Abstract
Steroids are perhydro-1,2-cyclopentanophenanthrene derivatives that are almost exclusively synthesised by eukaryotic organisms. Since the start of the Anthropocene, the presence of these molecules, as well as related synthetic compounds (ethinylestradiol, dexamethasone, and others), has increased in different habitats due to farm and municipal effluents and discharge from the pharmaceutical industry. In addition, the highly hydrophobic nature of these molecules, as well as the absence of functional groups, makes them highly resistant to biodegradation. However, some environmental bacteria are able to modify or mineralise these compounds. Although steroid-metabolising bacteria have been isolated since the beginning of the 20th century, the genetics and catabolic pathways used have only been characterised in model organisms in the last few decades. Here, the metabolic alternatives used by different bacteria to metabolise steroids (e.g., cholesterol, bile acids, testosterone, and other steroid hormones), as well as the organisation and conservation of the genes involved, are reviewed.
1. Introduction
Steroids are tetracyclic triterpenoid lipids containing a perhydro-1,2-cyclopentanophenanthrene structure, and include sterols, bile acids, steroid hormones, cardenolides, sapogenins, saponins, and vitamin D derivatives. Classically, they have been studied based on their physiological role. In mammals, the steroid hormones, bile acids, and other essential steroids are produced from cholesterol (Figure 1), a molecule that is also an important component of membranes, maintaining their fluidity, and participating in cell differentiation and proliferation. The major constituents of plant sterols are sitosterol, stigmasterol, campesterol, and brassicasterol, while in yeast and filamentous fungi, ergosterol is an important component of cell walls [1,2,3].
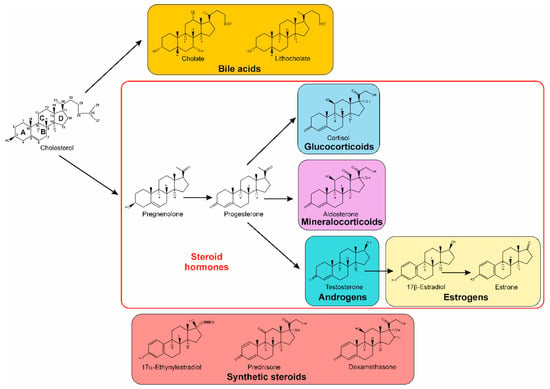
Figure 1.
Chemical structure of cholesterol and some of its mammalian derivatives, and selected synthetic steroids.
It is generally accepted that the biosynthesis of steroids is one of the hallmarks of the evolutionary progression of eukaryotes [4]. However, reports on the biosynthesis of sterols and modified sterols in methanotrophic bacteria (Methylococcus capsulatus, Methylosphaera hansonii, and Methylobacterium organophilum), Gemmata obscuriglobus, Eudoraea adriatica, and in a variety of myxobacteria have been published, though there is limited information about the metabolic machinery involved [5,6,7,8,9,10,11,12,13,14,15].
From an ecological point of view, the release into the environment of vertebrate steroids, such as androgens, estrogens, progestogens, cholesterol, and bile acids, during excreta or decomposition of biomass is constantly occurring [16,17,18,19,20,21,22]. In addition, not only natural steroids are released. Along with antibiotics, synthetic steroids represent a significant sector of the global pharmaceutical market, and they have found their way into environmental niches, as in the case of synthetic hormones (ethynylestradiol and anti-inflammatory drugs like dexamethasone) (Figure 1) [19,20,23,24,25,26,27]. With their widespread presence, they can affect endocrine activity, even at low concentrations, with potential adverse effects for both the environment and human health. As such, there is an increasing need for new approaches in the bioremediation of steroids from the environment.
Although steroid synthesis is mostly restricted to eukaryotic organisms [4], the biodegradation of these compounds seems to be carried out exclusively by bacteria. Even so, steroids are highly recalcitrant to microbial degradation because of the low number of functional groups present in their structure and their extremely low solubility in water. Nevertheless, the persistence of these molecules in the environment has triggered a response by some bacteria, either efficiently detoxifying these hydrophobic molecules or metabolising their carbon and energy-rich scaffold. Thus, the isolation of microorganisms able to degrade or modify steroids has been reported from soil [28,29,30], and from freshwater [31] and marine [32,33,34] environments. This indicates the importance of bacteria in reducing the adverse impact of environmentally released steroids, as well as emphasising their importance in introducing these molecules into the carbon cycle. Moreover, for some pathogenic bacteria, such as Mycobacterium tuberculosis and Rhodococcus equii, catabolism of cholesterol has been identified as a trait involved in their pathogenicity and persistence in the host [35,36].
In 1913, Söhngen reported that a Mycobacterium strain could use phytosterols as a sole carbon and energy source [37]. Thereafter, many bacterial strains have been isolated that are able to degrade steroids, including species initially classified as belonging to genera Mycobacterium, Gordonia, Tsukamurella, Rhodococcus, Azotobacter, Nocardia, Flavobacterium, Arthrobacter, Bacillus, Brevibacterium, Corynebacterium, Streptomyces, Microbacterium, Serratia, Achromobacter, Protaminobacter, and Pseudomonas [29,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54]. However, it has not been until the last two decades of the 20th century that studies have been initiated to elucidate the physiological and metabolical determinants, and the mechanisms involved in the aerobic catabolism of different steroid molecules. These studies have included sterols, chiefly cholesterol, testosterone and 17β-estradiol, and bile acids, mainly cholic acid, using selected model organisms from Actinobacteria, comprising different strains of Mycobacterium, Rhodococcus, and Gordonia [36,55,56,57,58,59] (Table 1), and some selected Gram negative strains, the α-proteobacteria Comamonas testosteroni and a few Pseudomonas strains [28,29,60,61,62]. In addition, the catabolism of steroids under anoxic conditions has been studied in Sterolibacterium (β-proteobacteria) and in Steroidobacter (ϒ-proteobacteria) [63,64,65] (Table 1).

Table 1.
Model strains widely used in the studies that have allowed the characterization of the metabolic mechanisms used for steroid degradation. Steroids used in these studies are also indicated.
2. Uptake of Steroids
The catabolism of steroids implies their selective uptake and internalisation inside microbial cells. However, there is limited information about these processes. Regarding steroid uptake, given the physiological and structural surface differences between bacterial groups, its mechanisms in most bacteria have remained elusive. This is especially evident in Gram-negative bacteria, which possess an outer membrane where the lipopolysaccharide leaflet on the outer surface impedes access of steroids to the cytoplasm by passive diffusion [66].
In mycolic acid-containing Actinobacteria, the transport of steroids is carried out by different transport systems, depending on the particular steroid to be assimilated. The sterol uptake into cells of M. tuberculosis, M. smegmatis, Rhodococcus jostii, R. equi, and Gordonia cholesterolivorans is performed using a set of proteins encoded by the mce4 locus [35,67,68,69,70,71]. The ten genes contained in the mce4 locus, referred to in short-hand by the names yrbE4ABmce4ABCDEFmas4AB in Mycobacterium spp. [71,72,73], and supABmce4ABCDEFHI in Rhodococcus and Gordonia spp. [67,68,69] (Figure 2), codify multicomponent ATP-dependent sterol uptake systems. Thus, these genes are upregulated in M. tuberculosis, M. smegmatis, and R. jostii during growth on cholesterol [55,73]. The deletion of some or all of the genes of the cluster prevents mutants of M. smegmatis [72] and R. jostii [67] growing on cholesterol as the sole carbon and energy source. Surprisingly, analogous mutants of M. tuberculosis are still able to grow using cholesterol, suggesting such strains have an alternative cholesterol transport system [67].
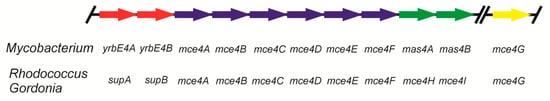
Figure 2.
Genetic organisation of the mce4 cluster involved in sterol uptake in Actinobacteria.
Further analysis revealed that yrbE4AB (supAB) encode transmembrane proteins similar to permease components of ABC (ATP-binding cassette) transporters [74]. These putative permease subunits should be anchored into the cytoplasmic membrane to facilitate cholesterol translocation across this cellular barrier. They are associated with Mkl ATPase, encoded by mceG, which is not linked with the cluster [35,75] (Figure 2). In M. tuberculosis, this protein acts as the ATPase component, not only with Mce4 proteins in the uptake of sterols, but also with Mce1, another transport system that is linked to fatty acid transport rather than to steroid catabolism [35,76]. Even so, both transport systems, Mce1 and Mce4, participate in mycobacterial infection [35,75,76,77,78]. Thus, MceG ATPase hydrolyses ATP, providing energy for the substrate import for both Mce4 in cholesterol uptake and for Mce1 in fatty acid internalisation [35,67]. Mce4ABCDEF are abundant in cell envelope protein fractions, hence their characterisation as cell wall-associated proteins. They probably facilitate cholesterol transport across the mycolic acid layer and/or pseudoperiplasmic space. Signal sequences for protein export via the general secretory pathway can be found in the amino acidic sequences of Mce4ABCDEF. Moreover, Mce4F has a putative transmembrane domain in the N-terminus of the protein, which probably allows for its embedding in the cytoplasmic membrane or cell wall [74,79,80]. In addition, Mas4AB (Mce4HI), both required for cholesterol import [74], have been suggested to be accessory subunits for their respective systems, with a putative function in stabilising or assembling Mce4ABCDEF complexes [76,81].
Other proteins, codified by genes not included in the mce4 operon, have been connected to cholesterol uptake in Mycobacterium. OmamA has been proposed to be a stabiliser of Mce1 and Mce4 complexes, playing a role in cholesterol and fatty acid uptake in this bacterium [81]. In the same way, LucA also participates in the uptake of both cholesterol and fatty acids, having a cell membrane or cell envelope location. LucA interacts with Mas4A, Mas4B, OmamA, and probably MceG, potentially protecting these proteins, and hence the uptake complexes, from proteolytic attack [76].
As previously indicated, the Mce4 system is required for the uptake of sterols in Actinobacteria. However, this system is not used by R. jostii when growing on cholic acid as the sole carbon and energy source [82]. The mutation of the gene coding the porin RjpA impairs growth on metabolising cholic acid, however, it had no effect when sterols were the molecules used to support growth, indicating the importance of this porin for cholic acid uptake [82]. Although the system for the entry of this bile acid through plasma membrane has not been identified, a proteomic study of cholic acid growing cells suggested there might be two systems, CamM, a major facilitator superfamily transporter, and CamABCD, an ATP-binding cassette (ABC) transporter [83]. However, a parallel study demonstrated that both systems act in the reassimilation of transiently accumulated cholic acid intermediates, previously secreted to the culture broth by bacteria during bile acid catabolism [84]. The accumulation of metabolic intermediates in culture broths by steroid-degrading bacteria has been documented [85]; Pseudomonas stutzeri strain Chol1 metabolising bile acids extracellularly accumulates intermediates, probably to regulate their intracellular levels or due to overflowing of the downstream metabolic pathway [85]. Later, during growth, these metabolites disappear from the culture broth, which implies: (i) the existence in these bacteria of an exclusion system, and (ii) a catabolic route involved in the reassimilation of these compounds [85].
In the anaerobic cholesterol-degrading Sterolibacterium denitrificans DSMZ 13999, it has been suggested that assimilation of cholesterol could proceed via direct adhesion, followed by outer membrane transport mediated by a FadL-like transport system. Once in the periplasm, initial reactions in cholesterol catabolism occur. Notably, the Fad-like system exhibited high substrate specificity for C27 sterols, but not for C19 androgens [86].
3. The Aerobic 9,10-seco Pathway
The most complete biochemical and genetic information about steroid catabolic pathways is based on the investigation of a few Actinobacteria and Proteobacteria species, which can mineralise steroids under aerobic conditions. Thus, testosterone and bile acid degradation have been studied in C. testosteroni strain TA441 and C. thiooxidans CNB-1 [60], P. stutzeri strain Chol1 [85,87], and Pseudomonas putida strain DOC21 [29,62]. Cholesterol degradation has been studied in R. jostii strain RHA1 [55], R. equi strain 103S [36], Rhodococcus rhodocrous strain DSM 43269 [88], Rhodococcus ruber strain Chol-4 [54], M. tuberculosis strain H37Rv [89,90], M. smegmatis strain mc2 155 [56,91], G. cholesterolivorans strain Chol-3 [53,69], and Gordonia neofelifaecis strain NRRL B-59395 [59,92]. Bile acid degradation has also been studied in R. jostii RHA1 [57,84]. In all these bacteria, the degradation of the chemical nucleus of the different steroids follows similar steps, using the 9,10-seco pathway. Results from the laboratory of Mohn [34,93] suggest that only Actinobacteria degrade sterols with the 9,10-seco pathway, while Proteobacteria degrade bile acids and other less structurally complex steroids using this pathway.
3.1. Oxidation of 3-hydroxyl-substituent from Sterols and Bile Acids
The degradation of steroid compounds by the 9,10-seco pathway is initiated by oxidative attack of 3-oxo steroid. Therefore, steroids, such as sterols or bile acids, carrying a 3-hydroxyl-substituent, need to be oxidised to the corresponding ketone as a preliminary step. However, in R. jostii strain RHA1 it has been reported that the catabolism of sterols starts through the ω-oxidation of the C17 side chain to the corresponding carboxylic acid, which is later catabolized through the classic pathway (see below). However, it is not clear if the formation of the carboxylic acid occurs before or simultaneously with the initial oxidation of the sterane rings [94].
For sterols, the initial reaction involves the oxidation of the 3β-hydroxy group, concomitantly with isomerisation of the C5–C6 double bond present in these molecules, to yield 3-keto-4-ene structures. Thus, considering cholesterol as a sterol model, the first reaction in its catabolism results in its transformation into cholest-4-en-3-one. This double step, oxidation of the hydroxyl group and isomerisation of the double bond, is catalysed either by a cholesterol oxidase requiring molecular oxygen or by NAD(P)-dependent 3-β-hydroxy-Δ(5)-steroid dehydrogenase (Figure 3) [95].
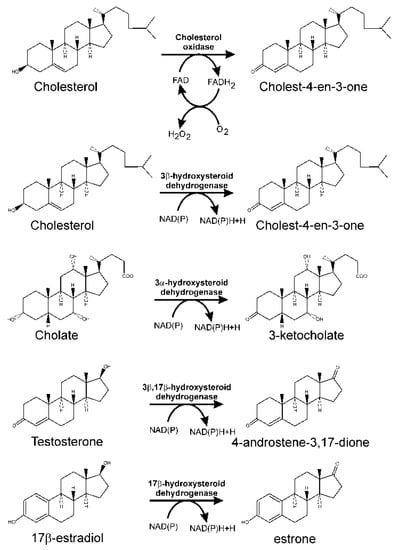
Figure 3.
Initial reactions catalysed by cholesterol oxidase and different hydroxysteroid dehydrogenases in steroid degradation through the 9,10-seco pathway.
Most of the organisms that possess a cholesterol oxidase produce it as an extracellular form, either released into the culture broth or linked to the cell surface. Cholesterol oxidases are monomeric enzymes containing FAD, and belong to two different classes. Class I includes enzymes, belonging to the glucose/methanol/choline oxidoreductase family, which fix FAD into their active site cavity through non-covalent bonds [96]. However, Class II enzymes belong to the vanillyl-alcohol oxidase family, covalently binding the FAD cofactor [97].
A NAD(P)-dependent cholesterol dehydrogenase has been isolated from cells of Nocardia sp. Ch2-1 [98]. In M. tuberculosis, the formation of cholest-4-en-3-one is carried out by a 3-β-hydroxysteroid dehydrogenase (HsdD, Rv1106c) [99]. Although a gene for cholesterol oxidase (ChoD, Rv3409c) has also been found in this bacterium, its activity has not been confirmed in vitro, and its homology to many described cholesterol oxidases is rather limited. Moreover, its disruption does not result in a marked change in the ability of the mutant strain to grow using cholesterol [100].
In M. smegmatis, a constitutive cholesterol oxidase (Msmeg1604) similar to ChoD from M. tuberculosis, and two cholesterol inducible enzymes, Msmeg5228 (a cholesterol oxidase) and Msmeg5233 (a cholesterol dehydrogenase/isomerase), have been identified. Msmeg1604 does not seem to play a critical role in the mineralisation of cholesterol, since a specific mutation in this protein does not affected the production of cholest-4-en-3-one. However, an Msmeg5228 defective mutant shows a drastic reduction in the formation of this intermediate [56]. In addition, double mutants affecting HsdD and ChoD in M. tuberculosis, and Msmeg5228 and Msmeg5233 in M. smegmatis, are still able to grow on cholesterol, suggesting there are other dehydrogenase/isomerases that could replace them in the first reaction of the cholesterol degradation pathway [56,101].
In R. ruber, Chol-4, a constitutive extracellular cholesterol oxidase has been characterised [58]. However, although the deletion of this gene delayed bacterial growth when cholesterol was the sole carbon source, it did not completely prevent it, suggesting the existence of genes with overlapping activities in this bacterium [58].
Bile acids also have a hydroxyl group in the C3 position that should be oxidised to a 3-oxo group prior to oxidation of the steroid nucleus. However, this hydroxyl group is in an α-configuration, and its oxidation in those strains able to use bile acids as a carbon and energy source is performed by 3α-hydroxysteroid dehydrogenase, which is a NAD(P)-dependent enzyme belonging to the short chain dehydrogenase/reductase superfamily [102]. The first description of this kind of enzymes corresponds to a 3α-hydroxysteroid dehydrogenase/carbonyl reductase in C. testosteroni [103]. It was found to be functional as an oxidoreductase towards a variety of 3α-steroid substrates [104,105]. The enzyme also catalyses the reduction of non-steroidal aldehydes and ketones, and consequently, has been named 3α-hydroxysteroid dehydrogenase/carbonyl reductase [105]. Its gene has been cloned from C. testosteroni ATCC 11996 [106], and it is also involved in steroid catabolism in C. testosteroni strain TA441 [107].
