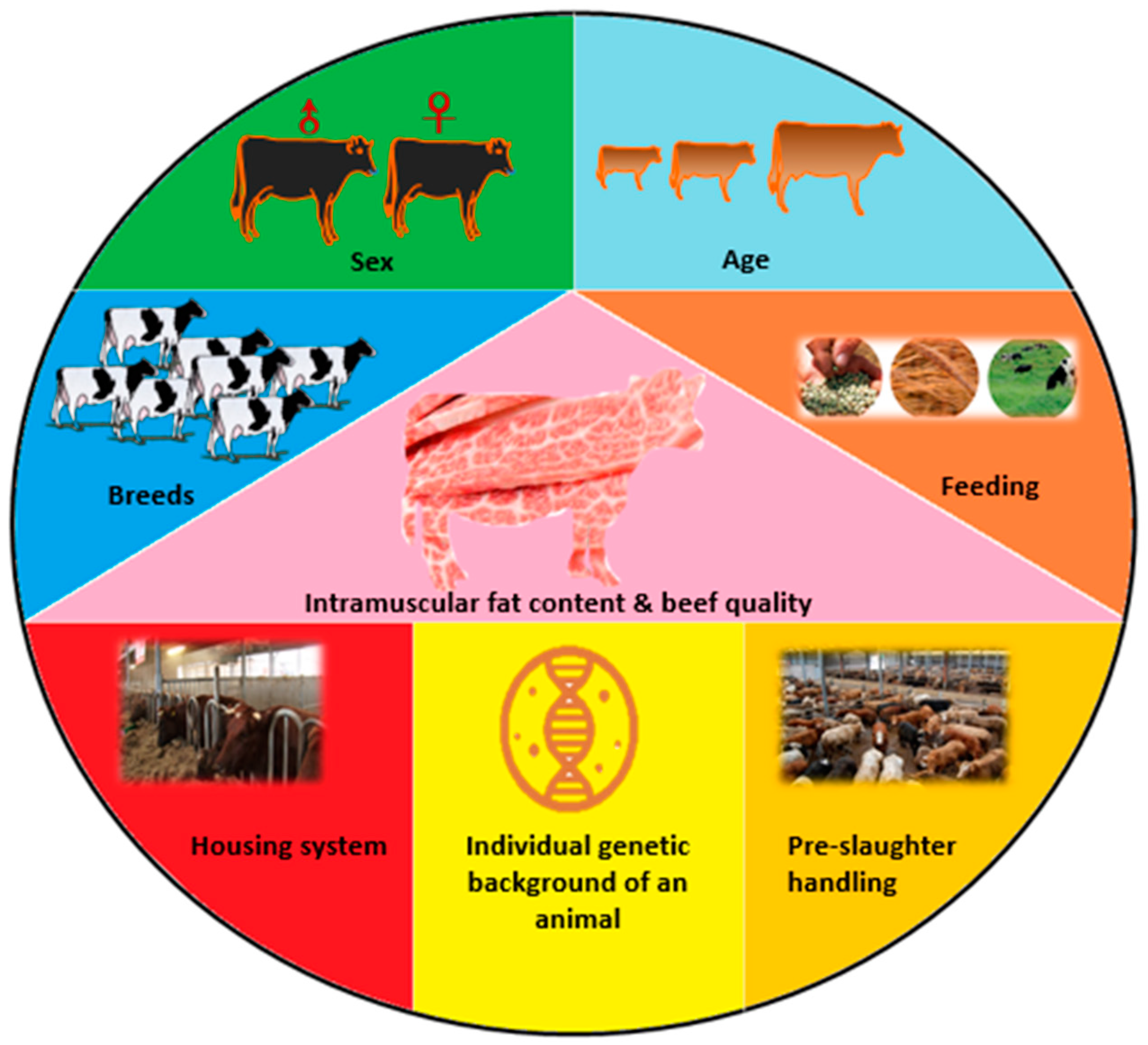
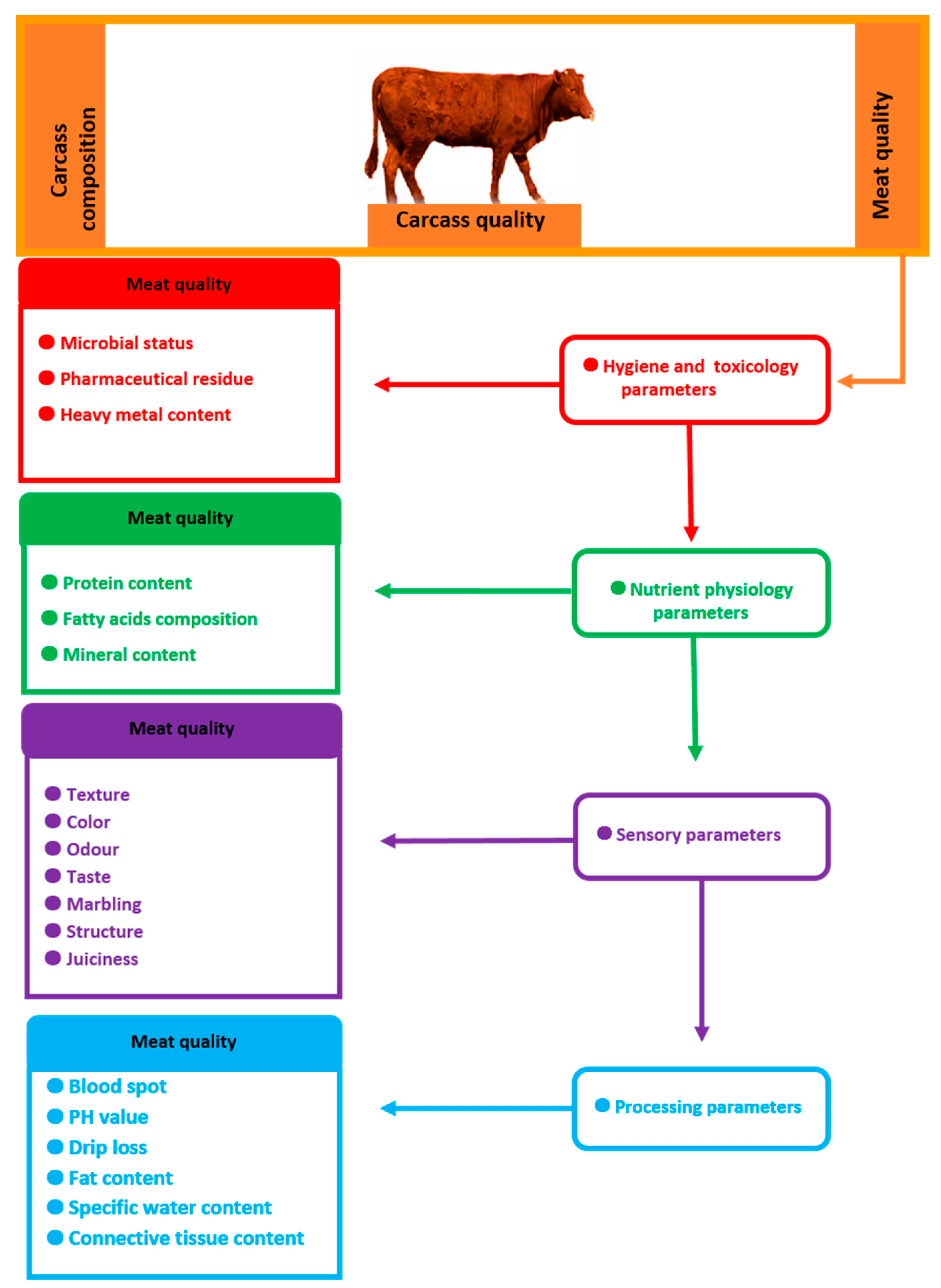
2. Effect of Some Slaughter Value Factors on Meat Quality
3. Most Important Beef Quality Traits
4. Gene Polymorphism vs. Slaughter Value and Beef Quality
4.1. The SIRT4 Gene
