Steroids as Environmental Compounds Recalcitrant to Degradation: Genetic Mechanisms of Bacterial Biodegradation Pathways
Abstract
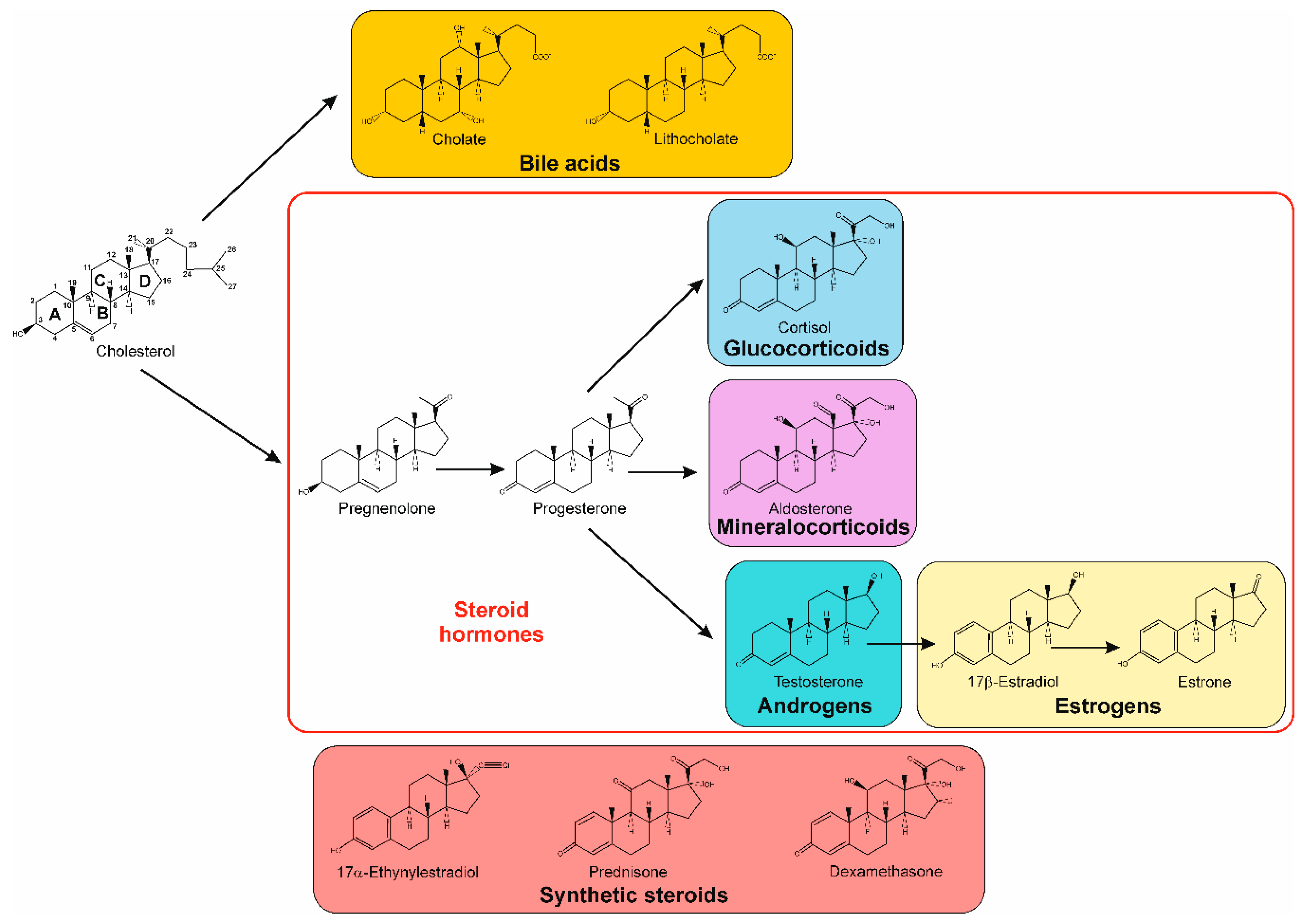
1. Introduction
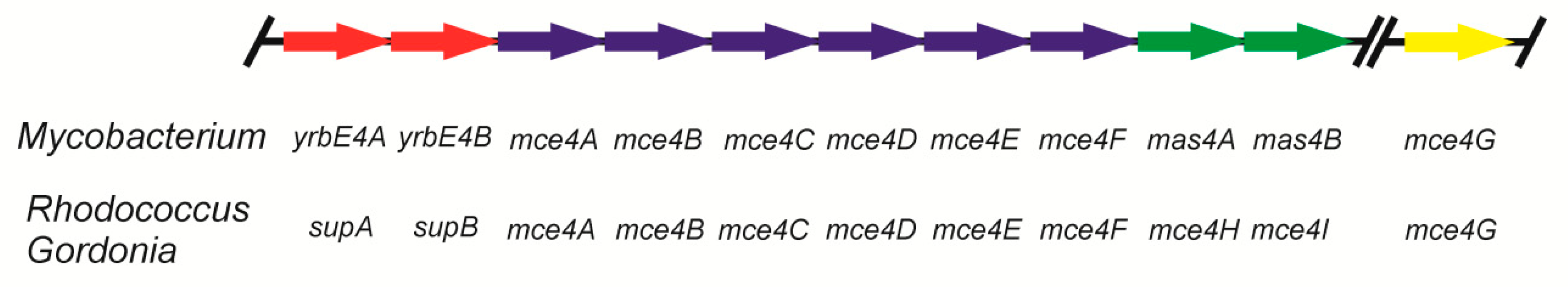
2. Uptake of Steroids
3. The Aerobic 9,10-seco Pathway
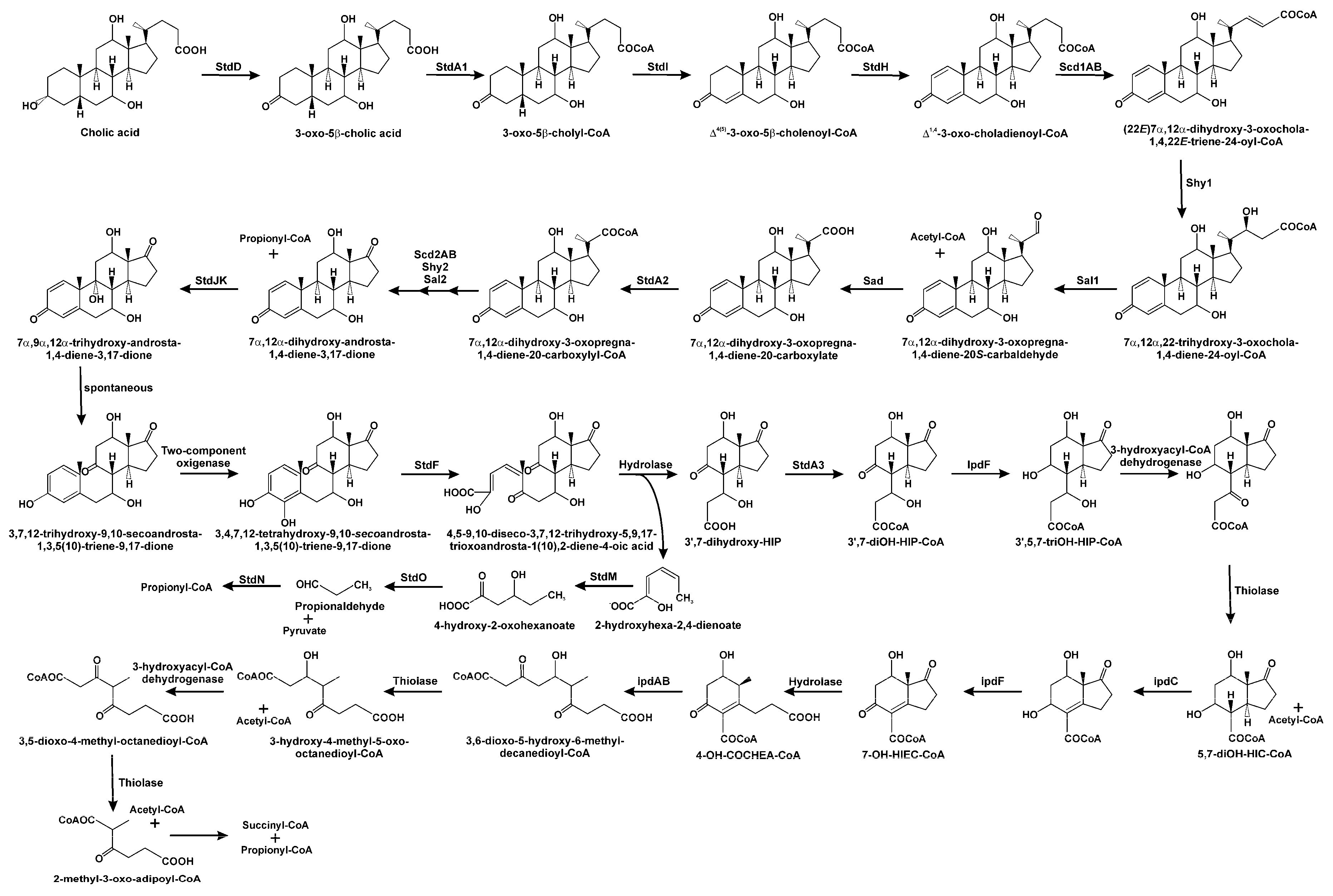
3.1. Oxidation of 3-hydroxyl-substituent from Sterols and Bile Acids
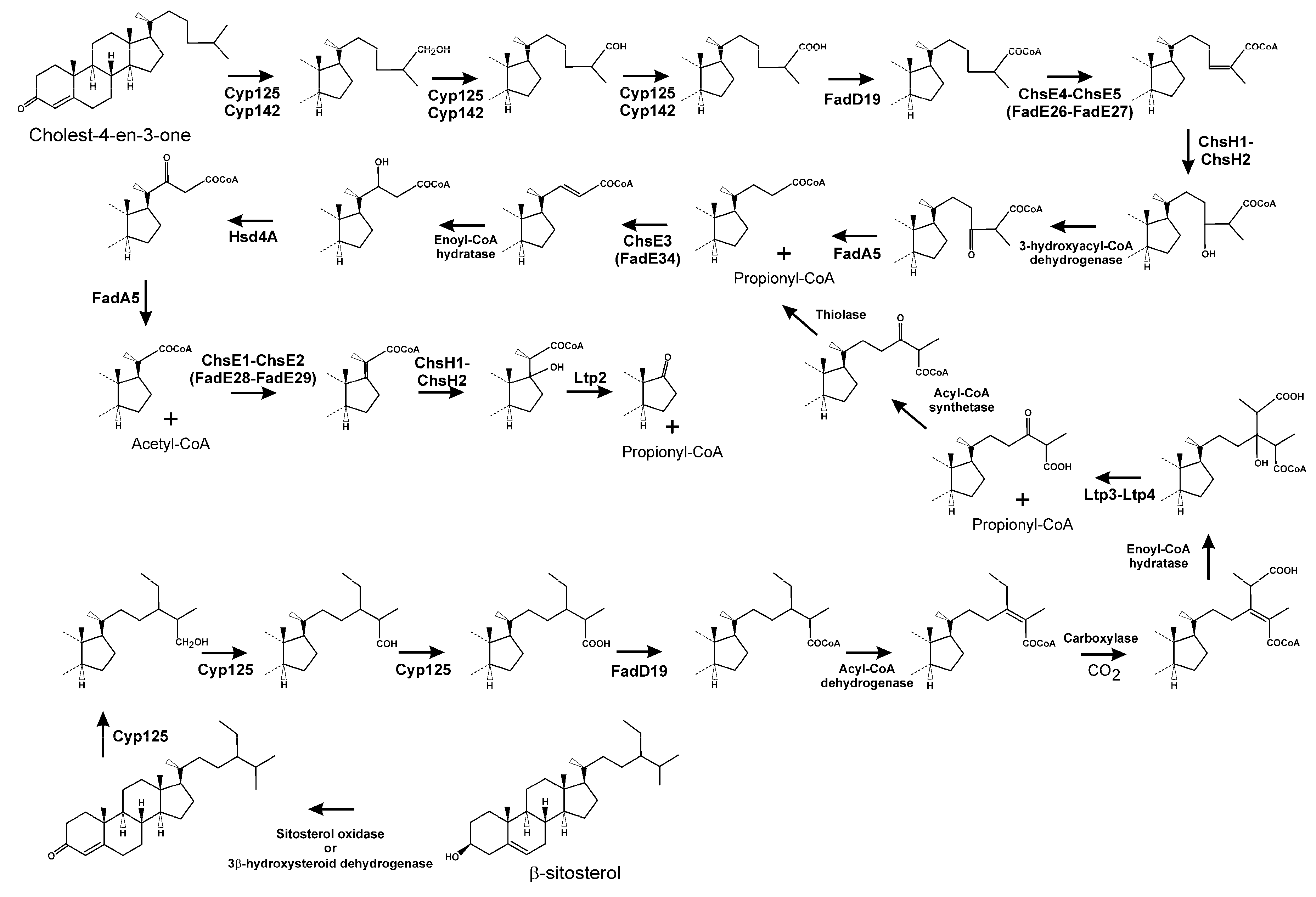
3.2. Side Chain Degradation of Sterols and Bile Acids
3.3. Testosterone Catabolism Convergence
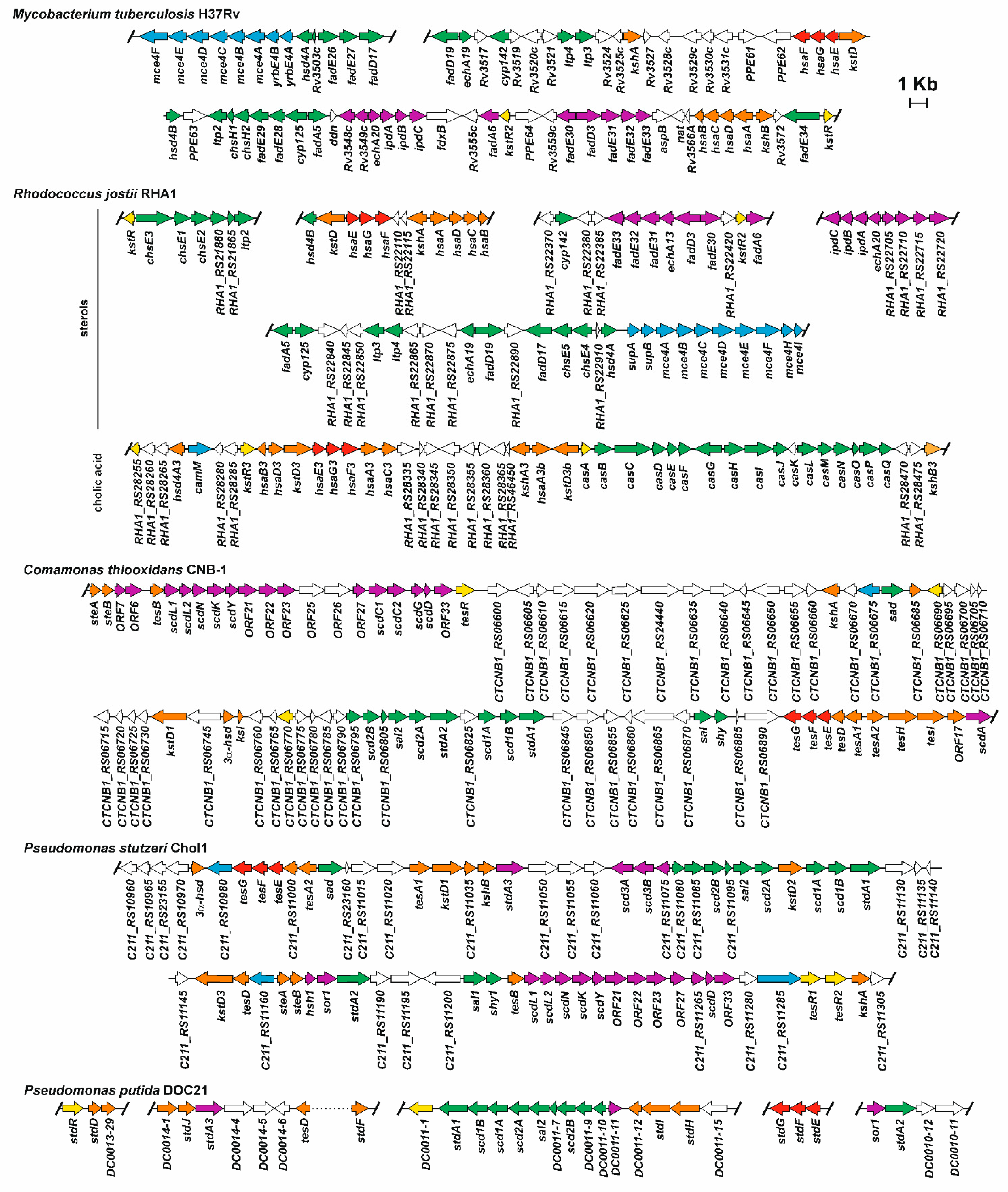
3.4. Oxidation of A/B Rings
3.5. Degradation of B/C Rings
3.6. Regulation of 9,10-seco Pathway
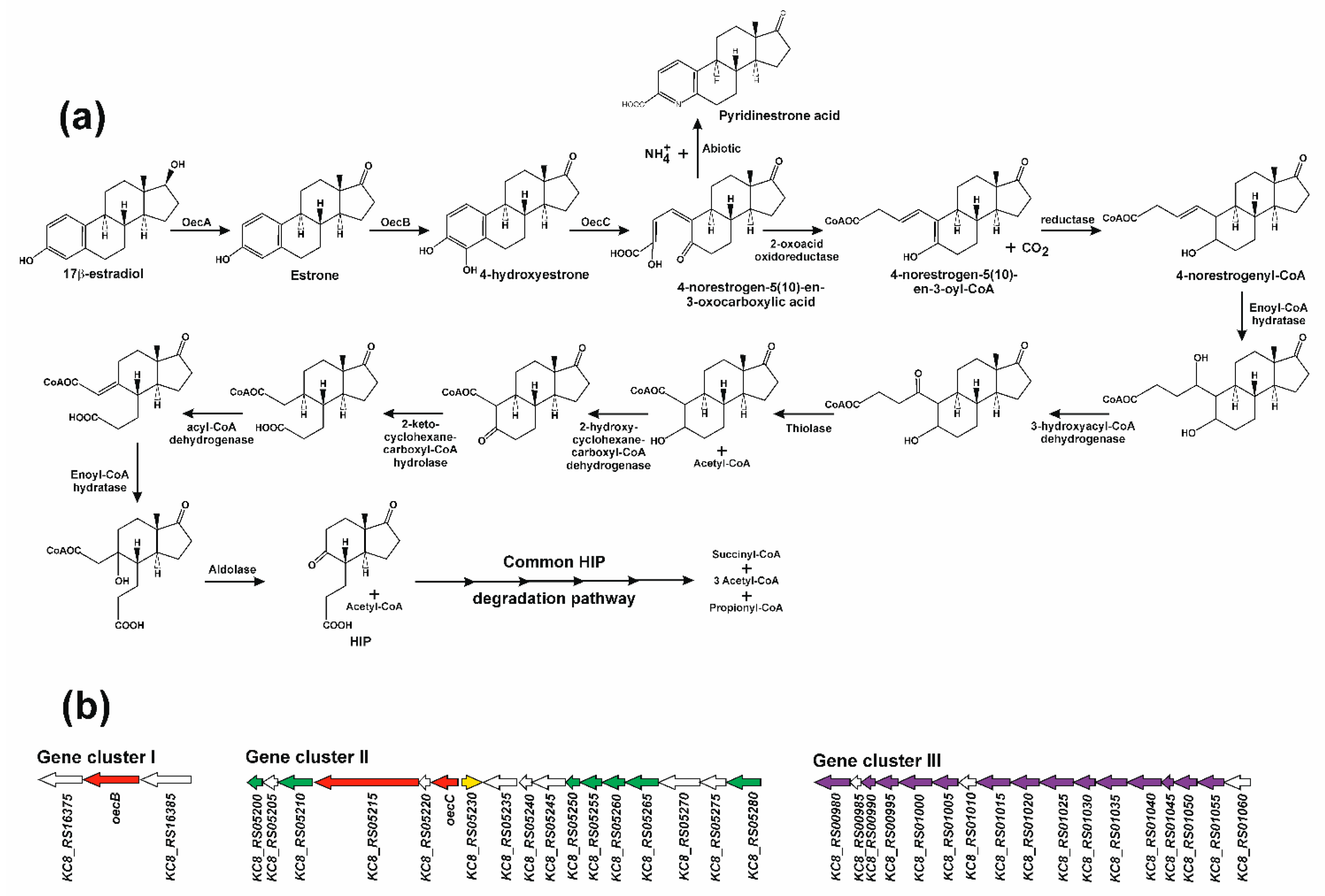
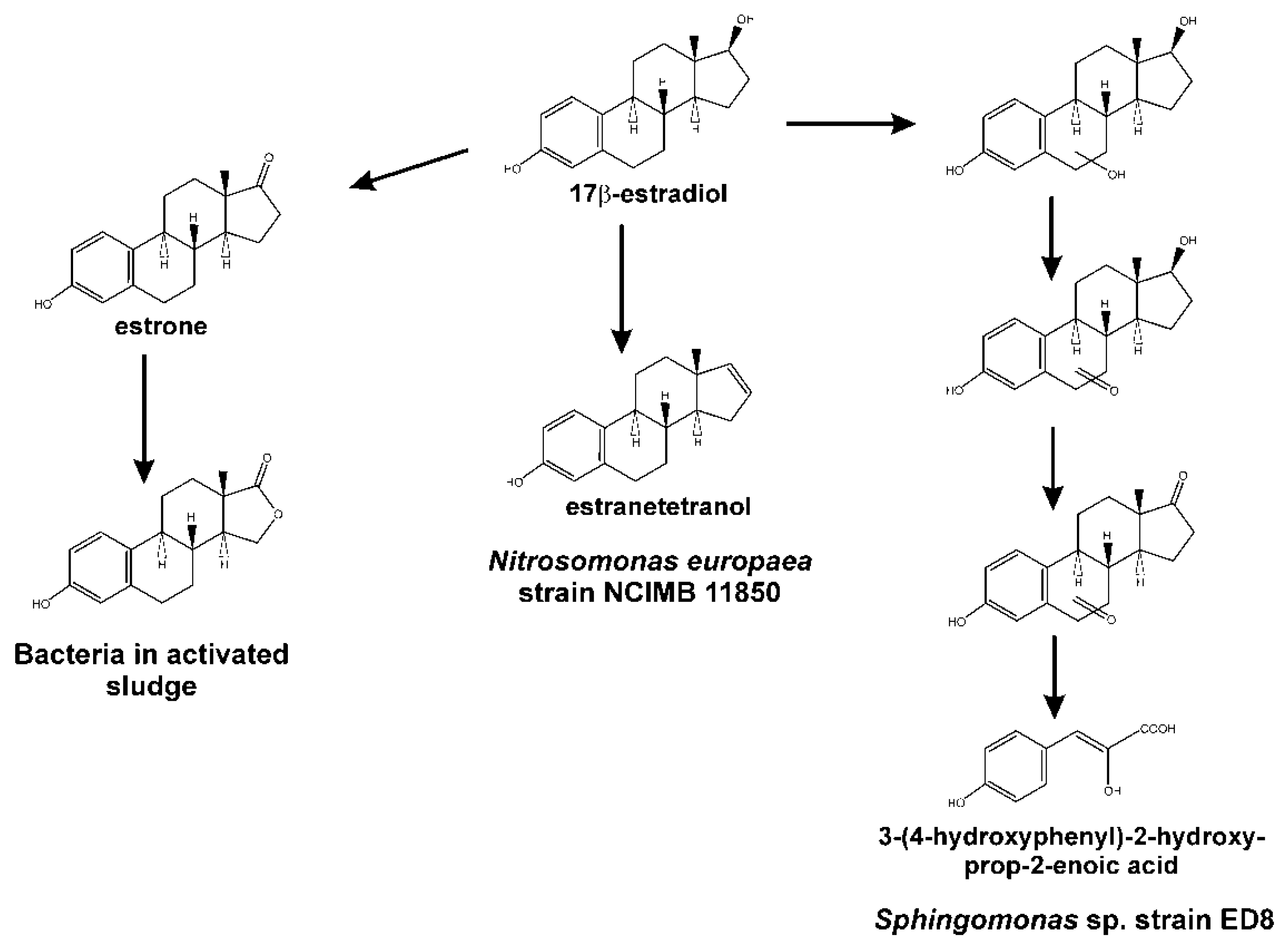
4. The 4,5-seco Pathway
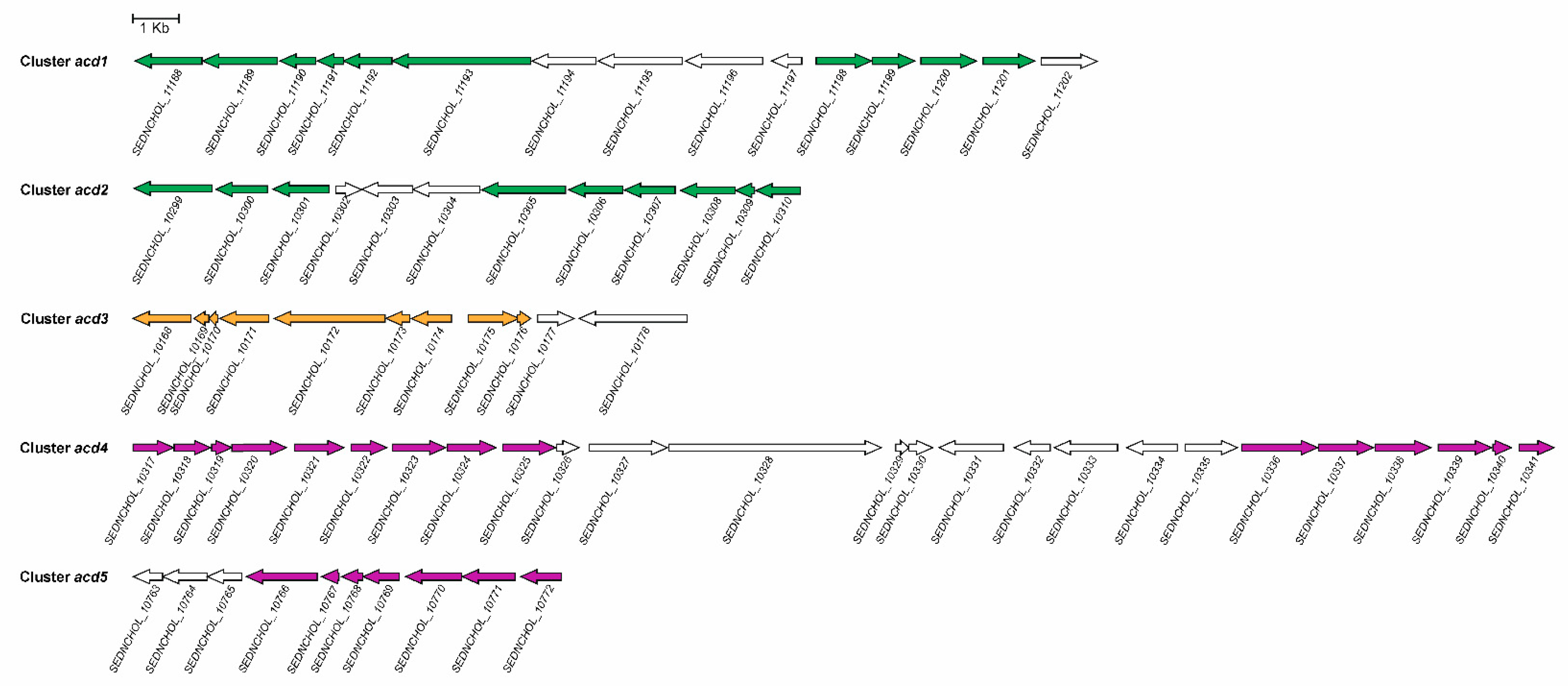
5. The Anaerobic 2,3-seco Pathway
6. Biotechnological Interest in Steroid-Degrading Microorganisms
Author Contributions
Funding
Conflicts of Interest
References
- Piironen, V.; Lindsay, D.G.; Miettinen, T.A.; Toivo, J.; Lampi, A.M. Plant sterols: Biosynthesis, biological function and their importance to human nutrition. J. Sci. Food Agric. 2000, 80, 939–966. [Google Scholar] [CrossRef]
- Fernandes, P.; Cabral, J.M.S. Phytosterols: Applications and recovery methods. Bioresour. Technol. 2007, 98, 2335–2350. [Google Scholar] [CrossRef] [PubMed]
- Wollam, J.; Antebi, A. Sterol regulation of metabolism, homeostasis, and development. Annu. Rev. Biochem. 2011, 80, 885–916. [Google Scholar] [CrossRef] [PubMed]
- Kodner, R.B.; Pearson, A.; Summons, R.E.; Knoll, A.H. Sterols in the red and green algae: Quantification, phylogeny, and relevance for the interpretation of geologic steranes. Geobiology 2008, 6, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Bird, C.W.; Lynch, J.M.; Pirt, F.J.; Reid, W.W. Steroids and squalene in Methylococcus capsulatus grown on methane. Nature 1971, 230, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Patt, T.E.; Hanson, R.S. Intracytoplasmic membrane, phospholipid, and sterol content of Methylobacterium organophilum cells grown under different conditions. J. Bacteriol. 1978, 134, 636–644. [Google Scholar]
- Kohl, W.; Gloe, A.; Reichenbach, H. Steroids from the myxobacterium Nannocystis exedens. J. Gen. Microbiol. 1983, 129, 1629–1635. [Google Scholar] [CrossRef]
- Schouten, S.; Bowman, J.P.; Rijpstra, W.I.C.; Damsté, J.S.S. Sterols in a psychrophilic methanotroph, Methylosphaera hansonii. FEMS Microbiol. Lett. 2000, 186, 193–195. [Google Scholar] [CrossRef]
- Bode, H.B.; Zeggel, B.; Silakowski, B.; Wenzel, S.C.; Reichenbach, H.; Müller, R. Steroid biosynthesis in prokaryotes: Identification of myxobacterial steroids and cloning of the first bacterial 2,3(S)-oxidosqualene cyclase from the mixobacterium Stigmatella aurantiaca. Mol. Microbiol. 2003, 47, 471–481. [Google Scholar] [CrossRef]
- Pearson, A.; Budin, M.; Brocks, J.J. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. USA 2003, 100, 15352–15357. [Google Scholar] [CrossRef]
- Lamb, D.C.; Jackson, C.J.; Warrilow, A.G.; Manning, N.J.; Kelly, D.E.; Kelly, S.L. Lanosterol biosynthesis in the prokaryote Methylococcus capsulatus. Insight into the evolution of sterol biosynthesis. Mol. Biol. Evol. 2007, 24, 1714–1721. [Google Scholar] [CrossRef] [PubMed]
- Gawas, D.; García, R.; Huch, V.; Müller, R. A highly conjugated dihydroxylated C28 steroid from a myxobacterium. J. Nat. Prod. 2011, 74, 1281–1283. [Google Scholar] [CrossRef] [PubMed]
- García, R.; Gemperlein, K.; Müller, R. Minicystis rosea gen. Nov., sp. Nov., a polyunsaturated fatty acid-rich and steroid-producing soil myxobacterium. Int. J. Syst. Evol. Microbiol. 2014, 64, 3733–3742. [Google Scholar] [CrossRef] [PubMed]
- Banta, A.B.; Wei, J.H.; Welander, P.V. A distinc pathway for tetrahymanol synthesis in bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 13478–13483. [Google Scholar] [CrossRef] [PubMed]
- Banta, A.B.; Wei, J.H.; Gill, C.C.C.; Giner, J.-L.; Welander, P.V. Synthesis of arborane triterpenols by a bacterial oxidosqualene cyclase. Proc. Natl. Acad. Sci. USA 2017, 114, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Lange, I.G.; Daxenberger, A.; Schiffer, B.; Witters, H.; Ibarreta, D.; Meyer, H.D.D. Sex hormones originating from different livestock production systems: Fate and potential disrupting activity in the environment. Anal. Chim. Acta 2002, 473, 27–37. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Froehner, S.; Martins, R.F.; Errera, M.R. Assessment of fecal sterols in Barigui River sediments in Curitiba, Brazil. Environ. Monit. Assess. 2009, 157, 591–600. [Google Scholar] [CrossRef]
- Chang, H.S.; Choo, K.H.; Lee, B.; Choi, S.J. The methods for identification, analysis, and removal of endocrine disrupting compounds (EDCs) in water. J. Hazard. Mater. 2009, 172, 1–12. [Google Scholar] [CrossRef]
- Santos, L.H.; Araújo, A.N.; Fachini, A.; Pena, A.; Delerue-Matos, C.; Montenegro, M.C. Ecotoxicological aspects related to the presence of pharmaceuticals in the aquatic environment. J. Hazard. Mater. 2010, 175, 45–95. [Google Scholar] [CrossRef]
- Prost, K.; Birk, J.J.; Lehndorff, E.; Gerlach, R.; Amelung, W. Steroid biomarkers revisited-improved source identification of faecal remains in archaeological soil material. PLoS ONE 2017, 12, e0164882. [Google Scholar] [CrossRef] [PubMed]
- Matić Bujagić, I.; Grujić, S.; Laušević, M.; Hofmann, T.; Micić, V. Emerging contaminants in sediment core from the Iron Gate I Reservoir on the Danube River. Sci. Total Environ. 2019, 662, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Gagné, F.; Blaise, C.; André, C. Occurrence of pharmaceutical products in a municipal effluent and toxicity to rainbow trout (Oncorhynchus mykiss) hepatocytes. Ecotoxicol. Environ. Saf. 2006, 64, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Crane, M.; Watts, C.; Boucard, T. Chronic aquatic environmental risks from exposure to human pharmaceuticals. Sci. Total Environ. 2006, 367, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Galli, R.; Braun, C. Integrative risk assessment of endocrine disruptors in Switzerland. Chimia 2008, 62, 417–423. [Google Scholar] [CrossRef]
- Tong, W.-Y.; Dong, X. Microbial biotransformation: Recent developments on steroid drugs. Recent Pat. Biotechnol. 2009, 3, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Runnalls, T.J.; Margiotta-Casaluci, L.; Kugathas, S.; Sumpter, J.P. Pharmaceuticals in the aquatic environment: Steroids and anti-steroids as high priorities for research. Hum. Ecol. Risk Assess. 2010, 16, 1318–1338. [Google Scholar] [CrossRef]
- Philipp, B.; Erdbrink, H.; Suter, M.J.; Schink, B. Degradation of and sensitivity to cholate in Pseudomonas sp. strain Chol1. Arch. Microbiol. 2006, 185, 192–201. [Google Scholar] [CrossRef]
- Merino, E.; Barrientos, A.; Rodríguez, J.; Naharro, G.; Luengo, J.M.; Olivera, E.R. Isolation of cholesterol- and deoxycholate-degrading bacteria from soil samples: Evidence of a common pathway. Appl. Microbiol. Biotechnol. 2013, 97, 891–904. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, W.; Cao, Z.; Xu, B.; Wang, G.; Luo, M. High correlation between genotypes and phenotypes of environmental bacteria Comamonas testosteroni strains. BMC Genom. 2015, 16, 110. [Google Scholar] [CrossRef]