NAD-dependent 3α-hydroxysteroid dehydrogenase activity has been detected in P. stutzeri Chol1 cell-free extracts when this bacterium was cultured in a medium containing cholic acid as the carbon source [85]. Moreover, the encoding gene has been annotated in the draft genome of this bacterium within the 79-Kb gene cluster containing the ORFs required for steroid assimilation (Figure 4) [108].
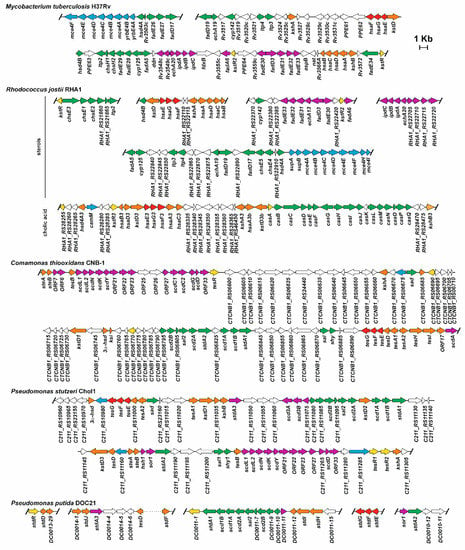
Figure 4.
Genetic organisation of the genes encoding the 9,10-seco pathway involved in cholesterol (Mycobacterium tuberculosis H37Rv, Rhodococcus jostii RHA1), or in cholic acid and testosterone (R. jostii RHA1, Comamonas thiooxidans CNB-1 (formerly, Comamonas testosteroni CNB-2), Pseudomonas stutzeri Chol1 (formerly Pseudomonas sp. Chol1), and Pseudomonas putida DOC21 catabolism. Genes coding enzymes involved in cholesterol or bile acids side chain degradation are shown in green; genes participating in ring A/B degradation are in orange; in purple are shown genes coding for ring C/D degradation; blue color indicates genes coding transport systems.
In P. putida DOC21, a Tn5 insertion in a gene encoding a 3α-hydroxysteroid dehydrogenase revealed that this mutant was unable to further metabolise any of the tested bile acids, cholic, lithocholic, chenodeoxycholic, ursodeoxycholic, and deoxycholic acid [62].
3.2. Side Chain Degradation of Sterols and Bile Acids
After this initial step, the catabolism of steroids proceeds through two sub-pathways, which involve C17 side chain cleavage and/or steroid nucleus oxidation. Degradation of the alkane side chain of cholesterol has been proposed to proceed through a β-oxidation-like process analogous to that of the catabolism of fatty acyl-CoA in human mitochondria and peroxisomes (Figure 5) [109,110,111]. Thus, cholest-4-en-3-one is used as substrate by Cyp125 P450 cytochrome, catalysing the oxidation of the C26 or C27 terminal methyl group [94,112,113,114], followed by further oxidation to provide the initial carboxylate. Although in M. tuberculosis strain CDC1551 this protein is absolutely required for bacterial growth on cholesterol as the sole carbon source [115], in M. tuberculosis strain H37Rv, loss of this activity is compensated for by another P450 cytochrome, Cyp142 [116]. However, substrate specificities of the two P450 cytochromes are different, with Cyp125 being specific for cholesterol, and although able to oxidise cholesteryl sulfate at a low rate, it is unable to oxidise cholesteryl propionate. By contrast, Cyp142 can efficiently metabolise cholesteryl-sulfate as well as cholesteryl-propionate. This suggests that Cyp142 enzymes may play an important role during M. tuberculosis infection, by providing access to additional reservoirs of esterified intracellular cholesterol that would not otherwise be available to the pathogen [117].
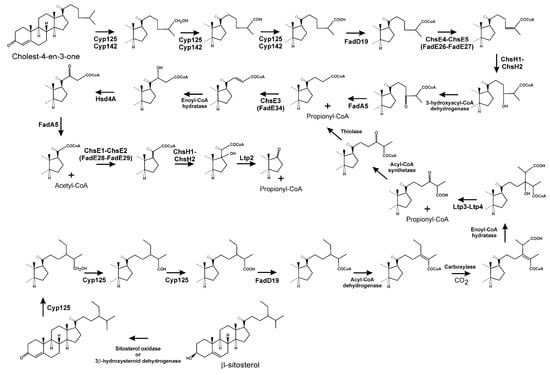
Figure 5.
Catabolism of the cholesterol side chain and C24-branched chain of β-sitosterol in Actinobacteria.
The carboxyl-functionalised side chain is then converted to its CoA thioester by FadD19, producing 3-oxocholest-4-en-26-oyl-CoA (Figure 5) [118,119]. Later, an α2β2 heterotetrameric acyl-CoA dehydrogenase, ChsE4-ChsE5, also named FadE26-FadE27, catalyses the α,β-unsaturation of this acyl-CoA thioester, to form 3-oxocholest-4,24-dien-26-oyl-CoA (Figure 5) [90,120,121]. Notably, those human and bacterial acyl-CoA dehydrogenases involved in β-oxidation form α4 homotetramers or α2 homodimers [122], in contrast to the unusual quaternary structure of acyl-CoA dehydrogenases acting in cholesterol side chain catabolism. 3-Oxocholest-4,24-dien-26-oyl-CoA is the substrate of a MaoC-like enoyl-CoA hydratase, ChsH1-ChsH2, being converted into 24-hydroxy-3-oxocholest-4-en-26-oyl-CoA (Figure 5) [123]. Transformation of this compound to 3,24-dioxocholest-4-en-26-oyl-CoA should be carried out by a β-hydroxyacyl-CoA dehydrogenase. Although there is no experimental evidence, it has been proposed that Hsd4A in M. tuberculosis could be the enzyme involved in this process [124,125]. Next, FadA5, a thiolase, catalyses the cleavage of this last CoA-esterified oxo-derivative, forming 3-oxochol-4-en-24-oyl-CoA and propionyl-CoA (Figure 5) [126,127]. FadA5 has been categorised as a member of the trifunctional enzyme-like thiolases, type-1 class, which contains a predicted binding site for a bulky fatty acid tail [128,129]. At this point, a second β-oxidation cycle starts, with the introduction of a trans double-bond in 3-oxochol-4-en-24-oyl-CoA by ChsE3 (FadE34), followed by the hydration of the double bond by an as yet unknown enoyl-CoA hydratase, and then a dehydrogenation performed by HsdA4, generating 3,22-dioxochol-4-en-24-oyl-CoA [130]. Finally, FadA5 acts again, releasing acetyl-CoA and 3-oxo-pregne-20-carboxyl-CoA (Figure 5) [124,126,127].
FadA5 has been proposed as the thiolase involved in finishing the first two β-oxidation-like rounds. However, although the gene encoding this activity has been shown to be upregulated in the presence of cholesterol in M. tuberculosis strain H37Rv, the fadA5 homologue in R. jostii strain RHA1 was not similarly upregulated [55]. Conversely, when fadA5 was inactivated in a R. rhodochrous DSM 43269 derivative, the resulting mutant was not impaired in cholesterol or β-sitosterol side chain degradation, indicating that FadA5 is not essential for the degradation of the sterol side chain in this strain [126,131].
The elimination of the side chain ends by a mechanism resembling a new β-oxidation cycle (Figure 5), starting with dehydrogenation of 3-oxo-4-pregnene-20-carboxyl-CoA to 3-oxo-4,17-pregnadiene-20-carboxyl-CoA in a process catalysed by another heterotetrameric α2β2 acyl-CoA dehydrogenase complex, ChsE1-ChsE2 (FadE28-FadE29; Figure 5) [90,120,121]. This last compound is then the substrate of an enoyl-CoA hydratase (ChsH1-ChsH2) [90], and the hydrated molecule undergoes an aldol-lyase cleavage reaction catalysed by Lpt2, producing androst-4-en-3,17-dione (AD) and releasing another propionyl-CoA molecule (Figure 5) [90,109,110,132]. Ltp2, at the protein sequence level, is more related to thiolases and acetoacetyl-CoA synthases than to aldolases from other metabolic pathways. It has been shown that Lpt2 interacts with ChsH2, forming a complex ChsH1-ChsH2-Ltp2, catalysing the last two steps of side chain removal from cholesterol [132]. In summary, the complete metabolism of the cholesterol side chain results in a 17-keto steroid intermediate, AD, as well as one acetyl-CoA and two propionyl-CoA molecules.
Removal of the C17 side chains of β-sitosterol and campesterol are processes less characterised than cholesterol side chain degradation. Both β-sitosterol and campesterol contain branched side chains that impede the first β-oxidation round of the side chain; therefore, these need to be eliminated to allow full degradation of the side chain. Initially, the C24-branched side chain is oxidised at position C26 by Cyp125, followed by CoA activation by FadD19 (Figure 5) [118]. Next, the bond between C24/C25 is desaturated. Removal of the C24-branches then starts by carboxylation of the C28 carbon, followed by a hydration reaction of the double bond, and finally the release of acetyl-CoA, from campesterol, or propionyl-CoA, from β-sitosterol, via cleavage of the C24–C25 bond by a heteromeric aldol-lyase (Ltp3-Ltp4) (Figure 5) [131]. It is evident that after aldolytic cleavage of the branch, thioesterification of the resulting carboxylate should be mandatory for degradation of the side chain (Figure 5). At the end of side chain degradation from β-sitosterol and campesterol, AD is formed.
Catabolism of the side chain of bile acids is a process in which not all the enzymatic steps are fully characterised. After oxidation of their hydroxy-group at C3 to an oxo-group in these carboxylated molecules, the degradation of the lateral chain begins. Firstly, the carboxy group from the side chain in C17 is activated to a coenzyme A thioester through a reaction catalysed by a bile acid-CoA ligase (StdA1 in P. putida DOC21 and CasG in R. jostii; Figure 6) [56,62,118]. Once activated, a β-oxidation-like process occurs, although it is different to the two initial rounds in the cholesterol side chain degradation. In the first place, a dehydrogenation of C2/C3 of the acyl-CoA side chain occurs. Based on bioinformatics analysis of clusters coding for bile acid catabolism in P. stutzeri Chol1, the participation of a heteromeric acyl-CoA dehydrogenase, Scd1AB, has been proposed [108]. In a second step, the hydration of the α,β-double bond takes place. The introduction of a hydroxy group at the third carbon of the lateral chain in P. stutzeri Chol1 is catalysed by the enoyl-CoA hydratase Shy1 (Figure 6) [87]. Until this step, the process is identical to a canonical β-oxidation. However, the next enzymatic step is not the expected oxidation of the hydroxy function to a keto group, followed by a thiolytic cleavage mediated by the introduction of a CoA molecule; instead, it is an aldolic cleavage of the C–C bond, catalysed by Sal1, generating an aldehyde and releasing an acetyl-CoA molecule (Figure 6) [87,133]. Thus, cleavage of these two carbons in unbranched side chains of bile acids proceeds through a retroaldol reaction, as a retro-Claisen reaction, instead of the typical reaction catalysed by thiolases. The resulting aldehyde is then oxidised to the corresponding carboxylic acid by a specific aldehyde dehydrogenase (Sad in P. stutzeri Chol1; Figure 6) [87], and later, a second acyl-CoA synthetase (StdA2 in P. putida DOC21 and CasI in R. jostii) activates it to a CoA derivative [56,62,119]. The subsequent degradation of the remaining side chain is believed to occur through a similar mechanism, where an acyl-CoA dehydrogenase, putatively Scd2AB, introduces an α,β-desaturation in the CoA-activated C3 side chain (Figure 6) [85,108]. Hydration of the double bond takes place, resulting in the formation of a hydroxyl group at C17, and then an aldolytic cleavage of the molecule occurs, yielding a molecule of propionyl-CoA and a steroid derivative with a keto function at C17 (Figure 6). The enzymes catalysing these two steps have not been fully characterised yet [108].

Figure 6.
Cholic acid metabolism in Pseudomonas putida DOC21 and P. stutzeri Chol1 through the 9,10-seco pathway. Preliminary evidence suggests that oxidation of the A-ring occurs simultaneously with C17 side chain degradation. Hydroxyl groups at C7 and C12 are maintained during degradation of the molecule, the affecting 3aα-H-4α(3′-propanoate)7a-β-methylhexahydro-1,5- indanedione-hydroxylated derivatives metabolism. 3′,7-diOH-HIP, 3aα-H-4α(3′(R)-hydroxy-3′propanoate)-7-hydroxy-7aβ-methylhexahydro-1,5-indanedione; 3′,7-diOH-HIP-CoA, 3aα-H-4α(3′(R)-hydroxy-3′propanoyl-CoA)-7-hydroxy-7aβ-methylhexahydro- 1,5-indanedione; 3′,5,7-triOH-HIP-CoA, 3aα-H-(3′(R)-hydroxy-3′propanoyl-CoA)-5,7-dihydroxy-7aβ-methylhexahydro-1-indanone; 4OH-COCHEA-CoA, 2-(2-carboxyethyl)-4-hydroxy-3-methyl-6-oxocyclohex-1-ene-1-carboxyl-CoA; 7-OH-HIEC-CoA, (7aS)-7a-methyl-7-hydroxy-1,5-dioxo-2,3,5,6,7,7a-hexahydro-1H-indene-4-carboxyl-CoA; 5,7-diOH-HIC-CoA, 3aα-H-4α(3′-carboxyl-CoA)-5,7-dihydroxy-7aβ-methylhexahydro-1-indenone.
3.3. Testosterone Catabolism Convergence
For the catabolic convergence of testosterone into this pathway, the 17β-hydroxy substituent needs to be oxidised to a 17-oxo-derivative. The oxidation of this substituent to a keto group produces androst-4-en-3,17-dione [134]. Currently, the best characterised bacterial enzyme performing this reaction is 3β,17β-hydroxysteroid dehydrogenase (3,17β-HSD) from C. testosteroni, encoded by the gene βhsd (Figure 3). This NAD(H)-dependent enzyme belongs to the short-chain dehydrogenases/reductases superfamily. It was first purified from strain ATCC11996, and was initially defined as 3β-HSD; however, the cloned enzyme was revealed to act on both the 3β-hydroxyl and 17β-hydroxyl groups of androgens, estrogens, and iso-bile acids [134,135,136]. Horinouchi et al. [107] suggested that the main role of 3,17β-HSD in C. testosteroni TA441 cells is 3β-dehydrogenation, and there is at least one more dehydrogenase acting on the 17β-hydroxyl group, since a knock out mutation in the coding gene did not prevent its growth on testosterone. Thus, C. testosteroni may have more than one enzyme with 17β-dehydrogenating/hydrogenating activities, to deal with intermediate compounds having considerable structural differences [107]. However, the degradation of testosterone by the 3,17β-HSD mutant is highly affected. Moreover, 3,17β-HSD gene expression is highly induced by testosterone, but not by estradiol and cholesterol, suggesting that this enzyme is a key component in the degradation of testosterone [137].
In P. putida DOC21, a microorganism isolated from soil based on its capacity to degrade bile acids and testosterone, a gene encoding 17β-hydroxysteroid dehydrogenase has been identified (unpublished results). This enzyme only has the ability to oxidise the 17β-hydroxy-group of testosterone, because, although this bacterium efficiently metabolises testosterone and androstanolone, it is unable to metabolise closely related compounds with a 3β-hydroxy group (i.e., trans-androsterone) [29]. However, current studies are being performed to more completely characterise this enzyme. Other bacterial 17β-hydroxysteroid dehydrogenases have been identified; however, they have been linked to estrogen degradation (Figure 3) through the 4,5-seco pathway (see below).
3.4. Oxidation of A/B Rings
Because of the initial processing of the steroid nucleus and elimination of the C17 side-chain, a common intermediate, AD, arises from sterols and testosterone. Starting from this compound, a common pathway is used, under aerobic conditions, for the complete assimilation of carbon atoms with the concomitant production of metabolic energy (Figure 7). This starts with the transformation of AD into androst-1,4-dien-3,17-dione (ADD), by the desaturation of the bond between C1 and C2, with trans axial removal of the hydrogen atoms C1 (α) and C2 (β), in a reaction catalysed by a 3-ketosteroid-Δ1(2)-dehydrogenase [138,139]: TesH in C. testosteroni [140], StdH in P. putida DOC21 [141], KstD in M. tuberculosis and Rhodococcus erythropolis [142,143,144,145], and KsdD in M. smegmatis [146].

Figure 7.
Proposed 9,10-seco pathway from androst-4-en-3,17-dione to central metabolites, pyruvate, acetyl-CoA, propionyl-CoA, and succinyl-CoA. AD, androst-4-en-3,17-dione; ADD, androst-1,4-dien-3,17-dione; 9OH-AD, 9-hydroxy-androst-4-en-3,17-dione; 9OH-ADD, 9-hydroxy-androst-1,4-dien-3,17-dione; 3-HAS, 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione; 3,4-DHSA, 3,4-dihydroxy-9,10- secoandrosta-1,3,5(10)-triene-9,17-dione; 4,9-DSHA, 4,5,9,10-diseco-3-hydroxy-5-9-17- trioxoandrosta-1(10),2-diene-4-oic acid; HHD, 2-hydroxy-2,4-hexadienoic acid; HIP, 3aα-H-4α(3′-propanoate)7a-β-methylhexahydro-1,5-indanedione; 5OH-HIP-CoA, 3aα-H-4α(3′-propanoyl-CoA)-5-hydroxy-7a-β-methylhexahydro-1-indanone; 5OH-HIC-CoA, 3aα-H-4α(3′-carboxyl-CoA)-5-hydroxy-7a-β-methylhexahydro-1-indanone; HIEC-CoA, (7aS)-7a-methyl-1,5-dioxo-2,3,5,6,7,7a-hexahydro-1H-indene-4-carboxyl-CoA; COCHEA-CoA, 2-(2-carboxyethyl)-3-methyl-6-oxocyclohex-1-ene-1-carboxyl-CoA; MOODA-CoA, 4-methyl-5-oxo- octanedioyl-CoA. Actinobacterial enzymes from cholesterol metabolism are indicated in red, actinobacterial enzymes involved in cholic acid catabolism are indicated in orange, and those from catabolism of testosterone and cholic acid from Comamonas spp. are written in blue.
