Variability in Assembly of Degradation Operons for Naphthalene and its derivative, Carbaryl, Suggests Mobilization through Horizontal Gene Transfer
Abstract
1. Introduction
2. Microbial Adaptation to Aromatics and Xenobiotics
3. Horizontal Gene Transfer Elements Involved in Catabolism of Aromatics
4. Assembly of Naphthalene Degradation Pathway
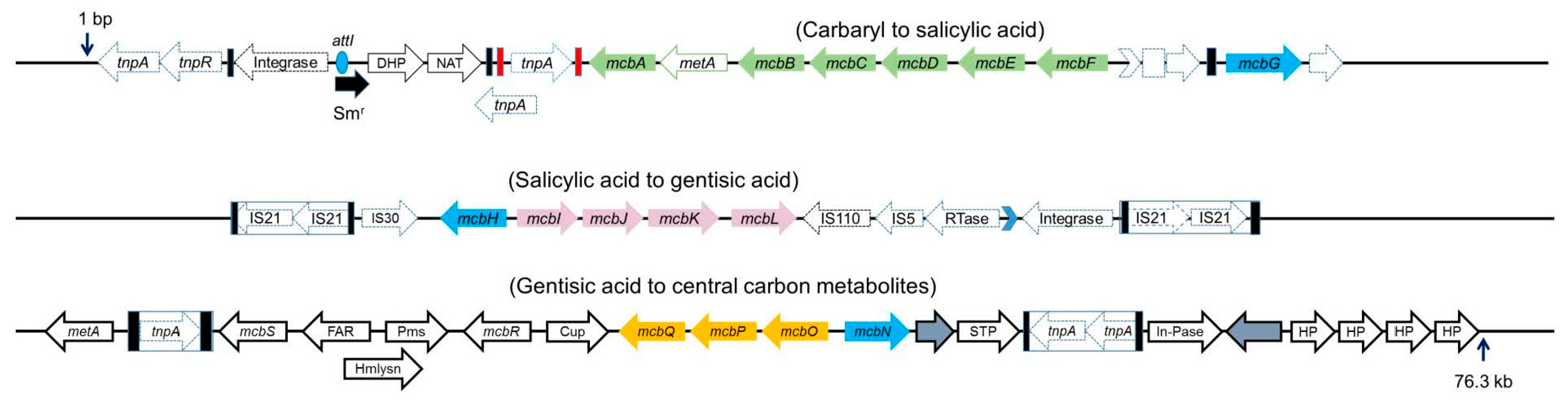
5. Assembly of Carbaryl Degradation Pathway
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Danzo, B.J. Environmental xenobiotics may disrupt normal endocrine function by interfering with the binding of physiological ligands to steroid receptors and binding proteins. Environ. Health Perspect. 1997, 105, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, S.; Kawanishi, S. Double base lesions of DNA by a metabolite of carcinogenic benzo[a]pyrene. Biochem. Biophys. Res. Commun. 2002, 290, 778–782. [Google Scholar] [CrossRef] [PubMed]
- Conney, A.H. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: GHA Clowes memorial lecture. Cancer Res. 1982, 42, 4875–4917. [Google Scholar]
- Stegeman, J.J.; Lech, J.J. Cytochrome P-450 monooxygenase systems in aquatic species: Carcinogen metabolism and biomarkers for carcinogen and pollutant exposure. Environ. Health Perspect. 1991, 90, 101–109. [Google Scholar] [PubMed]
- Pashin, Y.V.; Bakhitova, L.M. Mutagenic and carcinogenic properties of polycyclic aromatic hydrocarbons. Environ. Health Perspect. 1979, 30, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Wild, S.R.; Jones, K.C. Polynuclear aromatic hydrocarbons in the United Kingdom environment: A preliminary source inventory and budget. Environ. Pollut. 1995, 88, 91–108. [Google Scholar] [CrossRef]
- Haritash, A.K.; Kaushik, C.P. Biodegradation aspects of polycyclic aromatic hydrocarbons (PAHs): A review. J. Hazard. Mater. 2009, 169, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Interactions of cyclic hydrocarbons with biological membranes. J. Biol. Chem. 1994, 269, 8022–8028. [Google Scholar] [PubMed]
- Hansen, B.H.; Altin, D.; Vang, S.H.; Nordtug, T.; Olsen, A.J. Effects of naphthalene on gene transcription in Calanus finmarchicus (Crustacea: Copepoda). Aquat. Toxicol. 2008, 86, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Elumalai, M.; Balasubramanian, M.P. Influence of naphthalene on esterase activity during vitellogenesis of marine edible crab, Scylla serrata. Bull. Environ. Contam. Toxicol. 1999, 62, 743–748. [Google Scholar] [CrossRef]
- Valaes, T.; Doxiadis, S.A.; Fessas, P. Acute hemolysis due to naphthalene inhalation. J. Pediatr. 1963, 63, 904–915. [Google Scholar] [CrossRef]
- Gupta, R.; Singhal, P.C.; Muthusethupathy, M.A.; Malik, A.K.; Chugh, K.S. Cerebral oedema and renal failure following naphthalene poisoning. J. Assoc. Phys. India 1979, 27, 347–348. [Google Scholar]
- National Toxicology Program. Toxicology and carcinogenesis studies of naphthalene (CAS No. 91-20-3) in F344/N rats (inhalation studies). Natl. Toxicol. Program Tech. Rep. Ser. 2000, 500, 1–173. [Google Scholar]
- Smulders, C.J.; Bueters, T.J.; Van Kleef, R.G.; Vijverberg, H.P. Selective effects of carbamate pesticides on rat neuronal nicotinic acetylcholine receptors and rat brain acetylcholinesterase. Toxicol. Appl. Pharmacol. 2003, 193, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Moeller, F.E. Effect of pollen availability on poisoning of honey bees by Carbaryl applied to sweet corn. J. Econ. Entomol. 1971, 64, 1314–1315. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K.; Dhawan, A. Effect of Carbaryl on tissue composition, maturation, and breeding potential of Cirrhina mrigala (Ham.). Bull. Environ. Contam. Toxicol. 1996, 57, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Boone, M.D.; Semlitsch, R.D.; Little, E.E.; Doyle, M.C. Multiple stressors in amphibian communities: Effects of chemical contamination, bullfrogs, and fish. Ecol. Appl. 2007, 17, 291–301. [Google Scholar] [CrossRef]
- Eraslan, G.; Kanbur, M.; Silici, S. Effect of Carbaryl on some biochemical changes in rats: The ameliorative effect of bee pollen. Food Chem. Toxicol. 2009, 47, 86–91. [Google Scholar] [CrossRef]
- Bridges, C.M. Tadpole swimming performance and activity affected by acute exposure to sub lethal levels of Carbaryl. Environ. Toxicol. Chem. 1997, 16, 1935–1939. [Google Scholar] [CrossRef]
- Bulen, B.J.; Distel, C.A. Carbaryl concentration gradients in realistic environments and their influence on our understanding of the tadpole food web. Arch. Environ. Contam. Toxicol. 2011, 60, 343–350. [Google Scholar] [CrossRef]
- Lima, M.P.; Cardoso, D.N.; Soares, A.M.; Loureiro, S. Carbaryl toxicity prediction to soil organisms under high and low temperature regimes. Ecotoxicol. Environ. Saf. 2015, 114, 263–272. [Google Scholar] [CrossRef] [PubMed]
- USEPA/Office of Pesticides Programs. Interim Reregistration Eligibility Decision for Carbaryl. p.2. Available online: https://www3.epa.gov/pesticides/chemsearch/regactions/reregistration/iredPC-056801_22-Oct-04.pdf (accessed on 15 April 2019).