- Holert, J.; Yücel, O.; Suvekbala, V.; Kulić, Z.; Möller, H.; Philipp, B. Evidence of distinct pathways for bacterial degradation of the steroid compound cholate suggests the potential for metabolic interactions by interspecies cross-feeding. Environ. Microbiol. 2014, 16, 1424–1440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xiong, G.; Maser, E. Characterization of the steroid degrading bacterium S19-1 from the Baltic Sea at Kiel, Germany. Chem. Biol. Interact. 2011, 191, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Louvado, A.; Esteves, V.I.; Gomes, N.C.M.; Almeida, A.; Cunha, Â. Biodegradation of 17β-estradiol by bacteria isolated from deep sea sediments in aerobic and anaerobic media. J. Hazard. Mater. 2017, 323, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Holert, J.; Cardenas, E.; Bergstrand, L.H.; Zaikova, E.; Hahn, A.S.; Hallam, S.J.; Mohn, W.W. Metagenomes reveal global distribution of bacterial steroid catabolism in natural, engineered, and host environments. mBio 2018, 9, e02345-17. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.K.; Sassetti, C.M. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA 2008, 105, 4376–4380. [Google Scholar] [CrossRef] [PubMed]
- van der Geize, R.; Grommen, A.W.; Hessels, G.I.; Jacobs, A.A.; Dijkhuizen, L. The steroid catabolic pathway of the intracellular pathogen Rhodococcus equi is important for pathogenesis and a target for vaccine development. PLoS Pathog. 2011, 7, e1002181. [Google Scholar] [CrossRef]
- Söhngen, N.L. Benzin, petroleum, paraffinöl und paraffin als kohlenstoff- und energiequelle für mikroben. Zent. Bakteriol. Parasitenkd. Infekt. 1913, 27, 595–609. [Google Scholar]
- Tak, J.D. On bacteria decomposing cholesterol. Antonie Leeuwenhoek 1942, 8, 32–40. [Google Scholar] [CrossRef]
- Arnaudi, C. Flavobacterium dehydrogenans (Micrococcus dehydrogenans) und seine Fähigkeit zur oxidation von steroiden sowie substanzen aus der sexualhormonreibe. Zentr. Bakt. Parasitenk. II 1942, 105, 352–366. [Google Scholar]
- Turfitt, G.E. Microbiological agencies in the degradation of steroids: I. The cholesterol-decomposing organisms of soils. J. Bacteriol. 1944, 47, 487–493. [Google Scholar]
- Turfitt, G.E. The microbiological degradation of steroids 4. Fission of the steroid molecule. Biochem. J. 1948, 42, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Horvath, J.; Kramli, A. Microbiological oxidation of cholesterol with Azotobacter. Nature 1947, 160, 639. [Google Scholar] [CrossRef] [PubMed]
- Whitmarsh, J.M. Intermediates of microbiological metabolism of cholesterol. Biochem. J. 1964, 90, 23–24. [Google Scholar]
- Brown, R.L.; Peterson, G.E. Cholesterol oxidation by soil Actinomycetes. J. Gen. Microbiol. 1966, 45, 441–450. [Google Scholar] [CrossRef][Green Version]
- Arima, K.; Nagasawa, M.; Bae, M.; Tamura, G. Microbial transformation of sterols. Part I. Decomposition of cholesterol by microorganisms. Agric. Biol. Chem. 1969, 33, 1636–1643. [Google Scholar]
- Chipley, J.R.; Dreyfuss, M.S.; Smucker, R.A. Cholesterol metabolism by Mycobacterium. Microbios 1975, 12, 199–207. [Google Scholar] [PubMed]
- Martin, C.K.A. Microbial transformation of β-sitosterol by Nocardia sp. M29. Eur. J. Appl. Microbiol. 1976, 2, 243–255. [Google Scholar] [CrossRef]
- Ferreira, N.P.; Tracey, R.P. Numerical taxonomy of cholesterol-degrading soil bacteria. J. Appl. Microbiol. 1984, 57, 429–446. [Google Scholar] [CrossRef]
- Mahato, S.B.; Banerjee, S.; Sahu, N.P. Metabolism of progesterone and testosterone by a Bacillus sp. Steroids 1984, 43, 545–558. [Google Scholar] [CrossRef]
- Mahato, S.B.; Banerjee, S. Metabolism of 11-deoxycortisol by a Bacillus species. J. Steroid Biochem. 1986, 25, 995–999. [Google Scholar] [CrossRef]
- Yoshimoto, T.; Nagai, F.; Fujimoto, J.; Watanabe, K.; Mizukoshi, H.; Makino, T.; Kimura, K.; Saino, H.; Sawada, H.; Omura, H. Degradation of strogens by Rhodococcus zopfii and Rhodococcus equi isolates from activated sludge in wastewater treatment plants. Appl. Environ. Microbiol. 2004, 70, 5283–5289. [Google Scholar] [CrossRef] [PubMed]
- Fahrbach, M.; Kuever, J.; Meinke, R.; Kämpfer, P.; Hollender, J. Denitratisoma oestradiolicum gen. nov., sp. nov., a 17beta-oestradiol-degrading, denitrifying betaproteobacterium. Int. J. Syst. Evol. Microbiol. 2006, 56, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Drzyzga, O.; Navarro Llorens, J.M.; Fernández de Las Heras, L.; García Fernández, E.; Perera, J. Gordonia cholesterolivorans sp. nov., a cholesterol-degrading actinomycete isolated from sewage sludge. Int. J. Syst. Evol. Microbiol. 2009, 59, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Fernández de las Heras, L.; García Fernández, E.; Navarro Llorens, J.M.; Perera, J.; Drzyzga, O. Morphological, physiological, and molecular characterization of a newly isolated steroid-degrading Actinomycete, identified as Rhodococcus ruber strain Chol-4. Curr. Microbiol. 2009, 59, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Van der Geize, R.; Yam, K.; Heuser, T.; Wilbrink, M.H.; Hara, H.; Anderton, M.C.; Sim, E.; Dijkhuizen, L.; Davies, J.E.; Mohn, W.W.; et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. USA 2007, 104, 1947–1952. [Google Scholar] [CrossRef] [PubMed]
- Uhía, I.; Galán, B.; Morales, V.; García, J.L. Initial step in the catabolism of cholesterol by Mycobacterium smegmatis mc2 155. Environ. Microbiol. 2011, 13, 943–959. [Google Scholar] [CrossRef]
- Mohn, W.W.; Wilbrink, M.H.; Casabon, I.; Stewart, G.R.; Liu, J.; van der Geize, R.; Eltis, L.D. A gene cluster encoding cholate catabolism in Rhodococcus spp. J. Bacteriol. 2012, 194, 6712–6719. [Google Scholar] [CrossRef]
- Fernández de las Heras, L.; Perera, J.; Navarro Llorens, J.M. Cholesterol to cholestenone oxidation by ChoG, the main extracellular oxidase of Rhodococcus ruber strain Chol-4. J. Steroid Biochem. Mol. Biol. 2013, 139, 33–44. [Google Scholar] [CrossRef]
- Liu, Y.; Ge, F.; Chen, G.; Li, W.; Ma, P.; Zhang, G.; Zeng, L. Gordonia neofelifaecis sp. nov., a cholesterol side-chain-cleaving actinomycete isolated from the faeces of Neofelis nebulosa. Int. J. Syst. Evol. Microbiol. 2011, 61, 165–169. [Google Scholar] [CrossRef]
- Horinouchi, M.; Hayashi, T.; Kudo, T. Steroid degradation in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 2012, 129, 4–14. [Google Scholar] [CrossRef]
- Holert, J.; Alam, I.; Larsen, M.; Antunes, A.; Bajic, V.B.; Stingl, U.; Philipp, B. Genome sequence of Pseudomonas sp. strain Chol1, a model organism for the degradation of bile acids and other steroid compounds. Genome Announc. 2013, 1, e00014-12. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, A.; Merino, E.; Casabon, I.; Rodríguez, J.; Crowe, A.M.; Holert, J.; Philipp, B.; Eltis, L.D.; Olivera, E.R.; Luengo, J.M. Functional analyses of three acyl-CoA synthetases involved in bile acid degradation in Pseudomonas putida DOC21. Environ. Microbiol. 2015, 17, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Tarlera, S.; Denner, E.B.M. Sterolibacterium denitrificans gen. nov., sp. nov., a novel cholesterol-oxidizing, denitrifying member of the β-Proteobacteria. Int. J. Syst. Evol. Microbiol. 2003, 53, 1085–1091. [Google Scholar] [PubMed]