For the degradation of bile acids in P. putida DOC21, concomitant with the elimination of the C17 side chain, oxidation of the ring A starts with desaturation at C4 (catalysed by a 3-ketosteroid-Δ4(5α)-dehydrogenase, StdI) and at C1 (catalysed by StdH; Figure 6) [62,141]. After the elimination of the acyl-side chain and the introduction of two double bonds in the A ring of the cholic acid molecule, 7α,12α-dihydroxy-androsta-1,4-diene-3,17-dione is obtained (Figure 6). Notably, these hydroxyl substituents are maintained during the catabolic process (see below). However, the degradation of lithocholic acid (lacking the hydroxy groups at C7 and C12) results in ADD, which is a catabolite of convergence when testosterone and AD degradation occurs. Thus, ADD, as well as the hydroxylated derivatives generated from other bile acids, seems to represent a convergence point for the degradative pathway of steroids in Pseudomonas species.
Moreover, a 3-ketosteroid-Δ4(5a)-dehydrogenase, TesI, has been identified in C. testosteroni strains ATCC17410 and TA441 (Figure 4) [107,147]. This enzyme is needed by this bacterium to efficiently catabolise androsterone, androstanolone, androstenedione, and bile acids. Along with its gene sequence, this enzyme has also been characterised in R. jostii RHA1 [148]. It is interesting to note that stdI and stdH from P. putida DOC21, as well as tesH and tesI from C. testosteroni TA441, are adjacent to each other in their respective genomes, being part of the steroid-degrading gene clusters (Figure 4) [60,141].
Once ADD, or the hydroxylated derivatives originating from catabolism of different bile acids catabolism, has been synthesised, a monooxygenase/reductase complex (3-ketosteroid 9α-hydroxylase) introduces an α-hydroxy group at C9 with KshAB (Figure 7) in R. erythropolis [149,150] and in M. tuberculosis [151,152]. The resulting 9-hydroxy-androst-1,4-dien-3,17-dione is an unstable molecule that undergoes abiotic cleavage of the B-ring and aromatisation of the A-ring to form 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione (Figure 7).
While the formation of 9α-hydroxy-4-androstene-3,17-dione, and later desaturation in the A ring has been described, it is worth mentioning that the order of desaturation and hydroxylation reactions in Actinobacteria is unclear [149,150,151]. Moreover, the introduction of desaturation between C1 and C2 has been proposed to occur at different stages of side chain degradation in some Actinobacteria, i.e., in R. ruber Chol4, R. erythropolis SQ1, and different strains of Nocardia, Arthrobacter, and Mycobacterium [153,154]. In fact, different 3-ketosteroid-Δ1(2)-dehydrogenase paralogs showing differences in their substrate specificities have been characterised in some of these strains. For example, in R. ruber Chol-4, three different KstD isoenzymes (KstD1, KstD2, and KstD3) differing in their respective substrate profiles have been described, with KstD2 being the isoenzyme mainly involved in AD degradation in this strain [155]. In Mycobacterium neoaurum ATCC 25795, three different paralogs of this dehydrogenase have also been identified. In this strain, KstD1 showed a higher affinity for 9α-hydroxy-4-androstene-3,17-dione, while KstD3 preferred AD [156].
In Actinobacteria, it is not surprising to find different paralog genes coding KshA subunits, ranging from one to six. In M. tuberculosis H37Rv, only one kshA homolog has been found, in Mycobacterium spp. VKM Ac-1815D and 1816D, two different versions have been reported, and in Mycobacterium VKM Ac-1817, five different paralogs have been described [157,158]. In R. rhodochrous DSM43269 and R. erythropolis SQ1 chromosomes, five different paralogs of kshA have been identified [157,159], and the coded proteins showed different substrate specificities. Thus, KshA5 from R. rhodochrous appears to have the broadest substrate range, but without a clear substrate preference. By contrast, KshA1 seems to be specifically involved in cholic acid catabolism [159]. The reductase component of 3-ketosteroid 9α-hydroxylase in Actinobacteria, KshB, is generally present as a single copy gene, with some exceptions for those bacteria able to degrade sterols and bile acids, suggesting that each copy of this gene could be involved in the specific degradation of a particular compound.
Conversely, steroid-degrading Proteobacteria seem to only have single copies of 3-ketosteroid-Δ1(2)-dehydrogenase and both subunits of 3-ketosteroid 9α-hydroxylase, suggesting a possible reason for their rare ability to catabolise a wide range of steroids. Although it has been proposed that the multiplicity of genes encoding 3-ketosteroid-Δ1(2)-dehydrogenase and the oxygenase subunits of 3-ketosteroid 9α-hydroxylase in some Actinobacteria could represent an environmental advantage, their role could also be in the maintaining metabolic flux throughout the pathway, avoiding the accumulation of intermediates, and so facilitating a dynamic and fine-tuned steroid catabolism.
Catabolism continues through the hydroxylation of the aromatised A-ring in 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione by a two-component oxygenase, TesA1A2 in C. testosteroni [160], and HsaAB in M. tuberculosis and R. jostii RHA1 [161], leading to the formation of a catecholic derivative, 3,4-dihydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione (Figure 7). In this type of monooxygenase, the reductase utilises NADH to reduce a flavin, which is then transferred to the oxygenase. The dihydroxylated ring is then opened by meta-cleavage by an extradiol dioxygenase (TesB in C. testosteroni [162], and HsaC in M. tuberculosis [163] and R. jostii RHA1 [55]) that introduces two oxygen atoms. In P. putida DOC21 the coding gene for this meta-cleavage dioxygenase, stdF, was identified by Tn5 transposon mutagenesis and was sequenced [29]. The product of the cleavage, 4,5,9,10-diseco-3-hydroxy-5-9-17-trioxoandrosta- 1(10),2-diene-4-oic acid, is hydrolysed by TesD in C. testosteroni [164], and by HsaD in M. tuberculosis [165,166] and R. jostii [55], yielding 3aα-H-4α(3′-propanoate)7a-β-methylhexahydro-1,5-indanedione (HIP) and 2-hydroxy-2,4-hexadienoic acid (Figure 7).
In C. testosteroni, the catabolism of 2-hydroxy-2,4-hexadienoic acid is carried out by the products of the genes tesEFG [167]. Thus, 2-hydroxy-2,4-hexadienoic acid is the substrate of the hydratase TesE leading to the formation of 4-hydroxy-2-oxohexanoate, which then undergoes an aldol cleavage catalysed by TesG aldolase, yielding pyruvate and propionaldehyde. Finally, propionaldehyde is the substrate of an acylating aldehyde dehydrogenase, TesF, producing propionyl-CoA, which enters into the central metabolism (Figure 7). Orthologous genes have been identified in the M. tuberculosis H37Rv genome, hsaEFG, and it has been proposed that the aldolase and dehydrogenase, HsaF and HsaG, interact as a complex to efficiently perform the catalysis [168].
3.5. Degradation of B/C Rings
The degradation of HIP starts with its CoA thioesterification by a specific acyl-CoA synthetase (Figure 7), StdA3 in P. putida DOC21 [62], ScdA (ORF18) in C. testosteroni [169] and FadD3 in Actinobacteria [119,170]. It has been proposed that in M. tuberculosis, HIP-CoA is the substrate of IpdF (ScdG/ORF31 in C. testosteroni), which reduces the 5′-keto substituent to a hydroxyl group, leading to the formation of 5-hydroxy-HIP (Figure 7). This compound is then subjected to a β-oxidative process for the elimination of the propionyl-CoA side chain, yielding 3aα-H-4α(3′-carboxyl-CoA)-5-hydroxy-7aβ-methylhexahydro-1-indanone (Figure 7). The enzymes involved in this β-oxidation in M. tuberculosis are unknown, with the exception of FadE30, which has been proposed to be the acyl-CoA dehydrogenase involved in this process [36,171]. However, in C. testosteroni, it has been proposed that this enzymatic reaction is carried out by a heteromeric acyl-CoA dehydrogenase, ScdC1C2 (ORF28,30), and the product of this reaction is the substrate of hydration of the double bond, in a reaction catalysed by an enoyl-CoA hydratase, ScdD (ORF32; Figure 7) [172,173,174]. (7aS)-7a-methyl-1,5-dioxo-2,3,5,6,7,7a-hexahydro-1H-indene-carboxyl-CoA, in which the rings C and D remain intact, is then produced by two reactions catalysed by IpdC in M. tuberculosis (ScdK/ORF4 in C. testosteroni), which introduces a double bond in the C ring, and IpdF in M. tuberculosis (ScdG/ORF31 in C. testosteroni), which oxidises the 5-OH group (Figure 7) [171,172,173,174]. However, the order of these two reactions has not been determined. Ring D is now opened, previously to ring C, through a hydrolytic reaction mediated by a crotonase, EchA20 (ScdY/ORF5 in C. testosteroni) [171,174], producing (R)-2-(2-carboxyethyl)-3-methyl-6-oxocyclohex-1-ene- 1-carboxyl-CoA. This molecule is the substrate for the hydrolytic cleavage of ring C, catalysed by IpdAB (ScdL1L2/ORF1,2 in C. testosteroni) by a retro-Claisen hydrolysis (Figure 7) [175,176]. The product of the opening of both the C and D rings is the substrate of a thiolase (putatively Fad6), resulting in a molecule of acetyl-CoA and 4-methyl-5-oxo-octanedioyl-CoA (Figure 7) [171,177]. It has been proposed that this last intermediate undergoes a β-oxidation process, starting with a desaturation catalysed by an acyl-CoA dehydrogenase, FadE32, or by the heteromeric Fad31-FadE32 in Mycobacterium [171]. In C. testosteroni, the enoyl-CoA hydratase involved in the next reaction of this β-oxidation process, ScdN (ORF3), has been identified [177]. As final products of the β-oxidation, a molecule of acetyl-CoA would be released together with 2-methyl-β-ketoadipyl-CoA, which could be cleaved to propionyl-CoA and succinyl-CoA (Figure 7) by a mechanism analogous to the final step in the catabolism of aromatic compounds through the β-ketoadipate pathway [178].
It is important to mention that in the degradation of bile acids in P. putida DOC21, the hydroxyl substituents present at C7 and/or C12 in the different bile acids are maintained during the degradative process. Thus, a P. putida DOC21 mutant lacking StdA3, the ATP-depending acyl-CoA synthetase activating HIP, when cultured in the presence of chenodeoxycholic acid accumulates 3′(R)-hydroxy-HIP in the culture broth. When deoxycholic acid is used, 7β-hydroxy-HIP accumulation is observed, and in the presence of cholic acid, 3′(R),7β-dihydroxy-HIP is accumulated [62]. For R. jostii RHA1, it has been proposed that echA13, a gene included in the HIP catabolic gene cluster, could codify a function for the removal of the hydroxyl group at position 7β [57]. On the other hand, maintaining the hydroxyl group in the 3′ position in the aliphatic chain of HIP could potentially generate, after CoA thiosterification of the molecule, a C3-hydroxy-intermediate from the first β-oxidation round, avoiding the need for an acyl-CoA dehydrogenase, FadE30, and enoyl-CoA hydratase, in the formation of 3aα-H-4α(3′-carboxyl-CoA)-5-hydroxy-7aβ-methylhexahydro-1-indanone.
A bioinformatics approach, using selected characteristic genes from this pathway described from model organisms against genomes deposited in public databases, made it possible to identify 265 putative steroid degraders within only the Actinobacteria and Proteobacteria, from different habitats, including eukaryote hosts, soil, and aquatic environments [93]. This study allowed for a comparison of the organisation of the genetic clusters encoding this pathway in different organisms, and with different steroid use profiles (sterols, testosterone and bile acids). This study showed that most of the genes coding the whole 9,10-seco pathway are randomly located in different low number large clusters in the bacterial chromosome. However, there are some exceptions, such as M. tuberculosis H37Rv, where the predicted catabolic genes are mainly located in a single cluster (80 genes in a region of about 50 kb; Figure 4) [55,93]. Rhodococcus spp. have their cholic acid catabolism genes grouped in a separate cluster not close to the cholesterol catabolism genes, and lacking C/D-rings catabolic genes (i.e., R. jostii RHA1, Figure 4). Even so, the R. equii genome has a single cluster containing the putative cholic acid and cholesterol catabolic genes [93]. In general, the location of these clusters is chromosomal, however, Bergstrand et al. [93] identified some exceptions, where the genes are located on plasmids, such as the putative cholic acid A/B-ring degradation genes located on pRHL1 from R. jostii, and a gene cluster encoding putative enzymes involved in testosterone and/or cholic acid catabolism in two strains belonging to the genus Novosphingobium. The association of genes coding this pathway opens the possibility of the horizontal transfer of catabolism of steroids between bacteria.
Genes involved in the catabolism of cholesterol through the 9,10-seco pathway are part of the core genome characterising most of the genera of the Corynebacterinae. However, the cholic acid pathway, although conserved in Rhodococcus spp., is seldom found in closely related genera. This restricted distribution of the actinobacterial bile acid pathway suggests that its origin could be through the duplication of a pre-existing cholesterol pathway in an ancestor of the genus Rhodococcus. This could be the reason why genes encoding A/B-ring degradation are found in gene clusters for both cholesterol and bile acid pathways. Surprisingly, genes for C/D-ring degradation are found only in cholesterol pathway clusters, indicating that they have not been duplicated, or have been lost through the evolution of the strains.
Proteobacteria are unable to degrade sterols with alkyl side chains [29,60]. This could be due to the absence of Cyp125 and/or Cyp142 orthologs allowing side chain functionalisation and degradation, as well as the lack of a mce-like transport system for sterol uptake. The wide distribution of 9,10-seco pathway genes in Actinobacteria contrasts with the scarcity of androgen/bile acids pathway genes in Proteobacteria genomes. However, an exception to this could be the genus Comamonas, in which this pathway seems to be part of the genome core [93].
Although the bile acid and the androgen catabolic pathways of Proteobacteria are highly similar to the proposed pathways for actinobacterial cholesterol and cholic acid catabolism, the encoding genes show low sequence similarity. This could be explained by: (i) convergent evolution, where these genes evolved independently in both taxonomic groups [179]; or (ii) divergent evolution, with the genes having an actinobacterial origin, and being disseminated through horizontal gene transfer to the other taxon [93]. Although some interesting articles assess the phylogeny of some critical genes from these pathways [93,141,158], complementary studies will be necessary to clarify this issue.
3.6. Regulation of 9,10-seco Pathway
In M. tuberculosis, the regulation of cholesterol degradation is carried out by two TetR-like transcriptional repressors, KstR and KstR2 [73,180]. These repressors regulate the expression of particular genes after binding specific intermediates from cholesterol degradation. Thus, KstR unlocks the expression of the genes encoding the membrane transport system of cholesterol, as well as enzymes involved in side chain catabolism and the opening and removal of steroidal A and B rings through binding to 3-hydroxy-cholest-5-en-26-oyl-CoA [181]. This regulatory protein is not only involved in cholesterol catabolism, as the KstR regulon comprises 74 genes in M. tuberculosis, some of them involved in growth on palmitate, suggesting that KstR may control metabolism of lipids in this bacterium [73]. KstR may act as a de-repressor by binding to molecules other than 3-hydroxy-cholest-5-en-26-oyl-CoA. Moreover, in M. smegmatis, it has been demonstrated that cholesterol and its first catabolic intermediate, cholest-4-en-3-one, were unable to induce the release of KstR from proposed promoter regions in the regulon. Furthermore, in M. smegmatis, this regulator remains unaffected by other cholesterol catabolic intermediates, such as AD and ADD [182]. By contrast, KstR2 also downregulates the pool of genes needed for C and D ring degradation, and de-represses their expression by binding HIP-CoA as a ligand [183,184].
The regulation of the 9,10-seco pathway mediated steroid catabolism in Proteobacteria has mainly been studied in C. testosteroni. A gene called tesR in strain TA441, and teiR in strain ATCC11996, in the steroid degradation gene clusters, encode a transcriptional regulator needed for induction of 3,17β-HSD dehydrogenase, 3α-hydroxysteroid dehydrogenase, 3-ketosteroid-Δ1(2)-dehydrogenase, 3-ketosteroid-Δ4(5α)-dehydrogenase and most of the identified steroid degradation genes [185,186,187,188]. It has been reported that TesR/TeiR is a membrane protein with a polar location in cells involved in chemotaxis, and mediates steroid sensing and metabolism via kinase activity, which likely triggers a cascade of phosphorylation events that induce the expression of steroid catabolising enzymes [187].
In addition, two negative regulator genes, repA and repB, have been identified close to hsdA, the gene coding for 3α-hydroxysteroid dehydrogenase. The products of these genes repress hsdA expression: (i) at the transcriptional level, by binding to hsdA promoter sequences in response to the presence of steroids (RepA) [189]; and (ii) by interfering with its translation, by binding to the mRNA of 3α-hydroxysteroid dehydrogenase (RepB) [190]. Moreover, HsdR, a LysR-type transcriptional repressor, activates transcription of the hsdA gene in C. testosterone, dependent on decreased repression by RepA [191,192].
By contrast, in C. testosteroni ATCC11996, a complex network regulating the expression of the 3,17β-HSD gene, βhsd, has been proposed, with several transcriptional repressors, PhaR [193], LuxR [194], TetR [195], and BRP [196] having been described. In knock-out mutants for phaR, luxR, tetR, and brp, the basal expression levels of βhsd did not increase with reference to the wild type strain; however, on the addition of testosterone there was a several-fold increase in expression levels compared to the wild type, suggesting that these regulators function as repressors of βhsd expression [193,194,195,196]. This implies the existence of a complex controlling βhsd expression, which is regulated by testosterone. In addition, the expression of 3α-HSD was also increased in the brp knock-out mutant, indicating that BRP represses the expression of both hsdA and βhsd in the presence of testosterone [196].
