Activating Mutations in PTPN11 and KRAS in Canine Histiocytic Sarcomas
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. DNA Sample Acquisition
2.3. Methods for PTPN11 and KRAS Mutation Identification and Analysis
2.4. Statistical Methods
3. Results
3.1. Cases Characteristics
3.2. PTPN11 Mutation Status
3.3. KRAS Mutation Status
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bruner, S.R. Updates in therapeutics for veterinary dermatology. Vet. Clin. N. Am. Small Anim. Pract. 2006, 36, 39–58, vi. [Google Scholar] [CrossRef] [PubMed]
- Abadie, J.; Hedan, B.; Cadieu, E.; De Brito, C.; Devauchelle, P.; Bourgain, C.; Parker, H.G.; Vaysse, A.; Margaritte-Jeannin, P.; Galibert, F.; et al. Epidemiology, pathology, and genetics of histiocytic sarcoma in the Bernese mountain dog breed. J. Hered. 2009, 100 (Suppl. 1), S19–S27. [Google Scholar] [CrossRef] [PubMed]
- Dervisis, N.G.; Kiupel, M.; Qin, Q.; Cesario, L. Clinical prognostic factors in canine histiocytic sarcoma. Vet. Comp. Oncol. 2017, 15, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- Dobson, J.; Hoather, T.; McKinley, T.J.; Wood, J.L. Mortality in a cohort of flat-coated retrievers in the UK. Vet. Comp. Oncol. 2009, 7, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Fidel, J.; Schiller, I.; Hauser, B.; Jausi, Y.; Rohrer-Bley, C.; Roos, M.; Kaser-Hotz, B. Histiocytic sarcomas in flat-coated retrievers: A summary of 37 cases (November 1998-March 2005). Vet. Comp. Oncol. 2006, 4, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Affolter, V.K.; Moore, P.F. Localized and disseminated histiocytic sarcoma of dendritic cell origin in dogs. Vet. Pathol. 2002, 39, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Craig, L.E.; Julian, M.E.; Ferracone, J.D. The diagnosis and prognosis of synovial tumors in dogs: 35 cases. Vet. Pathol. 2002, 39, 66–73. [Google Scholar] [CrossRef]
- Skorupski, K.A.; Clifford, C.A.; Paoloni, M.C.; Lara-Garcia, A.; Barber, L.; Kent, M.S.; LeBlanc, A.K.; Sabhlok, A.; Mauldin, E.A.; Shofer, F.S.; et al. CCNU for the treatment of dogs with histiocytic sarcoma. J. Vet. Intern. Med. 2007, 21, 121–126. [Google Scholar] [CrossRef]
- Ruple, A.; Morley, P.S. Risk Factors Associated with Development of Histiocytic Sarcoma in Bernese Mountain Dogs. J. Vet. Intern. Med. 2016, 30, 1197–1203. [Google Scholar] [CrossRef]
- Klahn, S.L.; Kitchell, B.E.; Dervisis, N.G. Evaluation and comparison of outcomes in dogs with periarticular and nonperiarticular histiocytic sarcoma. J. Am. Vet. Med. Assoc. 2011, 239, 90–96. [Google Scholar] [CrossRef]
- Cannon, C.; Borgatti, A.; Henson, M.; Husbands, B. Evaluation of a combination chemotherapy protocol including lomustine and doxorubicin in canine histiocytic sarcoma. J. Small Anim. Pract. 2015, 56, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Rassnick, K.M.; Al-Sarraf, R.; Bailey, D.B.; Chretin, J.D.; Phillips, B.; Zwhalen, C.H. Phase II open-label study of single-agent hydroxyurea for treatment of mast cell tumours in dogs. Vet. Comp. Oncol. 2010, 8, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Hedan, B.; Thomas, R.; Motsinger-Reif, A.; Abadie, J.; Andre, C.; Cullen, J.; Breen, M. Molecular cytogenetic characterization of canine histiocytic sarcoma: A spontaneous model for human histiocytic cancer identifies deletion of tumor suppressor genes and highlights influence of genetic background on tumor behavior. BMC Cancer 2011, 11, 201. [Google Scholar] [CrossRef] [PubMed]