- Chiang, Y.R.; Ismail, W.; Heintz, D.; Schaeffer, C.; Van Dorsselaer, A.; Fuchs, G. Study of anoxic and oxic cholesterol metabolism by Sterolibacterium denitrificans. J. Bacteriol. 2008, 190, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Fahrbach, M.; Kuever, J.; Remesch, M.; Huber, B.E.; Kämpfer, P.; Dott, W.; Hollender, J. Steroidobacter denitrificans gen. nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 2008, 58, 2215–2223. [Google Scholar] [CrossRef] [PubMed]
- Plésiat, P.; Nikaido, H. Outer membranes of gram-negative bacteria are permeable to steroid probes. Mol. Microbiol. 1992, 6, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Mohn, W.W.; van der Geize, R.; Stewart, G.R.; Okamoto, S.; Liu, J.; Dijkhuizen, L.; Eltis, L.D. The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 2008, 283, 35368–35374. [Google Scholar] [CrossRef] [PubMed]
- Van der Geize, R.; de Jong, W.; Hessels, G.I.; Grommen, A.W.F.; Jacobs, A.A.C.; Dijkhuizen, L. A novel method to generate unmarked gene deletions in the intracellular pathogen Rhodococcus equi using 5-fluorocytosine conditional lethality. Nucleic Acids Res. 2008, 36, e151. [Google Scholar] [CrossRef]
- Drzyzga, O.; Fernández de las Heras, L.; Morales, V.; Navarro Llorens, J.M.; Perera, J. Cholesterol degradation by Gordonia cholesterolivorans. Appl. Environ. Microbiol. 2011, 77, 4802–4810. [Google Scholar] [CrossRef]
- Klepp, L.I.; Forrellad, M.A.; Osella, A.V.; Blanco, F.C.; Stella, E.J.; Bianco, M.V.; de la Paz Santangelo, M.; Sassetti, C.; Jackson, M.; Cataldi, A.A.; et al. Impact of the deletion of the six mce operons in Mycobacterium smegmatis. Microbes Infect. 2012, 14, 590–599. [Google Scholar] [CrossRef][Green Version]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., 3rd; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, J.; Papavinasasundaram, K.; Galán, B.; Sassetti, C.M.; García, J.L. Molecular and functional analysis of the mce4 operon in Mycobacterium smegmatis. Environ. Microbiol. 2017, 19, 3689–3699. [Google Scholar] [CrossRef] [PubMed]
- Kendall, S.L.; Withers, M.; Soffair, C.N.; Moreland, N.J.; Gurcha, S.; Sidders, B.; Frita, R.; ten Bokum, A.; Besra, G.S.; Lott, J.S.; et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol. Microbiol. 2007, 65, 684–699. [Google Scholar] [CrossRef] [PubMed]
- Casali, N.; Riley, L.W. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genom. 2007, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.M.; Pandey, A.K.; Capite, N.; Fortune, S.M.; Rubin, E.J.; Sassetti, C.M. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc. Natl. Acad. Sci. USA 2006, 103, 11760–11765. [Google Scholar] [CrossRef] [PubMed]
- Nazarova, E.V.; Montague, C.R.; La, T.; Wilburn, K.M.; Sukumar, N.; Lee, W.; Caldwell, S.; Russell, D.G.; VanderVen, B.C. Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. eLife 2017, 6, e26969. [Google Scholar] [CrossRef] [PubMed]
- Sassetti, C.M.; Rubin, E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 2003, 100, 12989–12994. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Magallanes, V.; Stadthagen-Gomez, G.; Rauzier, J.; Barreiro, L.B.; Tailleux, L.; Boudou, F.; Griffin, R.; Nigou, J.; Jackson, M.; Gicquel, B.; et al. Signature-tagged transposon mutagenesis identifies novel Mycobacterium tuberculosis genes involved in the parasitism of human macrophages. Infect. Immun. 2007, 75, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Sandie, R.; Wang, Y.; Andrade-Navarro, M.A.; Niederweis, M. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis 2008, 88, 526–544. [Google Scholar] [CrossRef] [PubMed]
- Feltcher, M.E.; Gunawardena, H.P.; Zulauf, K.E.; Malik, S.; Griffin, J.E.; Sassetti, C.M.; Chen, X.; Braunstein, M. Label-free Quantitative Proteomics reveals a role for the Mycobacterium tuberculosis SecA2 pathway in exporting solute binding proteins and Mce transporters to the cell wall. Mol. Cell. Proteom. 2015, 14, 1501–1516. [Google Scholar] [CrossRef] [PubMed]
- Perkowski, E.F.; Miller, B.K.; McCann, J.R.; Sullivan, J.T.; Malik, S.; Allen, I.C.; Godfrey, V.; Hayden, J.D.; Braunstein, M. An orphaned Mce-associated membrane protein of Mycobacterium tuberculosis is a virulence factor that stabilizes Mce transporters. Mol. Microbiol. 2016, 100, 90–107. [Google Scholar] [CrossRef] [PubMed]
- Somalinga, V.; Mohn, W.W. Rhodococcus jostii porin A (RjpA) functions in cholate uptake. Appl. Environ. Microbiol. 2013, 79, 6191–6193. [Google Scholar] [CrossRef] [PubMed]
- Haußmann, U.; Wolters, D.A.; Fränzel, B.; Eltis, L.D.; Poetsch, A. Physiological adaptation of the Rhodococcus jostii RHA1 membrane proteome to steroids as growth substrates. J. Proteome Res. 2013, 12, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Swain, K.; Casabon, I.; Eltis, L.D.; Mohn, W.W. Two transporters essential for reassimilation of novel cholate metabolites by Rhodococcus jostii RHA1. J. Bacteriol. 2012, 194, 6720–6727. [Google Scholar] [CrossRef] [PubMed]
- Birkenmaier, A.; Holert, J.; Erdbrink, H.; Moeller, H.M.; Friemel, A.; Schoenenberger, R.; Suter, M.J.; Klebensberger, J.; Philipp, B. Biochemical and genetic investigation of initial reactions in aerobic degradation of the bile acid cholate in Pseudomonas sp. strain Chol1. J. Bacteriol. 2007, 189, 7165–7173. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Wang, P.H.; Ismail, W.; Tsai, Y.W.; El Nayal, A.; Yang, C.Y.; Yang, F.C.; Wang, C.H.; Chiang, Y.R. Substrate uptake and subcellular compartmentation of anoxic cholesterol catabolism in Sterolibacterium denitrificans. J. Biol. Chem. 2015, 290, 1155–1169. [Google Scholar] [CrossRef] [PubMed]
- Holert, J.; Jagmann, N.; Philipp, B. The essential function of genes for a hydratase and an aldehyde dehydrogenase for growth of Pseudomonas sp. strain Chol1 with the steroid compound cholate indicates an aldolytic reaction step for deacetylation of the side chain. J. Bacteriol. 2013, 195, 3371–3380. [Google Scholar] [CrossRef]
- Petrusma, M.; Dijkhuizen, L.; van der Geize, R. Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl. Environ. Microbiol. 2009, 75, 5300–5307. [Google Scholar] [CrossRef]
- Griffin, J.E.; Gawronski, J.D.; Dejesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef]
- Thomas, S.T.; VanderVen, B.C.; Sherman, D.R.; Russell, D.G.; Sampson, N.S. Pathway profiling in Mycobacterium tuberculosis: Elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J. Biol. Chem. 2011, 286, 43668–43678. [Google Scholar] [CrossRef]
- Uhía, I.; Galán, B.; Kendall, S.L.; Stoker, N.G.; García, J.L. Cholesterol metabolism in Mycobacterium smegmatis. Environ. Microbiol. Rep. 2012, 4, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ge, F.; Zhang, Q.; Ren, Y.; Yuan, J.; He, J.; Li, W.; Chen, G.; Zhang, G.; Zhuang, Y.; et al. Identification of gene expression profiles in the actinomycete Gordonia neofelifaecis grown with different steroids. Genome 2014, 57, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Bergstrand, L.H.; Cardenas, E.; Holert, J.; Van Hamme, J.D.; Mohn, W.W. Delineation of steroid-degrading microorganisms through comparative genomic analysis. mBio 2016, 7, e00166-16. [Google Scholar] [CrossRef] [PubMed]