4. The 4,5-seco Pathway
Compared with the 9,10-seco pathway for sterols, bile acids, and androgen aerobic degradation, current knowledge on estrogen degradation pathways is very limited. The partial characterisation of the aerobic degradation of 17β-estradiol by Sphingomonas sp. strain KC8, an obligate aerobic alpha-proteobacterium isolated from wastewater [197], has allowed a new pathway for aerobic estrogen degradation, the 4,5-seco pathway, to be proposed [198].
Sphingomonas KC8 codes for a gene for 3β,17β-hydroxysteroid dehydrogenase (oecA) that is responsible for the transformation of 17-hydroxy from 17β-estradiol and testosterone to their respective oxo-derivatives. This gene was similarly expressed when this strain was cultured on 17β-estradiol and testosterone, although testosterone is degraded by this strain through the 9,10-seco pathway [199]. OecA converts 17β-estradiol into estrone (Figure 8). However, this gene does not cluster with other steroid-degrading genes in the genome of Sphingomonas KC8. An ortholog of this gene has been characterised in P. putida SJTE-1, a strain isolated from sludge, which can use 17β-estradiol as a sole carbon source [200,201].

Figure 8.
Metabolism of 17β-estradiol in Sphingomonas sp. KC8. (a) Reactions of the 4,5-seco pathway. HIP, 3aα-H-4α(3′-propanoate)7a-β-methylhexahydro-1,5-indanedione. (b) Genetic organisation of the three clusters codifying the enzymes required for 17β-estradiol assimilation (Genome Accession NZ_CP016306). Annotation from the genome: KC8_RS16375, putative dioxygenase; KC8_RS16385, Rieske (2Fe 2S) protein; KC8_RS05200, MaoC dehydratase; KC8_RS05205, hypothetical protein; KC8_RS05210, acyl-CoA dehydrogenase; KC8_RS05215, ferredoxin oxidoreductase; KC8_RS05220, VOC family protein; KC8_RS05230, TetR transcriptional regulator; KC8_RS05235, cytochrome P450; KC8_RS05240, hypothetical protein; KC8_RS05245, lipid-transfer protein; KC8_RS05250, MaoC dehydratase; KC8_RS05255 and KC8_RS05260, enoyl-CoA hydratases; KC8_RS05265, acetyl-CoA acetyltransferase; KC8_RS05270, 3-hydroxy-3-mthylglutaryl-CoA synthase; KC8_RS05275, Short-chain oxidoreductase; KC8_RS05280, acyl-CoA dehydrogenase; KC8_RS00980, CoA acyltransferase; KC8_RS00985, steroid Δ-isomerase; KC8_RS00990, MaoC dehydratase; KC8_RS00995, short-chain dehydrogenase/reductase; KC8_RS01000, acyl-CoA dehydrogenase; KC8_RS01005, short-chain oxidoreductase; KC8_RS01010, phenylacetic acid degradation protein PaaY; KC8_RS01015, acetyl-CoA acetyltransferase; KC8_RS01020, acyl-CoA dehydrogenase; KC8_RS01025, acyl-CoA dehydrogenase; KC8_RS01030, enoyl-CoA hydratase; KC8_RS01035, monooxygenase; KC8_RS01040, lipid-transfer protein (Ltp); KC8_RS01045, thiolase; KC8_RS01050, CoA transferase, β-subunit; KC8_RS01055, CoA transferase, α-subunit; KC8_RS01060, meta-dioxygenase. Genes encoding key enzymes for A ring degradation are depicted in red; genes coding β-oxidation related enzymes putatively catalyzing degradation of A/B ringare indicated in green; genes for C/D-ring degradation are depicted in purple.
Additionally, other genetic clusters involved in estrogen catabolism have been identified in the genome of Sphingomonas KC8 (Figure 8). In gene cluster I, an oecB gene encodes a flavin-dependent estrone 4-hydroxylase, which introduces a hydroxyl group at C4 in the aromatic ring, yielding 4-hydroxyestrone. In gene cluster II, oecC codes for a 4-hydroxyestrone 4,5-dioxygenase that catalyses the meta-cleavage of the aromatic ring, creating 4-norestrogen-5(10)-en-3-oxo-carboxylic acid. This meta-cleavage product is unstable and undergoes an abiotic recyclisation, producing pyridinestrone acid, a dead-end byproduct that accumulates in the culture broth of Sphingomonas KC8 when grown on 17β-estradiol (Figure 8) [198]. However, the metabolic pathway continues by using a 2-oxoacid oxidoreductase that catalyses the decarboxylation of this meta-cleavage product, as well as the thioesterification of the resulting molecule with CoA, to produce 4-norestrogen-5(10)-en-3-oyl-CoA (Figure 8). The gene encoding this 2-oxoacid oxidoreductase is also found in gene cluster II. This cluster also includes genes encoding β-oxidation-like enzymes. Metabolic intermediates in estrone degradation have allowed the identification of products of these genes involved in the transformation of the cleavage product from 4-hydroxyestrone to HIP (Figure 8). In addition, the existence of a gene encoding for a 3-hydroxy-3-methylglutaryl-CoA synthase-like protein in this cluster suggests its involvement in estrogen catabolism [202].
In summary, oecA and genes from clusters I and II are likely to be involved in estrogen A/B-ring degradation to HIP in Sphingomonas KC8. In the genome of this strain, a third cluster (cluster III) related to steroid degradation has also been identified (Figure 8). It contains genes similar to those proposed in C. testosteroni for C/D-ring degradation. Thus, orthologs of echA20, ipdB, and ipdA from M. tuberculosis have been found (Figure 8), and they are expressed during the aerobic growth of Sphingomonas KC8 on testosterone and 17β-estradiol, being involved in the transformation of HIP into general catabolites [198,202]. Similar clusters have been identified in other estrogen-degrading aerobes, such as Altererythrobacter estronivorus MH-B5 [203] and Novosphingobium tardaugens NBRC 16725 [204].
It has been proposed that the 4,5-seco pathway is highly prevalent, since: (i) the dead-end product pyridinestrone is found in wastewater treatment plants exposed to estrogens, and (ii) the gene oecC has been identified in bacteria isolated from these environments [198,205]. Alternative estrogen degradation pathways and metabolites have been proposed (Figure 9), although there is no biochemical or genetic evidence identifying their significance. Thus, in Sphingomonas sp. strain ED8, a pathway has been proposed, involving hydroxylation at different positions of the saturated ring of 17β-estradiol. This is supported by the detection of hydroxyl-17β-estradiol, keto-17β-estradiol, keto-estrone and 3-(4-hydroxyphenyl)-2-hydroxy-prop-2-enoate in the culture broth of this bacterium when 7β-estradiol was used as the sole carbon source (Figure 9). The appearance of this last compound suggested that catabolism of 17β-estradiol through this pathway takes place by opening the rings B, C, or D [206]. Another metabolic alternative for 17β-estradiol mineralisation has been proposed in Nitrosomonas europaea, based on the appearance of estra-1,3,5(10),16-tetraen-3-ol (estratetraenol) after dehydratation of ring D at C17 position (Figure 9). It was suggested that this strain could further metabolise this intermediate to non-estrogenic compounds [207]. The finding, in activated sludge, of a new intermediate from 17β-estradiol derived from estrone and containing a lactone in ring D, opens the possibility of a new estrogen catabolic pathway used for the mineralisation of this kind of steroid (Figure 9) [208].

Figure 9.
Metabolic alternatives to the 4,5-seco pathway proposed for estrogen mineralisation/biotransformation in different bacterial species.
5. The Anaerobic 2,3-seco Pathway
Several denitrifying Proteobacteria that degrade steroids, cholesterol, and testosterone under anaerobic conditions have been characterised [63,65,209]. Among them, Sterolibacterium denitrificans (Stl. denitrificans) and Steroidobacter denitrificans (Std. denitrificans) have been used as model organisms for studying anaerobic steroid metabolism [63,65]. The pathway involved in anoxic steroid catabolism, although having some similarities with the 9,10-seco pathway, uses dioxygen-independent reactions to degrade the steroidal core. It is referred to as the 2,3-seco pathway [210,211,212]. Similar to the aerobic metabolism performed through the 9,10-seco pathway, the anaerobic degradation of cholesterol by Stl. denitrificans is initiated by the oxidation of ring A using AcmA, a dehydrogenase/isomerase belonging to the short-chain dehydrogenase/reductase superfamily, yielding 4-cholesten-3-one (Figure 10) [64,213].
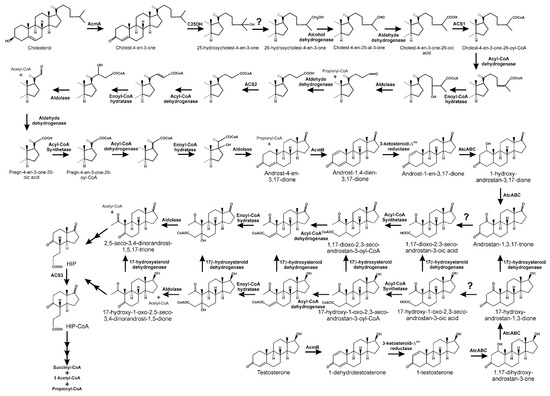
Figure 10.
Anaerobic catabolism of cholesterol, in Sterolibacterium denitrificans, and testosterone, in Steroidobacter denitrificans, by the 2,3-seco pathway. Putative points of convergence between both metabolic mechanisms are also suggested.
In a second step, the anaerobic degradation of cholesterol C17 side chain starts with an oxygen-independent water-dependent hydroxylation at the tertiary C25 atom of the side chain of C27 esteroid substrates, resulting in the formation of a tertiary alcohol, 25-hydroxy-4-cholesten-3-one (Figure 10) [213]. This hydroxylation is carried out by a molybdopterin-containing enzyme, C25DH, belonging to the dimethyl sulfoxide dehydrogenase molybdoenzyme family. This enzyme consists of three different subunits, an α-subunit containing the molybdo-bis(pyranopterin guanine dinucleotide) cofactor in the active site and an iron–sulfur cluster [4Fe-4S], associated with electron-transfer machinery composed of β-subunits that contain three [4Fe-4S] and one [3Fe-4S] iron–sulfur clusters, and ϒ-subunits associated with one heme b. Notably, the genome of Stl. denitrificans Chol1-S contains seven paralogs that code for putative α-subunits of S25DH-like dehydrogenases, although there are few genes putatively encoding the β- and ϒ-subunits, indicating that molybdoenzymes share common βϒ-components, but have different α-subunits [214,215]. Stl. denitrificans is able to not only degrade cholesterol, but also β-sitosterol, stigmasterol, and ergosterol. Surprisingly, S25DH used in cholesterol catabolism is unable to hydroxylate any of the 4-en-3-one analogues of these sterols [214,216]. However, several S25DHs carrying different α-subunits have been functionally characterised, and are able to activate those sterols [217].
The next step in the anoxic degradation of cholesterol involves an unprecedented isomerisation of the hydroxyl group from the tertiary C25 to the primary C26, catalysed by a yet unknown enzyme (Figure 10) [86,218]. Further degradation proceeds via oxidation of C26 primary alcohol to a carboxylate by the action of a putative cholesterol-induced alcohol dehydrogenase and aldehyde dehydrogenase (Figure 10) [216]. This carboxylic derivative is activated to a C26-oyl-CoA thioesterifed molecule by a specific ATP-depending acyl-CoA synthetase, followed by a modified β-oxidation-like reaction sequence, with some steps similar to those of bile acid catabolism through the 9,10-seco pathway, yielding AD, two propionyl-CoAs, and one acetyl-CoA (Figure 10) [216,219]. Thus, in the genome of Stl. denitrificans, two gene clusters, acd1 and acd2, contain the genes that, after induction by cholesterol, code for functions needed for cholesterol side chain elimination (Figure 11). Amazingly, although more of the intermediates of side chain degradation have been identified, no 3-oxoacyl-CoA intermediates have been found, which correlates with the lacking of genes specifically involved in the thiolytic β-oxidative processes (3-hydroxyacyl-CoA dehydrogenases and thiolases). Alternatively, the two aldolases and an aldehyde dehydrogenase in the acd1 and acd2 clusters indicate that the release of propionyl-CoA and acetyl-CoA molecules from degradation of the side chain occurs via aldolytic cleavage [216,219].
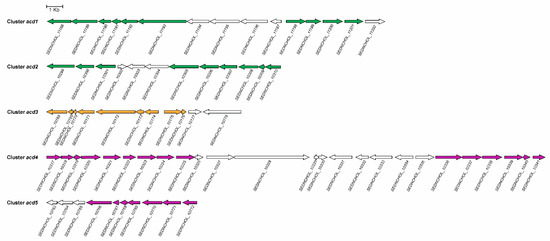
Figure 11.
Genetic organisation of the genes encoding 2,3-seco pathway functions for cholesterol degradation in Sterolibacterium denitrificans. Annotation in the genome (accession LT837803): SEDNCHOL_11188, aldehyde dehydrogenase; SEDNCHOL_11189, acyl-CoA synthetase 1 (ACS1); SEDNCHOL_11190, short-chain dehydrogenase; SEDNCHOL_11191, SEDNCHOL_11192, and SEDNCHOL_11193 steroid C25 dehydrogenase, γ-, β- and α- subunit, respectively; SEDNCHOL_11194 and SEDNCHOL_11195, proteins with unknown functions; SEDNCHOL_11196, AcmB; SEDNCHOL_11197, putative transcriptional regulatory protein; SEDNCHOL_11198, putative aldolase; SEDNCHOL_11199, enoyl-CoA hydratase; SEDNCHOL_11200 and SEDNCHOL_11201, acyl-CoA dehydrogenases; SEDNCHOL_11202, oxidoreductase; SEDNCHOL_10299, acyl-CoA synthetase 2 (ACS2); SEDNCHOL_10300 and SEDNCHOL_10301, acyl-CoA dehydrogenases; SEDNCHOL_10302, protein of unknown function; SEDNCHOL_10303, putative metallo-β-lactamase; SEDNCHOL_10304, phytoene dehydrogenase-like protein; SEDNCHOL_10305, putative C22 acyl-CoA synthetase; SEDNCHOL_10306 and SEDNCHOL_10307, acyl-CoA dehydrogenases; SEDNCHOL_10308, aldolase; SEDNCHOL_10309 and SEDNCHOL_10310, enoyl-CoA hydratases; SEDNCHOL_10168, CoA transferase; SEDNCHOL_10169, plasmid stabilization system; SEDNCHOL_10170 and SEDNCHOL_10171, proteins of unknown function; SEDNCHOL_10172, SEDNCHOL_10173, and SEDNCHOL_10174, A, B, and C subunits of AtcABC, respectively; SEDNCHOL_10175 and SEDNCHOL_10176, proteins of unknown function; SEDNCHOL_10177; putative electron-transfer flavoprotein, β-subunit; SEDNCHOL_10178, protein of unknown function; SEDNCHOL_10317, SEDNCHOL_10318, and SEDNCHOL_10321, IpdABC-like proteins; SEDNCHOL_10319, probably subunit of benzoylsuccinyl-CoA thiolase; SEDNCHOL_10320, Propanoyl-CoA C-acyltransferase; SEDNCHOL_10322, enoyl-CoA hydratase; SEDNCHOL_10323 and SEDNCHOL_10324, acyl-CoA dehydrogenases; SEDNCHOL_10325, thiolase; SEDNCHOL_10326, MarR family transcriptional regulator; SEDNCHOL_10327, outer membrane protein; SEDNCHOL_10328, putative Filamentous hemagglutinin family N-terminal domain containing protein; SEDNCHOL_10329, protein of unknown function; SEDNCHOL_10330, N-acetyltransferase; SEDNCHOL_10331, RseB; SEDNCHOL_10332, enoyl-CoA hydratase; SEDNCHOL_10333 and SEDNCHOL_10334, proteins of unknown function; SEDNCHOL_10335, CoA transferase; SEDNCHOL_10336, putative IpdC; SEDNCHOL_10337 and SEDNCHOL_10338, CoA transferases; SEDNCHOL_10339, acetyl-CoA acetyltransferase; SEDNCHOL_10340, protein of unknown function; SEDNCHOL_10341, enoyl-CoA hydratase; SEDNCHOL_10763, putative 2-phospho-L-lactate guanylyltransferase; SEDNCHOL_10764, 2-phospho-L-lactate transferase; SEDNCHOL_10765, Coenzyme F420:L-glutamate ligase; SEDNCHOL_10766, acyl-CoA synthetase 3 (ACS3); SEDNCHOL_10767, steroid Δ–isomerase; SEDNCHOL_10768, enoyl-CoA hydratase; SEDNCHOL_10769, short chain dehydrogenase; SEDNCHOL_10770 and SEDNCHOL_10771, acyl-CoA dehydrogenases; SEDNCHOL_10772, short chain alcohol dehydrogenase. Genes encoding enzymes involved in cholesterol side chain degradation are shown in green; genes participating in A ring degradation are shown in orange; and in purple is shown genes coding for ring C/D degradation.
In vitro assays using cell-free extracts indicate the existence of an FAD-containing 4-cholesten-3-one-Δ1(2)-dehydrogenase (AcmB) in Stl. denitrificans, catalysing Δ1(2)-desaturation of 4-cholesten-3-one to 1,4-cholestadien-3-one (Figure 10) [220]. However, the topological location of AcmB on the cytoplasmic side of the inner membrane and 4-cholesten-3-one in the cellular periplasm puts this proposed reaction in doubt. Thus, AcmB may be a 3-ketosteroid-Δ1(2)-dehydrogenase, with AD, obtained after complete removal of the lateral side chain from 4-cholesten-3-one, being its physiological substrate, leading to the formation of ADD. This later compound is then specifically reduced to 1-androsten-3,17-dione, which acts as a substrate for AtcABC, a bifunctional molybdopterin-containing hydratase/dehydrogenase, which introduces a water molecule to the double bond between C1 and C2, leading to the formation of a hydroxyl group at C1. Later, this hydroxyl group is oxidised to an oxo-group, releasing androstan-1,3,17-trione (Figure 10) [212]. The A ring of this compound is then cleaved, resulting in 1,17-dioxo-2,3-seco-androstan-3-oic acid, which would be activated by a yet unknown acyl-CoA synthetase (Figure 10) [86,212,218]. It has been suggested that acetyl-CoA could be released from 1,17-dioxo-2,3-seco-androstan-3-oyl-CoA through an aldolytic cleavage, to produce 2,5-seco-3,4-dinorandrost-1,5,17-trione [213]. Although the mechanism for cleavage of the B-ring remains unknown, HIP, the predicted product generated from 2,5-seco-3,4-dinorandrost-1,5,17-trione, has been identified (Figure 10) [216].