- Shearin, A.L.; Hedan, B.; Cadieu, E.; Erich, S.A.; Schmidt, E.V.; Faden, D.L.; Cullen, J.; Abadie, J.; Kwon, E.M.; Grone, A.; et al. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol. 2012, 21, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Hof, P.; Pluskey, S.; Dhe-Paganon, S.; Eck, M.J.; Shoelson, S.E. Crystal structure of the tyrosine phosphatase SHP-2. Cell 1998, 92, 441–450. [Google Scholar] [CrossRef]
- Tartaglia, M.; Niemeyer, C.M.; Fragale, A.; Song, X.; Buechner, J.; Jung, A.; Hahlen, K.; Hasle, H.; Licht, J.D.; Gelb, B.D. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat. Genet. 2003, 34, 148–150. [Google Scholar] [CrossRef] [PubMed]
- Loh, M.L.; Vattikuti, S.; Schubbert, S.; Reynolds, M.G.; Carlson, E.; Lieuw, K.H.; Cheng, J.W.; Lee, C.M.; Stokoe, D.; Bonifas, J.M.; et al. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood 2004, 103, 2325–2331. [Google Scholar] [CrossRef] [PubMed]
- Bentires-Alj, M.; Paez, J.G.; David, F.S.; Keilhack, H.; Halmos, B.; Naoki, K.; Maris, J.M.; Richardson, A.; Bardelli, A.; Sugarbaker, D.J.; et al. Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004, 64, 8816–8820. [Google Scholar] [CrossRef] [PubMed]
- Niihori, T.; Aoki, Y.; Ohashi, H.; Kurosawa, K.; Kondoh, T.; Ishikiriyama, S.; Kawame, H.; Kamasaki, H.; Yamanaka, T.; Takada, F.; et al. Functional analysis of PTPN11/SHP-2 mutants identified in Noonan syndrome and childhood leukemia. J. Hum. Genet. 2005, 50, 192–202. [Google Scholar] [CrossRef]
- Yu, W.M.; Daino, H.; Chen, J.; Bunting, K.D.; Qu, C.K. Effects of a leukemia-associated gain-of-function mutation of SHP-2 phosphatase on interleukin-3 signaling. J. Biol. Chem. 2006, 281, 5426–5434. [Google Scholar] [CrossRef] [PubMed]
- Frankson, R.; Yu, Z.H.; Bai, Y.; Li, Q.; Zhang, R.Y.; Zhang, Z.Y. Therapeutic Targeting of Oncogenic Tyrosine Phosphatases. Cancer Res. 2017, 77, 5701–5705. [Google Scholar] [CrossRef] [PubMed]
- Voruz, S.; Cairoli, A.; Naveiras, O.; de Leval, L.; Missiaglia, E.; Homicsko, K.; Michielin, O.; Blum, S. Response to MEK inhibition with trametinib and tyrosine kinase inhibition with imatinib in multifocal histiocytic sarcoma. Haematologica 2018, 103, e39–e41. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Tomaszewicz, K.; Hutchinson, L.; Hornick, J.L.; Woda, B.; Yu, H. Somatic mutations in histiocytic sarcoma identified by next generation sequencing. Virchows Arch. 2016, 469, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Batra, S.; Martin, S.C.; Nassiri, M.; Qureshi, A.; Markel, T.A. Histiocytic Sarcoma Associated with Coombs Negative Acute Hemolytic Anemia: A Rare Presentation. Case Rep. Oncol. Med. 2016, 2016, 3179147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shibani, A.; Sadikovic, B.; Howlett, C.J.; Ang, L.C. An aggressive multifocal primary CNS histiocytosis with PTPN11 (Shp2) mutation. Neuropathol. Appl. Neurobiol. 2018, 44, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, E.S.; World Health Organization. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues; IARC Press: Lyon, France, 2001; 351p. [Google Scholar]
- Jaffe, E.S. Histiocytoses of lymph nodes: biology and differential diagnosis. Semin. Diagn. Pathol. 1988, 5, 376–390. [Google Scholar] [PubMed]
- Vos, J.A.; Abbondanzo, S.L.; Barekman, C.L.; Andriko, J.W.; Miettinen, M.; Aguilera, N.S. Histiocytic sarcoma: a study of five cases including the histiocyte marker CD163. Mod. Pathol. 2005, 18, 693–704. [Google Scholar] [CrossRef]