- Rosłoniec, K.Z.; Wilbrink, M.H.; Capyk, J.K.; Mohn, W.W.; Ostendorf, M.; van der Geize, R.; Dijkhuizen, L.; Eltis, L.D. Cytochrome P450 125 (CYP125) catalyses C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii RHA1. Mol. Microbiol. 2009, 74, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Kreit, J. Microbial catabolism of sterols: Focus on the enzymes that transform the sterol 3β-hydroxy-5-en into 3-keto-4-en. FEMS Microbiol. Lett. 2017, 364, fnx007. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Vrielink, A.; Brick, P.; Blow, D.M. Crystal structure of cholesterol oxidase complexed with a steroid substrate: Implications for flavin adenine dinucleotide dependent alcohol oxidases. Biochemistry 1993, 32, 11507–11515. [Google Scholar] [CrossRef]
- Coulombe, R.; Yue, K.Q.; Ghisla, S.; Vrielink, A. Oxygen access to the active site of cholesterol oxidase through a narrow channel is gated by an Arg-Glu pair. J. Biol. Chem. 2001, 276, 30435–30441. [Google Scholar] [CrossRef]
- Horinouchi, S.; Ishizuka, H.; Beppu, T. Cloning, nucleotide sequence, and transcriptional analysis of the NAD(P)-dependent cholesterol dehydrogenase gene from a Nocardia sp. and its hyperexpression in Streptomyces spp. Appl. Environ. Microbiol. 1991, 57, 1386–1393. [Google Scholar]
- Yang, X.; Dubnau, E.; Smith, I.; Sampson, N.S. Rv1106c from Mycobacterium tuberculosis is a 3β-hydroxysteroid dehydrogenase. Biochemistry 2007, 46, 9058–9067. [Google Scholar] [CrossRef]
- Yang, X.; Gao, J.; Smith, I.; Dubnau, E.; Sampsonn, N.S. Cholesterol is not an essential source of nutrition for Mycobacterium tuberculosis during infection. J. Bacteriol. 2011, 193, 1473–1476. [Google Scholar] [CrossRef]
- Brzostek, A.; Rumijowska-Galewicz, A.; Dziadek, B.; Wojcik, E.A.; Dziadek, J. ChoD and HsdD can be dispensable for cholesterol degradation in mycobacteria. J. Steroid Biochem. Mol. Biol. 2013, 134, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, F.; Maser, E. Carbonyl reductases and pluripotent hydroxysteroid dehydrogenases of the short-chain dehydrogenase/reductase superfamily. Drug Metab. Rev. 2007, 39, 87–144. [Google Scholar] [CrossRef] [PubMed]
- Marcus, P.I.; Talalay, P. Induction and purification of α- and β-hydroxysteroid dehydrogenases. J. Biol. Chem. 1956, 218, 661–674. [Google Scholar] [PubMed]
- Oppermann, U.C.T.; Maser, E. Characterization of a 3 α-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium Comamonas testosteroni. Eur. J. Biochem. 1996, 241, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Oppermann, U.C.T.; Belai, I.; Maser, E. Antibiotic resistance and enhanced insecticide catabolism as consequences of steroid induction in the Gram-negative bacterium Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 1996, 58, 217–223. [Google Scholar] [CrossRef]
- Möbus, E.; Maser, E. Molecular cloning, overexpression, and characterization of steroid-inducible 3α-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni. A novel member of the short-chain dehydrogenase/reductase superfamily. J. Biol. Chem. 1998, 273, 30888–30896. [Google Scholar] [CrossRef]
- Horinouchi, M.; Kurita, T.; Hayashi, T.; Kudo, T. Steroid degradation genes in Comamonas testosteroni TA441: Isolation of genes encoding a Δ4(5)-isomerase and 3α- and 3β-dehydrogenases and evidence for a 100 kb steroid degradation gene hot spot. J. Steroid Biochem. Mol. Biol. 2010, 122, 253–263. [Google Scholar] [CrossRef]
- Holert, J.; Yücel, O.; Jagmann, N.; Prestel, A.; Möller, H.M.; Philipp, B. Identification of bypass reactions leading to the formation of one central steroid degradation intermediate in metabolism of different bile salts in Pseudomonas sp. strain Chol1. Environ. Microbiol. 2016, 18, 3373–3389. [Google Scholar] [CrossRef]
- Shi, C.J.; Tai, H.H.; Tsong, Y.Y. The mechanism of microbial conversion of cholesterol into 17-keto steroids. J. Am. Chem. Soc. 1967, 89, 1957–1958. [Google Scholar]
- Shi, C.J.; Tai, H.H.; Tsong, Y.Y.; Lee, S.S.; Coombe, R.G. Mechanisms of steroid oxidation by microorganisms 0.14. Pathway of cholesterol side-chain degradation. Biochemistry 1968, 7, 808–818. [Google Scholar]
- Reddy, J.K.; Hashimoto, T. Peroxisomal β-oxidation and peroxisome proliferator activated receptor α: An adaptive metabolic system. Annu. Rev. Nutr. 2001, 21, 193–230. [Google Scholar] [CrossRef] [PubMed]
- Capyk, J.K.; Kalscheuer, R.; Stewart, G.R.; Liu, J.; Kwon, H.; Zhao, R.; Okamoto, S.; Jacobs, W.R., Jr.; Eltis, L.D.; Mohn, W.W. Mycobacterial cytochrome p450 125 (cyp125) catalyzes the terminal hydroxylation of C27 steroids. J. Biol. Chem. 2009, 284, 35534–35542. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, H.; Guan, S.; Johnston, J.B.; Chow, E.D.; Kells, P.M.; Burlingame, A.L.; Cox, J.S.; Podust, L.M.; de Montellano, P.R. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Mol. Microbiol. 2010, 77, 730–742. [Google Scholar] [CrossRef] [PubMed]
- McLean, K.J.; Lafite, P.; Levy, C.; Cheesman, M.R.; Mast, N.; Pikuleva, I.A.; Leys, D.; Munro, A.W. The structure of Mycobacterium tuberculosis CYP125: Molecular basis for cholesterol binding in a P450 needed for host infection. J. Biol. Chem. 2009, 284, 35524–35533. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, H.; Johnston, J.B.; de Montellano, P.R. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends Microbiol. 2011, 19, 530–539. [Google Scholar] [CrossRef]
- Johnston, J.B.; Ouellet, H.; Ortiz de Montellano, P.R. Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses. J. Biol. Chem. 2010, 285, 36352–36360. [Google Scholar] [CrossRef]
- Frank, D.J.; Madrona, Y.; Ortiz de Montellano, P.R. Cholesterol ester oxidation by mycobacterial cytochrome P450. J. Biol. Chem. 2014, 289, 30417–30425. [Google Scholar] [CrossRef]
- Wilbrink, M.H.; Petrusma, M.; Dijkhuizen, L.; van der Geize, R. FadD19 of Rhodococcus rhodochrous DSM43269, a steroid-coenzyme A ligase essential for degradation of C-24 branched sterol side chains. Appl. Environ. Microbiol. 2011, 77, 4455–4464. [Google Scholar] [CrossRef]
- Casabon, I.; Swain, K.; Crowe, A.M.; Eltis, L.D.; Mohn, W.W. Actinobacterial acyl coenzyme A synthetases involved in steroid side-chain catabolism. J. Bacteriol. 2014, 196, 579–587. [Google Scholar] [CrossRef]
- Thomas, S.T.; Sampson, N.S. Mycobacterium tuberculosis utilizes a unique heterotetrameric structure for dehydrogenation of the cholesterol side chain. Biochemistry 2013, 52, 2895–2904. [Google Scholar] [CrossRef]
- Yang, M.; Lu, R.; Guja, K.E.; Wipperman, M.F.; St Clair, J.R.; Bonds, A.C.; Garcia-Diaz, M.; Sampson, N.S. Unraveling cholesterol catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 α2β2 acyl-CoA dehydrogenase initiates β-oxidation of 3-oxo-cholest-4-en-26-oyl CoA. ACS Infect. Dis. 2015, 1, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Battaile, K.P. Burning fat: The structural basis of fatty acid β-oxidation. Curr. Opin. Struct. Biol. 2002, 12, 721–728. [Google Scholar] [CrossRef]