In Stl. denitrificans, genes involved in ring A cleavage have been found in the cluster acd3 (Figure 11), although no candidates encoding a B-ring cleaving hydrolase or its degradation to HIP have been identified. Clusters acd4 and acd5 contain all the genes required for the integration of HIP into the central metabolism (Figure 11) [216].
The anaerobic degradation of the androgen testosterone has mainly been studied in model microorganism Std. denitrificans DSMZ 18526 [210,211,221,222,223]. The pathway used by this denitrifying strain generates analogous intermediates to those of the 2,3-seco pathway involved in cholesterol catabolism in Stl. denitrificans, and common steps in these two pathways are catalysed by orthologous enzymes (Figure 10) [212].
Thus, testosterone is initially transformed into 1-dehydrotestosterone by a 3-ketosteroid Δ1(2)-dehydrogenase/reductase (Figure 10). This compound is then transformed to 1-testosterone in a process catalysed by a 3-ketosteroid Δ4-dehydrogenase/reductase (Figure 10). AtcABC, the analogous bifunctional molybdoenzyme, in this case a 1-testosterone hydratase/dehydrogenase, catalyses the C1-C2 hydration reaction and the subsequent oxidation, leading to the formation of 17-hydroxy-androstan-1,3-one (Figure 10) [211,212]. It has been proposed that the process continues through the hydrolytic cleavage of this last compound, giving rise to 17-hydroxy-1-oxo-2,3-seco-androstan-3-oic acid (Figure 10). However, the hydrolase that would catalyse this process remains unidentified. Taking into account that 17-hydroxy-2,5-seco-3,4-dinorandrost-1,5-dione has been identified in cultures of Std. denitrificans growing in media containing testosterone as a carbon source, the metabolic steps for the transformation of 17-hydroxy-1-oxo-2,3-seco-androstan-3-oic acid into this compound have been proposed. Thus, the acid is first activated to a coenzyme A thioester, and after the introduction of a double bond and its hydration, a retroaldolic reaction, a molecule of acetyl-CoA is released, producing 17-hydroxy-2,5-seco-3,4-dinorandrost-1,5-dione (Figure 10). Notably, the use of acyl-CoA dehydrogenase inhibitors impairs the biotransformation of 17-hydroxy-1-oxo-2,3-seco-androstan-3-oic acid to 17-hydroxy-2,5-seco-3,4-dinorandrost-1,5-dione, reinforcing the participation of a β-oxidation-like mechanism in the processing of the A-ring cleavage product [211]. It has been proposed that, as occurs in cholesterol degradation, this compound will be converted to HIP.
It has to be noted that proposed intermediates in testosterone anaerobic catabolism conserve a hydroxyl group at C17 until their convergence at the HIP level. However, this only reflects the fact that these compounds are more prominent in the culture broth than their 17-keto structures, which appear at lower levels. This has resulted in a proposal that a 17β-hydroxysteroid dehydrogenase, or a group of enzymes with this activity, could interconvert the different 17β-hydroxyl compounds and their 17-keto intermediates [211,223]. Undoubtedly, if the catabolism of steroids through this pathway occurs by the formation of HIP at some stage, the keto present in the D-ring should be evident.
Taking into account that the aerobic and anaerobic degradation of steroids converge at HIP, it might be expected that the same metabolic mechanisms are used for its subsequent degradation. This hypothesis is reinforced by the conservation of genes in the genomes of microbial steroid degraders [216,219].
6. Biotechnological Interest in Steroid-Degrading Microorganisms
In addition to the relevance that steroid-degrading microorganisms have for the environment and maintaining the carbon cycle, their potential use in the pharmaceutical industry has to be highlighted. Classical pharmaceutical production processes for steroids have been carried out by extraction from plant or animal sources, by full organic synthesis, or by a combination of chemical and enzymatic synthesis. With exceptions, extraction from animals or plants is an inefficient large-scale production method. Full chemical and chemical-enzymatic syntheses are often multistage and very expensive in time, labor, and energy, as well as posing a potential risk for the environment.
As an alternative, the bioconversion of steroids from low-cost precursors (i.e., phytosterols) with genetically tailored microorganisms is becoming the method of choice for industrial production. The development of these biotechnological approaches, which are more cost-effective and environmentally friendly, have been stimulated due to the growing demand for steroid pharmaceuticals. This approach is classically developed with heterologous gene expressing transgenic bacteria, yeast, and fungi, or with specific strains empirically showing a specific reaction. Steroid degrading strains, or specific mutants tailored from them, has given rise to a new source of syntons (mainly AD, ADD, or 9-hydroxy-derivatives) or genes/enzymes useful for steroid transformation.
As far as we know, today there have been no studies focused in the development of biodegradative strategies for environmental steroid amendment. Despite the great interest that the elimination of steroids can have due to the risk that the release of these compounds to the environment has for flora and wild animal reproduction, and the potential effect on human beings, the biotechnological approaches to these applications have not been developed yet.
Although our knowledge about the metabolic mechanisms for catabolism of sterols, bile acids, testosterone, or 17β-estradiol has increased in recent years, some black holes need to be clarified. The transport of steroids across cell walls and membranes in some of the bacterial groups able to metabolize steroids, rate-limiting steps in the pathways, or global regulation and integration of these pathways at intermediate metabolism should be more deeply elucidated. Moreover, there are many steroid compounds released to the environment for which it is not yet known how they could be integrated into carbon cycles.
Study of metagenomics communities will allow the identification of novel steroid biodegraders. The isolation and characterization of individual microorganisms with the ability for steroid degradation will increase the pool of genes, enzymes, and metabolic strategies for environmental steroid amendment. It is to be expected that this set of knowledge, together with technologies developed in other disciplines, will allow the design of new bacteria, or communities of them, suitable for use in the biodegradation of these molecules. Thus, for instance, system biology will allow the design of interacting communities of microorganisms for the biodegradation of different steroids or the integration of new metabolic circuits inside a bacteria; metabolomics will permit the improvement of metabolic fluxes, ensuring a high efficiency in the catabolism of steroids; and synthetic biology would allow the design and expression of improved or new activities based on mammalian, fungi, or gut microbiota steroid metabolism. In sum, the open future for a biodegradative approach to remove environmentally released steroids is expected to be a promising reality.
Author Contributions
Both authors contributed similarly to the organization and writing of the manuscript.
Funding
Our research on the biodegradation of steroid compounds by bacteria has been supported by the Ministerio de Economía y Competitividad (Madrid, España), grants BIO2012-39695-C02-02, RTC2014-2249-1 and BIO2015-66960-C3-3-R, and by grants from the Junta de Castilla y León (Consejería de Educación, Valladolid, España; LE246A11-2 and LE029P17).
Conflicts of Interest
The authors declare no conflict of interest. Those funding our research had no role in the design of the study, in the collection, analyses, or interpretation of data, or in the writing of the manuscript.
References
- Piironen, V.; Lindsay, D.G.; Miettinen, T.A.; Toivo, J.; Lampi, A.M. Plant sterols: Biosynthesis, biological function and their importance to human nutrition. J. Sci. Food Agric. 2000, 80, 939–966. [Google Scholar] [CrossRef]
- Fernandes, P.; Cabral, J.M.S. Phytosterols: Applications and recovery methods. Bioresour. Technol. 2007, 98, 2335–2350. [Google Scholar] [CrossRef] [PubMed]
- Wollam, J.; Antebi, A. Sterol regulation of metabolism, homeostasis, and development. Annu. Rev. Biochem. 2011, 80, 885–916. [Google Scholar] [CrossRef] [PubMed]
- Kodner, R.B.; Pearson, A.; Summons, R.E.; Knoll, A.H. Sterols in the red and green algae: Quantification, phylogeny, and relevance for the interpretation of geologic steranes. Geobiology 2008, 6, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Bird, C.W.; Lynch, J.M.; Pirt, F.J.; Reid, W.W. Steroids and squalene in Methylococcus capsulatus grown on methane. Nature 1971, 230, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Patt, T.E.; Hanson, R.S. Intracytoplasmic membrane, phospholipid, and sterol content of Methylobacterium organophilum cells grown under different conditions. J. Bacteriol. 1978, 134, 636–644. [Google Scholar]
- Kohl, W.; Gloe, A.; Reichenbach, H. Steroids from the myxobacterium Nannocystis exedens. J. Gen. Microbiol. 1983, 129, 1629–1635. [Google Scholar] [CrossRef]
- Schouten, S.; Bowman, J.P.; Rijpstra, W.I.C.; Damsté, J.S.S. Sterols in a psychrophilic methanotroph, Methylosphaera hansonii. FEMS Microbiol. Lett. 2000, 186, 193–195. [Google Scholar] [CrossRef]
- Bode, H.B.; Zeggel, B.; Silakowski, B.; Wenzel, S.C.; Reichenbach, H.; Müller, R. Steroid biosynthesis in prokaryotes: Identification of myxobacterial steroids and cloning of the first bacterial 2,3(S)-oxidosqualene cyclase from the mixobacterium Stigmatella aurantiaca. Mol. Microbiol. 2003, 47, 471–481. [Google Scholar] [CrossRef]
- Pearson, A.; Budin, M.; Brocks, J.J. Phylogenetic and biochemical evidence for sterol synthesis in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. USA 2003, 100, 15352–15357. [Google Scholar] [CrossRef]
- Lamb, D.C.; Jackson, C.J.; Warrilow, A.G.; Manning, N.J.; Kelly, D.E.; Kelly, S.L. Lanosterol biosynthesis in the prokaryote Methylococcus capsulatus. Insight into the evolution of sterol biosynthesis. Mol. Biol. Evol. 2007, 24, 1714–1721. [Google Scholar] [CrossRef] [PubMed]
- Gawas, D.; García, R.; Huch, V.; Müller, R. A highly conjugated dihydroxylated C28 steroid from a myxobacterium. J. Nat. Prod. 2011, 74, 1281–1283. [Google Scholar] [CrossRef] [PubMed]
- García, R.; Gemperlein, K.; Müller, R. Minicystis rosea gen. Nov., sp. Nov., a polyunsaturated fatty acid-rich and steroid-producing soil myxobacterium. Int. J. Syst. Evol. Microbiol. 2014, 64, 3733–3742. [Google Scholar] [CrossRef] [PubMed]
- Banta, A.B.; Wei, J.H.; Welander, P.V. A distinc pathway for tetrahymanol synthesis in bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 13478–13483. [Google Scholar] [CrossRef] [PubMed]
- Banta, A.B.; Wei, J.H.; Gill, C.C.C.; Giner, J.-L.; Welander, P.V. Synthesis of arborane triterpenols by a bacterial oxidosqualene cyclase. Proc. Natl. Acad. Sci. USA 2017, 114, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Lange, I.G.; Daxenberger, A.; Schiffer, B.; Witters, H.; Ibarreta, D.; Meyer, H.D.D. Sex hormones originating from different livestock production systems: Fate and potential disrupting activity in the environment. Anal. Chim. Acta 2002, 473, 27–37. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.-J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- Froehner, S.; Martins, R.F.; Errera, M.R. Assessment of fecal sterols in Barigui River sediments in Curitiba, Brazil. Environ. Monit. Assess. 2009, 157, 591–600. [Google Scholar] [CrossRef]
- Chang, H.S.; Choo, K.H.; Lee, B.; Choi, S.J. The methods for identification, analysis, and removal of endocrine disrupting compounds (EDCs) in water. J. Hazard. Mater. 2009, 172, 1–12. [Google Scholar] [CrossRef]
- Santos, L.H.; Araújo, A.N.; Fachini, A.; Pena, A.; Delerue-Matos, C.; Montenegro, M.C. Ecotoxicological aspects related to the presence of pharmaceuticals in the aquatic environment. J. Hazard. Mater. 2010, 175, 45–95. [Google Scholar] [CrossRef]
- Prost, K.; Birk, J.J.; Lehndorff, E.; Gerlach, R.; Amelung, W. Steroid biomarkers revisited-improved source identification of faecal remains in archaeological soil material. PLoS ONE 2017, 12, e0164882. [Google Scholar] [CrossRef] [PubMed]
- Matić Bujagić, I.; Grujić, S.; Laušević, M.; Hofmann, T.; Micić, V. Emerging contaminants in sediment core from the Iron Gate I Reservoir on the Danube River. Sci. Total Environ. 2019, 662, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Gagné, F.; Blaise, C.; André, C. Occurrence of pharmaceutical products in a municipal effluent and toxicity to rainbow trout (Oncorhynchus mykiss) hepatocytes. Ecotoxicol. Environ. Saf. 2006, 64, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Crane, M.; Watts, C.; Boucard, T. Chronic aquatic environmental risks from exposure to human pharmaceuticals. Sci. Total Environ. 2006, 367, 23–41. [Google Scholar] [CrossRef] [PubMed]
- Galli, R.; Braun, C. Integrative risk assessment of endocrine disruptors in Switzerland. Chimia 2008, 62, 417–423. [Google Scholar] [CrossRef]
- Tong, W.-Y.; Dong, X. Microbial biotransformation: Recent developments on steroid drugs. Recent Pat. Biotechnol. 2009, 3, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Runnalls, T.J.; Margiotta-Casaluci, L.; Kugathas, S.; Sumpter, J.P. Pharmaceuticals in the aquatic environment: Steroids and anti-steroids as high priorities for research. Hum. Ecol. Risk Assess. 2010, 16, 1318–1338. [Google Scholar] [CrossRef]
- Philipp, B.; Erdbrink, H.; Suter, M.J.; Schink, B. Degradation of and sensitivity to cholate in Pseudomonas sp. strain Chol1. Arch. Microbiol. 2006, 185, 192–201. [Google Scholar] [CrossRef]
- Merino, E.; Barrientos, A.; Rodríguez, J.; Naharro, G.; Luengo, J.M.; Olivera, E.R. Isolation of cholesterol- and deoxycholate-degrading bacteria from soil samples: Evidence of a common pathway. Appl. Microbiol. Biotechnol. 2013, 97, 891–904. [Google Scholar] [CrossRef]
- Liu, L.; Zhu, W.; Cao, Z.; Xu, B.; Wang, G.; Luo, M. High correlation between genotypes and phenotypes of environmental bacteria Comamonas testosteroni strains. BMC Genom. 2015, 16, 110. [Google Scholar] [CrossRef]
- Holert, J.; Yücel, O.; Suvekbala, V.; Kulić, Z.; Möller, H.; Philipp, B. Evidence of distinct pathways for bacterial degradation of the steroid compound cholate suggests the potential for metabolic interactions by interspecies cross-feeding. Environ. Microbiol. 2014, 16, 1424–1440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xiong, G.; Maser, E. Characterization of the steroid degrading bacterium S19-1 from the Baltic Sea at Kiel, Germany. Chem. Biol. Interact. 2011, 191, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Louvado, A.; Esteves, V.I.; Gomes, N.C.M.; Almeida, A.; Cunha, Â. Biodegradation of 17β-estradiol by bacteria isolated from deep sea sediments in aerobic and anaerobic media. J. Hazard. Mater. 2017, 323, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Holert, J.; Cardenas, E.; Bergstrand, L.H.; Zaikova, E.; Hahn, A.S.; Hallam, S.J.; Mohn, W.W. Metagenomes reveal global distribution of bacterial steroid catabolism in natural, engineered, and host environments. mBio 2018, 9, e02345-17. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.K.; Sassetti, C.M. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA 2008, 105, 4376–4380. [Google Scholar] [CrossRef] [PubMed]
