Characterization of Aminoacyl-tRNA Synthetases in Chromerids
Abstract
1. Introduction
2. Material and Methods
2.1. Gene Identification and Model Assessment
2.2. Localization Prediction
2.3. Molecular Phylogeny
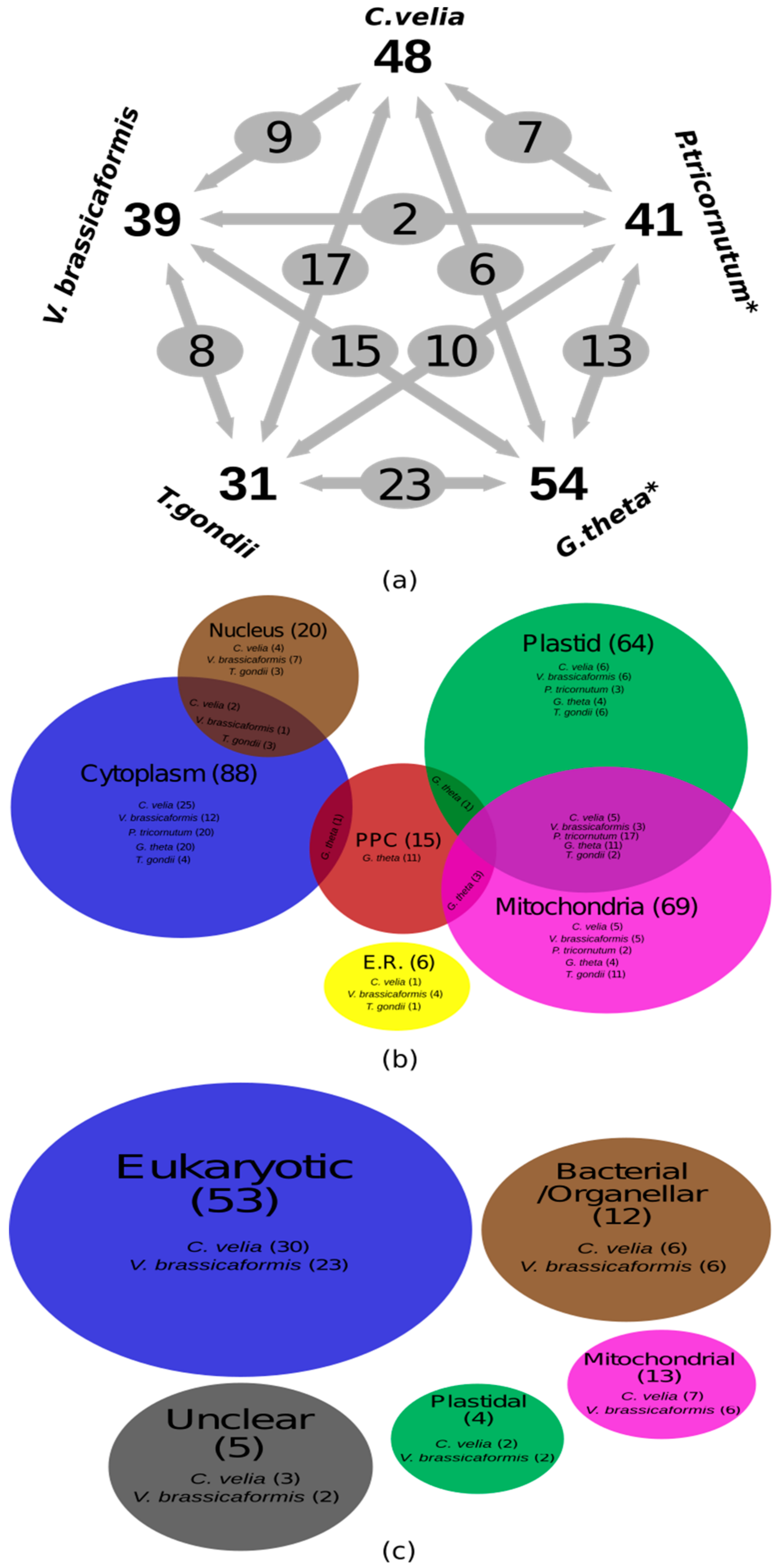
3. Results and Discussion
Gene Identification and in Silico Localization of Gene Products
4. Phylogenetic Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moore, R.B.; Oborník, M.; Janouškovec, J.; Chrudimský, T.; Vancová, M.; Green, D.H.; Wright, S.W.; Davies, N.W.; Bolch, C.J.S.; Heimann, K.; et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature 2008, 451, 959–963. [Google Scholar] [CrossRef] [PubMed]
- Oborník, M.; Modrý, D.; Lukeš, M.; Černotíková-Stříbrná, E.; Cihlář, J.; Tesařová, M.; Kotabová, E.; Vancová, M.; Prášil, O.; Lukeš, J. Morphology, Ultrastructure and Life Cycle of Vitrella brassicaformis n. sp., n. gen., a Novel Chromerid from the Great Barrier Reef. Protist 2012, 163, 306–323. [Google Scholar] [CrossRef] [PubMed]
- Cumbo, V.R.; Baird, A.H.; Moore, R.B.; Negri, A.P.; Neilan, B.A.; Salih, A.; van Oppen, M.J.H.; Wang, Y.; Marquis, C.P. Chromera velia is endosymbiotic in larvae of the reef corals Acropora digitifera and A. tenuis. Protist 2013, 164, 237–244. [Google Scholar] [CrossRef]
- Janouškovec, J.; Horák, A.; Barott, K.L.; Rohwer, F.L.; Keeling, P.J. Environmental distribution of coral-associated relatives of apicomplexan parasites. ISME J. 2013, 7, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Füssy, Z.; Oborník, M. Chromerids and Their Plastids. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2017; Volume 84, pp. 187–218. ISBN 9780128026519. [Google Scholar]
- McFadden, G.I.; Waller, R.F. Plastids in parasites of humans. Bioessays 1997, 19, 1033–1040. [Google Scholar] [CrossRef]
- Woo, Y.H.; Ansari, H.; Otto, T.D.; Linger, C.M.K.; Olisko, M.K.; Michálek, J.; Saxena, A.; Shanmugam, D.; Tayyrov, A.; Veluchamy, A.; et al. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. eLife 2015, 4, e06974. [Google Scholar] [CrossRef] [PubMed]
- Flegontov, P.; Michálek, J.; Janouškovec, J.; Lai, D.H.; Jirků, M.; Hajdušková, E.; Tomčala, A.; Otto, T.D.; Keeling, P.J.; Pain, A.; et al. Divergent mitochondrial respiratory chains in phototrophic relatives of apicomplexan parasites. Mol. Biol. Evol. 2015, 32, 1115–1131. [Google Scholar] [CrossRef] [PubMed]
- Oborník, M.; Janouškovec, J.; Chrudimský, T.; Lukeš, J. Evolution of the apicoplast and its hosts: From heterotrophy to autotrophy and back again. Int. J. Parasitol. 2009, 39, 1–12. [Google Scholar] [CrossRef]
- Oborník, M.; Lukeš, J. The organellar genomes of Chromera and Vitrella, the phototrophic relatives of apicomplexan parasites. Annu. Rev. Microbiol. 2015, 69, 129–144. [Google Scholar] [CrossRef]
- Oborník, M.; Lukeš, J. Cell Biology of Chromerids: Autotrophic Relatives to Apicomplexan Parasites. In International Review of Cell and Molecular Biology; Academic Press: Cambridge, MA, USA, 2013; Volume 306, pp. 333–369. [Google Scholar]