- Yang, M.; Guja, K.E.; Thomas, S.T.; Garcia-Diaz, M.; Sampson, N.S. A distinct MaoC-like enoyl-CoA hydratase architecture mediates cholesterol catabolism in Mycobacterium tuberculosis. ACS Chem. Biol. 2014, 9, 2632–2645. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.E.; Pandey, A.K.; Gilmore, S.A.; Mizrahi, V.; McKinney, J.D.; Bertozzi, C.R.; Sassetti, C.M. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 2012, 19, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Wipperman, M.F.; Yang, M.; Thomas, S.T.; Sampson, N.S. Shrinking the FadE proteome of Mycobacterium tuberculosis: Insights into cholesterol metabolism through identification of an α2β2 heterotetrameric acyl coenzyme A dehydrogenase family. J. Bacteriol. 2013, 195, 4331–4341. [Google Scholar] [CrossRef] [PubMed]
- Nesbitt, N.M.; Yang, X.; Fontán, P.; Kolesnikova, I.; Smith, I.; Sampson, N.S.; Dubnau, E. A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol. Infect. Immun. 2010, 78, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, C.M.; Lu, R.; Nesbitt, N.M.; Schiebel, J.; Sampson, N.S.; Kisker, C. FadA5 a thiolase from Mycobacterium tuberculosis: A steroid-binding pocket reveals the potential for drug development against tuberculosis. Structure 2015, 23, 21–33. [Google Scholar] [CrossRef]
- Lu, R.; Schaefer, C.M.; Nesbitt, N.M.; Kuper, J.; Kisker, C.; Sampson, N.S. Catabolism of the cholesterol side chain in Mycobacterium tuberculosis is controlled by a Redox-Sensitive Thiol Switch. ACS Infect. Dis. 2017, 3, 666–675. [Google Scholar] [CrossRef]
- Anbazhagan, P.; Harijan, R.K.; Kiema, T.R.; Janardan, N.; Murthy, M.R.; Michels, P.A.; Juffer, A.H.; Wierenga, R.K. Phylogenetic relationships and classification of thiolases and thiolase-like proteins of Mycobacterium tuberculosis and Mycobacterium smegmatis. Tuberculosis 2014, 94, 405–412. [Google Scholar] [CrossRef]
- Xu, L.Q.; Liu, Y.J.; Yao, K.; Liu, H.H.; Tao, X.Y.; Wang, F.Q.; Wei, D.Z. Unraveling and engineering the production of 23,24-bisnorcholenic steroids in sterol metabolism. Sci. Rep. 2016, 6, 21928. [Google Scholar] [CrossRef]
- Wilbrink, M.H.; van der Geize, R.; Dijkhuizen, L. Molecular characterization of lpt3 and lpt4, essential for C24-branched chain sterol-side-chain degradation in Rhodococcus rhodochrous DSM 43269. Microbiology 2012, 158, 3054–3062. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.; Hood, L.; Seah, S.Y.K. Characterization of an aldolase involved in cholesterol side chain degradation in Mycobacterium tuberculosis. J. Bacteriol. 2018, 200, e00512-17. [Google Scholar] [CrossRef] [PubMed]
- Holert, J.; Kulić, Ž.; Yücel, O.; Suvekbala, V.; Suter, M.J.; Möller, H.M.; Philipp, B. Degradation of the acyl side chain of the steroid compound cholate in Pseudomonas sp. strain Chol1 proceeds via an aldehyde intermediate. J. Bacteriol. 2013, 195, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Genti-Raimondi, S.; Tolmasky, M.E.; Patrito, L.C.; Flury, A.; Actis, L.A. Molecular cloning and expression of the β-hydroxysteroid dehydrogenase gene from Pseudomonas testosteroni. Gene 1991, 105, 43–49. [Google Scholar] [CrossRef]
- Schultz, R.M.; Groman, E.V.; Engel, L.L. 3(17)β-Hydroxysteroid dehydrogenase of Pseudomonas testosteroni. A convenient purification and demonstration of multiple molecular forms. J. Biol. Chem. 1977, 252, 3775–3783. [Google Scholar] [PubMed]
- Minard, P.; Legoy, M.D.; Thomas, D. 3β,17β-hydroxysteroid dehydrogenase of Pseudomonas testosteroni. Kinetic evidence for the bifunctional activity at a common catalytic site. FEBS Lett. 1985, 188, 85–90. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, C.; Wang, B.; Li, Y.; Zhang, H. Characterization of 3,17β-hydroxysteroid dehydrogenase in Comamonas testosteroni. Chem. Biol. Interact. 2015, 234, 221–228. [Google Scholar] [CrossRef]
- Itagaki, E.; Matushita, H.; Hatta, T. Steroid transhydrogenase activity of 3-ketosteroid-Δ1-dehydrogenase from Nocardia corallina. J. Biochem. 1990, 108, 122–127. [Google Scholar] [CrossRef]
- Rohman, A.; van Oosterwijk, N.; Thunnissen, A.M.W.H.; Dijkstra, B.W. Crystal structure and site-directed mutagenesis of 3-ketosteroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 explain its catalytic mechanism. J. Biol. Chem. 2013, 288, 35559–35568. [Google Scholar] [CrossRef]
- Horinouchi, M.; Hayashi, T.; Yamamoto, T.; Kudo, T. A new bacterial steroid degradation gene cluster in Comamonas testosteroni TA441 which consists of aromatic-compound degradation genes for seco-steroids and 3-ketosteroid dehydrogenase genes. Appl. Environ. Microbiol. 2003, 69, 4421–4430. [Google Scholar] [CrossRef]
- Olivera, E.R.; de la Torre, M.; Barrientos, A.; Luengo, J.M. Steroid catabolism in bacteria: Genetic and functional analyses of stdH and stdJ in Pseudomonas putida DOC21. Can. J. Biotechnol. 2018, 2, 88–99. [Google Scholar] [CrossRef]
- Brzostek, A.; Pawelczyk, J.; Rumijowska-Galewicz, A.; Dziadek, B.; Dziadek, J. Mycobacterium tuberculosis is able to accumulate and utilize cholesterol. J. Bacteriol. 2009, 191, 6584–6591. [Google Scholar] [CrossRef] [PubMed]
- Knol, J.; Bodewits, K.; Hessels, G.I.; Dijkhuizen, L.; van der Geize, R. 3-Keto-5α-steroid Δ(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem. J. 2008, 410, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Van der Geize, R.; Hessels, G.I.; Dijkhuizen, L. Molecular and functional characterization of the kstD2 gene of Rhodococcus erythropolis SQ1 encoding a second 3-ketosteroid Δ(1)-dehydrogenase isoenzyme. Microbiology 2002, 148, 3285–3292. [Google Scholar] [CrossRef] [PubMed]
- van der Geize, R.; Hessels, G.I.; van Gerwen, R.; van der Meijden, P.; Dijkhuizen, L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Δ1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 2001, 205, 197–202. [Google Scholar] [CrossRef]
- Brzostek, A.; Sliwiński, T.; Rumijowska-Galewicz, A.; Korycka-Machała, M.; Dziadek, J. Identification and targeted disruption of the gene encoding the main 3-ketosteroid dehydrogenase in Mycobacterium smegmatis. Microbiology 2005, 151, 2393–2402. [Google Scholar] [CrossRef]
- Florin, C.; Kohler, M.; Grandguillot, M.; Plesiat, P. Comamonas testosteroni 3-ketosteroid-Δ4(5α)-dehydrogenase: Gene and protein characterization. J. Bacteriol. 1996, 178, 3322–3330. [Google Scholar] [CrossRef][Green Version]
- van Oosterwijk, N.; Knol, J.; Dijkhuizen, L.; van der Geize, R.; Dijkstra, B.W. Structure and catalytic mechanism of 3-ketosteroid-Delta4-(5α)-dehydrogenase from Rhodococcus jostii RHA1 genome. J. Biol. Chem. 2012, 287, 30975–30983. [Google Scholar] [CrossRef]
- van der Geize, R.; Hessels, G.I.; van Gerwen, R.; van der Meijden, P.; Dijkhuizen, L. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9alpha-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol. Microbiol. 2002, 45, 1007–1018. [Google Scholar] [CrossRef]
- van der Geize, R.; Hessels, G.I.; Nienhuis-Kuiper, M.; Dijkhuizen, L. Characterization of a second Rhodococcus erythropolis SQ1 3-ketosteroid 9alpha-hydroxylase activity comprising a terminal oxygenase homologue, KshA2, active with oxygenase-reductase component KshB. Appl. Environ. Microbiol. 2008, 74, 7197–7203. [Google Scholar] [CrossRef]