- Gibson, D.T.; Subramanian, V. Microbial degradation of aromatic hydrocarbons. In Microbial Degradation of Organic Compounds; Gibson, D.T., Ed.; Microbiology Series; Marcel Dekker Inc.: New York, NY, USA; Basel, Switzerland, 1984; Volume 13, pp. 181–252. [Google Scholar]
- Singleton, I. Microbial metabolism of xenobiotics: Fundamental and applied research. J. Chem. Technol. Biotech. Int. Res. Process Environ. Clean Technol. 1994, 59, 9–23. [Google Scholar] [CrossRef]
- Nojiri, H.; Shintani, M.; Omori, T. Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl. Microbiol. Biotechnol. 2004, 64, 154–174. [Google Scholar] [CrossRef]
- Furukawa, K.; Matsumura, F. Microbial metabolism of polychlorinated biphenyls. Relative degradability of polychlorinated biphenyl components by Alkaligenes species. J. Agric. Food Chem. 1976, 24, 251–256. [Google Scholar] [CrossRef]
- Chatterjee, D.K.; Kellogg, S.T.; Hamada, S.; Chakrabarty, A.M. Plasmid specifying total degradation of 3-chlorobenzoate by a modified ortho pathway. J. Bacteriol. 1981, 146, 639–646. [Google Scholar]
- Baggi, G.; Barbieri, P.; Galli, E.; Tollari, S. Isolation of a Pseudomonas stutzeri strain that degrades o-xylene. Appl. Environ. Microbiol. 1987, 53, 2129–2132. [Google Scholar]
- Mahajan, M.C.; Phale, P.S.; Vaidyanathan, C.S. Evidence for the involvement of multiple pathways in the biodegradation of 1-and 2-methylnaphthalene by Pseudomonas putida CSV86. Arch. Microbiol. 1994, 161, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Swetha, V.P.; Phale, P.S. Metabolism of Carbaryl via 1,2-dihydroxynaphthalene by soil isolates Pseudomonas sp. strains C4, C5, and C6. Appl. Environ. Microbiol. 2005, 71, 5951–5956. [Google Scholar] [CrossRef] [PubMed]
- Vamsee-Krishna, C.; Mohan, Y.; Phale, P.S. Biodegradation of phthalate isomers by Pseudomonas aeruginosa PP4, Pseudomonas sp. PPD and Acinetobacter lwoffii ISP4. Appl. Microbiol. Biotechnol. 2006, 72, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- John, R.C.; Essien, J.P.; Akpan, S.B.; Okpokwasili, G.C. Polycyclic aromatic hydrocarbon-degrading bacteria from aviation fuel spill site at Ibeno, Nigeria. Bull. Environ. Contam. Toxicol. 2012, 88, 1014–1019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jiang, X.; Lu, L.; Xiao, W. Biodegradation of polychlorinated biphenyls (PCBs) by the novel identified cyanobacterium Anabaena PD-1. PLoS ONE 2015, 10, e0131450. [Google Scholar] [CrossRef] [PubMed]
- Kanaly, R.A.; Harayama, S. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 2000, 182, 2059–2067. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.R. The biodegradation of aromatic hydrocarbons by bacteria. Biodegradation 1990, 1, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Samanta, S.K.; Singh, O.V.; Jain, R.K. Polycyclic aromatic hydrocarbons: Environmental pollution and bioremediation. Trends Biotechnol. 2002, 20, 243–248. [Google Scholar] [CrossRef]
- González-Gaya, B.; Martínez-Varela, A.; Vila-Costa, M.; Casal, P.; Cerro-Gálvez, E.; Berrojalbiz, N.; Jiménez, B. Biodegradation as an important sink of aromatic hydrocarbons in the oceans. Nat. Geosci. 2019, 12, 119. [Google Scholar] [CrossRef]
- Srivastava, S.; Kumar, M. Biodegradation of polycyclic aromatic hydrocarbons (PAHs): A sustainable approach. In Sustainable Green Technologies for Environmental Management; Shah, S., Ramanan, V., Prasad, R., Eds.; Springer: Singapore, 2019; pp. 111–139. [Google Scholar]
- Ladino-Orjuela, G.; Gomes, E.; da Silva, R.; Salt, C.; Parsons, J.R. Metabolic pathways for degradation of aromatic hydrocarbons by bacteria. In Reviews of Environmental Contamination and Toxicology; de Voogt, P., Ed.; Springer: Cham, Switzerland, 2016; Volume 237, pp. 105–121. [Google Scholar]