- van der Geize, R.; Grommen, A.W.; Hessels, G.I.; Jacobs, A.A.; Dijkhuizen, L. The steroid catabolic pathway of the intracellular pathogen Rhodococcus equi is important for pathogenesis and a target for vaccine development. PLoS Pathog. 2011, 7, e1002181. [Google Scholar] [CrossRef]
- Söhngen, N.L. Benzin, petroleum, paraffinöl und paraffin als kohlenstoff- und energiequelle für mikroben. Zent. Bakteriol. Parasitenkd. Infekt. 1913, 27, 595–609. [Google Scholar]
- Tak, J.D. On bacteria decomposing cholesterol. Antonie Leeuwenhoek 1942, 8, 32–40. [Google Scholar] [CrossRef]
- Arnaudi, C. Flavobacterium dehydrogenans (Micrococcus dehydrogenans) und seine Fähigkeit zur oxidation von steroiden sowie substanzen aus der sexualhormonreibe. Zentr. Bakt. Parasitenk. II 1942, 105, 352–366. [Google Scholar]
- Turfitt, G.E. Microbiological agencies in the degradation of steroids: I. The cholesterol-decomposing organisms of soils. J. Bacteriol. 1944, 47, 487–493. [Google Scholar]
- Turfitt, G.E. The microbiological degradation of steroids 4. Fission of the steroid molecule. Biochem. J. 1948, 42, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Horvath, J.; Kramli, A. Microbiological oxidation of cholesterol with Azotobacter. Nature 1947, 160, 639. [Google Scholar] [CrossRef] [PubMed]
- Whitmarsh, J.M. Intermediates of microbiological metabolism of cholesterol. Biochem. J. 1964, 90, 23–24. [Google Scholar]
- Brown, R.L.; Peterson, G.E. Cholesterol oxidation by soil Actinomycetes. J. Gen. Microbiol. 1966, 45, 441–450. [Google Scholar] [CrossRef][Green Version]
- Arima, K.; Nagasawa, M.; Bae, M.; Tamura, G. Microbial transformation of sterols. Part I. Decomposition of cholesterol by microorganisms. Agric. Biol. Chem. 1969, 33, 1636–1643. [Google Scholar]
- Chipley, J.R.; Dreyfuss, M.S.; Smucker, R.A. Cholesterol metabolism by Mycobacterium. Microbios 1975, 12, 199–207. [Google Scholar] [PubMed]
- Martin, C.K.A. Microbial transformation of β-sitosterol by Nocardia sp. M29. Eur. J. Appl. Microbiol. 1976, 2, 243–255. [Google Scholar] [CrossRef]
- Ferreira, N.P.; Tracey, R.P. Numerical taxonomy of cholesterol-degrading soil bacteria. J. Appl. Microbiol. 1984, 57, 429–446. [Google Scholar] [CrossRef]
- Mahato, S.B.; Banerjee, S.; Sahu, N.P. Metabolism of progesterone and testosterone by a Bacillus sp. Steroids 1984, 43, 545–558. [Google Scholar] [CrossRef]
- Mahato, S.B.; Banerjee, S. Metabolism of 11-deoxycortisol by a Bacillus species. J. Steroid Biochem. 1986, 25, 995–999. [Google Scholar] [CrossRef]
- Yoshimoto, T.; Nagai, F.; Fujimoto, J.; Watanabe, K.; Mizukoshi, H.; Makino, T.; Kimura, K.; Saino, H.; Sawada, H.; Omura, H. Degradation of strogens by Rhodococcus zopfii and Rhodococcus equi isolates from activated sludge in wastewater treatment plants. Appl. Environ. Microbiol. 2004, 70, 5283–5289. [Google Scholar] [CrossRef] [PubMed]
- Fahrbach, M.; Kuever, J.; Meinke, R.; Kämpfer, P.; Hollender, J. Denitratisoma oestradiolicum gen. nov., sp. nov., a 17beta-oestradiol-degrading, denitrifying betaproteobacterium. Int. J. Syst. Evol. Microbiol. 2006, 56, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Drzyzga, O.; Navarro Llorens, J.M.; Fernández de Las Heras, L.; García Fernández, E.; Perera, J. Gordonia cholesterolivorans sp. nov., a cholesterol-degrading actinomycete isolated from sewage sludge. Int. J. Syst. Evol. Microbiol. 2009, 59, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Fernández de las Heras, L.; García Fernández, E.; Navarro Llorens, J.M.; Perera, J.; Drzyzga, O. Morphological, physiological, and molecular characterization of a newly isolated steroid-degrading Actinomycete, identified as Rhodococcus ruber strain Chol-4. Curr. Microbiol. 2009, 59, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Van der Geize, R.; Yam, K.; Heuser, T.; Wilbrink, M.H.; Hara, H.; Anderton, M.C.; Sim, E.; Dijkhuizen, L.; Davies, J.E.; Mohn, W.W.; et al. A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. USA 2007, 104, 1947–1952. [Google Scholar] [CrossRef] [PubMed]
- Uhía, I.; Galán, B.; Morales, V.; García, J.L. Initial step in the catabolism of cholesterol by Mycobacterium smegmatis mc2 155. Environ. Microbiol. 2011, 13, 943–959. [Google Scholar] [CrossRef]
- Mohn, W.W.; Wilbrink, M.H.; Casabon, I.; Stewart, G.R.; Liu, J.; van der Geize, R.; Eltis, L.D. A gene cluster encoding cholate catabolism in Rhodococcus spp. J. Bacteriol. 2012, 194, 6712–6719. [Google Scholar] [CrossRef]
- Fernández de las Heras, L.; Perera, J.; Navarro Llorens, J.M. Cholesterol to cholestenone oxidation by ChoG, the main extracellular oxidase of Rhodococcus ruber strain Chol-4. J. Steroid Biochem. Mol. Biol. 2013, 139, 33–44. [Google Scholar] [CrossRef]
- Liu, Y.; Ge, F.; Chen, G.; Li, W.; Ma, P.; Zhang, G.; Zeng, L. Gordonia neofelifaecis sp. nov., a cholesterol side-chain-cleaving actinomycete isolated from the faeces of Neofelis nebulosa. Int. J. Syst. Evol. Microbiol. 2011, 61, 165–169. [Google Scholar] [CrossRef]
- Horinouchi, M.; Hayashi, T.; Kudo, T. Steroid degradation in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 2012, 129, 4–14. [Google Scholar] [CrossRef]
- Holert, J.; Alam, I.; Larsen, M.; Antunes, A.; Bajic, V.B.; Stingl, U.; Philipp, B. Genome sequence of Pseudomonas sp. strain Chol1, a model organism for the degradation of bile acids and other steroid compounds. Genome Announc. 2013, 1, e00014-12. [Google Scholar] [CrossRef] [PubMed]
- Barrientos, A.; Merino, E.; Casabon, I.; Rodríguez, J.; Crowe, A.M.; Holert, J.; Philipp, B.; Eltis, L.D.; Olivera, E.R.; Luengo, J.M. Functional analyses of three acyl-CoA synthetases involved in bile acid degradation in Pseudomonas putida DOC21. Environ. Microbiol. 2015, 17, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Tarlera, S.; Denner, E.B.M. Sterolibacterium denitrificans gen. nov., sp. nov., a novel cholesterol-oxidizing, denitrifying member of the β-Proteobacteria. Int. J. Syst. Evol. Microbiol. 2003, 53, 1085–1091. [Google Scholar] [PubMed]
- Chiang, Y.R.; Ismail, W.; Heintz, D.; Schaeffer, C.; Van Dorsselaer, A.; Fuchs, G. Study of anoxic and oxic cholesterol metabolism by Sterolibacterium denitrificans. J. Bacteriol. 2008, 190, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Fahrbach, M.; Kuever, J.; Remesch, M.; Huber, B.E.; Kämpfer, P.; Dott, W.; Hollender, J. Steroidobacter denitrificans gen. nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 2008, 58, 2215–2223. [Google Scholar] [CrossRef] [PubMed]
- Plésiat, P.; Nikaido, H. Outer membranes of gram-negative bacteria are permeable to steroid probes. Mol. Microbiol. 1992, 6, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Mohn, W.W.; van der Geize, R.; Stewart, G.R.; Okamoto, S.; Liu, J.; Dijkhuizen, L.; Eltis, L.D. The actinobacterial mce4 locus encodes a steroid transporter. J. Biol. Chem. 2008, 283, 35368–35374. [Google Scholar] [CrossRef] [PubMed]
- Van der Geize, R.; de Jong, W.; Hessels, G.I.; Grommen, A.W.F.; Jacobs, A.A.C.; Dijkhuizen, L. A novel method to generate unmarked gene deletions in the intracellular pathogen Rhodococcus equi using 5-fluorocytosine conditional lethality. Nucleic Acids Res. 2008, 36, e151. [Google Scholar] [CrossRef]
- Drzyzga, O.; Fernández de las Heras, L.; Morales, V.; Navarro Llorens, J.M.; Perera, J. Cholesterol degradation by Gordonia cholesterolivorans. Appl. Environ. Microbiol. 2011, 77, 4802–4810. [Google Scholar] [CrossRef]
- Klepp, L.I.; Forrellad, M.A.; Osella, A.V.; Blanco, F.C.; Stella, E.J.; Bianco, M.V.; de la Paz Santangelo, M.; Sassetti, C.; Jackson, M.; Cataldi, A.A.; et al. Impact of the deletion of the six mce operons in Mycobacterium smegmatis. Microbes Infect. 2012, 14, 590–599. [Google Scholar] [CrossRef][Green Version]
- Cole, S.T.; Brosch, R.; Parkhill, J.; Garnier, T.; Churcher, C.; Harris, D.; Gordon, S.V.; Eiglmeier, K.; Gas, S.; Barry, C.E., 3rd; et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998, 393, 537–544. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, J.; Papavinasasundaram, K.; Galán, B.; Sassetti, C.M.; García, J.L. Molecular and functional analysis of the mce4 operon in Mycobacterium smegmatis. Environ. Microbiol. 2017, 19, 3689–3699. [Google Scholar] [CrossRef] [PubMed]
- Kendall, S.L.; Withers, M.; Soffair, C.N.; Moreland, N.J.; Gurcha, S.; Sidders, B.; Frita, R.; ten Bokum, A.; Besra, G.S.; Lott, J.S.; et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol. Microbiol. 2007, 65, 684–699. [Google Scholar] [CrossRef] [PubMed]
- Casali, N.; Riley, L.W. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genom. 2007, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.M.; Pandey, A.K.; Capite, N.; Fortune, S.M.; Rubin, E.J.; Sassetti, C.M. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc. Natl. Acad. Sci. USA 2006, 103, 11760–11765. [Google Scholar] [CrossRef] [PubMed]
- Nazarova, E.V.; Montague, C.R.; La, T.; Wilburn, K.M.; Sukumar, N.; Lee, W.; Caldwell, S.; Russell, D.G.; VanderVen, B.C. Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. eLife 2017, 6, e26969. [Google Scholar] [CrossRef] [PubMed]
- Sassetti, C.M.; Rubin, E.J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 2003, 100, 12989–12994. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Magallanes, V.; Stadthagen-Gomez, G.; Rauzier, J.; Barreiro, L.B.; Tailleux, L.; Boudou, F.; Griffin, R.; Nigou, J.; Jackson, M.; Gicquel, B.; et al. Signature-tagged transposon mutagenesis identifies novel Mycobacterium tuberculosis genes involved in the parasitism of human macrophages. Infect. Immun. 2007, 75, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Sandie, R.; Wang, Y.; Andrade-Navarro, M.A.; Niederweis, M. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis 2008, 88, 526–544. [Google Scholar] [CrossRef] [PubMed]
- Feltcher, M.E.; Gunawardena, H.P.; Zulauf, K.E.; Malik, S.; Griffin, J.E.; Sassetti, C.M.; Chen, X.; Braunstein, M. Label-free Quantitative Proteomics reveals a role for the Mycobacterium tuberculosis SecA2 pathway in exporting solute binding proteins and Mce transporters to the cell wall. Mol. Cell. Proteom. 2015, 14, 1501–1516. [Google Scholar] [CrossRef] [PubMed]
- Perkowski, E.F.; Miller, B.K.; McCann, J.R.; Sullivan, J.T.; Malik, S.; Allen, I.C.; Godfrey, V.; Hayden, J.D.; Braunstein, M. An orphaned Mce-associated membrane protein of Mycobacterium tuberculosis is a virulence factor that stabilizes Mce transporters. Mol. Microbiol. 2016, 100, 90–107. [Google Scholar] [CrossRef] [PubMed]
- Somalinga, V.; Mohn, W.W. Rhodococcus jostii porin A (RjpA) functions in cholate uptake. Appl. Environ. Microbiol. 2013, 79, 6191–6193. [Google Scholar] [CrossRef] [PubMed]
- Haußmann, U.; Wolters, D.A.; Fränzel, B.; Eltis, L.D.; Poetsch, A. Physiological adaptation of the Rhodococcus jostii RHA1 membrane proteome to steroids as growth substrates. J. Proteome Res. 2013, 12, 1188–1198. [Google Scholar] [CrossRef] [PubMed]
- Swain, K.; Casabon, I.; Eltis, L.D.; Mohn, W.W. Two transporters essential for reassimilation of novel cholate metabolites by Rhodococcus jostii RHA1. J. Bacteriol. 2012, 194, 6720–6727. [Google Scholar] [CrossRef] [PubMed]
- Birkenmaier, A.; Holert, J.; Erdbrink, H.; Moeller, H.M.; Friemel, A.; Schoenenberger, R.; Suter, M.J.; Klebensberger, J.; Philipp, B. Biochemical and genetic investigation of initial reactions in aerobic degradation of the bile acid cholate in Pseudomonas sp. strain Chol1. J. Bacteriol. 2007, 189, 7165–7173. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.W.; Wang, P.H.; Ismail, W.; Tsai, Y.W.; El Nayal, A.; Yang, C.Y.; Yang, F.C.; Wang, C.H.; Chiang, Y.R. Substrate uptake and subcellular compartmentation of anoxic cholesterol catabolism in Sterolibacterium denitrificans. J. Biol. Chem. 2015, 290, 1155–1169. [Google Scholar] [CrossRef] [PubMed]
- Holert, J.; Jagmann, N.; Philipp, B. The essential function of genes for a hydratase and an aldehyde dehydrogenase for growth of Pseudomonas sp. strain Chol1 with the steroid compound cholate indicates an aldolytic reaction step for deacetylation of the side chain. J. Bacteriol. 2013, 195, 3371–3380. [Google Scholar] [CrossRef]
- Petrusma, M.; Dijkhuizen, L.; van der Geize, R. Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl. Environ. Microbiol. 2009, 75, 5300–5307. [Google Scholar] [CrossRef]
- Griffin, J.E.; Gawronski, J.D.; Dejesus, M.A.; Ioerger, T.R.; Akerley, B.J.; Sassetti, C.M. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011, 7, e1002251. [Google Scholar] [CrossRef]
- Thomas, S.T.; VanderVen, B.C.; Sherman, D.R.; Russell, D.G.; Sampson, N.S. Pathway profiling in Mycobacterium tuberculosis: Elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J. Biol. Chem. 2011, 286, 43668–43678. [Google Scholar] [CrossRef]
- Uhía, I.; Galán, B.; Kendall, S.L.; Stoker, N.G.; García, J.L. Cholesterol metabolism in Mycobacterium smegmatis. Environ. Microbiol. Rep. 2012, 4, 168–182. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ge, F.; Zhang, Q.; Ren, Y.; Yuan, J.; He, J.; Li, W.; Chen, G.; Zhang, G.; Zhuang, Y.; et al. Identification of gene expression profiles in the actinomycete Gordonia neofelifaecis grown with different steroids. Genome 2014, 57, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Bergstrand, L.H.; Cardenas, E.; Holert, J.; Van Hamme, J.D.; Mohn, W.W. Delineation of steroid-degrading microorganisms through comparative genomic analysis. mBio 2016, 7, e00166-16. [Google Scholar] [CrossRef] [PubMed]
- Rosłoniec, K.Z.; Wilbrink, M.H.; Capyk, J.K.; Mohn, W.W.; Ostendorf, M.; van der Geize, R.; Dijkhuizen, L.; Eltis, L.D. Cytochrome P450 125 (CYP125) catalyses C26-hydroxylation to initiate sterol side-chain degradation in Rhodococcus jostii RHA1. Mol. Microbiol. 2009, 74, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Kreit, J. Microbial catabolism of sterols: Focus on the enzymes that transform the sterol 3β-hydroxy-5-en into 3-keto-4-en. FEMS Microbiol. Lett. 2017, 364, fnx007. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Vrielink, A.; Brick, P.; Blow, D.M. Crystal structure of cholesterol oxidase complexed with a steroid substrate: Implications for flavin adenine dinucleotide dependent alcohol oxidases. Biochemistry 1993, 32, 11507–11515. [Google Scholar] [CrossRef]
- Coulombe, R.; Yue, K.Q.; Ghisla, S.; Vrielink, A. Oxygen access to the active site of cholesterol oxidase through a narrow channel is gated by an Arg-Glu pair. J. Biol. Chem. 2001, 276, 30435–30441. [Google Scholar] [CrossRef]
- Horinouchi, S.; Ishizuka, H.; Beppu, T. Cloning, nucleotide sequence, and transcriptional analysis of the NAD(P)-dependent cholesterol dehydrogenase gene from a Nocardia sp. and its hyperexpression in Streptomyces spp. Appl. Environ. Microbiol. 1991, 57, 1386–1393. [Google Scholar]
- Yang, X.; Dubnau, E.; Smith, I.; Sampson, N.S. Rv1106c from Mycobacterium tuberculosis is a 3β-hydroxysteroid dehydrogenase. Biochemistry 2007, 46, 9058–9067. [Google Scholar] [CrossRef]
- Yang, X.; Gao, J.; Smith, I.; Dubnau, E.; Sampsonn, N.S. Cholesterol is not an essential source of nutrition for Mycobacterium tuberculosis during infection. J. Bacteriol. 2011, 193, 1473–1476. [Google Scholar] [CrossRef]
- Brzostek, A.; Rumijowska-Galewicz, A.; Dziadek, B.; Wojcik, E.A.; Dziadek, J. ChoD and HsdD can be dispensable for cholesterol degradation in mycobacteria. J. Steroid Biochem. Mol. Biol. 2013, 134, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, F.; Maser, E. Carbonyl reductases and pluripotent hydroxysteroid dehydrogenases of the short-chain dehydrogenase/reductase superfamily. Drug Metab. Rev. 2007, 39, 87–144. [Google Scholar] [CrossRef] [PubMed]
- Marcus, P.I.; Talalay, P. Induction and purification of α- and β-hydroxysteroid dehydrogenases. J. Biol. Chem. 1956, 218, 661–674. [Google Scholar] [PubMed]