- Capyk, J.K.; D’Angelo, I.; Strynadka, N.C.; Eltis, L.D. Characterization of 3-ketosteroid 9{α}-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J. Biol. Chem. 2009, 284, 9937–9946. [Google Scholar] [CrossRef] [PubMed]
- Capyk, J.K.; Casabon, I.; Gruninger, R.; Strynadka, N.C.; Eltis, L.D. Activity of 3-ketosteroid 9α-hydroxylase (KshAB) indicates cholesterol side chain and ring degradation occur simultaneously in Mycobacterium tuberculosis. J. Biol. Chem. 2011, 286, 40717–40724. [Google Scholar] [CrossRef] [PubMed]
- García, J.L.; Uhía, I.; Galán, B. Catabolism and biotechnological applications of cholesterol degrading bacteria. Microb. Biotechnol. 2012, 5, 679–699. [Google Scholar] [CrossRef] [PubMed]
- Donova, M.; Gulevskaya, S.; Dovbnya, D.; Puntus, I. Mycobacterium sp. mutant strain producing 9α-hydroxy androstenedione from sitosterol. Appl. Microbiol. Biotechnol. 2005, 67, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Guevara, G.; Fernández de las Heras, L.; Perera, J.; Navarro Llorens, J.M. Functional differentiation of 3-ketosteroid Δ1-dehydrogenase isoenzymes in Rhodococcus ruber strain Chol-4. Microb. Cell Fact. 2017, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Xu, L.Q.; Wang, F.Q.; Wei, D.Z. Characterization and engineering of 3-ketosteroid Δ1-dehydrogenase and 3-ketosteroid 9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9a-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab. Eng. 2014, 24, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; van der Geize, R.; Besra, G.S.; Gurcha, A.; Liu, M.; Rohde, M.; Singh, M.; Coares, A. 3-Ketosteroid 9α-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 2010, 75, 107–121. [Google Scholar] [CrossRef]
- Bragin, E.Y.; Shtratnikova, V.Y.; Dovbnya, D.V.; Schelkunov, M.I.; Pekov, Y.A.; Malakho, S.G.; Egoroba, O.V.; Ivashina, T.V.; Sokolov, S.L.; Ashapkin, V.V.; et al. Comparative analysis of genes encoding key steroid core oxidation enzymes in fast-growing Mycobacterium spp. Strains. J. Steroid Biochem. Mol. Biol. 2013, 138, 41–53. [Google Scholar] [CrossRef]
- Petrusma, M.; Hessels, G.; Dijkhuizen, L.; van der Geize, R. Multiplicity of 3-ketosteroid-9α-hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids. J. Bacteriol. 2011, 193, 3931–3940. [Google Scholar] [CrossRef]
- Horinouchi, M.; Hayashi, T.; Kudo, T. The genes encoding the hydroxylase of 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione in steroid degradation in Comamonas testosteroni TA441. J. Steroid. Biochem. Mol. Biol. 2004, 92, 143–154. [Google Scholar] [CrossRef]
- Dresen, C.; Lin, L.Y.; D’Angelo, I.; Tocheva, E.I.; Strynadka, N.; Eltis, L.D. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J. Biol. Chem. 2010, 285, 22264–22275. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Yamamoto, T.; Taguchi, K.; Arai, H.; Kudo, T. Meta-cleavage enzyme gene tesB is necessary for testosterone degradation in Comamonas testosteroni TA441. Microbiology 2001, 147, 3367–3375. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yam, K.C.; D’Angelo, I.; Kalscheuer, R.; Zhu, H.; Wang, J.X.; Snieckus, V.; Ly, L.H.; Converse, P.J.; Jacobs, W.R. Jr.; Strynadka, N.; et al. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 2009, 5, e1000344. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Hayashi, T.; Koshino, H.; Yamamoto, T.; Kudo, T. Gene encoding the hydrolase for the product of the meta-cleavage reaction in testosterone degradation by Comamonas testosteroni. Appl. Environ. Microbiol. 2003, 69, 2139–2152. [Google Scholar] [CrossRef] [PubMed]
- Lack, N.; Lowe, E.D.; Liu, J.; Eltis, L.D.; Noble, M.E.; Sim, E.; Westwood, I.M. Structure of HsaD, a steroid-degrading hydrolase, from Mycobacterium tuberculosis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Lack, N.A.; Yam, K.C.; Lowe, E.D.; Horsman, G.P.; Owen, R.L.; Sim, E.; Eltis, L.D. Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism. J. Biol. Chem. 2010, 285, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Hayashi, T.; Koshino, H.; Kurita, T.; Kudo, T. Identification of 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid, 4-hydroxy-2-oxohexanoic acid, and 2-hydroxyhexa-2,4-dienoic acid and related enzymes involved in testosterone degradation in Comamonas testosteroni TA441. Appl. Environ. Microbiol. 2005, 71, 5275–5281. [Google Scholar] [CrossRef] [PubMed]
- Carere, J.; McKenna, S.E.; Kimber, M.S.; Seah, S.Y. characterization of an aldolase_dehydrogenase complex from the cholesterol degrading pathway of Mycobacterium tuberculosis. Biochemistry 2013, 52, 3502–3511. [Google Scholar] [CrossRef]
- Horinouchi, M.; Hayashi, T.; Koshino, H.; Kudo, T. ORF18 disrupted mutant of Comamonas testosteroni TA441 accumulates significant amounts of 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid and its derivatives after incubation with steroids. J. Steroid Biochem. Mol. Biol. 2006, 101, 78–84. [Google Scholar] [CrossRef]
- Casabon, I.; Crowe, A.M.; Liu, J.; Eltis, L.D. FadD3 is an acyl-CoA synthetase that initiates catabolism of cholesterol rings C and D in actinobacteria. Mol. Microbiol. 2013, 87, 269–283. [Google Scholar] [CrossRef]
- Crowe, A.M.; Casabon, I.; Brown, K.L.; Liu, J.; Lian, J.; Rogalski, J.C.; Hurst, T.E.; Snieckus, V.; Foster, L.J.; Eltis, L.D. Catabolism of the last two steroid rings in Mycobacterium tuberculosis and other bacteria. mBio 2017, 8, e00321-17. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Hayashi, T.; Koshino, H.; Kudo, T. Identification of 9alpha-hydroxy-17-oxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid in steroid degradation by Comamonas testosteroni TA441 and its conversion to the corresponding 6-en-5-oyl Coenzyme A (CoA) involving open reading frame 28 (ORF28)- and ORF30-encoded acyl-CoA dehydrogenases. J. Bacteriol. 2014, 196, 3598–3608. [Google Scholar]
- Horinouchi, M.; Hayashi, T.; Koshino, H.; Malon, M.; Hirota, H.; Kudo, T. Identification of 9α-hydroxy-17-oxo-1,2,3,4,10,19-hexanorandrost-6-en-5-oic acid and β-oxidation products of the C-17 side chain in cholic acid degradation by Comamonas testosteroni TA441. J. Steroid Biochem. Biol. Mol. 2014, 143, 306–322. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Koshino, H.; Malon, M.; Hirota, H.; Hayashi, T. Steroid degradation in Comamonas testosteroni TA441: Identification of metabolites and the genes involved in the reactions necessary before D-ring cleavage. Appl. Environ. Microbiol. 2018, 84, e01324-18. [Google Scholar] [CrossRef] [PubMed]
- Crowe, A.M.; Workman, S.D.; Watanabe, N.; Worrall, L.J.; Strynadka, N.C.J.; Eltis, L.D. IpdAB, a virulence factor in Mycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase. Proc. Natl. Acad. Sci. USA 2018, 115, E3378–E3387. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Koshino, H.; Malon, M.; Hirota, H.; Hayashi, T. Identification of 9-oxo-1,2,3,4,5,6,10,19-octanor-13,17-secoandrost-8(14)-ene-7,17-dioic acid as a metabolite of steroid degradation in Comamonas testosteroni TA441 and the genes involved in the conversion. J. Steroid Biochem. Mol. Biol. 2019, 185, 268–276. [Google Scholar] [CrossRef]