- Liang, D.W.; Zhang, T.; Fang, H.H.; He, J. Phthalates biodegradation in the environment. Appl. Microbiol. Biotechnol. 2008, 80, 183. [Google Scholar] [CrossRef]
- Kumar, V.; Sharma, N.; Maitra, S.S. Comparative study on the degradation of dibutyl phthalate by two newly isolated Pseudomonas sp. V21b and Comamonas sp. 51F. Biotechnol. Rep. 2017, 15, 1–10. [Google Scholar] [CrossRef]
- Basu, A.; Dixit, S.S.; Phale, P.S. Metabolism of benzyl alcohol via catechol ortho-pathway in methylnaphthalene-degrading Pseudomonas putida CSV86. Appl. Microbiol. Biotechnol. 2003, 62, 579–585. [Google Scholar] [CrossRef]
- Chapalamadugu, S.; Chaudhry, G.R. Hydrolysis of Carbaryl by a Pseudomonas sp. and construction of a microbial consortium that completely metabolizes Carbaryl. Appl. Environ. Microbiol. 1991, 57, 744–750. [Google Scholar]
- Deng, Y.; Yang, F.; Deng, C.; Yang, J.; Jia, J.; Yuan, H. Biodegradation of BTEX aromatics by a haloduric microbial consortium enriched from a sediment of Bohai Sea, China. Appl. Biochem. Biotechnol. 2017, 183, 893–905. [Google Scholar] [CrossRef]
- Zafra, G.; Absalón, Á.E.; Anducho-Reyes, M.Á.; Fernandez, F.J.; Cortés-Espinosa, D.V. Construction of PAH-degrading mixed microbial consortia by induced selection in soil. Chemosphere 2017, 172, 120–126. [Google Scholar] [CrossRef]
- Vaidya, S.; Devpura, N.; Jain, K.; Madamwar, D. Degradation of chrysene by enriched bacterial consortium. Front. Microbiol. 2018, 9, 1333. [Google Scholar] [CrossRef]
- Cerniglia, C.E.; Freeman, J.P.; Althaus, J.R.; van Baalen, C. Metabolism and toxicity of 1-and 2-methylnaphthalene and their derivatives in cyanobacteria. Arch. Microbiol. 1983, 136, 177–183. [Google Scholar] [CrossRef]
- Cerniglia, C.E. Microbial metabolism of polycyclic aromatic hydrocarbons. Adv. Appl. Microbiol. 1984, 30, 31–71. [Google Scholar]
- Phale, P.S.; Mahajan, M.C.; Vaidyanathan, C.S. A pathway for biodegradation of 1-naphthoic acid by Pseudomonas maltophilia CSV89. Arch. Microbiol. 1995, 163, 42–47. [Google Scholar] [CrossRef]
- Vamsee-Krishna, C.; Phale, P.S. Bacterial degradation of phthalate isomers and their esters. Indian J. Microbiol. 2008, 48, 19–34. [Google Scholar] [CrossRef]
- Vamsee-Krishna, C.; Phale, P.S. Bypassing isophthalate inhibition by modulating glutamate dehydrogenase (GDH): Purification and kinetic characterization of NADP-GDHs from isophthalate-degrading Pseudomonas aeruginosa strain PP4 and Acinetobacter lwoffii strain ISP4. J. Bacteriol. 2010, 192, 801–806. [Google Scholar] [CrossRef]
- Arber, W. Genetic variation: Molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 2000, 24, 1–7. [Google Scholar] [CrossRef]
- Hacker, J.; Carniel, E. Ecological fitness, genomic islands and bacterial pathogenicity: A Darwinian view of the evolution of microbes. EMBO Rep. 2001, 2, 376–381. [Google Scholar] [CrossRef]
- van der Meer, J.R.; Sentchilo, V. Genomic islands and the evolution of catabolic pathways in bacteria. Curr. Opin. Biotechnol. 2003, 14, 248–254. [Google Scholar] [CrossRef]
- Springael, D.; Top, E.M. Horizontal gene transfer and microbial adaptation to xenobiotics: New types of mobile genetic elements and lessons from ecological studies. Trends Microbiol. 2004, 12, 53–58. [Google Scholar] [CrossRef]
- Van Der Meer, J.R.; De Vos, W.M.; Harayama, S.; Zehnder, A.J. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Mol. Biol. Rev. 1992, 56, 677–694. [Google Scholar]
- Nagata, Y.; Kato, H.; Ohtsubo, Y.; Tsuda, M. Mobile genetic elements involved in the evolution of bacteria that degrade recalcitrant xenobiotic compounds. In DNA Traffic in the Environment; Hiromi, N., Taku, O., Eds.; Springer: Singapore, 2019; pp. 215–244. [Google Scholar]