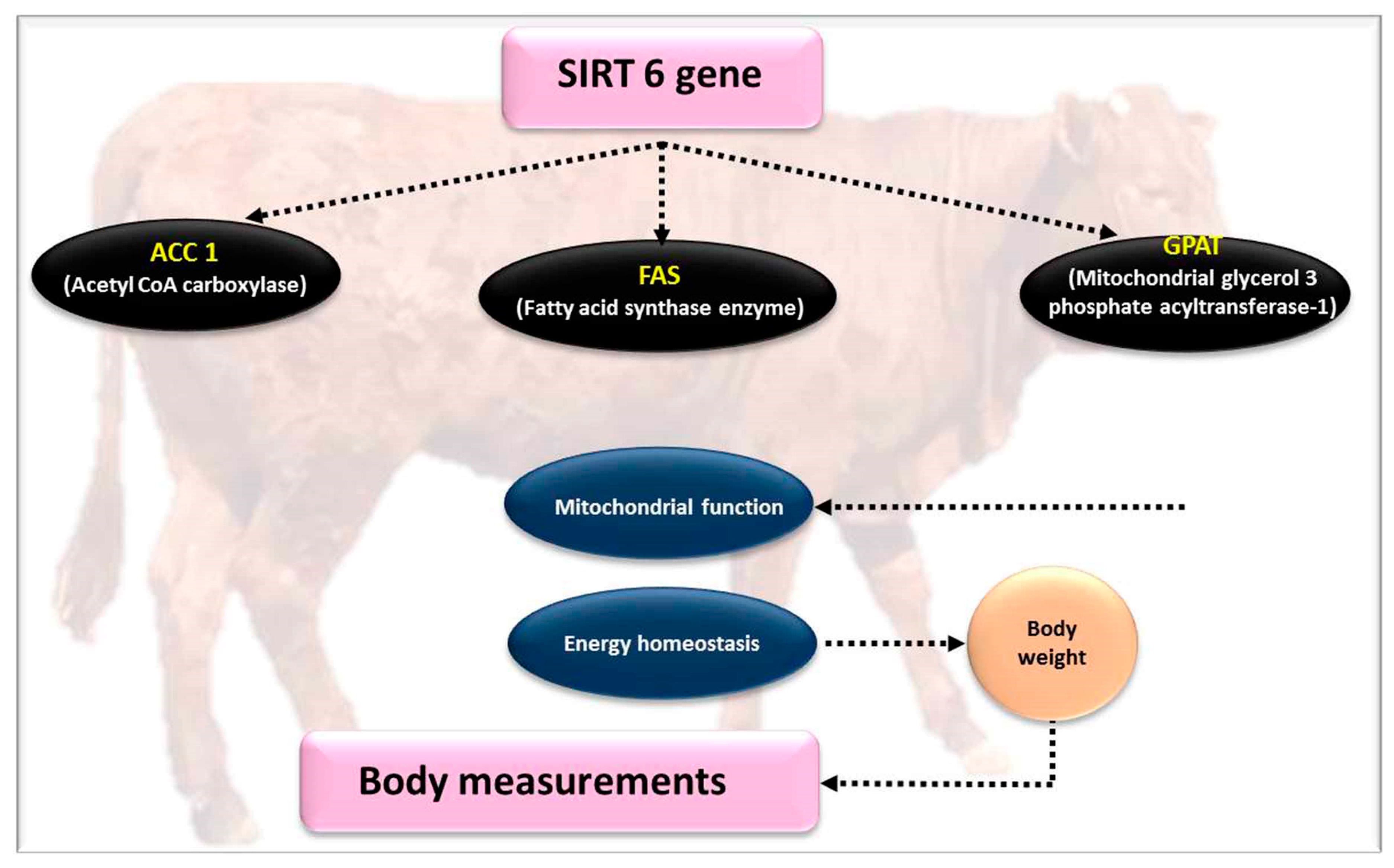
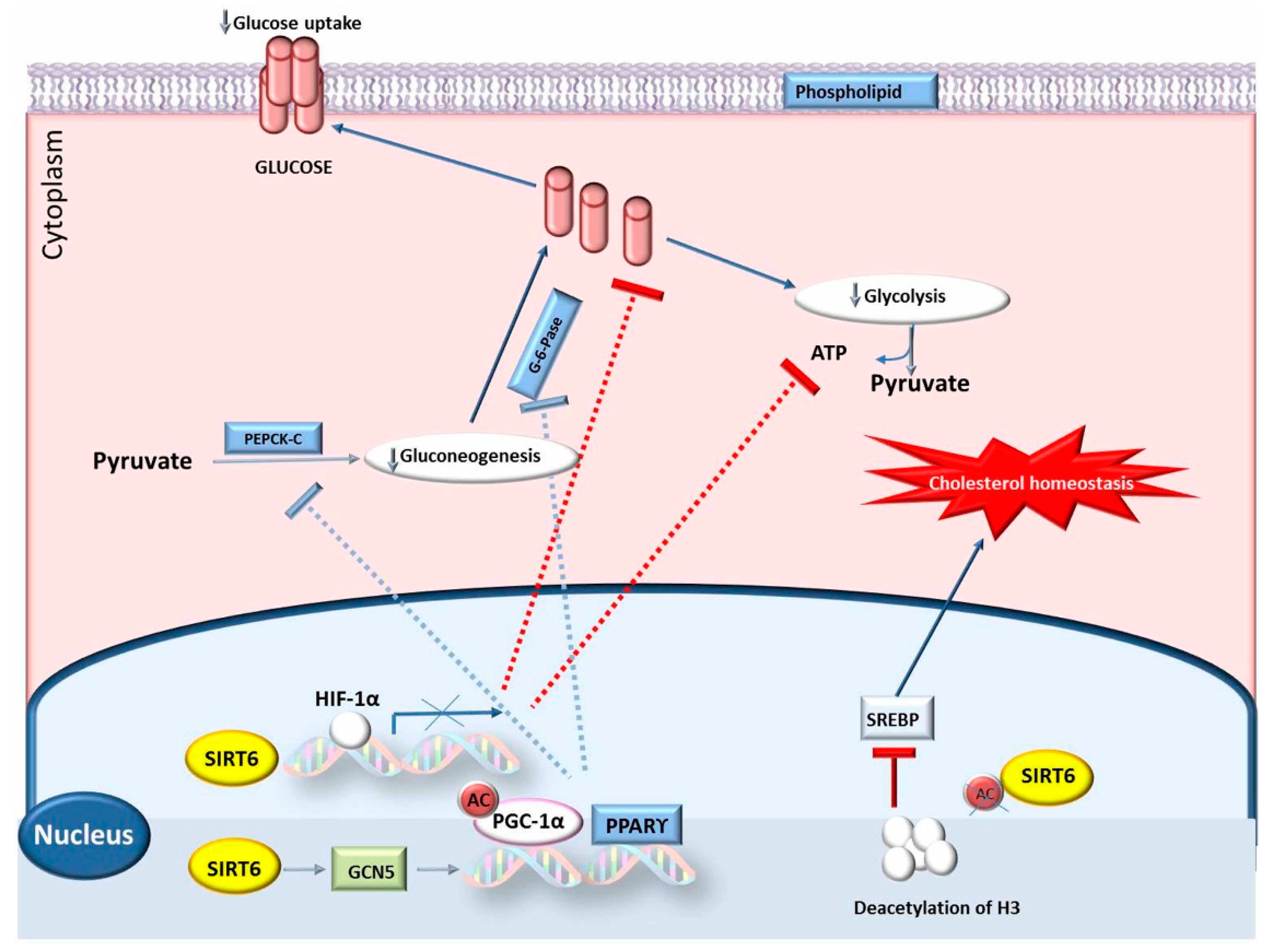
4.2. SIRT6 Gene
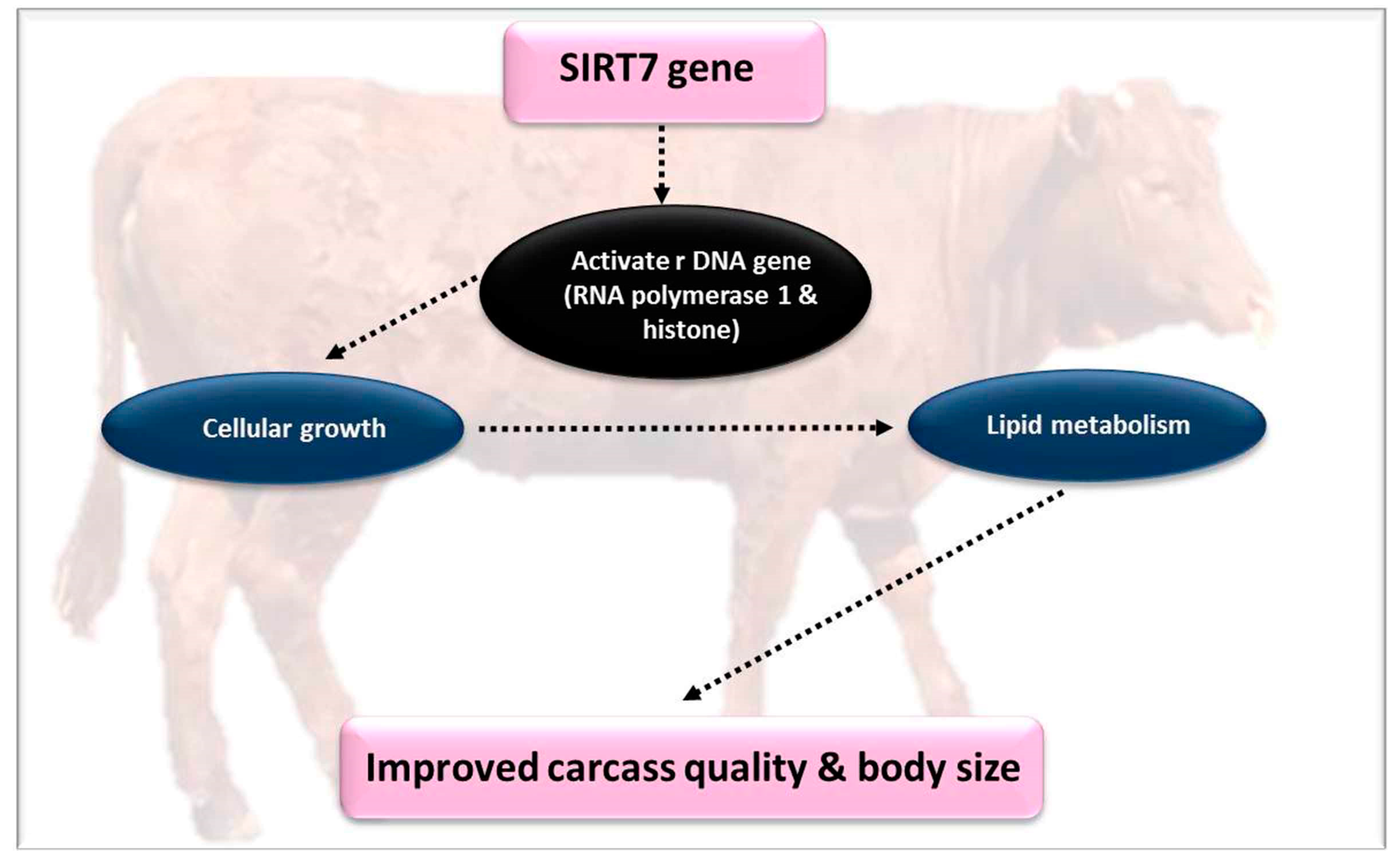
4.3. Silent Information Regulators 7 (SIRT7)
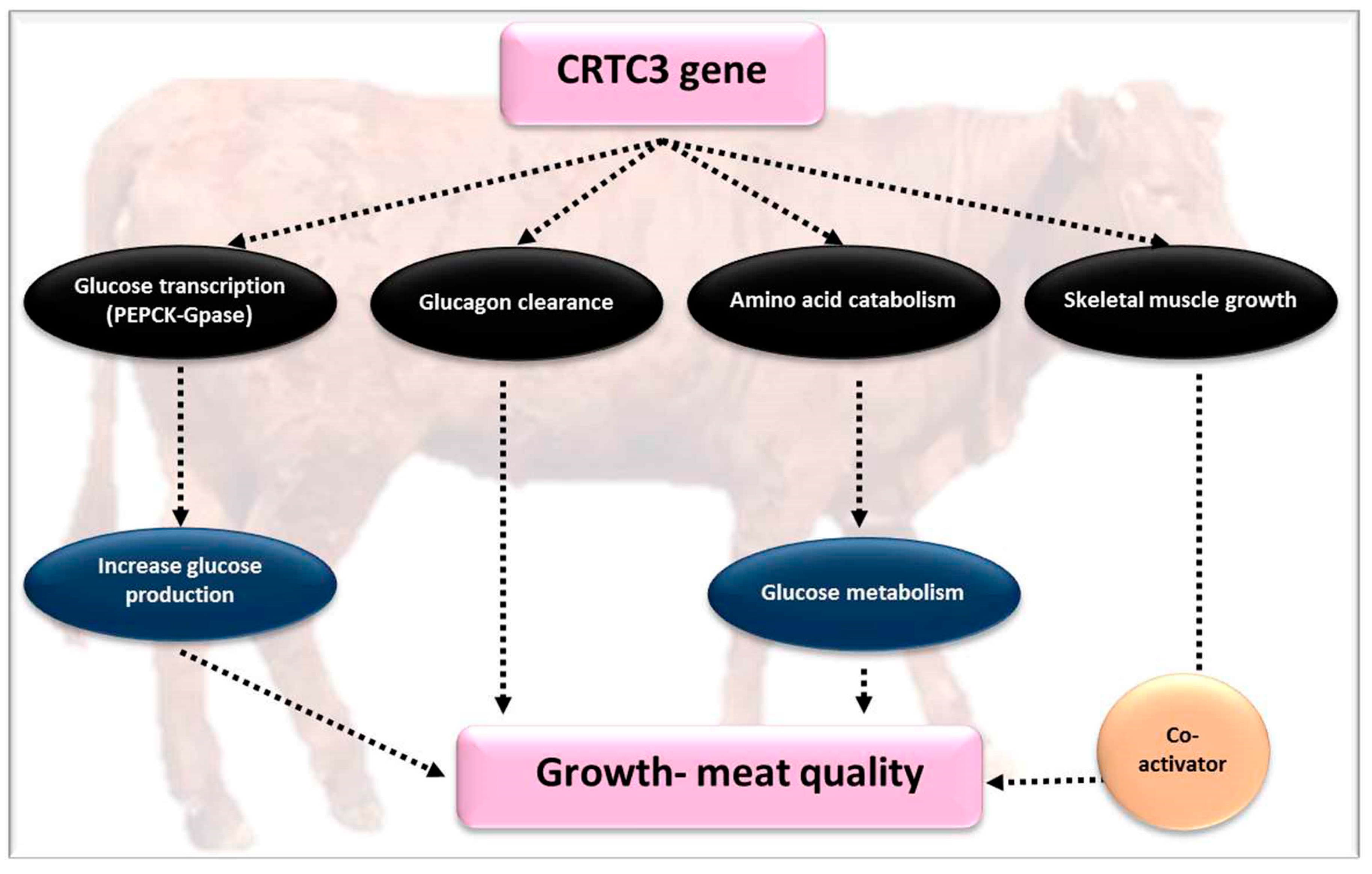
4.4. CREB-Regulated Transcription Coactivator 3 (CRTC3)