- Oborník, M.; Kručinská, J.; Esson, H. Life cycles of chromerids resemble those of colpodellids and apicomplexan parasites. Perspect. Phycol. 2016, 3, 21–27. [Google Scholar] [CrossRef]
- Janouškovec, J.; Tikhonenkov, D.V.; Burki, F.; Howe, A.T.; Kolísko, M.; Mylnikov, A.P.; Keeling, P.J. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc. Natl. Acad. Sci. USA 2015, 112, 10200–10207. [Google Scholar] [CrossRef] [PubMed]
- Füssy, Z.; Oborník, M. Reductive Evolution of Apicomplexan Parasites from Phototrophic Ancestors. In Evolutionary Biology: Self/Nonself Evolution, Species and Complex Traits Evolution, Methods and Concepts; Springer: Cham, Switzerland, 2017; pp. 217–236. [Google Scholar]
- Janouškovec, J.; Horák, A.; Oborník, M.; Lukeš, J.; Keeling, P.J. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl. Acad. Sci. USA 2010, 107, 10949–10954. [Google Scholar] [CrossRef] [PubMed]
- Sobotka, R.; Esson, H.J.; Koník, P.; Trsková, E.; Moravcová, L.; Horák, A.; Dufková, P.; Oborník, M. Extensive gain and loss of photosystem I subunits in chromerid algae, photosynthetic relatives of apicomplexans. Sci. Rep. 2017, 7, 13214. [Google Scholar] [CrossRef] [PubMed]
- Füssy, Z.; Faitová, T.; Oborník, M. Subcellular compartments interplay for carbon and nitrogen allocation in Chromera velia and Vitrella brassicaformis. Genome Biol. Evol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Ševcíková, T.; Horák, A.; Klimeš, V.; Zbránková, V.; Demir-Hilton, E.; Sudek, S.; Jenkins, J.; Schmutz, J.; Pribyl, P.; Fousek, J.; et al. Updating algal evolutionary relationships through plastid genome sequencing: Did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci. Rep. 2015, 5, 10134. [Google Scholar] [CrossRef]
- Kořený, L.; Sobotka, R.; Janouškovec, J.; Keeling, P.J.; Oborník, M. Tetrapyrrole Synthesis of Photosynthetic Chromerids Is Likely Homologous to the Unusual Pathway of Apicomplexan Parasites. Plant Cell 2011, 23, 3454–3462. [Google Scholar] [CrossRef] [PubMed][Green Version]
- van Dooren, G.G.; Kennedy, A.T.; McFadden, G.I. The Use and Abuse of Heme in Apicomplexan Parasites. Antioxid. Redox Signal. 2012, 17, 634–656. [Google Scholar] [CrossRef] [PubMed]
- Patron, N.J.; Waller, R.F.; Archibald, J.M.; Keeling, P.J. Complex protein targeting to dinoflagellate plastids. J. Mol. Biol. 2005, 348, 1015–1024. [Google Scholar] [CrossRef]
- Slamovits, C.H.; Saldarriaga, J.F.; Larocque, A.; Keeling, P.J. The Highly Reduced and Fragmented Mitochondrial Genome of the Early-branching Dinoflagellate Oxyrrhis marina Shares Characteristics with both Apicomplexan and Dinoflagellate Mitochondrial Genomes. J. Mol. Biol. 2007, 372, 356–368. [Google Scholar] [CrossRef]
- Nash, E.A.; Nisbet, R.E.R.; Barbrook, A.C.; Howe, C.J. Dinoflagellates: A mitochondrial genome all at sea. Trends Genet. 2008, 24, 328–335. [Google Scholar] [CrossRef]