- Johnsen, A.R.; Wick, L.Y.; Harms, H. Principles of microbial PAH-degradation in soil. Environ. Pollut. 2005, 133, 71–84. [Google Scholar] [CrossRef]
- Chakrabarty, A.M. Genetic basis of the biodegradation of salicylate in Pseudomonas. J. Bacteriol. 1972, 112, 815–823. [Google Scholar]
- Chakrabarty, A.M.; Chou, G.; Gunsalus, I.C. Genetic regulation and extra chromosomal nature of octane degradative pathway in Pseudomonas. Proc. Natl. Acad. Sci. USA 1973, 70, 1137–1140. [Google Scholar] [CrossRef]
- Dunn, N.W.; Gunsalus, I.C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J. Bacteriol. 1973, 114, 974–979. [Google Scholar]
- Rheinwald, J.G.; Chakrabarty, A.M.; Gunsalus, I.C. A transmissible plasmid controlling camphor oxidation in Pseudomonas putida. Proc. Natl. Acad. Sci. USA 1973, 70, 885–889. [Google Scholar] [CrossRef]
- Worsey, M.J.; Williams, P.A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: Evidence for a new function of the TOL plasmid. J. Bacteriol. 1975, 124, 7–13. [Google Scholar]
- Mulbry, W.W.; Karns, J.S.; Kearney, P.C.; Nelson, J.O.; McDaniel, C.S.; Wild, J.R. Identification of a plasmid-borne parathion hydrolase gene from Flavobacterium sp. by southern hybridization with opd from Pseudomonas diminuta. Appl. Environ. Microbiol. 1986, 51, 926–930. [Google Scholar]
- Hayatsu, M.; Hirano, M.; Tokuda, S. Involvement of two plasmids in fenitrothion degradation by Burkholderia sp. strain NF100. Appl. Environ. Microbiol. 2000, 66, 1737–1740. [Google Scholar] [CrossRef]
- Wu, S.J.; Hu, Z.H.; Zhang, L.L.; Yu, X.; Chen, J.M. A novel dichloromethane-degrading Lysinibacillus sphaericus strain wh22 and its degradative plasmid. Appl. Microbiol. Biotechnol. 2009, 82, 731–740. [Google Scholar] [CrossRef]
- Feng, X.; Ou, L.T.; Ogram, A. Plasmid-mediated mineralization of carbofuran by Sphingomonas sp. strain CF06. Appl. Environ. Microbiol. 1997, 63, 1332–1337. [Google Scholar]
- Maeda, K.; Nojiri, H.; Shinatani, M.; Yoshida, T.; Habe, H.; Omori, T. Complete nucleotide sequence of carbazole/dioxin degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J. Mol. Biol. 2003, 326, 21–33. [Google Scholar] [CrossRef]
- Martinez, B.; Tomkins, J.; Wackett, L.P.; Wing, R.; Sadowsky, M.J. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol. 2001, 183, 5684–5697. [Google Scholar] [CrossRef]
- Sota, M.; Kawasaki, H.; Tsuda, M. Structure of haloacetate catabolic IncP-1β plasmid pUO1 and genetic mobility of its residing haloacetate-catabolic transposon. J. Bacteriol. 2003, 185, 6741–6745. [Google Scholar] [CrossRef]
- Tsuda, M.; Iino, T. Identification and characterization of Tn4653, a transposon covering the toluene transposon Tn4651 on TOL plasmid pWW0. Mol. Gen. Genet. 1988, 213, 72–77. [Google Scholar] [CrossRef]
- Tsuda, M.; Iino, T. Naphthalene degrading genes on plasmid NAH7 are on a defective transposon. Mol. Gen. Genet. 1990, 223, 33–39. [Google Scholar] [CrossRef]
- Obayori, O.S.; Salam, L.B. Degradation of polycyclic aromatic hydrocarbons: Role of plasmids. Sci. Res. Essays 2010, 5, 4093–4106. [Google Scholar]
- Weightman, A.J.; Topping, A.W.; Hill, K.E.; Lee, L.L.; Sakai, K.; Slater, J.H.; Thomas, A.W. Transposition of DEH, a broad-host-range transposon flanked by ISPpu12, in Pseudomonas putida is associated with genomic rearrangements and dehalogenase gene silencing. J. Bacteriol. 2002, 184, 6581–6591. [Google Scholar] [CrossRef]
- Williams, P.A.; Jones, R.M.; Shaw, L.E. A third transposable element, ISPpu12, from the toluene-xylene catabolic plasmid pWW0 of Pseudomonas putida mt-2. J. Bacteriol. 2002, 184, 6572–6580. [Google Scholar] [CrossRef]