4.5. The α/β Hydrolase Domain-Containing Protein 5 (ABHD5)
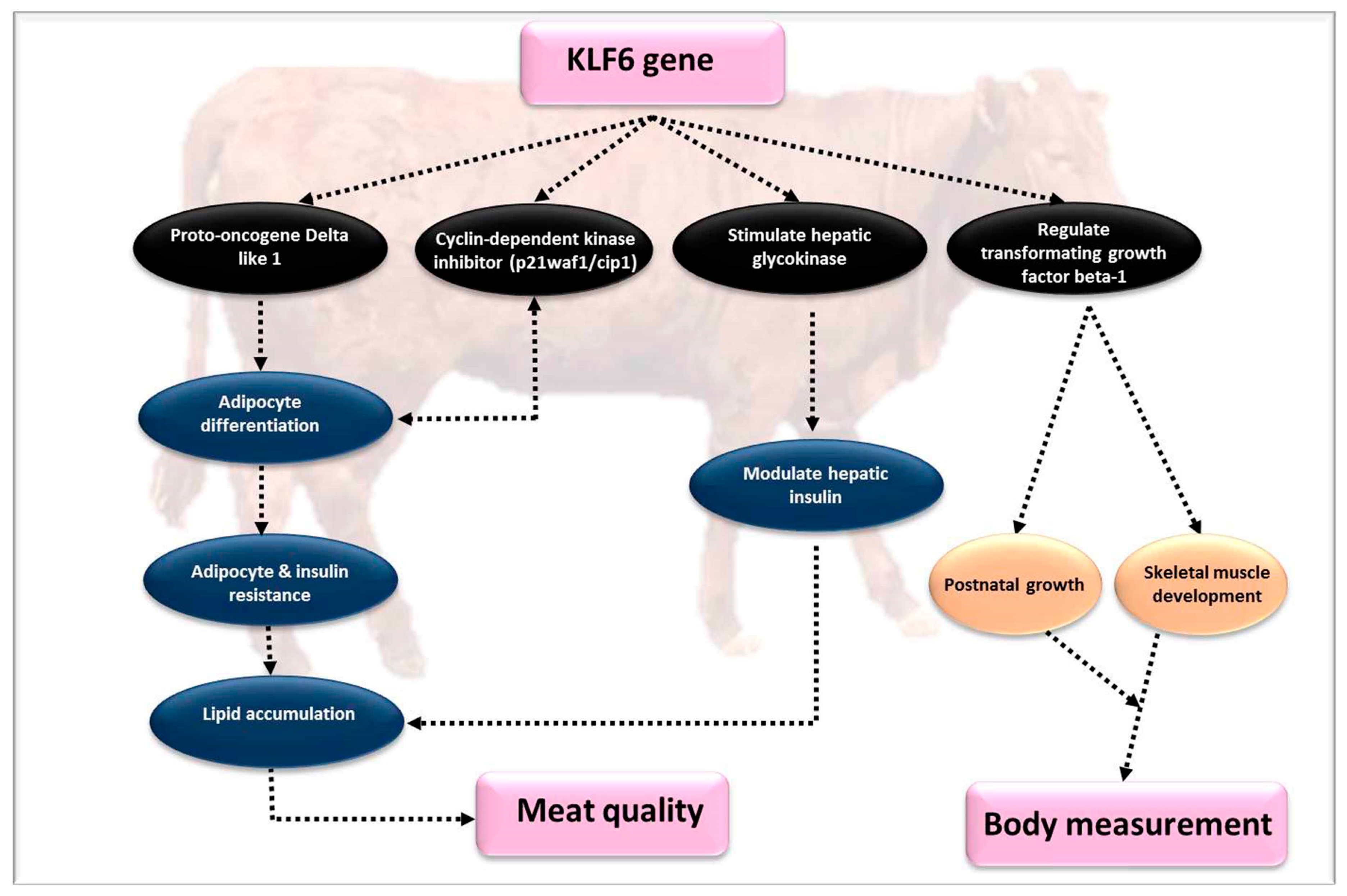
4.6. Kruppel-Like Factor 6 (KLF6)
4.7. Heart Type Fatty Acid Binding Protein (H-FABP)
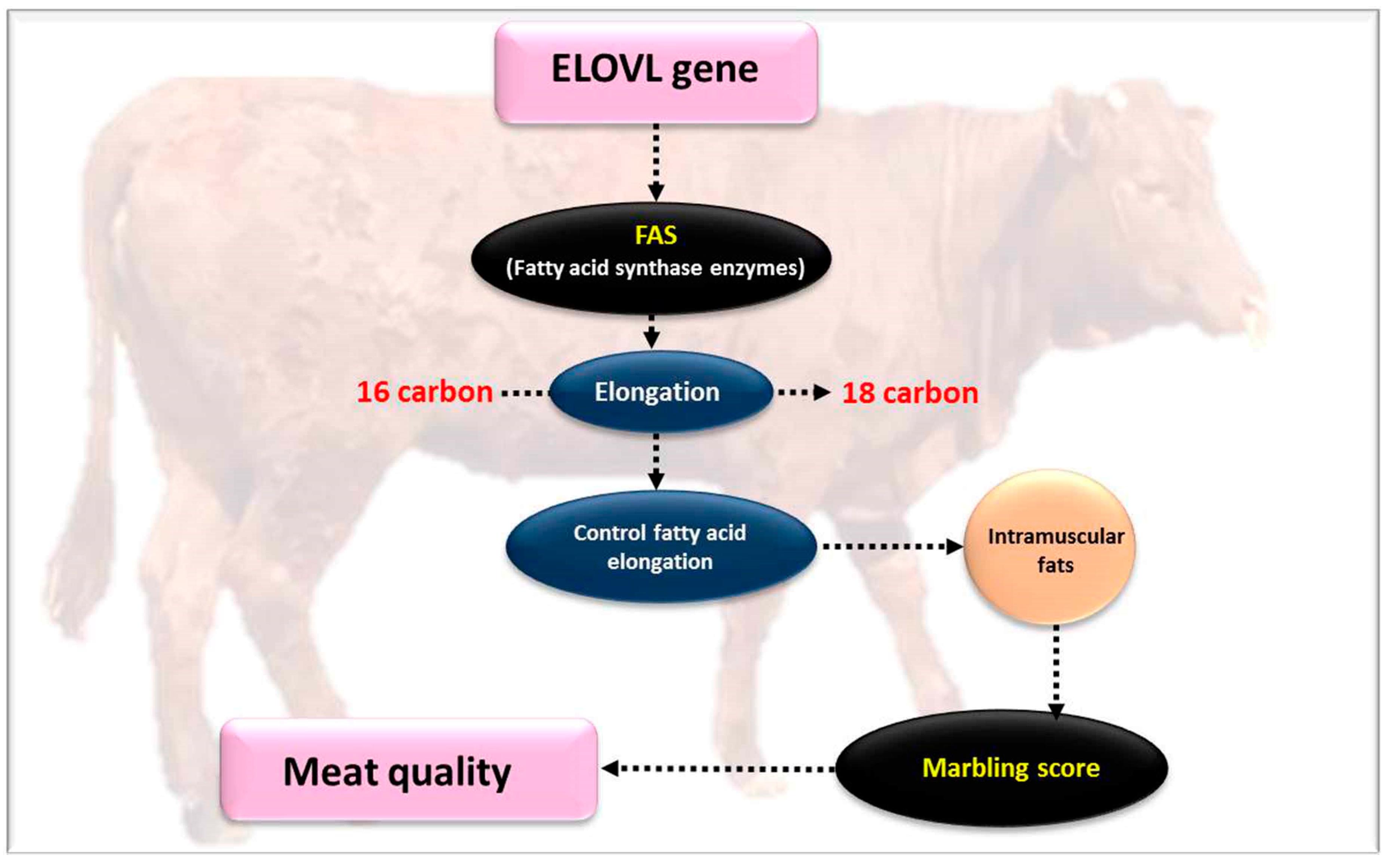
4.8. Very Long Chain Fatty Acids Protein 5 (ELOVL5)
4.9. Very Long Chain Fatty Acids Protein 6 (ELOVL6)
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Q&A on the Carcinogenicity of the Consumption of Red Meat and Processed Meat. Available online: https://www.who.int/features/qa/cancer-red-meat/en/ (accessed on 7 August 2019).