- Waller, R.F.; Jackson, C.J. Dinoflagellate mitochondrial genomes: Stretching the rules of molecular biology. BioEssays 2009, 31, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Waller, R.F.; Kořený, L. Plastid Complexity in Dinoflagellates: A Picture of Gains, Losses, Replacements and Revisions. In Advances in Botanical Research; Academic Press: Cambridge, MA, USA, 2017; Volume 84, pp. 105–143. ISBN 9780128026519. [Google Scholar]
- John, U.; Lu, Y.; Wohlrab, S.; Groth, M.; Janouškovec, J.; Kohli, G.S.; Mark, F.C.; Bickmeyer, U.; Farhat, S.; Felder, M.; et al. An aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome. Sci. Adv. 2019, 6, eaav1110. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.L.; Du, D.H.; Tan, M.; Lei, H.Y.; Ruan, L.L.; Eriani, G.; Wang, E.D. Role of tRNA amino acid-accepting end in aminoacylation and its quality control. Nucleic Acids Res. 2011, 20, 8857–8868. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.-I.; Chen, Y.-W.; Wu, Y.-H.; Chang, C.-Y.; Wang, T.-L.; Wang, C.-C. Functional Substitution of a Eukaryotic Glycyl-tRNA Synthetase with an Evolutionarily Unrelated Bacterial Cognate Enzyme. PLoS ONE 2014, 9, e94659. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carter, C.W., Jr. Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 1993, 62, 715–748. [Google Scholar] [CrossRef] [PubMed]
- Giege, R.; Sissler, M.; Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998, 26, 5017–5035. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, P.R.; Söll, D. Aminoacyl-tRNA Synthetases: General Features and Recognition of Transfer RNAs. Annu. Rev. Biochem. 1979, 48, 601–648. [Google Scholar] [CrossRef]
- Chaliotis, A.; Vlastaridis, P.; Mossialos, D.; Ibba, M.; Becker, H.D.; Stathopoulos, C.; Amoutzias, G.D. The complex evolutionary history of aminoacyl-tRNA synthetases. Nucleic Acids Res. 2017, 45, 1059–1068. [Google Scholar] [CrossRef]
- Eriani, G.; Delarue, M.; Poch, O.; Gangloff, J.; Moras, D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 1990, 347, 203–206. [Google Scholar] [CrossRef]
- Ludmerer, S.W.; Schimmel, P. Gene for yeast glutamine tRNA synthetase encodes a large amino-terminal extension and provides a strong confirmation of the signature sequence for a group of the aminoacyl-tRNA synthetases. J. Biol. Chem. 1987, 262, 10801–10806. [Google Scholar]
- Mazauric, M.H.; Keith, G.; Logan, D.; Kreutzer, R.; Giegé, R.; Kern, D. Glycyl-tRNA synthetase from Thermus thermophilus--wide structural divergence with other prokaryotic glycyl-tRNA synthetases and functional inter-relation with prokaryotic and eukaryotic glycylation systems. Eur. J. Biochem. 1998, 251, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Ostrem, D.L.; Berg, P. Glycyl transfer ribonucleic acid synthetase from Escherichia coli: Purification, properties, and substrate binding. Biochemistry 1974, 13, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Surguchov, A.P.; Surguchov, I.G. Two Enzymically Active Forms of Glycyl-tRNA Synthetase from Bacillus brevis Purification and Properties. Eur. J. Biochem. 