- Tsuda, M.; Genka, H. Identification and characterization of Tn4656, a novel class II transposon carrying a set of toluene degrading genes from TOL plasmid pWW53. J. Bacteriol. 2001, 183, 6215–6224. [Google Scholar] [CrossRef]
- Grindley, N.D.; Reed, R.R. Transpositional recombination in prokaryotes. Annu. Rev. Biochem. 1985, 54, 863–896. [Google Scholar] [CrossRef]
- van der Meer, J.R.; Zehnder, A.J.; de Vos, W.M. Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain P51. J. Bacteriol. 1991, 173, 7077–7083. [Google Scholar] [CrossRef][Green Version]
- Tsuda, M.; Minegishi, K.I.; Iino, T. Toluene transposons Tn4651 and Tn4653 are class II transposons. J. Bacteriol. 1989, 171, 1386–1393. [Google Scholar] [CrossRef]
- Burrus, V.; Pavlovic, G.; Decaris, B.; Guédon, G. Conjugative transposons: The tip of the iceberg. Mol. Microbiol. 2002, 46, 601–610. [Google Scholar] [CrossRef]
- Hentschel, U.; Hacker, J. Pathogenicity islands: The tip of the iceberg. Microb. Infect. 2001, 3, 545–548. [Google Scholar] [CrossRef]
- Blum, G.; Ott, M.; Lischewski, A.; Ritter, A.; Imrich, H.; Tschäpe, H.; Hacker, J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 1994, 62, 606–614. [Google Scholar]
- Yamaguchi, T.; Nishifuji, K.; Sasaki, M.; Fudaba, Y.; Aepfelbacher, M.; Takata, T.; Ohara, M.; Komatsuzawa, H.; Amagai, M.; Sugai, M. Identification of the Staphylococcus aureus etd pathogenicity island which encodes a novel exfoliative toxin, ETD, and EDIN-B. Infect. Immun. 2002, 70, 5835–5845. [Google Scholar] [CrossRef]
- Parreira, V.R.; Gyles, C.L. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect. Immun. 2003, 71, 5087–5096. [Google Scholar] [CrossRef]
- Luck, S.N.; Turner, S.A.; Rajakumar, K.; Sakellaris, H.; Adler, B. Ferric dicitrate transport system (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 2001, 69, 6012–6021. [Google Scholar] [CrossRef]
- Novick, R.P. Mobile genetic elements and bacterial toxinoses: The superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 2003, 49, 93–105. [Google Scholar] [CrossRef]
- Inouye, S.; Sunshine, M.G.; Six, E.W.; Inouye, M. Retronphage phi R73: An E. coli phage that contains a retroelement and integrates into a tRNA gene. Science 1991, 252, 969–971. [Google Scholar] [CrossRef]
- Cheetham, B.F.; Katz, M.E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 1995, 18, 201–208. [Google Scholar] [CrossRef]
- Gaillard, M.; Vallaeys, T.; Vorhölter, F.J.; Minoia, M.; Werlen, C.; Sentchilo, V.; Pühler, A.; van der Meer, J.R. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 2006, 188, 1999–2013. [Google Scholar] [CrossRef]
- Zamarro, M.T.; Martín-Moldes, Z.; Díaz, E. The ICEXTD of Azoarcus sp. CIB, an integrative and conjugative element with aerobic and anaerobic catabolic properties. Environ. Microbiol. 2016, 18, 5018–5031. [Google Scholar] [CrossRef]
- Paliwal, V.; Raju, S.C.; Modak, A.; Phale, P.S.; Purohit, H.J. Pseudomonas putida CSV86: A candidate genome for genetic bioaugmentation. PLoS ONE 2014, 9, e84000. [Google Scholar] [CrossRef]
- Obi, C.C.; Vayla, S.; De Gannes, V.; Berres, M.E.; Walker, J.; Pavelec, D.; Hyman, J.; Hickey, W.J. The integrative conjugative element clc (ICEclc) of Pseudomonas aeruginosa JB2. Front. Microbiol. 2018, 9, 1532. [Google Scholar] [CrossRef]
- Ohtsubo, Y.; Ishibashi, Y.; Naganawa, H.; Hirokawa, S.; Atobe, S.; Nagata, Y.; Tsuda, M. Conjugal transfer of polychlorinated biphenyl/biphenyl degradation genes in Acidovorax sp. strain KKS102, which are located on an integrative and conjugative element. J. Bacteriol. 2012, 194, 4237–4248. [Google Scholar] [CrossRef]