- Abd El-Hack, M.E.; Abdelnour, S.A.; Swelum, A.A.; Arif, M. The application of gene marker-assisted selection and proteomics for the best meat quality criteria and body measurements in Qinchuan cattle breed. Mol. Boil. Rep. 2018, 45, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.H.A.; Khan, R.; Schreurs, N.M.; Guo, H.; Gui, L.S.; Mei, C.; Zan, L. Expression of the bovine KLF6 gene polymorphisms and their association with carcass and body measures in Qinchuan cattle (Bos Taurus). Genomic 2019. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, L.D.; Sørensen, A.C.; Berg, P. Marker-assisted selection can reduce true as well as pedigree-estimated inbreeding. J. Dairy Sci. 2009, 92, 2214–2223. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.H.A.; Gui, L.; Khan, R.; Schreurs, N.M.; Wang, X.; Wu, C.; Mei, L.; Wang, X.; Ma, D.; Wei, H.; et al. Association between FASN gene polymorphisms ultrasound carcass traits and intramuscular fat in Qinchuan cattle. Gene 2018, 645, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Boucher, C.A.; Carey, N.; Edwards, Y.H.; Siciliano, M.J.; Johnson, K.J. Cloning of the human SIX1 gene and its assignment to chromosome 14. Genomics 1996, 33, 140–142. [Google Scholar] [CrossRef] [PubMed]
- D’Andre Hirwa, C.; Wallace, P.; Shen, X.; Nie, Q.; Yang, G.; Zhang, X. Genes related to economically important traits in beef cattle. Asian J. Anim. Sci. 2011, 5, 34–45. [Google Scholar]
- Boukha, A.; Bonfatti, V.; Cecchinato, A.; Albera, A.; Gallo, L.; Carnier, P.; Bittante, G. Genetic parameters of carcass and meat quality traits of double muscled Piemontese cattle. Meat Sci. 2011, 89, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.U.; Zhang, R.; Hu, X.; Li, N. Application of genomic technologies to the improvement of meat quality of farm animals. Meat Sci. 2007, 77, 36–45. [Google Scholar] [CrossRef]
- Ribeca, C.; Bonfatti, V.; Cecchinato, A.; Albera, A.; Gallo, L.; Carnier, P. Effect of polymorphisms in candidate genes on carcass and meat quality traits in double muscled Piemontese cattle. Meat Sci. 2014, 96, 1376–1383. [Google Scholar] [CrossRef]
- Gui, L.; Wu, H.; Raza, S.H.A.; Schreurs, N.M.; Shah, M.A. The effect of haplotypes in the promoter region of SIRT4 gene on the ultrasound traits in Qinchuan cattle. Trop. Anim. Health Prod. 2019, 1–6. [Google Scholar] [CrossRef]
- Luft, F.C. Are you certain about SIRT? J. Mol. Med. 2014, 92, 305–306. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Silberman, D.M.; Ross, K.; Sande, P.H.; Kubota, S.; Ramaswamy, S.; Apte, R.S.; Mostoslavsky, R. SIRT6 is required for normal retinal function. PLoS ONE 2014, 9, e98831. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kazgan, N. Mammalian sirtuins and energy metabolism. Int. J. Biol. Sci. 2011, 7, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Xin, X.; Wang, J.; Hong, J.; Zan, L. Expression analysis, single nucleotide polymorphisms within SIRT4 and SIRT7 genes and their association with body size and meat quality traits in Qinchuan cattle. J. Integr. Agric. 2016, 15, 2819–2826. [Google Scholar] [CrossRef]
- Wang, X.; Khana, R.; Razaa, S.H.A.; Lia, A.; Zhanga, Y.; Lianga, C.; Yanga, W.; Wua, S.; Zan, L. Molecular characterization of ABHD5 gene promoter in intramuscular preadipocytes of Qinchuan cattle: Roles of Evi1 and C/EBP α. Gene 2019, 690, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Junjvlieke, Z.; Mei, C.G.; Khan, R.; Zhang, W.Z.; Hong, J.Y.; Wang, L.; Zan, L.S. Transcriptional regulation of bovine elongation of very long chain fatty acids protein 6 in lipid metabolism and adipocyte proliferation. J. Cell Biochem. 2019, 120, 13932–13943. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Bach, A.; Velarde, A.; Devant, M. Association between animal, transportation, slaughterhouse practices, and meat pH in beef. Meat Sci. 2008, 78, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Węglarz, A. Effect of pre-slaughter housing of different cattle categories on beef quality. Anim. Sci. Pap. Rep. 2011, 29, 43–52. [Google Scholar]
- Chambaz, A.; Scheeder, M.R.L.; Kreuzer, M.; Dufer, P.A. Meat quality of Angus, Simmental, Charolais and Limousin steers compared at the same level of intramuscular fat. Meat Sci. 2003, 63, 491–500. [Google Scholar] [CrossRef]
- Keane, M.G.; Dunne, P.G.; Kenny, D.A.; Berry, D.P. Effect of genetic merit for carcass weight, breed type and slaughter weight on performance and carcass traits of beef x diary steers. Animal 2011, 5, 182–194. [Google Scholar] [CrossRef]
- Magolski, J.D.; Buchanan, D.S.; Maddock-Carlin, K.R.; Anderson, V.L.; Newman, D.J.; Berg, E.P. Relationship between commercially available DNA analysis and phenotypic observations on beef quality and tenderness. Meat Sci. 2013, 95, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.S. Trends in skeletal muscle biology and underestanding of toughness in beef. Aust. J. Agric. Res. 1999, 50, 1105–1129. [Google Scholar] [CrossRef]
- Juszczuk-Kubiak, E.; Rosochacki, S.J.; Wicińska, K.; Słoniewski, K.; Łyczyński, A.; Sakowski, T. Association of the SNP in the 3’-UTR region of the small subunit of bovine calcium-activated neutral protease (CAPN1S) with the activity of the calpain II and meat quality traits of Fresian bulls. Eurobiotech I Międzynarodowa Konferencja oraz Targi. Biotechnologia w Rolnictwie 2007, 4, 25–27. [Google Scholar]
- Resurreccion, A.V.A. Sensory aspects of consumer choices for meat and meat products. Meat Sci. 2004, 66, 11–20. [Google Scholar] [CrossRef]
- Purslow, P.P. Intramuscular connective tissue and its role in meat quality. Meat Sci. 2005, 70, 435447. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.C.; Savell, J. Perimysium thickness as an indicator of beef tenderness. Meat Sci. 2004, 67, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Kołczak, T.; Pałka, K.; Pośpiech, E. Changes in collagen solubility of raw and roasted bovine psoas major and minor and semitendinosus muscles during cold storage. Pol. J. Food Nutr. Sci. 2003, 12, 57–61. [Google Scholar]
- Maddock, K.R.; Huff-Lonergan, E.; Rowe, L.J.; Lonergan, S.M. Effect of pH and ionic strength on μ-and m-calpain inhibition by calpastatin. J. Anim. Sci. 2005, 83, 1370–1376. [Google Scholar] [CrossRef] [PubMed]
- Allais, S.; Journaux, L.; Levéziel, H.; Payet-Duprat, N.; Raynaud, P.; Hocquette, J.F.; Lepetit, J.; Rousset, S.; Denoyelle, C.; Bernard-Capel, C.; et al. Effects of polymorphisms in the calpastatin and μ-calpain genes on meat tenderness in 3 French beef breeds. J. Anim. Sci. 2011, 89, 1–11. [Google Scholar] [CrossRef]
- Reardon, W.; Mullen, A.M.; Sweeney, T.; Hamill, R.M. Association of polymorphisms in candidate genes with colour, water-holding capacity, and composition traits in bovine m. longissimus and m. semimembranosus. Meat Sci. 2010, 86, 270–275. [Google Scholar] [CrossRef]
- Laurent, G.; German, N.J.; Saha, A.K.; de Boer, V.C.J.; Davies, M.; Koves, T.R.; Dephoure, N.; Fischer, F.; Boanca, G.; Vaitheesvaran, B. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol. Cell 2013, 50, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Nasrin, N.; Wu, X.; Fortier, E.; Feng, Y.; Bare, O.C.; Chen, S.; Ren, X.; Wu, Z.; Streeper, R.S.; Bordone, L. SIRT 4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J. Biol. Chem. 2010, 285, 31995–32002. [Google Scholar] [CrossRef] [PubMed]
- Laurent, G.; de Boer, V.C.J.; Finley, L.W.S.; Sweeney, M.; Lu, H.; Schug, T.T.; Cen, Y.; Jeong, S.M.; Li, X.; Sauve, A.A. SIRT4 represses PPARα activity to suppress hepatic fat oxidation. Mol. Cell Biol. 2013, 33, 4552–4561. [Google Scholar] [CrossRef] [PubMed]