1975, 54, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Kern, D.; Giegé, R.; Ebel, J.P. Purification and some properties of alanyl- and leucyl-tRNA synthetases from baker’s yeast. BBA Sect. Nucleic Acids Protein Synth. 1981, 653, 83–90. [Google Scholar] [CrossRef]
- Shiba, K.; Schimmel, P.; Motegi, H.; Noda, T. Human glycyl-tRNA synthetase. Wide divergence of primary structure from bacterial counterpart and species-specific aminoacylation. J. Biol. Chem. 1994, 269, 30049–30055. [Google Scholar] [PubMed]
- Curnow, A.W.; Hong, K.W.; Yuan, R.; Kim, S.I.; Martins, O.; Winkler, W.; Henkin, T.M.; Söll, D. Glu-tRNAGln amidotransferase: A novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. USA 1997, 22, 11819–11826. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.D.; Kern, D. Thermus thermophilus: A link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl. Acad. Sci. USA 1998, 22, 12832–12837. [Google Scholar] [CrossRef]
- Ibba, M.; Curnow, A.W.; Söll, D. Aminoacyl-tRNA synthesis: Divergent routes to a common goal. Trends Biochem. Sci. 1997, 22, 39–42. [Google Scholar] [CrossRef]
- Leinfelder, W.; Zehelein, E.; Mandrandberthelot, M.; Bock, A. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature 1988, 331, 723–725. [Google Scholar] [CrossRef]
- Sauerwald, A.; Zhu, W.; Major, T.A.; Roy, H.; Palioura, S.; Jahn, D.; Whitman, W.B.; Yates, J.R.; Ibba, M.; Söll, D. RNA-dependent cysteine biosynthesis in archaea. Science 2005, 307, 1969–1972. [Google Scholar] [CrossRef]
- Sheppard, K.; Yuan, J.; Hohn, M.J.; Jester, B.; Devine, K.M.; Söll, D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008, 36, 1813–1825. [Google Scholar] [CrossRef] [PubMed]
- Gile, G.H.; Moog, D.; Slamovits, C.H.; Maier, U.G.; Archibald, J.M. Dual organellar targeting of aminoacyl-tRNA synthetases in diatoms and cryptophytes. Genome Biol. Evol. 2015, 7, 1728–1742. [Google Scholar] [CrossRef] [PubMed]
- Warrenfeltz, S.; Basenko, E.Y.; Crouch, K.; Harb, O.S.; Kissinger, J.C.; Roos, D.S.; Shanmugasundram, A.; Silva-Franco, F. EuPathDB: The eukaryotic pathogen genomics database resource. In Eukaryotic Genomic Databases; Humana Press: New York, NY, USA, 2018; Volume 1757, pp. 69–113. [Google Scholar]
- Woese, C.R.; Olsen, G.J.; Ibba, M.; Soll, D. Aminoacyl-tRNA Synthetases, the Genetic Code, and the Evolutionary Process. Microbiol. Mol. Biol. Rev. 2000, 64, 202–236. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R. Ancient horizontal gene transfer. Nat. Rev. Genet. 2003, 4, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Bendtsen, J.D.; Nielsen, H.; Von Heijne, G.; Brunak, S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Gruber, A.; Rocap, G.; Kroth, P.G.; Armbrust, E.V.; Mock, T. Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 2015, 81, 519–528. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Brunak, S.; von Heijne, G.; Nielsen, H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007, 2, 953–971. [Google Scholar] [CrossRef]