- Toussaint, A.; Merlin, C.; Monchy, S.; Benotmane, M.A.; Leplae, R.; Mergeay, M.; Springael, D. The biphenyl-and 4-chlorobiphenyl-catabolic transposon Tn4371, a member of a new family of genomic islands related to IncP and Ti plasmids. Appl. Environ. Microbiol. 2003, 69, 4837–4845. [Google Scholar] [CrossRef]
- Hickey, W.J.; Chen, S.; Zhao, J. The phn island: A new genomic island encoding catabolism of polynuclear aromatic hydrocarbons. Front. Microbiol. 2012, 3, 125. [Google Scholar] [CrossRef]
- Basu, A.; Phale, P.S. Conjugative transfer of preferential utilization of aromatic compounds from Pseudomonas putida CSV86. Biodegradation 2008, 19, 83–92. [Google Scholar] [CrossRef]
- Annweiler, E.; Richnow, H.H.; Antranikian, G.; Hebenbrock, S.; Garms, C.; Franke, S.; Francke, W.; Michaelis, W. Naphthalene degradation and incorporation of naphthalene-derived carbon into biomass by the thermophile Bacillus thermoleovorans. Appl. Environ. Microbiol. 2000, 66, 518–523. [Google Scholar] [CrossRef]
- Yen, K.M.; Gunsalus, I.C. Regulation of naphthalene catabolic genes of plasmid NAH7. J. Bacteriol. 1985, 162, 1008–1013. [Google Scholar]
- Sota, M.; Yano, H.; Ono, A.; Miyazaki, R.; Ishii, H.; Genka, H.; Top, E.M.; Tsuda, M. Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J. Bacteriol. 2006, 188, 4057–4067. [Google Scholar] [CrossRef]
- Shamsuzzaman, K.M.; Barnsley, E.A. The regulation of naphthalene oxygenase in pseudomonads. Microbiology 1974, 83, 165–170. [Google Scholar] [CrossRef][Green Version]
- Park, W.; Padmanabhan, P.; Padmanabhan, S.; Zylstra, G.J.; Madsen, E.L. nahR, encoding a LysR-type transcriptional regulator, is highly conserved among naphthalene-degrading bacteria isolated from a coal tar waste-contaminated site and in extracted community DNA. Microbiology 2002, 148, 2319–2329. [Google Scholar] [CrossRef][Green Version]
- Dennis, J.J.; Zylstra, G.J. Complete sequence and genetic organization of pDTG1, the 83 kilobase naphthalene degradation plasmid from Pseudomonas putida strain NCIB 9816-4. J. Mol. Biol. 2004, 341, 753–768. [Google Scholar] [CrossRef]
- Izmalkova, T.Y.; Sazonova, O.I.; Nagornih, M.O.; Sokolov, S.L.; Kosheleva, I.A.; Boronin, A.M. The organization of naphthalene degradation genes in Pseudomonas putida strain AK5. Res. Microbiol. 2013, 164, 244–253. [Google Scholar] [CrossRef]
- Zuniga, M.C.; Durham, D.R.; Welch, R.A. Plasmid-and chromosome-mediated dissimilation of naphthalene and salicylate in Pseudomonas putida PMD-1. J. Bacteriol. 1981, 147, 836–843. [Google Scholar]
- Bosch, R.; García-Valdés, E.; Moore, E.R. Genetic characterization and evolutionary implications of a chromosomally encoded naphthalene-degradation upper pathway from Pseudomonas stutzeri AN10. Gene 1999, 236, 149–157. [Google Scholar] [CrossRef]
- Bosch, R.; García-Valdés, E.; Moore, E.R. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene 2000, 245, 65–74. [Google Scholar] [CrossRef]
- Connors, M.A.; Barnsley, E.A. Naphthalene plasmids in pseudomonads. J. Bacteriol. 1982, 149, 1096–1101. [Google Scholar]
- Foght, J.M.; Westlake, D.W. Transposon and spontaneous deletion mutants of plasmid-borne genes encoding polycyclic aromatic hydrocarbon degradation by a strain of Pseudomonas fluorescens. Biodegradation 1996, 7, 353–366. [Google Scholar] [CrossRef]
- Li, W.; Shi, J.; Wang, X.; Han, Y.; Tong, W.; Ma, L.; Liu, B.; Cai, B. Complete nucleotide sequence and organization of the naphthalene catabolic plasmid pND6-1 from Pseudomonas sp. strain ND6. Gene 2004, 336, 231–240. [Google Scholar] [CrossRef]
- Heinaru, E.; Vedler, E.; Jutkina, J.; Aava, M.; Heinaru, A. Conjugal transfer and mobilization capacity of the completely sequenced naphthalene plasmid pNAH20 from multi plasmid strain Pseudomonas fluorescens PC20. FEMS Microbiol. Ecol. 2009, 70, 563–574. [Google Scholar] [CrossRef]