- Braidy, N.; Poljak, A.; Grant, R.; Jayasena, T.; Mansour, H.; Chan-Ling, T.; Smythe, G.; Sachdev, P.; Guillemin, G.J. Differential expression of sirtuins in the aging rat brain. Front. Cell Neurosci. 2015, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Michishita, E.; McCord, R.A.; Berber, E.; Kioi, M.; Padilla-Nash, H.; Damian, M.; Chang, H.Y. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452, 492. [Google Scholar] [CrossRef] [PubMed]
- Elhanati, S.; Kanfi, Y.; Varvak, A.; Roichman, A.; Carmel-Gross, I.; Barth, S.; Cohen, H.Y. Multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep. 2013, 4, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Jiang, B.; Zhang, Y.; Zan, L. Sequence variants in the bovine silent information regulator 6, their linkage and their associations with body measurements and carcass quality traits in Qinchuan cattle. Gene 2015, 559, 16–21. [Google Scholar] [CrossRef]
- Gui, L.S.; Raza, S.H.A.; Garcia, M.; Sun, Y.G.; Ullah, I.R.; Han, Y.C. Genetic variants in the SIRT6 transcriptional regulatory region affect gene activity and carcass quality traits in indigenous Chinese beef cattle (Bos taurus). BMC Genom. 2018, 19, 785. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, X.; Zang, R.; Cai, Y.; Cao, X.; Yang, J.; Li, J.; Lan, X.; Wu, J. Genetic variations in the sheep SIRT7 gene and their correlation with body size traits. Arch. Anim. Breed 2019, 62, 189–197. [Google Scholar] [CrossRef]
- Yamagata, K.; Yoshizawa, T. Transcriptional regulation of metabolism by SIRT1 and SIRT7. Int. Rev. Cell Mol. Biol. 2018, 335, 143–166. [Google Scholar]
- Shin, J.; He, M.; Liu, Y.; Paredes, S.; Villanova, L.; Brown, K.; Qiu, X.; Nabavi, N.; Mohrin, M.; Wojnoonski, K.; et al. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. 2013, 5, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Xiong, J.; Zhan, J.; Yuan, F.; Tang, M.; Zhang, C.; Cao, Z.; Chen, Y.; Lu, X.; Li, Y.; et al. Ubiquitin-specific peptidase 7 (USP7)-mediated deubiquitination of the histone deacetylase SIRT7 regulates gluconeogenesis. J. Biol. Chem. 2017, 292, 13296–13311. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Li, M.T.; Hou, T.Y.; Gao, T.; Zhu, W.G.; Yang, Y. Sirtuins in glucose and lipid metabolism. Oncotarget 2017, 8, 1845–1859. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.L. SIRT7 and hepatic lipid metabolism. Front. Cell. Dev. Biol. 2015, 3, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Altarejos, J.Y.; Montminy, M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 2011, 12, 141. [Google Scholar] [CrossRef] [PubMed]
- Bachman, E.S.; Dhillon, H.; Zhang, C.Y.; Cinti, S.; Bianco, A.C.; Kobilka, B.K.; Lowell, B.B. βAR signaling required for diet-induced thermogenesis and obesity resistance. Science 2002, 297, 843–845. [Google Scholar] [CrossRef]
- Wu, Z.; Huang, X.; Feng, Y.; Handschin, C.; Feng, Y.; Gullicksen, P.S.; Bare, O.; Labow, M.; Spiegelman, B.; Stevenson, S.C. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1 α transcription and mitochondrial biogenesis in muscle cells. Proc. Natl. Acad. Sci. USA 2006, 103, 14379–14384. [Google Scholar] [CrossRef] [PubMed]
- Than, T.A.; Lou, H.; Ji, C.; Win, S.; Kaplowitz, N. Role of cAMP-responsive elementbinding protein (CREB)-regulated transcription coactivator 3 (CRTC3) in the initiation of mitochondrial biogenesis and stress response in liver cells. J. Biol. Chem. 2011, 286, 22047–22054. [Google Scholar] [CrossRef]
- Wu, S.; Yue, N.; Raza, S.H.A.; Chengtu, Z.; Le, Z.; Gong, C.; Hongbao, W.; Nicola, S.; Linsen, Z. Genetic variants and haplotype combination in the bovine CRTC3 affected conformation traits in two Chinese native cattle breeds (Bos Taurus). Genomics 2018. [Google Scholar] [CrossRef]
- Bong, J.J.; Jeong, J.Y.; Rajasekar, P.; Cho, Y.M.; Kwon, E.G.; Kim, H.C.; Paek, B.H.; Baik, M. Differential expression of genes associated with lipid metabolism in longissimus dorsi of Korean bulls and steers. Meat Sci. 2012, 91, 284–293. [Google Scholar] [CrossRef]
- Zhao, S.M.; Ren, L.J.; Chen, L.; Zhang, X.; Cheng, M.L.; Li, W.Z.; Zhang, Y.Y.; Gao, S.Z. Differential expression of lipid metabolism related genes in porcine muscle tissue leading to different intramuscular fat deposition. Lipids 2009, 44, 1029. [Google Scholar] [CrossRef] [PubMed]
- Lass, A.; Zimmermann, R.; Haemmerle, G.; Riederer, M.; Schoiswohl, G.; Schweiger, M.; Kienesberger, P.; Strauss, J.G.; Gorkiewicz, G.; Zechner, R. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman syndrome. Cell Metab. 2006, 3, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Eichmann, T.O.; Kumari, M.; Haas, J.T.; Farese, R.V.; Zimmermann, R.; Lass, A.; Zechner, R. Studies on the substrate and stereo/regioselectivity of adipose triglyceride lipase, hormone-sensitive lipase, and diacylglycerol-O-acyltransferases. J. Biol. Chem. 2012, 287, 41446–41457. [Google Scholar] [CrossRef] [PubMed]
- Goshu, H.; Wu, X.; Chu, M.; Bao, P.; Ding, X.; Yan, P. Copy number variations of KLF6 modulate gene transcription and growth traits in Chinese Datong yak (Bos Grunniens). Animals 2018, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Yea, S.; Li, S.; Chen, Z.; Narla, G.; Banck, M.; Laborda, J.; Tan, S.; Friedman, J.M.; Friedman, S.L. Krüppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J. Biol. Chem. 2005, 280, 26941–26952. [Google Scholar] [CrossRef] [PubMed]
- Miele, L.; Beale, G.; Patman, G.; Nobili, V.; Leathart, J.; Grieco, A.; Abate, M.; Friedman, S.L.; Narla, G.; Bugianesi, E. The Kruppel-like factor 6 genotype is associated with fibrosis in nonalcoholic fatty liver disease. Gastroenterol 2008, 135, 282–291.e1. [Google Scholar] [CrossRef] [PubMed]
- Andreoli, V.; Gehrau, R.C.; Bocco, J.L. Biology of Krüppel-like factor 6 transcriptional regulator in cell life and death. IUBMB Life 2010, 62, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Dionyssiou, M.G.; Salma, J.; Bevzyuk, M.; Wales, S.; Zakharyan, L.; McDermott, J.C. Krüppel-like factor 6 (KLF6) promotes cell proliferation in skeletal myoblasts in response to TGF β/Smad3 signaling. Skelet. Muscle 2013, 3, 7. [Google Scholar] [CrossRef]
- Ito, G.; Uchiyama, M.; Kondo, M.; Mori, S.; Usami, N.; Maeda, O.; Kawabe, T.; Hasegawa, Y.; Shimokata, K.; Sekido, Y. Krüppel-like factor 6 is frequently downregulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res. 2004, 64, 3838–3843. [Google Scholar] [CrossRef]
- Zhang, J.; Cui, L.; Ma, J.; Chen, C.; Yang, B.; Huang, L. Transcriptome analyses reveal genes and pathways associated with fatty acid composition traits in pigs. Anim. Genet. 2017, 48, 645–652. [Google Scholar] [CrossRef]
- Shang, P.; Zhang, B.; Zhang, J.; Duan, M.; Wu, L.; Gong, X.; Tang, K.; Zhang, H.; Chamba, Y. Expression and singlenucleotide polymorphisms of the H-FABP gene in pigs. Gene 2019, 710, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Shearer, J.; Fueger, P.T.; Bracy, D.P.; Wasserman, D.H.; Rottman, J.N. Partial gene deletion of heart-type fatty acid-binding protein limits the severity of dietary-induced insulin resistance. Diabetes 2005, 54, 3133–3139. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Wu, L.; Wang, X.; Xin, Y.; Zan, L. Tissue expression analysis, cloning and characterization of the 5’-regulatory region of the bovine FABP3 gene. Mol. Biol. Rep. 2016, 43, 991–998. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.J.; Barcelo-Coblijn, G.; Binas, B.; Glatz, J.F. Heart fatty acid uptake is decreased in heart fatty acid-binding protein gene-ablated mice. J. Biol. Chem. 2004, 279, 34481–34488. [Google Scholar] [CrossRef] [PubMed]