- Horinouchi, M.; Malon, M.; Hirota, H.; Hayashi, T. Identification of 4-methyl-5-oxo-octane-1,8-dioic acid and the derivatives as metabolites of steroidal C,D-ring degradation in Comamonas testosteroni TA441. J. Steroid Biochem. Mol. Biol. 2019, 185, 277–286. [Google Scholar] [CrossRef]
- Harwood, C.S.; Parales, R.E. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996, 50, 553–590. [Google Scholar] [CrossRef]
- Chen, Y.L.; Wang, C.H.; Yang, F.C.; Ismail, W.; Wang, P.H.; Shih, C.J.; Wu, Y.C.; Chiang, Y.R. Identification of Comamonas testosteroni as an androgen degrader in sewage. Sci. Rep. 2016, 6, 35386. [Google Scholar] [CrossRef]
- Kendall, S.L.; Burgess, P.; Balhana, R.; Withers, M.; Ten Bokum, A.; Lott, J.S.; Gao, C.; Uhía-Castro, I.; Stocker, N.G. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: KstR and KstR2. Microbiology 2010, 156, 1362–1371. [Google Scholar] [CrossRef]
- Ho, N.A.T.; Dawes, S.S.; Crowe, A.M.; Casabon, I.; Gao, C.; Kendall, S.L.; Baker, E.N.; Eltis, L.D.; Lott, J.S. The structure of the transcriptional repressor KstR in complex with CoA thioester cholesterol metabolites sheds light on the regulation of cholesterol catabolism in Mycobacterium tuberculosis. J. Biol. Chem. 2016, 291, 7256–7266. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, E.; Medrano, F.J.; Galán, B.; García, J.L. Deciphering the transcriptional regulation of cholesterol catabolic pathway in mycobacteria: Identification of the inducer of KstR repressor. J. Biol. Chem. 2014, 289, 17576–17588. [Google Scholar] [CrossRef] [PubMed]
- Casabon, I.; Zhu, S.-H.; Otani, H.; Liu, J.; Mohn, W.W.; Eltis, L.D. Regulation of KstR2 regulon of Mycobacterium tuberculosis by a cholesterol catabolite. Mol. Microbiol. 2013, 89, 1201–1212. [Google Scholar] [CrossRef] [PubMed]
- Crowe, A.M.; Stogios, P.J.; Casabon, I.; Evdokimova, E.; Savchenko, A.; Eltis, L.D. Structural and functional characterization of a ketosteroid transcriptional regulator of Mycobacterium tuberculosis. J. Biol. Chem. 2015, 290, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Kurita, T.; Yamamoto, T.; Hatori, E.; Hayasi, T.; Kudo, T. Steroid degradation gene cluster of Comamonas testosteroni consisting of 18 putative genes from meta-cleavage enzyme gene tesB to regulator gene tesR. Biochem. Biophys. Res. Commun. 2004, 324, 597–604. [Google Scholar] [CrossRef]
- Pruneda-Paz, J.L.; Linares, M.; Cabrera, J.E.; Genti-Raimondi, S. TeiR, a LuxR-type transcriptional factor required for testosterone degradation in Comamonas testosteroni. J. Bacteriol. 2004, 186, 1430–1437. [Google Scholar] [CrossRef]
- Göhler, A.; Xiong, G.; Paulsen, S.; Trentmann, G.; Maser, E. Testosterone-inducible regulator is a kinase that drives steroid sensing and metabolism in Comamonas testosteroni. J. Biol. Chem. 2008, 283, 17380–17390. [Google Scholar] [CrossRef]
- Linares, M.; Pruneda-Paz, J.L.; Reyna, L.; Genti-Raimondi, S. Regulation of testosterone degradation in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 2008, 112, 145–150. [Google Scholar] [CrossRef]
- Xiong, G.; Maser, E. Regulation of the steroid-inducible 3α-hydroxysteroid dehydrogenase/carbonyl reductase gene in Comamonas testosteroni. J. Biol. Chem. 2001, 276, 9961–9970. [Google Scholar] [CrossRef]
- Xiong, G.; Martin, H.J.; Maser, E. Identification and characterization of a novel translational repressor of the steroid-inducible 3α-hydroxysteroid dehydrogenase/carbonyl reductase in Comamonas testosteroni. J. Biol. Chem. 2003, 278, 47400–47407. [Google Scholar] [CrossRef]
- Gong, W.; Xiong, G.; Maser, E. Identification and characterization of the LysR-type transcriptional regulator HsdR for steroid-inducible expression of the 3a-hydroxysteroid dehydrogenase/carbonyl reductase gene in Comamonas testosteroni. Appl. Environ. Microbiol. 2011, 78, 941–950. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gong, W.; Xiong, G.; Maser, E. Oligomerization and negative autoregulation of the LysR-type transcriptional regulator HsdR from Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 2012, 132, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiong, G.; Maser, E. A novel transcriptional repressor PhaR for the steroid-inducible expression of the 3,17β-hydroxysteroid dehydrogenase gene in Comamonas testosteroni ATCC11996. Chem. Biol. Interact. 2013, 202, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Pan, T.; Zhang, Y.; Xiong, G.; Yu, Y. Functional analysis of a novel repressor LuxR in Comamonas testosteroni. Chem. Biol. Interact. 2017, 276, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Huang, P.; Xiong, G.; Maser, E. Isolation and characterization of a repressor TetR for 3,17β-HSD expressional regulation in Comamonas testosteroni. Chem. Biol. Interact. 2015, 234, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Huang, P.; Xiong, G.; Maser, E. Identification and isolation of a regulator protein for 3,17β-HSD expression regulation in Comamonas testosteroni. Chem. Biol. Interact. 2015, 234, 197–204. [Google Scholar] [CrossRef]
- Yu, C.P.; Roh, H.; Chu, K.H. 17β-Estradiol-degrading bacteria isolated from activated sludge. Environ. Sci. Technol. 2007, 41, 486–492. [Google Scholar] [CrossRef]
- Chen, Y.L.; Yu, C.P.; Lee, T.H.; Goh, K.S.; Chu, K.H.; Wang, P.H.; Ismail, W.; Shih, C.J.; Chiang, Y.R. Biochemical mechanisms and catabolic enzymes involved in bacterial estrogen degradation pathways. Cell Chem. Biol. 2017, 24, 712–724. [Google Scholar] [CrossRef]
- Roh, H.; Chu, K.H. A 17beta-estradiol-utilizing bacterium, Sphingomonas strain KC8: Part I—Characterization and abundance in wastewater treatment plants. Environ. Sci. Technol. 2010, 44, 4943–4950. [Google Scholar] [CrossRef]
- Liang, R.; Liu, H.; Liu, J. Genome sequence of Pseudomonas putida strain SJTE-1, a bacterium capable of degrading estrogens and persistent organic pollutants. J. Bacteriol. 2012, 194, 4781–4782. [Google Scholar] [CrossRef]
- Wang, P.; Zheng, D.; Peng, W.; Wang, Y.; Wang, X.; Xiong, W.; Liang, R. Characterization of 17β-hydroxysteroid dehydrogenase and regulators involved in estrogen degradation in Pseudomonas putida SJTE-1. Appl. Microbiol. Biotechnol. 2019, 103, 2413–2425. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Lee, T.H.; Chen, Y.L.; Wang, Y.S.; Wang, P.H.; Yu, C.P.; Chu, K.H.; Chiang, Y.R. Metabolites involved in aerobic degradation of the A and B rings of estrogen. Appl. Environ. Microbiol. 2019, 85, e02223-18. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Me, C.; Hu, A.; Zhang, F.; Hu, H.; Yu, C.P. Altererythrobacter estronivorus sp. nov., an estrogen-degrading strain isolated from Yundang Lagoon of Xiamen City in China. Curr. Microbiol. 2016, 72, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Fujii, K.; Satomi, M.; Morita, N.; Motomura, T.; Tanaka, T.; Kikuchi, S. Novosphingobium tardaugens sp. nov., an oestradiol-degrading bacterium isolated from activated sludge of a sewage treatment plant in Tokyo. Int. J. Syst. Evol. Microbiol. 2003, 53, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Fu, H.-Y.; Lee, T.-H.; Shih, C.-J.; Huang, L.; Wang, Y.-S.; Ismail, W.; Chiang, Y.-R. Estrogen degraders and estrogen degradation pathway identified in an activated sludge. Appl. Environ. Microbiol. 2018, 84, e00001-18. [Google Scholar] [CrossRef] [PubMed]