- Kim, J.; Park, W. Genome analysis of naphthalene-degrading Pseudomonas sp. AS1 harboring the mega plasmid pAS1. J. Microbiol. Biotechnol. 2018, 28, 330–337. [Google Scholar] [CrossRef]
- Sud, R.K.; Sud, A.K.; Gupta, K.G. Degradation of Sevin (1-naphthyl N-methyl carbamate by Achromobacter sp. Arch. Mikrobiol. 1972, 87, 353–358. [Google Scholar] [CrossRef]
- Larkin, M.J.; Day, M.J. The metabolism of Carbaryl by three bacterial isolates, Pseudomonas spp. (NCIB 12042 & 12043) and Rhodococcus sp. (NCIB 12038) from garden soil. J. Appl. Bacteriol. 1986, 60, 233–242. [Google Scholar]
- Hayatsu, M.; Nagata, T. Purification and characterization of Carbaryl hydrolase from Blastobacter sp. strain M501. Appl. Environ. Microbiol. 1993, 59, 2121–2125. [Google Scholar]
- Hayatsu, M.; Hirano, M.; Nagata, T. Involvement of two plasmids in the degradation of Carbaryl by Arthrobacter sp. strain RC100. Appl. Environ. Microbiol. 1999, 65, 1015–1019. [Google Scholar]
- Hashimoto, M.; Fukui, M.; Hayano, K.; Hayatsu, M. Nucleotide sequence and genetic structure of a novel Carbaryl hydrolase gene (cehA) from Rhizobium sp. strain AC100. Appl. Environ. Microbiol. 2002, 68, 1220–1227. [Google Scholar] [CrossRef]
- Doddamani, H.P.; Ninnekar, H.Z. Biodegradation of Carbaryl by a Micrococcus species. Curr. Microbiol. 2001, 43, 69–73. [Google Scholar] [CrossRef]
- Seo, J.S.; Keum, Y.S.; Li, Q.X. Metabolomic and proteomic insights into Carbaryl catabolism by Burkholderia sp. C3 and degradation of ten N-methyl carbamates. Biodegradation 2013, 24, 795–811. [Google Scholar] [CrossRef]
- Trivedi, V.D.; Bharadwaj, A.; Varunjikar, M.S.; Singha, A.K.; Upadhyay, P.; Gautam, K.; Phale, P.S. Insights into metabolism and sodium chloride adaptability of Carbaryl degrading halotolerant Pseudomonas sp. strain C7. Arch. Microbiol. 2017, 199, 907–916. [Google Scholar] [CrossRef]
- Zhu, S.; Wang, H.; Jiang, W.; Yang, Z.; Zhou, Y.; He, J.; Hong, Q. Genome analysis of Carbaryl-degrading strain Pseudomonas putida XWY-1. Curr. Microbiol. 2019, 76, 927–929. [Google Scholar] [CrossRef]
- Cheesman, M.J.; Horne, I.; Weir, K.M.; Pandey, G.; Williams, M.R.; Scott, C.; Russell, R.J.; Oakeshott, J.G. Carbamate pesticides and their biological degradation: Prospects for enzymatic bioremediation. Ration. Environ. Manag. Agrochem. Risk Assess. Monit. Remedial Act. 2007, 966, 288–305. [Google Scholar]
- Singh, R.; Trivedi, V.D.; Phale, P.S. Metabolic regulation and chromosomal localization of Carbaryl degradation pathway in Pseudomonas sp. strains C4, C5 and C6. Arch. Microbiol. 2013, 195, 521–535. [Google Scholar] [CrossRef]
- Trivedi, V.D. Biochemical and Evolutionary Aspects of Carbaryl Metabolism in Pseudomonas sp. PhD Thesis, Indian Institute of Technology-Bombay, Mumbai, India, December 2016. [Google Scholar]
- Trivedi, V.D.; Jangir, P.K.; Sharma, R.; Phale, P.S. Insights into functional and evolutionary analysis of Carbaryl metabolic pathway from Pseudomonas sp. strain C5pp. Sci. Rep. 2016, 6, 38430. [Google Scholar] [CrossRef]
- Kamini; Sharma, R.; Punekar, N.S.; Phale, P.S. Carbaryl as a carbon and nitrogen source: An inducible methylamine metabolic pathway at the biochemical and molecular levels in Pseudomonas sp. strain C5pp. Appl. Environ. Microbiol. 2018, 84, e01866–e01918. [Google Scholar] [CrossRef]
- Kamini; Shetty, D.; Trivedi, V.D.; Varunjikar, M.; Phale, P.S. Compartmentalization of the Carbaryl degradation pathway: Molecular characterization of inducible periplasmic Carbaryl hydrolase from Pseudomonas spp. Appl. Environ. Microbiol. 2018, 84, e02115–e02117. [Google Scholar]
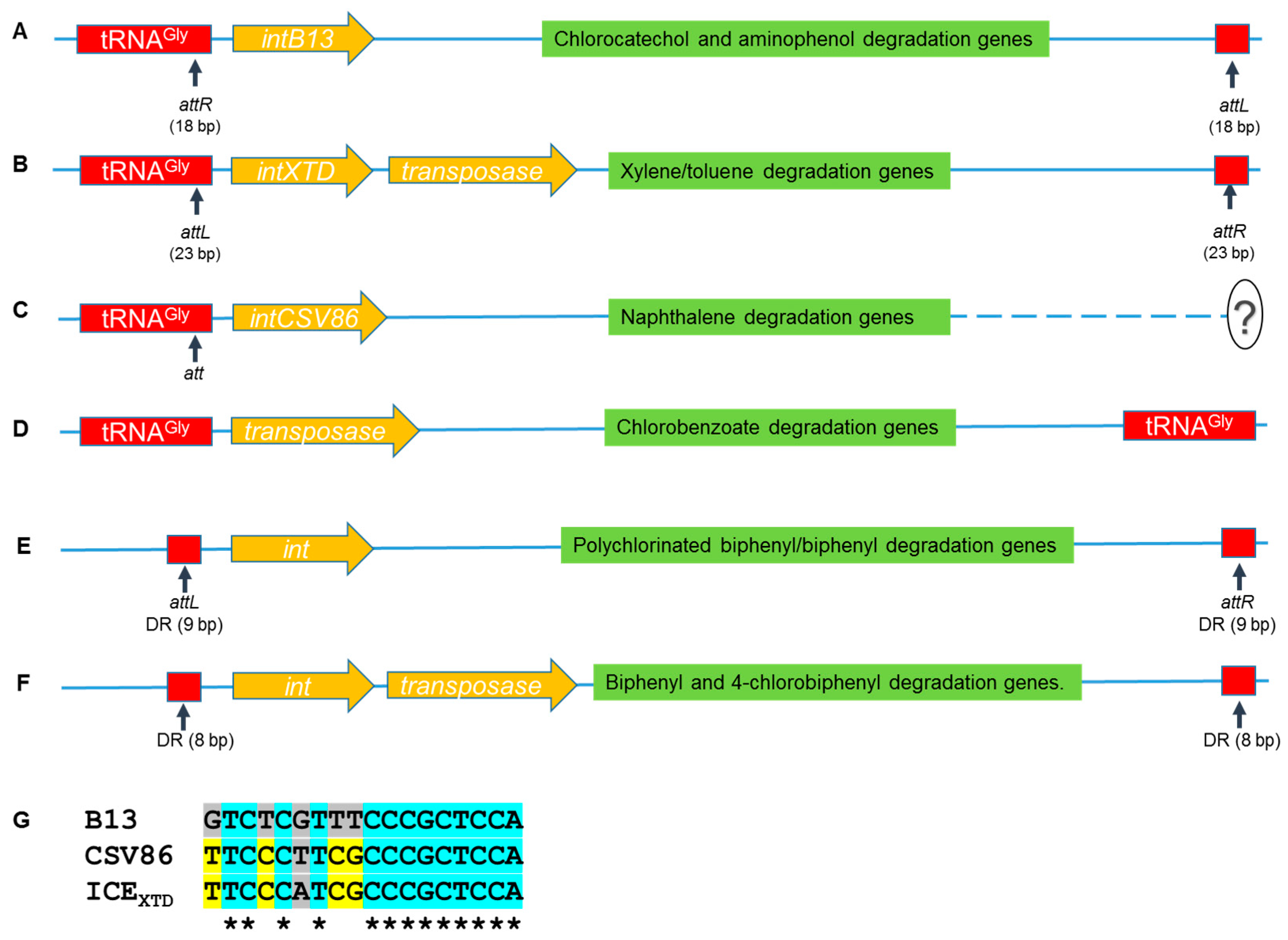
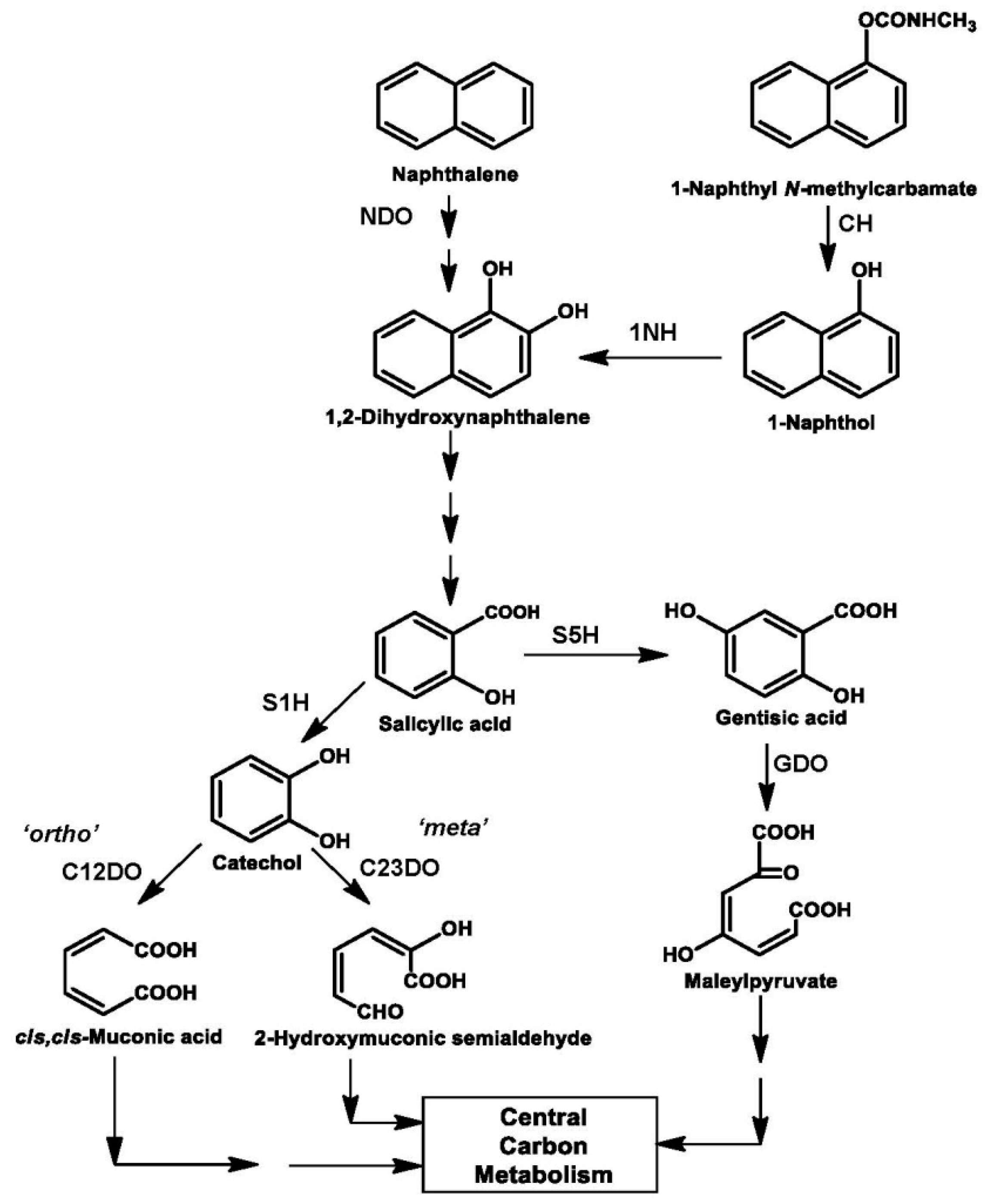
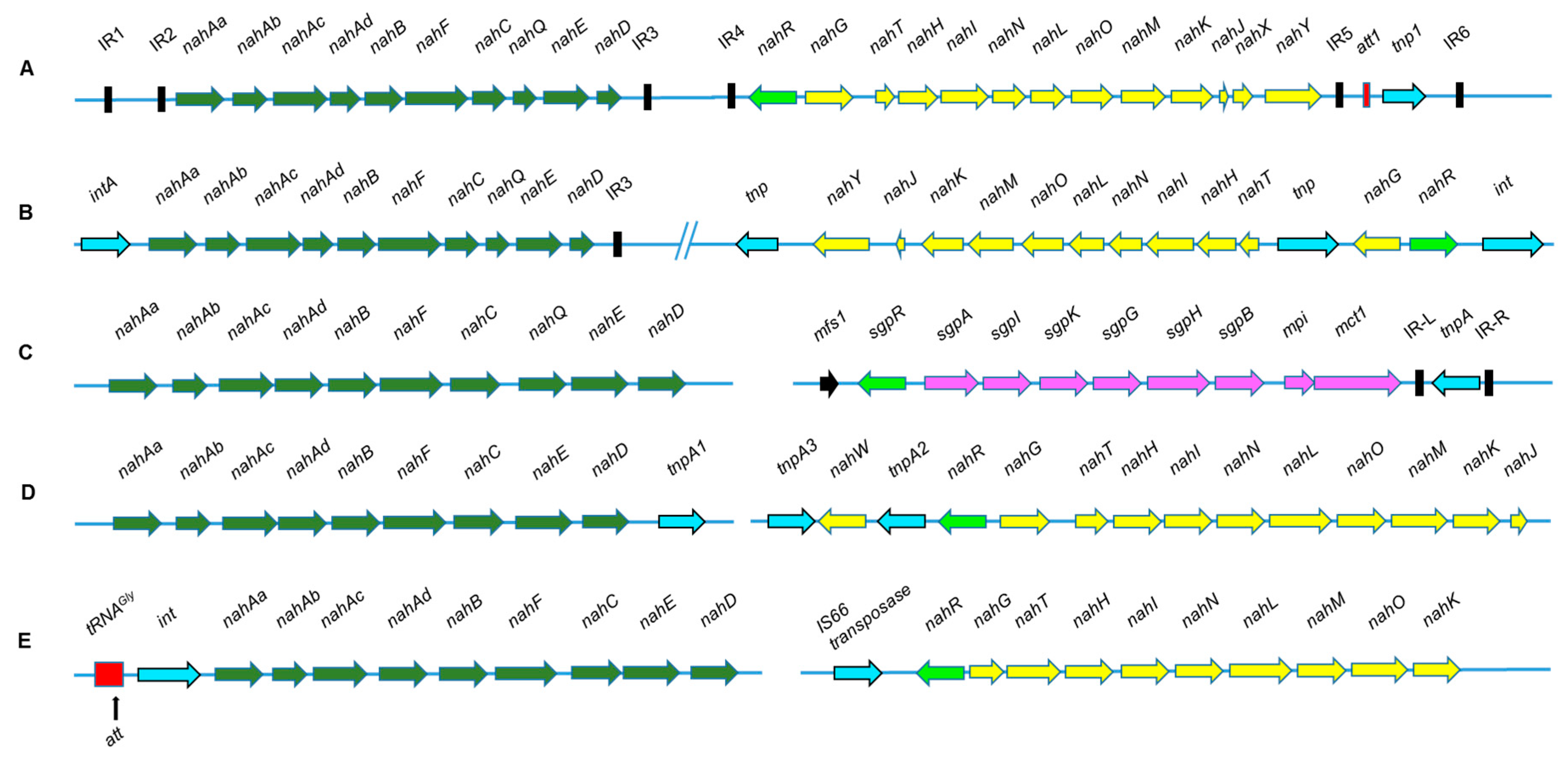