- Chao, Z.; Wang, F.; Deng, C.Y.; Wei, L.M.; Sun, R.P.; Liu, H.L.; Liu, Q.W.; Zheng, X.L. Distribution and linkage disequilibrium analysis of polymorphisms of MC4R, LEP, H-FABP genes in the different populations of pigs, associated with economic traits in DIV2 line. Mol. Biol. Rep. 2012, 39, 6329–6335. [Google Scholar] [CrossRef] [PubMed]
- Yi, B.; Wang, J.; Wang, S.; Yuan, D.; Sun, J.; Li, Z.; Mao, Y.; Hou, Q.; Liu, W. Overexpression of Banna mini-pig inbred line fatty acid binding protein 3 promotes adipogenesis in 3T3-L1 preadipocytes. Cell Biol. Int. 2014, 38, 918–923. [Google Scholar] [CrossRef] [PubMed]
- Green, C.D.; Ozguden-Akkoc, C.G.; Wang, Y.; Jump, D.B.; Olson, L.K. Role of fatty acid elongases in determination of de novo synthesized monounsaturated fatty acid species. J. Lipid. Res. 2010, 51, 1871–1877. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.A.; Hammer, R.E.; Horton, J.D. Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. J. Lipid Res. 2009, 50, 412–423. [Google Scholar] [CrossRef]
- Matsumoto, H.; Shimizu, Y.; Tanaka, A.; Nogi, T.; Tabuchi, I.; Oyama, K.; Taniguchi, M.; Mannen, H.; Sasazaki, S. The SNP in the promoter region of the bovine ELOVL5 gene influences economic traits including subcutaneous fat thickness. Mol. Biol. Rep. 2013, 40, 3231–3237. [Google Scholar] [CrossRef]
- Zhang, W.; Bin, Y.; Zhang, J.; Cui, L.; Ma, J.; Chen, C.; Ai, H.; Xiao, S.; Ren, J.; Huang, L. Genome-wide association studies for fatty acid metabolic traits in five divergent pig populations. Sci. Rep. 2016, 6, 24718. [Google Scholar] [CrossRef]
- Zhu, B.; Niu, H.; Zhang, W.; Wang, Z.; Liang, Y.; Guan, L.; Guo, P.; Chen, Y.; Zhang, L.; Guo, Y.; et al. Genome wide association study and genomic prediction for fatty acid composition in Chinese Simmental beef cattle using high density SNP array. BMC Genom. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.B.; Wu, M.; Zhu, J.J.; Zhang, C.H.; Yao, D.W.; Luo, J.; Loor, J.J. Fatty acid elongase 6 plays a role in the synthesis of long -chain fatty acids in goat mammary epithelial cells. J. Dairy Sci. 2017, 100, 4987–4995. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.S.; Oh, A.R.; Lee, H.J.; Ahn, Y.H.; Cha, J.Y. Hepatic Elovl6 gene expression is regulated by the synergistic action of ChREBP and SREBP -1c. Biochem. Biophys. Res. Commun. 2016, 478, 1060–1066. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Yu, J.H.; Kim, W.H.; Kim, J.W. Mouse elovl-6 promoter is an SREBP target. Biochem. Biophys. Res. Commun. 2008, 368, 261–266. [Google Scholar]
- Matsuzaka, T.; Shimano, H.; Yahagi, N.; Yoshikawa, T.; Amemiya-Kudo, M.; Hasty, A.H.; Okazaki, H.; Tamura, Y.; Iizuka, Y.; Ohashi, K.; et al. Cloning and characterization of a mammalian fatty acyl-CoA elongase as a lipogenic enzyme regulated by SREBPs. J. Lipid Res. 2002, 43, 911–920. [Google Scholar] [PubMed]
- Mariani, S.; Di Rocco, G.; Toietta, G.; Russo, M.A.; Petrangeli, E.; Salvatori, L. Sirtuins 1–7 expression in human adipose derived stem cells from subcutaneous and visceral fat depots: Influence of obesity and hypoxia. Endocrine 2017, 57, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Marcos, P.J.; Serrano, M. Sirt4: The glutamine gatekeeper. Cancer Cell 2013, 23, 427–428. [Google Scholar] [CrossRef] [PubMed]
- Pannek, M.; Simic, Z.; Fuszard, M.; Meleshin, M.; Rotili, D.; Mai, A.; Schutkowski, M.; Steegborn, C. Crystal structures of the mitochondrial deacylase Sirtuin 4 reveal isoform-specific acyl recognition and regulation features. Nature Commun. 2017, 8, 1513. [Google Scholar] [CrossRef] [PubMed]
- Acs, Z.; Bori, Z.; Takeda, M.; Osvath, P.; Berkes, I.; Taylor, A.W.; Yang, H.; Radak, Z. High altitude exposure alters gene expression levels of DNA repair enzymes, and modulates fatty acid metabolism by SIRT4 induction in human skeletal muscle. Res. Physiol. Neurobiol. 2014, 196, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Han, S.H. Potential role of sirtuin as a therapeutic target for neurodegenerative diseases. J. Clin. Neurol. 2009, 5, 1205. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Mucke, L. Paths of convergence: Sirtuins in aging and neurodegeneration. Neuron 2008, 58, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1 α. Cell 2010, 22, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Pastinen, T.; Hudson, T.J. Cis-acting regulatory variation in the human genome. Science 2004, 306, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.T.; Cheng, Y.S.; Huang, M.C. A novel nonsynonymous SNP of the COLX gene and its association with duck reproductive traits. Mol. Cell. Probes 2012, 26, 204–207. [Google Scholar] [CrossRef]
- Guarente, L. Sirtuins in aging and disease. In Cold Spring Harbor Symposia Quantitat Biology; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2007; Volume 72, pp. 483–488. [Google Scholar]
- Ford, E.; Voit, R.; Liszt, G.; Magin, C.; Grummt, I.; Guarente, L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006, 20, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Mu, P.; Liu, K.; Lin, Q.; Yang, W.; Liu, D.; Lin, Z.; Shao, W.; Ji, T. Sirtuin 7 promotes glioma proliferation and invasion through activation of the ERK/STAT3 signaling pathway. Oncol. Lett. 2019, 17, 1445–1452. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Xiong, Y.; Zhang, P.; Li, Z.; Jiang, Q.; Bi, P.; Yue, F.; Yang, G.; Wang, Y.; Liu, X. Lkb1 controls brown adipose tissue growth and thermogenesis by regulating the intracellular localization of CRTC3. Nat. Commun. 2016, 7, 12205. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Raza, S.H.A.; Junjvlieke, Z.; Xiaoyu, W.; Garcia, M.; Elnour, I.E.; Hongbao, W.; Linsen, Z. Function and Transcriptional Regulation of Bovine TORC2 Gene in Adipocytes: Roles of C/EBP, XBP1, INSM1 and ZNF263. Int. J. Mol. Sci. 2019, 20, 4338. [Google Scholar] [CrossRef]
- Iourgenko, V.; Zhang, W.; Mickanin, C.; Daly, I.; Jiang, C.; Hexham, J.M.; Orth, A.P.; Miraglia, L.; Meltzer, J.; Garza, D. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc. Natl. Acad. Sci. USA 2003, 100, 12147–12152. [Google Scholar] [CrossRef]
- Bittinger, M.A.; McWhinnie, E.; Meltzer, J.; Iourgenko, V.; Latario, B.; Liu, L.; Chen, C.H.; Song, C.; Garza, D.; Labow, M. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr. Biol. 2004, 14, 2156–2161. [Google Scholar]
- Screaton, R.A.; Conkright, M.D.; Katoh, Y.; Best, J.L.; Canettieri, G.; Jeffries, S.; Guzman, E.; Niessen, S.; Yates, J.R.; Takemori, H. The CREB coactivator TORC functions as a calcium-and cAMP-sensitive coincidence detector. Cell 2004, 119, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Haemmerle, G.; Lass, A.; Zimmermann, R.; Gorkiewicz, G.; Meyer, C.; Rozman, J.; Heldmaier, G.; Maier, R.; Theussl, C.; Eder, S. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 2006, 80, 734–737. [Google Scholar] [CrossRef]
- Lu, X.; Yang, X.; Liu, J. Differential control of ATGL-mediated lipid droplet degradation by CGI-58 and G0S2. Cell Cycle 2010, 9, 2791–2797. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lu, X.; Lombès, M.; Rha, G.B.; Chi, Y.I.; Guerin, T.M.; Smart, E.J.; Liu, J. The G0/G1 switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 2010, 11, 194–205. [Google Scholar] [CrossRef]
- Rowland, B.D.; Peeper, D.S. KLF4, p21 and context-dependent opposing forces in cancer. Nat. Rev. Cancer 2006, 6, 11. [Google Scholar] [CrossRef]