- Oppermann, U.C.T.; Maser, E. Characterization of a 3 α-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium Comamonas testosteroni. Eur. J. Biochem. 1996, 241, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Oppermann, U.C.T.; Belai, I.; Maser, E. Antibiotic resistance and enhanced insecticide catabolism as consequences of steroid induction in the Gram-negative bacterium Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 1996, 58, 217–223. [Google Scholar] [CrossRef]
- Möbus, E.; Maser, E. Molecular cloning, overexpression, and characterization of steroid-inducible 3α-hydroxysteroid dehydrogenase/carbonyl reductase from Comamonas testosteroni. A novel member of the short-chain dehydrogenase/reductase superfamily. J. Biol. Chem. 1998, 273, 30888–30896. [Google Scholar] [CrossRef]
- Horinouchi, M.; Kurita, T.; Hayashi, T.; Kudo, T. Steroid degradation genes in Comamonas testosteroni TA441: Isolation of genes encoding a Δ4(5)-isomerase and 3α- and 3β-dehydrogenases and evidence for a 100 kb steroid degradation gene hot spot. J. Steroid Biochem. Mol. Biol. 2010, 122, 253–263. [Google Scholar] [CrossRef]
- Holert, J.; Yücel, O.; Jagmann, N.; Prestel, A.; Möller, H.M.; Philipp, B. Identification of bypass reactions leading to the formation of one central steroid degradation intermediate in metabolism of different bile salts in Pseudomonas sp. strain Chol1. Environ. Microbiol. 2016, 18, 3373–3389. [Google Scholar] [CrossRef]
- Shi, C.J.; Tai, H.H.; Tsong, Y.Y. The mechanism of microbial conversion of cholesterol into 17-keto steroids. J. Am. Chem. Soc. 1967, 89, 1957–1958. [Google Scholar]
- Shi, C.J.; Tai, H.H.; Tsong, Y.Y.; Lee, S.S.; Coombe, R.G. Mechanisms of steroid oxidation by microorganisms 0.14. Pathway of cholesterol side-chain degradation. Biochemistry 1968, 7, 808–818. [Google Scholar]
- Reddy, J.K.; Hashimoto, T. Peroxisomal β-oxidation and peroxisome proliferator activated receptor α: An adaptive metabolic system. Annu. Rev. Nutr. 2001, 21, 193–230. [Google Scholar] [CrossRef] [PubMed]
- Capyk, J.K.; Kalscheuer, R.; Stewart, G.R.; Liu, J.; Kwon, H.; Zhao, R.; Okamoto, S.; Jacobs, W.R., Jr.; Eltis, L.D.; Mohn, W.W. Mycobacterial cytochrome p450 125 (cyp125) catalyzes the terminal hydroxylation of C27 steroids. J. Biol. Chem. 2009, 284, 35534–35542. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, H.; Guan, S.; Johnston, J.B.; Chow, E.D.; Kells, P.M.; Burlingame, A.L.; Cox, J.S.; Podust, L.M.; de Montellano, P.R. Mycobacterium tuberculosis CYP125A1, a steroid C27 monooxygenase that detoxifies intracellularly generated cholest-4-en-3-one. Mol. Microbiol. 2010, 77, 730–742. [Google Scholar] [CrossRef] [PubMed]
- McLean, K.J.; Lafite, P.; Levy, C.; Cheesman, M.R.; Mast, N.; Pikuleva, I.A.; Leys, D.; Munro, A.W. The structure of Mycobacterium tuberculosis CYP125: Molecular basis for cholesterol binding in a P450 needed for host infection. J. Biol. Chem. 2009, 284, 35524–35533. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, H.; Johnston, J.B.; de Montellano, P.R. Cholesterol catabolism as a therapeutic target in Mycobacterium tuberculosis. Trends Microbiol. 2011, 19, 530–539. [Google Scholar] [CrossRef]
- Johnston, J.B.; Ouellet, H.; Ortiz de Montellano, P.R. Functional redundancy of steroid C26-monooxygenase activity in Mycobacterium tuberculosis revealed by biochemical and genetic analyses. J. Biol. Chem. 2010, 285, 36352–36360. [Google Scholar] [CrossRef]
- Frank, D.J.; Madrona, Y.; Ortiz de Montellano, P.R. Cholesterol ester oxidation by mycobacterial cytochrome P450. J. Biol. Chem. 2014, 289, 30417–30425. [Google Scholar] [CrossRef]
- Wilbrink, M.H.; Petrusma, M.; Dijkhuizen, L.; van der Geize, R. FadD19 of Rhodococcus rhodochrous DSM43269, a steroid-coenzyme A ligase essential for degradation of C-24 branched sterol side chains. Appl. Environ. Microbiol. 2011, 77, 4455–4464. [Google Scholar] [CrossRef]
- Casabon, I.; Swain, K.; Crowe, A.M.; Eltis, L.D.; Mohn, W.W. Actinobacterial acyl coenzyme A synthetases involved in steroid side-chain catabolism. J. Bacteriol. 2014, 196, 579–587. [Google Scholar] [CrossRef]
- Thomas, S.T.; Sampson, N.S. Mycobacterium tuberculosis utilizes a unique heterotetrameric structure for dehydrogenation of the cholesterol side chain. Biochemistry 2013, 52, 2895–2904. [Google Scholar] [CrossRef]
- Yang, M.; Lu, R.; Guja, K.E.; Wipperman, M.F.; St Clair, J.R.; Bonds, A.C.; Garcia-Diaz, M.; Sampson, N.S. Unraveling cholesterol catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 α2β2 acyl-CoA dehydrogenase initiates β-oxidation of 3-oxo-cholest-4-en-26-oyl CoA. ACS Infect. Dis. 2015, 1, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Battaile, K.P. Burning fat: The structural basis of fatty acid β-oxidation. Curr. Opin. Struct. Biol. 2002, 12, 721–728. [Google Scholar] [CrossRef]
- Yang, M.; Guja, K.E.; Thomas, S.T.; Garcia-Diaz, M.; Sampson, N.S. A distinct MaoC-like enoyl-CoA hydratase architecture mediates cholesterol catabolism in Mycobacterium tuberculosis. ACS Chem. Biol. 2014, 9, 2632–2645. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.E.; Pandey, A.K.; Gilmore, S.A.; Mizrahi, V.; McKinney, J.D.; Bertozzi, C.R.; Sassetti, C.M. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 2012, 19, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Wipperman, M.F.; Yang, M.; Thomas, S.T.; Sampson, N.S. Shrinking the FadE proteome of Mycobacterium tuberculosis: Insights into cholesterol metabolism through identification of an α2β2 heterotetrameric acyl coenzyme A dehydrogenase family. J. Bacteriol. 2013, 195, 4331–4341. [Google Scholar] [CrossRef] [PubMed]
- Nesbitt, N.M.; Yang, X.; Fontán, P.; Kolesnikova, I.; Smith, I.; Sampson, N.S.; Dubnau, E. A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol. Infect. Immun. 2010, 78, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, C.M.; Lu, R.; Nesbitt, N.M.; Schiebel, J.; Sampson, N.S.; Kisker, C. FadA5 a thiolase from Mycobacterium tuberculosis: A steroid-binding pocket reveals the potential for drug development against tuberculosis. Structure 2015, 23, 21–33. [Google Scholar] [CrossRef]
- Lu, R.; Schaefer, C.M.; Nesbitt, N.M.; Kuper, J.; Kisker, C.; Sampson, N.S. Catabolism of the cholesterol side chain in Mycobacterium tuberculosis is controlled by a Redox-Sensitive Thiol Switch. ACS Infect. Dis. 2017, 3, 666–675. [Google Scholar] [CrossRef]
- Anbazhagan, P.; Harijan, R.K.; Kiema, T.R.; Janardan, N.; Murthy, M.R.; Michels, P.A.; Juffer, A.H.; Wierenga, R.K. Phylogenetic relationships and classification of thiolases and thiolase-like proteins of Mycobacterium tuberculosis and Mycobacterium smegmatis. Tuberculosis 2014, 94, 405–412. [Google Scholar] [CrossRef]
- Xu, L.Q.; Liu, Y.J.; Yao, K.; Liu, H.H.; Tao, X.Y.; Wang, F.Q.; Wei, D.Z. Unraveling and engineering the production of 23,24-bisnorcholenic steroids in sterol metabolism. Sci. Rep. 2016, 6, 21928. [Google Scholar] [CrossRef]
- Wilbrink, M.H.; van der Geize, R.; Dijkhuizen, L. Molecular characterization of lpt3 and lpt4, essential for C24-branched chain sterol-side-chain degradation in Rhodococcus rhodochrous DSM 43269. Microbiology 2012, 158, 3054–3062. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.; Hood, L.; Seah, S.Y.K. Characterization of an aldolase involved in cholesterol side chain degradation in Mycobacterium tuberculosis. J. Bacteriol. 2018, 200, e00512-17. [Google Scholar] [CrossRef] [PubMed]
- Holert, J.; Kulić, Ž.; Yücel, O.; Suvekbala, V.; Suter, M.J.; Möller, H.M.; Philipp, B. Degradation of the acyl side chain of the steroid compound cholate in Pseudomonas sp. strain Chol1 proceeds via an aldehyde intermediate. J. Bacteriol. 2013, 195, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Genti-Raimondi, S.; Tolmasky, M.E.; Patrito, L.C.; Flury, A.; Actis, L.A. Molecular cloning and expression of the β-hydroxysteroid dehydrogenase gene from Pseudomonas testosteroni. Gene 1991, 105, 43–49. [Google Scholar] [CrossRef]
- Schultz, R.M.; Groman, E.V.; Engel, L.L. 3(17)β-Hydroxysteroid dehydrogenase of Pseudomonas testosteroni. A convenient purification and demonstration of multiple molecular forms. J. Biol. Chem. 1977, 252, 3775–3783. [Google Scholar] [PubMed]
- Minard, P.; Legoy, M.D.; Thomas, D. 3β,17β-hydroxysteroid dehydrogenase of Pseudomonas testosteroni. Kinetic evidence for the bifunctional activity at a common catalytic site. FEBS Lett. 1985, 188, 85–90. [Google Scholar] [CrossRef]
- Yu, Y.; Liu, C.; Wang, B.; Li, Y.; Zhang, H. Characterization of 3,17β-hydroxysteroid dehydrogenase in Comamonas testosteroni. Chem. Biol. Interact. 2015, 234, 221–228. [Google Scholar] [CrossRef]
- Itagaki, E.; Matushita, H.; Hatta, T. Steroid transhydrogenase activity of 3-ketosteroid-Δ1-dehydrogenase from Nocardia corallina. J. Biochem. 1990, 108, 122–127. [Google Scholar] [CrossRef]
- Rohman, A.; van Oosterwijk, N.; Thunnissen, A.M.W.H.; Dijkstra, B.W. Crystal structure and site-directed mutagenesis of 3-ketosteroid Δ1-dehydrogenase from Rhodococcus erythropolis SQ1 explain its catalytic mechanism. J. Biol. Chem. 2013, 288, 35559–35568. [Google Scholar] [CrossRef]
- Horinouchi, M.; Hayashi, T.; Yamamoto, T.; Kudo, T. A new bacterial steroid degradation gene cluster in Comamonas testosteroni TA441 which consists of aromatic-compound degradation genes for seco-steroids and 3-ketosteroid dehydrogenase genes. Appl. Environ. Microbiol. 2003, 69, 4421–4430. [Google Scholar] [CrossRef]
- Olivera, E.R.; de la Torre, M.; Barrientos, A.; Luengo, J.M. Steroid catabolism in bacteria: Genetic and functional analyses of stdH and stdJ in Pseudomonas putida DOC21. Can. J. Biotechnol. 2018, 2, 88–99. [Google Scholar] [CrossRef]
- Brzostek, A.; Pawelczyk, J.; Rumijowska-Galewicz, A.; Dziadek, B.; Dziadek, J. Mycobacterium tuberculosis is able to accumulate and utilize cholesterol. J. Bacteriol. 2009, 191, 6584–6591. [Google Scholar] [CrossRef] [PubMed]
- Knol, J.; Bodewits, K.; Hessels, G.I.; Dijkhuizen, L.; van der Geize, R. 3-Keto-5α-steroid Δ(1)-dehydrogenase from Rhodococcus erythropolis SQ1 and its orthologue in Mycobacterium tuberculosis H37Rv are highly specific enzymes that function in cholesterol catabolism. Biochem. J. 2008, 410, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Van der Geize, R.; Hessels, G.I.; Dijkhuizen, L. Molecular and functional characterization of the kstD2 gene of Rhodococcus erythropolis SQ1 encoding a second 3-ketosteroid Δ(1)-dehydrogenase isoenzyme. Microbiology 2002, 148, 3285–3292. [Google Scholar] [CrossRef] [PubMed]
- van der Geize, R.; Hessels, G.I.; van Gerwen, R.; van der Meijden, P.; Dijkhuizen, L. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Δ1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 2001, 205, 197–202. [Google Scholar] [CrossRef]
- Brzostek, A.; Sliwiński, T.; Rumijowska-Galewicz, A.; Korycka-Machała, M.; Dziadek, J. Identification and targeted disruption of the gene encoding the main 3-ketosteroid dehydrogenase in Mycobacterium smegmatis. Microbiology 2005, 151, 2393–2402. [Google Scholar] [CrossRef]
- Florin, C.; Kohler, M.; Grandguillot, M.; Plesiat, P. Comamonas testosteroni 3-ketosteroid-Δ4(5α)-dehydrogenase: Gene and protein characterization. J. Bacteriol. 1996, 178, 3322–3330. [Google Scholar] [CrossRef][Green Version]
- van Oosterwijk, N.; Knol, J.; Dijkhuizen, L.; van der Geize, R.; Dijkstra, B.W. Structure and catalytic mechanism of 3-ketosteroid-Delta4-(5α)-dehydrogenase from Rhodococcus jostii RHA1 genome. J. Biol. Chem. 2012, 287, 30975–30983. [Google Scholar] [CrossRef]
- van der Geize, R.; Hessels, G.I.; van Gerwen, R.; van der Meijden, P.; Dijkhuizen, L. Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9alpha-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol. Microbiol. 2002, 45, 1007–1018. [Google Scholar] [CrossRef]
- van der Geize, R.; Hessels, G.I.; Nienhuis-Kuiper, M.; Dijkhuizen, L. Characterization of a second Rhodococcus erythropolis SQ1 3-ketosteroid 9alpha-hydroxylase activity comprising a terminal oxygenase homologue, KshA2, active with oxygenase-reductase component KshB. Appl. Environ. Microbiol. 2008, 74, 7197–7203. [Google Scholar] [CrossRef]
- Capyk, J.K.; D’Angelo, I.; Strynadka, N.C.; Eltis, L.D. Characterization of 3-ketosteroid 9{α}-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J. Biol. Chem. 2009, 284, 9937–9946. [Google Scholar] [CrossRef] [PubMed]
- Capyk, J.K.; Casabon, I.; Gruninger, R.; Strynadka, N.C.; Eltis, L.D. Activity of 3-ketosteroid 9α-hydroxylase (KshAB) indicates cholesterol side chain and ring degradation occur simultaneously in Mycobacterium tuberculosis. J. Biol. Chem. 2011, 286, 40717–40724. [Google Scholar] [CrossRef] [PubMed]
- García, J.L.; Uhía, I.; Galán, B. Catabolism and biotechnological applications of cholesterol degrading bacteria. Microb. Biotechnol. 2012, 5, 679–699. [Google Scholar] [CrossRef] [PubMed]
- Donova, M.; Gulevskaya, S.; Dovbnya, D.; Puntus, I. Mycobacterium sp. mutant strain producing 9α-hydroxy androstenedione from sitosterol. Appl. Microbiol. Biotechnol. 2005, 67, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Guevara, G.; Fernández de las Heras, L.; Perera, J.; Navarro Llorens, J.M. Functional differentiation of 3-ketosteroid Δ1-dehydrogenase isoenzymes in Rhodococcus ruber strain Chol-4. Microb. Cell Fact. 2017, 16, 42. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Xu, L.Q.; Wang, F.Q.; Wei, D.Z. Characterization and engineering of 3-ketosteroid Δ1-dehydrogenase and 3-ketosteroid 9α-hydroxylase in Mycobacterium neoaurum ATCC 25795 to produce 9a-hydroxy-4-androstene-3,17-dione through the catabolism of sterols. Metab. Eng. 2014, 24, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; van der Geize, R.; Besra, G.S.; Gurcha, A.; Liu, M.; Rohde, M.; Singh, M.; Coares, A. 3-Ketosteroid 9α-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 2010, 75, 107–121. [Google Scholar] [CrossRef]
- Bragin, E.Y.; Shtratnikova, V.Y.; Dovbnya, D.V.; Schelkunov, M.I.; Pekov, Y.A.; Malakho, S.G.; Egoroba, O.V.; Ivashina, T.V.; Sokolov, S.L.; Ashapkin, V.V.; et al. Comparative analysis of genes encoding key steroid core oxidation enzymes in fast-growing Mycobacterium spp. Strains. J. Steroid Biochem. Mol. Biol. 2013, 138, 41–53. [Google Scholar] [CrossRef]
- Petrusma, M.; Hessels, G.; Dijkhuizen, L.; van der Geize, R. Multiplicity of 3-ketosteroid-9α-hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids. J. Bacteriol. 2011, 193, 3931–3940. [Google Scholar] [CrossRef]
- Horinouchi, M.; Hayashi, T.; Kudo, T. The genes encoding the hydroxylase of 3-hydroxy-9,10-secoandrosta-1,3,5(10)-triene-9,17-dione in steroid degradation in Comamonas testosteroni TA441. J. Steroid. Biochem. Mol. Biol. 2004, 92, 143–154. [Google Scholar] [CrossRef]
- Dresen, C.; Lin, L.Y.; D’Angelo, I.; Tocheva, E.I.; Strynadka, N.; Eltis, L.D. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J. Biol. Chem. 2010, 285, 22264–22275. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Yamamoto, T.; Taguchi, K.; Arai, H.; Kudo, T. Meta-cleavage enzyme gene tesB is necessary for testosterone degradation in Comamonas testosteroni TA441. Microbiology 2001, 147, 3367–3375. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yam, K.C.; D’Angelo, I.; Kalscheuer, R.; Zhu, H.; Wang, J.X.; Snieckus, V.; Ly, L.H.; Converse, P.J.; Jacobs, W.R. Jr.; Strynadka, N.; et al. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 2009, 5, e1000344. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Hayashi, T.; Koshino, H.; Yamamoto, T.; Kudo, T. Gene encoding the hydrolase for the product of the meta-cleavage reaction in testosterone degradation by Comamonas testosteroni. Appl. Environ. Microbiol. 2003, 69, 2139–2152. [Google Scholar] [CrossRef] [PubMed]