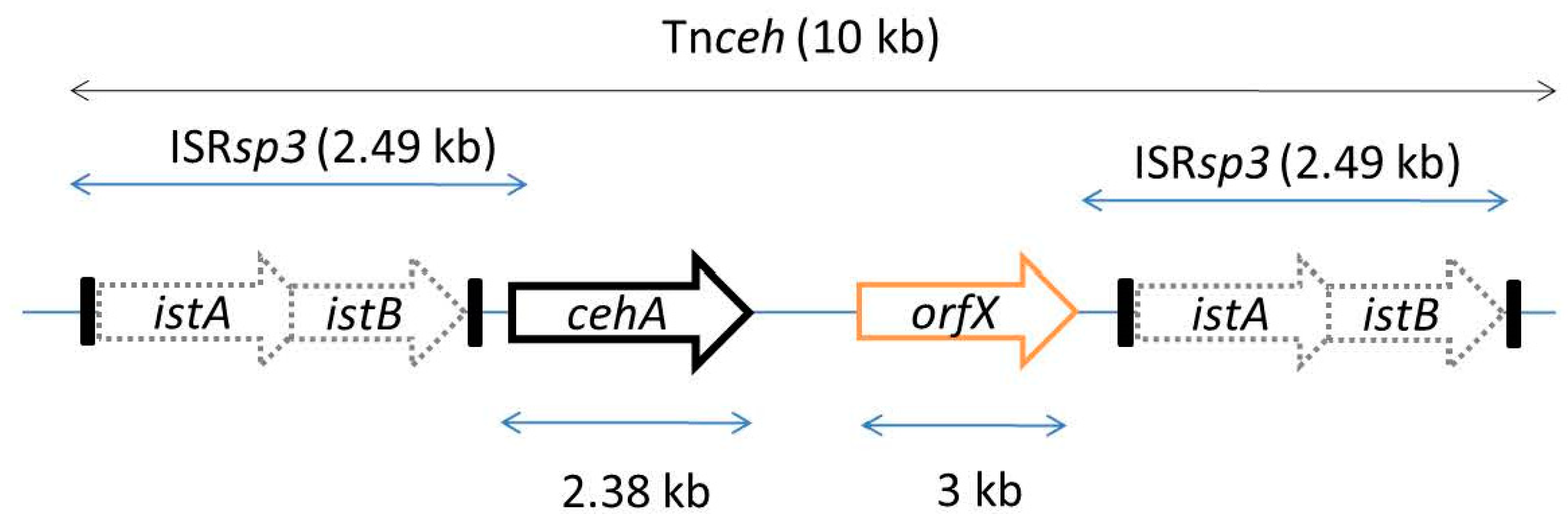
| Organism | Pathway, ring-cleavage mode | Chromosome/plasmid (kb); Operon size (kb) | References |
|---|---|---|---|
| A. Naphthalene degradation | |||
| Pseudomonas putida G7 | Catechol, meta | Plasmid, NAH7 (82); nah (10) and sal (8) | [99] |
| Pseudomonas putida strain NCIB 9816-4 | Catechol, meta | Plasmid, pDTG1 (88); nah (9.5) and sal (13.4) | [102] |
| Pseudomonas sp. strain ND6 | Catechol, meta | Plasmid, pND6-1 (102); nah (10) and sal (18) | [109] |
| Pseudomonas fluorescens strain PC20 | Catechol, meta | Plasmid, pNAH20 (88); nah (6) and sal (13) | [110] |
| Pseudomonas putida strain AK5 | Gentisic acid | Plasmid, pAK5 (-); nah (6.7) and sal (12.2) | [103] |
| Pseudomonas sp. AS1 | Catechol, ortho | Plasmid, pAS1 (82) | [111] |
| Pseudomonas putida strain PMD-1 | Catechol, meta | Chromosome and Plasmid, pMWD-1 | [104] |
| Pseudomonas stutzeri AN10 | Catechol, meta | Chromosome, nah (11.5) and sal (16) | [105,106] |
| Pseudomonas putida CSV86 | Catechol, meta | ICE, nah (8.2) and sal (9.8) | [91] |
| B. Carbaryl degradation | |||
| Achromobacter sp. | Hydroquinone, Catechol, | - * | [112] |
| Pseudomonas sp. NCIB 12043 | Gentisic acid | - | [113] |
| Pseudomonas sp. NCIB 12042 | Catechol, meta | - | [113] |
| Rhodococcus sp. NCIB 12038 | Gentisic acid | - | [113] |
| Consortia Pseudomonas spp. isolate 50581 and 50552 | Catechol | Plasmid, pCD1 (50) in isolate 50581 encodes Carbaryl hydrolase; Chromosome encodes degradative enzymes for 1-naphthol in isolate 50552 | [43] |
| Blastobacter sp. strain M501 | Hydrolysis to 1-naphthol | - | [114] |
| Sphingomonas sp. strain CF06 | Gentisic acid | Plasmids pCF01, pCF02, pCF03, pCF04, and pCF05, role of each plasmid is not clear | [67] |
| Arthrobacter sp. RC100 | Gentisic acid | Plasmid, pRC1 (110) encodes Carbaryl hydrolase; Plasmid, pRC2 (120) encodes enzymes for 1-naphthol to gentisic acid; chromosome encodes enzymes for utilization of gentisic acid | [115] |
| Rhizobium sp. strain AC100 | Partial hydrolysis to 1-naphthol | Plasmid, pAC200 encodes Carbaryl hydrolase | [116] |
| Micrococcus sp. | Gentisic acid | - | [117] |
| Pseudomonas sp. strain C4 | Gentisic acid | Chromosome | [30] |
| Pseudomonas sp. strain C5 | Gentisic acid | Chromosome | [30] |
| Pseudomonas sp. strain C6 | Gentisic acid | Chromosome | [30] |
| Burkholderia sp. C3 | Catechol and Gentisic acid | - | [118] |
| Pseudomonas sp. strain C7 | Gentisic acid | - | [119] |
| Pseudomonas putida XWY-1 | Gentisic acid | Plasmid, pXWY (400) encoding all enzymes of Carbaryl degradation | [120] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phale, P.S.; Shah, B.A.; Malhotra, H. Variability in Assembly of Degradation Operons for Naphthalene and its derivative, Carbaryl, Suggests Mobilization through Horizontal Gene Transfer. Genes 2019, 10, 569. https://doi.org/10.3390/genes10080569
Phale PS, Shah BA, Malhotra H. Variability in Assembly of Degradation Operons for Naphthalene and its derivative, Carbaryl, Suggests Mobilization through Horizontal Gene Transfer. Genes. 2019; 10(8):569. https://doi.org/10.3390/genes10080569
Chicago/Turabian StylePhale, Prashant S., Bhavik A. Shah, and Harshit Malhotra. 2019. "Variability in Assembly of Degradation Operons for Naphthalene and its derivative, Carbaryl, Suggests Mobilization through Horizontal Gene Transfer" Genes 10, no. 8: 569. https://doi.org/10.3390/genes10080569
APA StylePhale, P. S., Shah, B. A., & Malhotra, H. (2019). Variability in Assembly of Degradation Operons for Naphthalene and its derivative, Carbaryl, Suggests Mobilization through Horizontal Gene Transfer. Genes, 10(8), 569. https://doi.org/10.3390/genes10080569