- Kaczynski, J.; Cook, T.; Urrutia, R. Sp1-and Krüppel-like transcription factors. Genome Biol. 2003, 4, 206. [Google Scholar] [CrossRef] [PubMed]
- Ghaleb, A.M.; Katz, J.P.; Kaestner, K.H.; Du, J.X.; Yang, V.W. Krüppel-like factor 4 exhibits antiapoptotic activity following γ-radiation-induced DNA damage. Oncogene 2007, 26, 2365. [Google Scholar] [CrossRef]
- Suske, G.; Bruford, E.; Philipsen, S. Mammalian SP/KLF transcription factors: Bring in the family. Genomics 2005, 85, 551–556. [Google Scholar] [CrossRef]
- Inuzuka, H.; Nanbu-Wakao, R.; Masuho, Y.; Muramatsu, M.; Tojo, H.; Wakao, H. Differential regulation of immediate early gene expression in preadipocyte cells through multiple signaling pathways. Biochem. Biophys. Res. Commun. 1999, 265, 664–668. [Google Scholar] [CrossRef]
- Bechmann, L.P.; Gastaldelli, A.; Vetter, D.; Patman, G.L.; Pascoe, L.; Hannivoort, R.A.; Lee, U.E.; Fiel, I.; Muñoz, U.; Ciociaro, D. Glucokinase links Krüppel-like factor 6 to the regulation of hepatic insulin sensitivity in nonalcoholic fatty liver disease. Hepatology 2012, 55, 1083–1093. [Google Scholar] [CrossRef]
- Shioda, N.; Yamamoto, Y.; Watanabe, M.; Binas, B.; Owada, Y.; Fukunaga, K. Heart-type fatty acid binding protein regulates dopamine D2 receptor function in mouse brain. J. Neurosci. 2010, 30, 3146–3155. [Google Scholar] [CrossRef] [PubMed]
- Kusudo, T.; Kontani, Y.; Kataoka, N.; Ando, F.; Shimokata, H.; Yamashita, H. Fatty acid-binding protein 3 stimulates glucose uptake by facilitating AS160 phosphorylation in mouse muscle cells. Genes Cells 2011, 16, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.M.; Wang, H.; Zeng, Y.Q.; Chen, W. Developmental changes and effect on intramuscular fat content of H-FABP and A-FABP mRNA expression in pigs. J. Appl. Genet. 2013, 54, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Vergnes, L.; Chin, R.; Young, S.G.; Reue, K. Heart-type fatty acid-binding protein is essential for efficient brown adipose tissue fatty acid oxidation and cold tolerance. J. Biol. Chem. 2011, 286, 380–390. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Choi, Y.M.; Choe, J.H.; Kim, J.M.; Hong, K.C.; Park, H.C.; Kim, B.C. Association between polymorphisms of the heart fatty acid binding protein gene and intramuscular fat content, fatty acid composition, and meat quality in Berkshire breed. Meat Sci. 2010, 86, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, A.; Westerberg, R.; Jacobsson, A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006, 45, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Makino, A.; Hullin-Matsuda, F.; Kobayashi, T.; Furihata, M.; Chung, S.; Ashida, S.; Miki, T.; Fujioka, T.; Shuin, T.; et al. Novel lipogenic enzyme ELOVL7 is involved in prostate cancer growth through saturated long-chain fatty acid metabolism. Cancer Res. 2009, 69, 8133–8140. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, T.; Atsumi, A.; Matsumori, R.; Nie, T.; Shinozaki, H.; Suzuki-Kemuriyama, N.; Kuba, M.; Nakagawa, Y.; Ishii, K.; Shimada, M.; et al. Elovl6 promotes nonalcoholic steatohepatitis. Hepatlogy 2012, 56, 2199–2208. [Google Scholar] [CrossRef]
- Matsuzaka, T.; Shimano, H.; Yahagi, N.; Kato, T.; Atsumi, A.; Yamamoto, T.; Inoue, N.; Ishikawa, M.; Okada, S.; Ishigaki, N.; et al. Crucial role of a long -chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat. Med. 2007, 13, 1193–1202. [Google Scholar] [CrossRef]
- Senyilmaz, D.; Virtue, S.; Xu, X.; Tan, C.Y.; Griffin, J.L.; Miller, A.K.; Vidal-Puig, A.; Teleman, A.A. Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature 2015, 525, 124–128. [Google Scholar] [CrossRef]
- Chen, S.; He, H.; Liu, X.L. Tissue expression profiles and transcriptional regulation of elongase of very long chain fatty acid 6 in bovine mammary epithelial cells. PLoS ONE 2017, 12, e0175777. [Google Scholar] [CrossRef] [PubMed]


| Gene Type | Author(s) | Findings |
|---|---|---|
| The SIRT4 gene | [32] |
|
| [33] |
| |
| [34] |
| |
| [11] |
| |
| [35] |
| |
| SIRT6 gene | [36] |
|
| [37] |
| |
| [38] |
| |
| [39] |
| |
| [2] |
| |
| Silent information regulators 7 (SIRT7) | [40,41,42] |
|
| [43,44] |
| |
| [45] |
| |
| [57] |
| |
| [15] |
| |
| CREB-regulated transcription coactivator 3 (CRTC3) | [46] |
|
| [47,48,49] |
| |
| [46,48] |
| |
| [50] |
| |
| The α/β hydrolase domain containing 5 (ABHD5) | [51] |
|
| [52] |
| |
| [53] |
| |
| [54] |
| |
| [16] |
| |
| Kruppel-like factor 6 (KLF6) | [55] |
|
| [56] |
| |
| [57] |
| |
| [58,59] |
| |
| [3] |
| |
| [60] |
| |
| [56,60] |
| |
| Heart type fatty acid binding protein (H-FABP) | [61,62] |
|
| [63] |
| |
| [64] |
| |
| [11,65] |
| |
| [66] |
| |
| [67] |
| |
| Very long chain fatty acids protein 5 (ELOVL5) | [2,68] |
|
| [68] |
| |
| [69] |
| |
| [70] |
| |
| [71] |
| |
| [72] |
| |
| [70] |
| |
| Very long chain fatty acids protein 6 (ELOVL6) | [68] |
|
| [73] |
| |
| [74,75,76] |
|
| Gene | SNP | Site | Related Traits | References |
|---|---|---|---|---|
| SIRT4 | g.−311C > T and g.−771C | ND | Subcutaneous fat depths | [11] |
| g.−1022G > A | ND | Intramuscular fat content subcutaneous fat depth | [11] | |
| SIRT6 | g.8460GNA | ND | Body measurements | [38] |
| g.9429CNT | ND | Meat traits | [38] | |
| g.9735TNC | ND | Body measurements | [38] | |
| SIRT7 | g.3587C > T | Exon 6 | Body size and meat quality traits | [11,15] |
| g.3793T > C | Exon 7 | Body size and meat quality traits | [11,15] | |
| CREC3 | g.62652 A > G | Intron 3 | Loin muscle area | [50] |
| g.62730C > T | Exon 4 | BL, HH, RL, and HW | [50] | |
| g.66478G > C | Exon 6 | BL and CD | [50] | |
| g.91297C > T | Intron 13 | Body conformation | [50] | |
| ABHD5 | Evi1 and C/EBP α as a transcriptional factor | Transcriptional factor | Carcass quality traits | [16] |
| KLF6 | g.3332C > G | Exon2 | Body and carcass measurements | [3] |
| g.3413C > T | Exon2 | Body and carcass measurements | [3] | |
| g.3521G > A | Exon2 | Body and carcass measurements | [3] | |
| H-FABP | g.6643C > T | ND | Weight and body length | [11,65] |
| g.1375 C >G | ND | Lipid deposition | [62] | |
| ELOVL5 | g.−110T>C | ND | Monounsaturated fatty acid, SFA saturated fatty acid | [2,68,70] |
| ND | Subcutaneous fat thickness | [70] | ||
| Fatty acid profile | ||||
| ELOVL6 | ND | ND | Fatty acid profile in meat | [61] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raza, S.H.A.; Khan, R.; Abdelnour, S.A.; Abd El-Hack, M.E.; Khafaga, A.F.; Taha, A.; Ohran, H.; Mei, C.; Schreurs, N.M.; Zan, L. Advances of Molecular Markers and Their Application for Body Variables and Carcass Traits in Qinchuan Cattle. Genes 2019, 10, 717. https://doi.org/10.3390/genes10090717
Raza SHA, Khan R, Abdelnour SA, Abd El-Hack ME, Khafaga AF, Taha A, Ohran H, Mei C, Schreurs NM, Zan L. Advances of Molecular Markers and Their Application for Body Variables and Carcass Traits in Qinchuan Cattle. Genes. 2019; 10(9):717. https://doi.org/10.3390/genes10090717
Chicago/Turabian StyleRaza, Sayed Haidar Abbas, Rajwali Khan, Sameh A. Abdelnour, Mohamed E. Abd El-Hack, Asmaa F. Khafaga, Ayman Taha, Husein Ohran, Chugang Mei, Nicola M. Schreurs, and Linsen Zan. 2019. "Advances of Molecular Markers and Their Application for Body Variables and Carcass Traits in Qinchuan Cattle" Genes 10, no. 9: 717. https://doi.org/10.3390/genes10090717
APA StyleRaza, S. H. A., Khan, R., Abdelnour, S. A., Abd El-Hack, M. E., Khafaga, A. F., Taha, A., Ohran, H., Mei, C., Schreurs, N. M., & Zan, L. (2019). Advances of Molecular Markers and Their Application for Body Variables and Carcass Traits in Qinchuan Cattle. Genes, 10(9), 717. https://doi.org/10.3390/genes10090717