- Kurisu, F.; Ogura, M.; Saitoh, S.; Yamazoe, A.; Yagi, O. Degradation of natural estrogen and identification of the metabolites produced by soil isolates of Rhodococcus sp. and Sphingomonas sp. J. Biosci. Bioeng. 2010, 109, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Nakai, S.; Yamamura, A.; Tanaka, S.; Shi, J.; Nishikawa, M.; Nakashimada, Y.; Hosomi, M. Pathway of 17β-estradiol degradation by Nitrosomonas europaea and reduction in 17β-estradiol-derived estrogenic activity. Environ. Chem. Lett. 2011, 9, 1–6. [Google Scholar] [CrossRef]
- Lee, H.B.; Liu, D. Degradation of 17β-estradiol and its metabolites by sewage bacteria. Water Air Soil Pollut. 2002, 134, 353–368. [Google Scholar] [CrossRef]
- Harder, J.; Probian, C. Anaerobic mineralization of cholesterol by a novel type of denitrifying bacterium. Arch. Microbiol. 1997, 167, 269–274. [Google Scholar] [CrossRef]
- Wang, P.-H.; Leu, Y.-L.; Ismail, W.; Tang, S.-L.; Tsai, C.-Y.; Chen, H.-J.; Kao, A.-T.; Chiang, Y.-R. Anaerobic and aerobic cleavage of the steroid core ring structure by Steroidobacter denitrificans. J. Lipid Res. 2013, 54, 1493–1504. [Google Scholar] [CrossRef]
- Wang, P.-H.; Yu, C.-P.; Lee, T.-H.; Lin, C.-W.; Ismail, W.; Wey, S.-P.; Kuo, A.-T.; Chiang, Y.-R. Anoxic androgen degradation by the denitrifying bacterium Sterolibacterium denitrificans via the 2,3-seco pathway. Appl. Environ. Microbiol. 2014, 80, 3442–3452. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.C.; Chen, Y.L.; Tang, S.L.; Yu, C.P.; Wang, P.H.; Ismail, W.; Wang, C.H.; Ding, J.Y.; Yang, C.Y.; Yang, C.Y.; et al. Integrated multi-omics analyses reveal the biochemical mechanisms and phylogenetic relevance of anaerobic androgen biodegradation in the environment. ISME J. 2016, 10, 1967–1983. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.R.; Ismail, W.; Müller, M.; Fuchs, G. Initial steps in the anoxic metabolism of cholesterol by the denitrifying Sterolibacterium denitrificans. J. Biol. Chem. 2007, 282, 13240–13249. [Google Scholar] [CrossRef] [PubMed]
- Dermer, J.; Fuchs, G. Molybdoenzyme that catalyzes the anaerobic hydroxylation of a tertiary carbon atom in the side chain of cholesterol. J. Biol. Chem. 2012, 287, 36905–36916. [Google Scholar] [CrossRef] [PubMed]
- Rugor, A.; Wójcik-Augustyn, A.; Niedzialkowska, E.; Mordalski, S.; Staroń, J.; Bojarski, A.; Szaleniec, M. Reaction mechanism of sterol hydroxylation by steroid C25 dehydrogenase–Homology model, reactivity and isoenzymatic diversity. J. Inorg. Biochem. 2017, 173, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Warnke, M.; Jacoby, C.; Jung, T.; Agne, M.; Mergelsberg, M.; Starke, R.; Jehmlich, N.; von Bergen, M.; Richnow, H.H.; Brüls, T.; et al. A patchwork pathway for oxygenase-independent degradation of side chain containing steroids. Environ. Microbiol. 2017, 19, 4684–4699. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, C.; Eipper, J.; Warnke, M.; Tiedt, O.; Mergelsberg, M.; Stärk, H.J.; Daus, B.; Martín-Moldes, Z.; Zamarro, M.T.; Díaz, E.; et al. Four Molybdenum-dependent steroid C-25 hydroxylases: Heterologous overproduction, role in steroid degradation, and application for 25-hydroxyvitamin D3 synthesis. mBio 2018, 9, e00694-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.-H.; Lee, T.H.; Ismail, W.; Tsai, C.Y.; Lin, C.W.; Tsai, Y.W.; Chiang, Y.R. An oxygenase-independent cholesterol catabolic pathway operates under oxic conditions. PLoS ONE 2013, 8, e66675. [Google Scholar] [CrossRef] [PubMed]
- Warnke, M.; Jung, T.; Jacoby, C.; Agne, M.; Feller, F.M.; Philipp, B.; Seiche, W.; Breit, B.; Boll, M. Functional characterization of three specific acyl-coenzyme A synthetases involved in anaerobic cholesterol degradation in Sterolibacterium denitrificans Chol1S. Appl. Environ. Microbiol. 2018, 84, e02721-17. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.R.; Ismail, W.; Gallien, S.; Heintz, D.; Van Dorsselaer, A.; Fuchs, G. Cholest-4-en-3-one-Δ1-dehydrogenase, a flavoprotein catalyzing the second step in anoxic cholesterol metabolism. Appl. Environ. Microbiol. 2008, 74, 107–113. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chiang, Y.R.; Fang, J.Y.; Ismail, W.; Wang, P.H. Initial steps in anoxic testosterone degradation by Steroidobacter denitrificans. Microbiology 2010, 156, 2253–2259. [Google Scholar] [CrossRef] [PubMed]
- Fahrbach, M.; Krauss, M.; Preiss, A.; Kohler, H.P.; Hollender, J. Anaerobic testosterone degradation in Steroidobacter denitrificans—Identification of transformation products. Environ. Pollut. 2010, 158, 2572–2581. [Google Scholar] [CrossRef] [PubMed]
- Leu, Y.L.; Wang, P.H.; Shiao, M.S.; Ismail, W.; Chiang, Y.R. A novel testosterone catabolic pathway in bacteria. J. Bacteriol. 2011, 193, 4447–4455. [Google Scholar] [CrossRef] [PubMed][Green Version]



| Strain | Steroids Used | Reference Genome | ||
|---|---|---|---|---|
| 9,10-seco pathway | Mycobacterium | tuberculosis H37Rv | Cholesterol | NC_000962 |
| smegmatis mc2 155 | Cholesterol | NC_008596 | ||
| Rhodococcus | neoaurum ATCC 25795 | Cholesterol | NZ_JMDW00000000 | |
| β-sitosterol | ||||
| Stigmasterol | ||||
| Campesterol | ||||
| equi 103S | Cholesterol | NC_014659 | ||
| rhodochrous DSM43269 | Cholesterol | unpublished | ||
| β-sitosterol | ||||
| Campesterol | ||||
| jostii RHA1 | Cholesterol | NC_008268 | ||
| Cholic acid | ||||
| erythropolis SQ1 | Cholesterol | unpublished | ||
| Gordonia | neofelifaecis NRRL B-59395 | Cholesterol | NZ_AEUD00000000 | |
| cholesterolivorans Chol-3 | Cholesterol | unpublished | ||
| Ergosterol | ||||
| Stigmasterol | ||||
| Comamonas | testosteroni TA441 | Testosterone | NZ_CP006704 | |
| Cholic acid | ||||
| thiooxidans CNB-1 | Testosterone | NC_013446 | ||
| Pseudomonas | stutzeri Chol-1 | Cholic acid | NZ_AMSL00000000 | |
| putida DOC21 | Bile acids | unpublished | ||
| Testosterone | ||||
| 4,5-seco pathway | Sphingomonas | sp. KC8 | 17β-estradiol | NZ_AFMP01000000 |
| 2,3-seco pathway | Sterolibacterium | denitrificans Chol1S | Cholesterol | LT837803 |
| Steroidobacter | denitrificans DSMZ18526 | Testosterone | NZ_CP011971 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olivera, E.R.; Luengo, J.M. Steroids as Environmental Compounds Recalcitrant to Degradation: Genetic Mechanisms of Bacterial Biodegradation Pathways. Genes 2019, 10, 512. https://doi.org/10.3390/genes10070512
Olivera ER, Luengo JM. Steroids as Environmental Compounds Recalcitrant to Degradation: Genetic Mechanisms of Bacterial Biodegradation Pathways. Genes. 2019; 10(7):512. https://doi.org/10.3390/genes10070512
Chicago/Turabian StyleOlivera, Elías R., and José M. Luengo. 2019. "Steroids as Environmental Compounds Recalcitrant to Degradation: Genetic Mechanisms of Bacterial Biodegradation Pathways" Genes 10, no. 7: 512. https://doi.org/10.3390/genes10070512
APA StyleOlivera, E. R., & Luengo, J. M. (2019). Steroids as Environmental Compounds Recalcitrant to Degradation: Genetic Mechanisms of Bacterial Biodegradation Pathways. Genes, 10(7), 512. https://doi.org/10.3390/genes10070512