- Bannai, H.; Tamada, Y.; Maruyama, O.; Nakai, K.; Miyano, S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 2002, 18, 298–305. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Claros, M.G.; Vincens, P. Computational Method to Predict Mitochondrially Imported Proteins and their Targeting Sequences. Eur. J. Biochem. 1996, 241, 779–786. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest-HPC: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef] [PubMed]
- de Juan, D.; Pazos, F.; Valencia, A.; Evaluation, P.M.; Rehbein, P.; Schwalbe, H.; Jones, D.T.; Buchan, D.W.A.; Cozzetto, D.; Pontil, M.; et al. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2015, 21, 2104–2105. [Google Scholar]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Ludwig, T.; Meier, H. RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 2005, 21, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Hopper, A.K.; Martinis, S.A.; Schimmel, P.; Kisselev, L.; Wolfson, A.; Melton, D.A.; De Robertis, E.M.; Cortese, R.; Popenko, V.I.; Wolfe, C.L.; et al. Nuclear functions charge ahead. Science 1998, 282, 2003–2004. [Google Scholar] [CrossRef]
- Lund, E. Proofreading and Aminoacylation of tRNAs Before Export from the Nucleus. Science 1998, 282, 2082–2085. [Google Scholar] [CrossRef]
- Szymanski, M.; Deniziak, M.A.; Barciszewski, J. Aminoacyl-tRNA synthetases database. Nucleic Acids Res. 2001, 29, 288–290. [Google Scholar] [CrossRef] [PubMed]
- Nathanson, L.; Deutscher, M.P. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J. Biol. Chem. 2000, 275, 31559–31562. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.G.; Kang, Y.S.; Kim, E.K.; Park, S.G.; Kim, S. Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J. Cell Biol. 2000, 149, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Kaminska, M.; Havrylenko, S.; Decottignies, P.; Le Maréchal, P.; Negrutskii, B.; Mirande, M. Dynamic organization of aminoacyl-tRNA synthetase complexes in the cytoplasm of human cells. J. Biol. Chem. 2009, 284, 13746–13754. [Google Scholar] [CrossRef] [PubMed]
- Duchene, A.-M.; Giritch, A.; Hoffmann, B.; Cognat, V.; Lancelin, D.; Peeters, N.M.; Zaepfel, M.; Marechal-Drouard, L.; Small, I.D. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 16484–16489. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Batth, T.S.; Petzold, C.J.; Redding-Johanson, A.M.; Mukhopadhyay, A.; Verboom, R.; Meyer, E.H.; Millar, A.H.; Heazlewood, J.L. Analysis of the Arabidopsis cytosolic proteome highlights subcellular partitioning of central plant metabolism. J. Proteome Res. 2011, 10, 1571–1582. [Google Scholar] [CrossRef]