- Lack, N.; Lowe, E.D.; Liu, J.; Eltis, L.D.; Noble, M.E.; Sim, E.; Westwood, I.M. Structure of HsaD, a steroid-degrading hydrolase, from Mycobacterium tuberculosis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Lack, N.A.; Yam, K.C.; Lowe, E.D.; Horsman, G.P.; Owen, R.L.; Sim, E.; Eltis, L.D. Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism. J. Biol. Chem. 2010, 285, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Hayashi, T.; Koshino, H.; Kurita, T.; Kudo, T. Identification of 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid, 4-hydroxy-2-oxohexanoic acid, and 2-hydroxyhexa-2,4-dienoic acid and related enzymes involved in testosterone degradation in Comamonas testosteroni TA441. Appl. Environ. Microbiol. 2005, 71, 5275–5281. [Google Scholar] [CrossRef] [PubMed]
- Carere, J.; McKenna, S.E.; Kimber, M.S.; Seah, S.Y. characterization of an aldolase_dehydrogenase complex from the cholesterol degrading pathway of Mycobacterium tuberculosis. Biochemistry 2013, 52, 3502–3511. [Google Scholar] [CrossRef]
- Horinouchi, M.; Hayashi, T.; Koshino, H.; Kudo, T. ORF18 disrupted mutant of Comamonas testosteroni TA441 accumulates significant amounts of 9,17-dioxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid and its derivatives after incubation with steroids. J. Steroid Biochem. Mol. Biol. 2006, 101, 78–84. [Google Scholar] [CrossRef]
- Casabon, I.; Crowe, A.M.; Liu, J.; Eltis, L.D. FadD3 is an acyl-CoA synthetase that initiates catabolism of cholesterol rings C and D in actinobacteria. Mol. Microbiol. 2013, 87, 269–283. [Google Scholar] [CrossRef]
- Crowe, A.M.; Casabon, I.; Brown, K.L.; Liu, J.; Lian, J.; Rogalski, J.C.; Hurst, T.E.; Snieckus, V.; Foster, L.J.; Eltis, L.D. Catabolism of the last two steroid rings in Mycobacterium tuberculosis and other bacteria. mBio 2017, 8, e00321-17. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Hayashi, T.; Koshino, H.; Kudo, T. Identification of 9alpha-hydroxy-17-oxo-1,2,3,4,10,19-hexanorandrostan-5-oic acid in steroid degradation by Comamonas testosteroni TA441 and its conversion to the corresponding 6-en-5-oyl Coenzyme A (CoA) involving open reading frame 28 (ORF28)- and ORF30-encoded acyl-CoA dehydrogenases. J. Bacteriol. 2014, 196, 3598–3608. [Google Scholar]
- Horinouchi, M.; Hayashi, T.; Koshino, H.; Malon, M.; Hirota, H.; Kudo, T. Identification of 9α-hydroxy-17-oxo-1,2,3,4,10,19-hexanorandrost-6-en-5-oic acid and β-oxidation products of the C-17 side chain in cholic acid degradation by Comamonas testosteroni TA441. J. Steroid Biochem. Biol. Mol. 2014, 143, 306–322. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Koshino, H.; Malon, M.; Hirota, H.; Hayashi, T. Steroid degradation in Comamonas testosteroni TA441: Identification of metabolites and the genes involved in the reactions necessary before D-ring cleavage. Appl. Environ. Microbiol. 2018, 84, e01324-18. [Google Scholar] [CrossRef] [PubMed]
- Crowe, A.M.; Workman, S.D.; Watanabe, N.; Worrall, L.J.; Strynadka, N.C.J.; Eltis, L.D. IpdAB, a virulence factor in Mycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase. Proc. Natl. Acad. Sci. USA 2018, 115, E3378–E3387. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Koshino, H.; Malon, M.; Hirota, H.; Hayashi, T. Identification of 9-oxo-1,2,3,4,5,6,10,19-octanor-13,17-secoandrost-8(14)-ene-7,17-dioic acid as a metabolite of steroid degradation in Comamonas testosteroni TA441 and the genes involved in the conversion. J. Steroid Biochem. Mol. Biol. 2019, 185, 268–276. [Google Scholar] [CrossRef]
- Horinouchi, M.; Malon, M.; Hirota, H.; Hayashi, T. Identification of 4-methyl-5-oxo-octane-1,8-dioic acid and the derivatives as metabolites of steroidal C,D-ring degradation in Comamonas testosteroni TA441. J. Steroid Biochem. Mol. Biol. 2019, 185, 277–286. [Google Scholar] [CrossRef]
- Harwood, C.S.; Parales, R.E. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 1996, 50, 553–590. [Google Scholar] [CrossRef]
- Chen, Y.L.; Wang, C.H.; Yang, F.C.; Ismail, W.; Wang, P.H.; Shih, C.J.; Wu, Y.C.; Chiang, Y.R. Identification of Comamonas testosteroni as an androgen degrader in sewage. Sci. Rep. 2016, 6, 35386. [Google Scholar] [CrossRef]
- Kendall, S.L.; Burgess, P.; Balhana, R.; Withers, M.; Ten Bokum, A.; Lott, J.S.; Gao, C.; Uhía-Castro, I.; Stocker, N.G. Cholesterol utilization in mycobacteria is controlled by two TetR-type transcriptional regulators: KstR and KstR2. Microbiology 2010, 156, 1362–1371. [Google Scholar] [CrossRef]
- Ho, N.A.T.; Dawes, S.S.; Crowe, A.M.; Casabon, I.; Gao, C.; Kendall, S.L.; Baker, E.N.; Eltis, L.D.; Lott, J.S. The structure of the transcriptional repressor KstR in complex with CoA thioester cholesterol metabolites sheds light on the regulation of cholesterol catabolism in Mycobacterium tuberculosis. J. Biol. Chem. 2016, 291, 7256–7266. [Google Scholar] [CrossRef] [PubMed]
- García-Fernández, E.; Medrano, F.J.; Galán, B.; García, J.L. Deciphering the transcriptional regulation of cholesterol catabolic pathway in mycobacteria: Identification of the inducer of KstR repressor. J. Biol. Chem. 2014, 289, 17576–17588. [Google Scholar] [CrossRef] [PubMed]
- Casabon, I.; Zhu, S.-H.; Otani, H.; Liu, J.; Mohn, W.W.; Eltis, L.D. Regulation of KstR2 regulon of Mycobacterium tuberculosis by a cholesterol catabolite. Mol. Microbiol. 2013, 89, 1201–1212. [Google Scholar] [CrossRef] [PubMed]
- Crowe, A.M.; Stogios, P.J.; Casabon, I.; Evdokimova, E.; Savchenko, A.; Eltis, L.D. Structural and functional characterization of a ketosteroid transcriptional regulator of Mycobacterium tuberculosis. J. Biol. Chem. 2015, 290, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, M.; Kurita, T.; Yamamoto, T.; Hatori, E.; Hayasi, T.; Kudo, T. Steroid degradation gene cluster of Comamonas testosteroni consisting of 18 putative genes from meta-cleavage enzyme gene tesB to regulator gene tesR. Biochem. Biophys. Res. Commun. 2004, 324, 597–604. [Google Scholar] [CrossRef]
- Pruneda-Paz, J.L.; Linares, M.; Cabrera, J.E.; Genti-Raimondi, S. TeiR, a LuxR-type transcriptional factor required for testosterone degradation in Comamonas testosteroni. J. Bacteriol. 2004, 186, 1430–1437. [Google Scholar] [CrossRef]
- Göhler, A.; Xiong, G.; Paulsen, S.; Trentmann, G.; Maser, E. Testosterone-inducible regulator is a kinase that drives steroid sensing and metabolism in Comamonas testosteroni. J. Biol. Chem. 2008, 283, 17380–17390. [Google Scholar] [CrossRef]
- Linares, M.; Pruneda-Paz, J.L.; Reyna, L.; Genti-Raimondi, S. Regulation of testosterone degradation in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 2008, 112, 145–150. [Google Scholar] [CrossRef]
- Xiong, G.; Maser, E. Regulation of the steroid-inducible 3α-hydroxysteroid dehydrogenase/carbonyl reductase gene in Comamonas testosteroni. J. Biol. Chem. 2001, 276, 9961–9970. [Google Scholar] [CrossRef]
- Xiong, G.; Martin, H.J.; Maser, E. Identification and characterization of a novel translational repressor of the steroid-inducible 3α-hydroxysteroid dehydrogenase/carbonyl reductase in Comamonas testosteroni. J. Biol. Chem. 2003, 278, 47400–47407. [Google Scholar] [CrossRef]
- Gong, W.; Xiong, G.; Maser, E. Identification and characterization of the LysR-type transcriptional regulator HsdR for steroid-inducible expression of the 3a-hydroxysteroid dehydrogenase/carbonyl reductase gene in Comamonas testosteroni. Appl. Environ. Microbiol. 2011, 78, 941–950. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gong, W.; Xiong, G.; Maser, E. Oligomerization and negative autoregulation of the LysR-type transcriptional regulator HsdR from Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 2012, 132, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiong, G.; Maser, E. A novel transcriptional repressor PhaR for the steroid-inducible expression of the 3,17β-hydroxysteroid dehydrogenase gene in Comamonas testosteroni ATCC11996. Chem. Biol. Interact. 2013, 202, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Pan, T.; Zhang, Y.; Xiong, G.; Yu, Y. Functional analysis of a novel repressor LuxR in Comamonas testosteroni. Chem. Biol. Interact. 2017, 276, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Huang, P.; Xiong, G.; Maser, E. Isolation and characterization of a repressor TetR for 3,17β-HSD expressional regulation in Comamonas testosteroni. Chem. Biol. Interact. 2015, 234, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Huang, P.; Xiong, G.; Maser, E. Identification and isolation of a regulator protein for 3,17β-HSD expression regulation in Comamonas testosteroni. Chem. Biol. Interact. 2015, 234, 197–204. [Google Scholar] [CrossRef]
- Yu, C.P.; Roh, H.; Chu, K.H. 17β-Estradiol-degrading bacteria isolated from activated sludge. Environ. Sci. Technol. 2007, 41, 486–492. [Google Scholar] [CrossRef]
- Chen, Y.L.; Yu, C.P.; Lee, T.H.; Goh, K.S.; Chu, K.H.; Wang, P.H.; Ismail, W.; Shih, C.J.; Chiang, Y.R. Biochemical mechanisms and catabolic enzymes involved in bacterial estrogen degradation pathways. Cell Chem. Biol. 2017, 24, 712–724. [Google Scholar] [CrossRef]
- Roh, H.; Chu, K.H. A 17beta-estradiol-utilizing bacterium, Sphingomonas strain KC8: Part I—Characterization and abundance in wastewater treatment plants. Environ. Sci. Technol. 2010, 44, 4943–4950. [Google Scholar] [CrossRef]
- Liang, R.; Liu, H.; Liu, J. Genome sequence of Pseudomonas putida strain SJTE-1, a bacterium capable of degrading estrogens and persistent organic pollutants. J. Bacteriol. 2012, 194, 4781–4782. [Google Scholar] [CrossRef]
- Wang, P.; Zheng, D.; Peng, W.; Wang, Y.; Wang, X.; Xiong, W.; Liang, R. Characterization of 17β-hydroxysteroid dehydrogenase and regulators involved in estrogen degradation in Pseudomonas putida SJTE-1. Appl. Microbiol. Biotechnol. 2019, 103, 2413–2425. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Lee, T.H.; Chen, Y.L.; Wang, Y.S.; Wang, P.H.; Yu, C.P.; Chu, K.H.; Chiang, Y.R. Metabolites involved in aerobic degradation of the A and B rings of estrogen. Appl. Environ. Microbiol. 2019, 85, e02223-18. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Me, C.; Hu, A.; Zhang, F.; Hu, H.; Yu, C.P. Altererythrobacter estronivorus sp. nov., an estrogen-degrading strain isolated from Yundang Lagoon of Xiamen City in China. Curr. Microbiol. 2016, 72, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Fujii, K.; Satomi, M.; Morita, N.; Motomura, T.; Tanaka, T.; Kikuchi, S. Novosphingobium tardaugens sp. nov., an oestradiol-degrading bacterium isolated from activated sludge of a sewage treatment plant in Tokyo. Int. J. Syst. Evol. Microbiol. 2003, 53, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Fu, H.-Y.; Lee, T.-H.; Shih, C.-J.; Huang, L.; Wang, Y.-S.; Ismail, W.; Chiang, Y.-R. Estrogen degraders and estrogen degradation pathway identified in an activated sludge. Appl. Environ. Microbiol. 2018, 84, e00001-18. [Google Scholar] [CrossRef] [PubMed]
- Kurisu, F.; Ogura, M.; Saitoh, S.; Yamazoe, A.; Yagi, O. Degradation of natural estrogen and identification of the metabolites produced by soil isolates of Rhodococcus sp. and Sphingomonas sp. J. Biosci. Bioeng. 2010, 109, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Nakai, S.; Yamamura, A.; Tanaka, S.; Shi, J.; Nishikawa, M.; Nakashimada, Y.; Hosomi, M. Pathway of 17β-estradiol degradation by Nitrosomonas europaea and reduction in 17β-estradiol-derived estrogenic activity. Environ. Chem. Lett. 2011, 9, 1–6. [Google Scholar] [CrossRef]
- Lee, H.B.; Liu, D. Degradation of 17β-estradiol and its metabolites by sewage bacteria. Water Air Soil Pollut. 2002, 134, 353–368. [Google Scholar] [CrossRef]
- Harder, J.; Probian, C. Anaerobic mineralization of cholesterol by a novel type of denitrifying bacterium. Arch. Microbiol. 1997, 167, 269–274. [Google Scholar] [CrossRef]
- Wang, P.-H.; Leu, Y.-L.; Ismail, W.; Tang, S.-L.; Tsai, C.-Y.; Chen, H.-J.; Kao, A.-T.; Chiang, Y.-R. Anaerobic and aerobic cleavage of the steroid core ring structure by Steroidobacter denitrificans. J. Lipid Res. 2013, 54, 1493–1504. [Google Scholar] [CrossRef]
- Wang, P.-H.; Yu, C.-P.; Lee, T.-H.; Lin, C.-W.; Ismail, W.; Wey, S.-P.; Kuo, A.-T.; Chiang, Y.-R. Anoxic androgen degradation by the denitrifying bacterium Sterolibacterium denitrificans via the 2,3-seco pathway. Appl. Environ. Microbiol. 2014, 80, 3442–3452. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.C.; Chen, Y.L.; Tang, S.L.; Yu, C.P.; Wang, P.H.; Ismail, W.; Wang, C.H.; Ding, J.Y.; Yang, C.Y.; Yang, C.Y.; et al. Integrated multi-omics analyses reveal the biochemical mechanisms and phylogenetic relevance of anaerobic androgen biodegradation in the environment. ISME J. 2016, 10, 1967–1983. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.R.; Ismail, W.; Müller, M.; Fuchs, G. Initial steps in the anoxic metabolism of cholesterol by the denitrifying Sterolibacterium denitrificans. J. Biol. Chem. 2007, 282, 13240–13249. [Google Scholar] [CrossRef] [PubMed]
- Dermer, J.; Fuchs, G. Molybdoenzyme that catalyzes the anaerobic hydroxylation of a tertiary carbon atom in the side chain of cholesterol. J. Biol. Chem. 2012, 287, 36905–36916. [Google Scholar] [CrossRef] [PubMed]
- Rugor, A.; Wójcik-Augustyn, A.; Niedzialkowska, E.; Mordalski, S.; Staroń, J.; Bojarski, A.; Szaleniec, M. Reaction mechanism of sterol hydroxylation by steroid C25 dehydrogenase–Homology model, reactivity and isoenzymatic diversity. J. Inorg. Biochem. 2017, 173, 28–43. [Google Scholar] [CrossRef] [PubMed]
- Warnke, M.; Jacoby, C.; Jung, T.; Agne, M.; Mergelsberg, M.; Starke, R.; Jehmlich, N.; von Bergen, M.; Richnow, H.H.; Brüls, T.; et al. A patchwork pathway for oxygenase-independent degradation of side chain containing steroids. Environ. Microbiol. 2017, 19, 4684–4699. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, C.; Eipper, J.; Warnke, M.; Tiedt, O.; Mergelsberg, M.; Stärk, H.J.; Daus, B.; Martín-Moldes, Z.; Zamarro, M.T.; Díaz, E.; et al. Four Molybdenum-dependent steroid C-25 hydroxylases: Heterologous overproduction, role in steroid degradation, and application for 25-hydroxyvitamin D3 synthesis. mBio 2018, 9, e00694-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.-H.; Lee, T.H.; Ismail, W.; Tsai, C.Y.; Lin, C.W.; Tsai, Y.W.; Chiang, Y.R. An oxygenase-independent cholesterol catabolic pathway operates under oxic conditions. PLoS ONE 2013, 8, e66675. [Google Scholar] [CrossRef] [PubMed]
- Warnke, M.; Jung, T.; Jacoby, C.; Agne, M.; Feller, F.M.; Philipp, B.; Seiche, W.; Breit, B.; Boll, M. Functional characterization of three specific acyl-coenzyme A synthetases involved in anaerobic cholesterol degradation in Sterolibacterium denitrificans Chol1S. Appl. Environ. Microbiol. 2018, 84, e02721-17. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.R.; Ismail, W.; Gallien, S.; Heintz, D.; Van Dorsselaer, A.; Fuchs, G. Cholest-4-en-3-one-Δ1-dehydrogenase, a flavoprotein catalyzing the second step in anoxic cholesterol metabolism. Appl. Environ. Microbiol. 2008, 74, 107–113. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chiang, Y.R.; Fang, J.Y.; Ismail, W.; Wang, P.H. Initial steps in anoxic testosterone degradation by Steroidobacter denitrificans. Microbiology 2010, 156, 2253–2259. [Google Scholar] [CrossRef] [PubMed]
- Fahrbach, M.; Krauss, M.; Preiss, A.; Kohler, H.P.; Hollender, J. Anaerobic testosterone degradation in Steroidobacter denitrificans—Identification of transformation products. Environ. Pollut. 2010, 158, 2572–2581. [Google Scholar] [CrossRef] [PubMed]
- Leu, Y.L.; Wang, P.H.; Shiao, M.S.; Ismail, W.; Chiang, Y.R. A novel testosterone catabolic pathway in bacteria. J. Bacteriol. 2011, 193, 4447–4455. [Google Scholar] [CrossRef] [PubMed][Green Version]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).