- Araiso, Y.; Huot, J.L.; Sekiguchi, T.; Frechin, M.; Fischer, F.; Enkler, L.; Senger, B.; Ishitani, R.; Becker, H.D.; Nureki, O. Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases. Nucleic Acids Res. 2014, 42, 6052–6063. [Google Scholar] [CrossRef]
- Frechin, M.; Duchêne, A.-M.; Becker, H.D. Translating organellar glutamine codons: A case by case scenario? RNA Biol. 2009, 6, 31–34. [Google Scholar] [CrossRef][Green Version]
- Freist, W.; Logan, D.T.; Gauss, D.H. Glycyl-tRNA synthetase. Biol. Chem. Hoppe Seyler 1996, 377, 343–356. [Google Scholar]
- Hipps, D.; Shiba, K.; Henderson, B.; Schimmel, P. Operational RNA code for amino acids: Species-specific aminoacylation of minihelices switched by a single nucleotide. Proc. Natl. Acad. Sci. USA 1995, 92, 5550–5552. [Google Scholar] [CrossRef]
- Duchêne, A.M.; Peeters, N.; Dietrich, A.; Cosset, A.; Small, I.D.; Wintz, H. Overlapping Destinations for Two Dual Targeted Glycyl-tRNA Synthetases in Arabidopsis thaliana and Phaseolus vulgaris. J. Biol. Chem. 2001, 276, 15275–15283. [Google Scholar] [CrossRef] [PubMed]
- Halary, S.; McInerney, J.O.; Lopez, P.; Bapteste, E. EGN: A wizard for construction of gene and genome similarity networks. BMC Evol. Biol. 2013, 13, 146. [Google Scholar] [CrossRef] [PubMed]
- Brindefalk, B.; Viklund, J.; Larsson, D.; Thollesson, M.; Andersson, S.G.E. Origin and evolution of the mitochondrial aminoacyl-tRNA synthetases. Mol. Biol. Evol. 2007, 24, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 2005, 33, 4626–4638. [Google Scholar] [CrossRef] [PubMed]
- Ribas de Pouplana, L.; Brown, J.R.; Schimmel, P. Structure-based phylogeny of class IIa tRNA synthetases in relation to an unusual biochemistry. J. Mol. Evol. 2001, 53, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Roy, H.; Ling, J.; Alfonzo, J.; Ibba, M. Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J. Biol. Chem. 2005, 280, 38186–38192. [Google Scholar] [CrossRef]

| AaRSs | Chromera velia | Vitrella brassicaformis | ||||
|---|---|---|---|---|---|---|
| 48 | 39 | |||||
| Gene | Localization | Origin | Gene | Localization | Origin | |
| ArgRS | cvel_7363 | Cyto.|Nucl. | Bact./Orga. | vbra_19912 | Nucl. | Plas. |
| cvel_22194 | Cyto. | Euka. | ||||
| cvel_3008 | Mito. | Plas. | ||||
| CysRS | cvel_18990 | Cyto | Euka. | vbra_21787 | Cyto. | Euka. |
| cvel_1412 | Plas. | Bact./Orga. | vbra_7006 | Mito. | Bact./Orga. | |
| GluRS | cvel_31597 | Plas. | Bact./Orga. | vbra_21912 | Plas. | Bact./Orga. |
| GlnRS | cvel_8110 | Cyto. | Mito. | vbra_11768 | Cyto. | Mito. |
| cvel_1112 | Cyto. | Mito. | vbra_19782 | Mito. | Bact./Orga. | |
| cvel_874 | Mito. | Bact./Orga. | ||||
| LeuRS | cvel_7809 | Plas.|Mito. | Bact./Orga. | vbra_9619 | Plas.|Mito. | Bact./Orga. |
| cvel_16973 | Cyto. | Euka. | vbra_9824 | Nucl. | Euka. | |
| IleRS | cvel_28423 | Plas. | Euka. | vbra_7770 | Cyto. | Euka. |
| cvel_28875 | Cyto. | Euka. | vbra_11931 | Endo. | Euka. | |
| MetRS | cvel_21221 | Cyto. | Euka. | vbra_1113 | Plas. | Mito. |
| cvel_7165 | Plas.|Mito. | Mito. | vbra_13101 | Cyto. | Euka. | |
| TrpRS | cvel_7641 | Cyto. | Euka. | vbra_16713 | Mito. | Euka. |
| cvel_20758 | Plas.|Mito. | Plas. | vbra_20646 | Endo. | Plas. | |
| TyrRS | cvel_7146 | Plas. | Mito. | vbra_8727 | Endo. | Mito. |
| cvel_4070 | Cyto. | Euka. | vbra_13490 | Cyto. | Euka. | |
| ValRS | cvel_6889 | Nucl. | Euka. | vbra_14431 | Plas. | Euka. |
| cvel_16754 | Cyto. | Euka. | vbra_19209 | Cyto. | Euka. | |
| cvel_9091 | Cyto. | Euka. | ||||
| AlaRS | cvel_11640 | Nucl. | Bact./Orga. | vbra_6631 | Nucl. | Euka. |
| cvel_19371 | Mito. | Euka. | vbra_13386 | Mito. | Bact./Orga. | |
| cvel_21018 | Nucl. | Euka. | ||||
| AspRS | cvel_29834 | Cyto. | Euka. | vbra_13004 | Mito. | Euka. |
| cvel_4486 | Cyto. | Euka. | vbra_6695 | Plas. | Euka. | |
| cvel_13271 | Plas. | Euka. | ||||
| AsnRS | cvel_14360 | Cyto. | Uncl. | vbra_5336 | Plas.|Mito. | Euka. |
| cvel_7405 | Mito. | Euka. | vbra_5330 | Mito. | Euka. | |
| cvel_23534 | Cyto. | Euka. | ||||
| GlyRS | cvel_30153 | Cyto. | Euka. | vbra_22902 | Cyto. | Euka. |
| cvel_8554 | Cyto. | Euka. | vbra_14962 | Plas.|Mito. | Bact./Orga. | |
| HisRS | cvel_21705 | Cyto. | Euka. | vbra_20528 | Plas. | Euka. |
| cvel_3311(2) | (2) Plas. | Euka. | vbra_6931 | Cyto. | Euka. | |
| LysRS | cvel_9397 | Mito. | Mito. | vbra_6201 | Cyto. | Euka. |
| cvel_20258 | Cyto. | Euka. | vbra_4678 | Nucl. | Mito. | |
| cvel_7175 | Cyto.|Nul. | Euka. | ||||
| α-PheRS | cvel_9563 | Plas.|Mito. | Uncl. | vbra_15842 | Cyto.|Nucl. | Uncl. |
| cvel_5984 (3) | (3) Cyto. | Euka. | vbra_3529 | Nucl. | Euka. | |
| β-PheRS | cvel_22325 | Cyto. | Euka. | vbra_20531 | Cyto. | Euka. |
| ProRS | cvel_26651 | Cyto. | Uncl. | vbra_17540 | Cyto. | Euka. |
| cvel_17161 | Cyto. | Euka. | vbra_8238 | Endo. | Uncl. | |
| SerRS | cvel_28365 | Cyto. | Euka. | vbra_2397 | Cyto. | Euka. |
| cvel_4888 | Nucl. | Euka. | vbra_7841 | Nucl. | Mito. | |
| cvel_18368 | Endo. | Mito. | ||||
| ThrRS | cvel_8549 | Plas.|Mito. | Mito. | vbra_13128 | Nucl. | Euka. |
| cvel_11445 | Cyto. | Euka. | vbra_14881 | Plas. | Mito. | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharaf, A.; Gruber, A.; Jiroutová, K.; Oborník, M. Characterization of Aminoacyl-tRNA Synthetases in Chromerids. Genes 2019, 10, 582. https://doi.org/10.3390/genes10080582
Sharaf A, Gruber A, Jiroutová K, Oborník M. Characterization of Aminoacyl-tRNA Synthetases in Chromerids. Genes. 2019; 10(8):582. https://doi.org/10.3390/genes10080582
Chicago/Turabian StyleSharaf, Abdoallah, Ansgar Gruber, Kateřina Jiroutová, and Miroslav Oborník. 2019. "Characterization of Aminoacyl-tRNA Synthetases in Chromerids" Genes 10, no. 8: 582. https://doi.org/10.3390/genes10080582
APA StyleSharaf, A., Gruber, A., Jiroutová, K., & Oborník, M. (2019). Characterization of Aminoacyl-tRNA Synthetases in Chromerids. Genes, 10(8), 582. https://doi.org/10.3390/genes10080582