- Kommalapati, A.; Tella, S.H.; Durkin, M.; Go, R.S.; Goyal, G. Histiocytic sarcoma: a population-based analysis of incidence, demographic disparities, and long-term outcomes. Blood 2018, 131, 265–268. [Google Scholar] [CrossRef]
- Thaiwong, T.; Sirivisoot, S.; Takada, M.; Yuzbasiyan-Gurkan, V.; Kiupel, M. Gain-of-function mutation in PTPN11 in histiocytic sarcomas of Bernese Mountain Dogs. Vet. Comp. Oncol. 2018, 16, 220–228. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Takada, M.; Hix, J.M.L.; Corner, S.; Schall, P.Z.; Kiupel, M.; Yuzbasiyan-Gurkan, V. Targeting MEK in a Translational Model of Histiocytic Sarcoma. Mol. Cancer Ther. 2018, 17, 2439–2450. [Google Scholar] [CrossRef] [PubMed]
- Wellman, M.L.; Krakowka, S.; Jacobs, R.M.; Kociba, G.J. A macrophage-monocyte cell line from a dog with malignant histiocytosis. Vitr. Cell. Dev. Biol. J. Tissue Cult. Assoc. 1988, 24, 223–229. [Google Scholar]
- Heinrich, F.; Contioso, V.B.; Stein, V.M.; Carlson, R.; Tipold, A.; Ulrich, R.; Puff, C.; Baumgartner, W.; Spitzbarth, I. Passage-dependent morphological and phenotypical changes of a canine histiocytic sarcoma cell line (DH82 cells). Vet. Immunol. Immunopathol. 2015, 163, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Takada, M.; Parys, M.; Gregory-Bryson, E.; Vilar Saavedra, P.; Kiupel, M.; Yuzbasiyan-Gurkan, V. A novel canine histiocytic sarcoma cell line: initial characterization and utilization for drug screening studies. BMC Cancer 2018, 18, 237. [Google Scholar] [CrossRef] [PubMed]
- Lowry, R. VassarStats: Website for Statistical Computation. Available online: http://vassarstats.net/index.html (accessed on 20 April 2018).
- Neel, B.G.; Gu, H.; Pao, L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem. Sci. 2003, 28, 284–293. [Google Scholar] [CrossRef]
- Qu, C.K.; Nguyen, S.; Chen, J.; Feng, G.S. Requirement of Shp-2 tyrosine phosphatase in lymphoid and hematopoietic cell development. Blood 2001, 97, 911–914. [Google Scholar] [CrossRef] [PubMed]
- Constantino-Casas, F.; Mayhew, D.; Hoather, T.M.; Dobson, J.M. The clinical presentation and histopathologic-immunohistochemical classification of histiocytic sarcomas in the Flat Coated Retriever. Vet. Pathol. 2011, 48, 764–771. [Google Scholar] [CrossRef]
- Dobson, J.M. Breed-predispositions to cancer in pedigree dogs. ISRN Vet. Sci. 2013, 2013, 941275. [Google Scholar] [CrossRef]
- Morris, J.S.; Bostock, D.E.; McInnes, E.F.; Hoather, T.M.; Dobson, J.M. Histopathological survey of neoplasms in flat-coated retrievers, 1990 to 1998. Vet. Rec. 2000, 147, 291–295. [Google Scholar] [CrossRef]
- Tartaglia, M.; Mehler, E.L.; Goldberg, R.; Zampino, G.; Brunner, H.G.; Kremer, H.; van der Burgt, I.; Crosby, A.H.; Ion, A.; Jeffery, S.; et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 2001, 29, 465–468. [Google Scholar] [CrossRef]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, V.; Griffin, G.K.; Jacobsen, E.D.; Fletcher, C.D.M.; Sholl, L.M.; Hornick, J.L. Identification of diverse activating mutations of the RAS-MAPK pathway in histiocytic sarcoma. Mod. Pathol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Diamond, E.L.; Durham, B.H.; Ulaner, G.A.; Drill, E.; Buthorn, J.; Ki, M.; Bitner, L.; Cho, H.; Young, R.J.; Francis, J.H.; et al. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature 2019, 567, 521–524. [Google Scholar] [CrossRef] [PubMed]
- Takada, M.; Smyth, L.A.; Hix, J.M.; Corner, S.M.; O’Reilly, S.; Kiupel, M.; Yuzbasiyan-Gurkan, V. Development of an Orthotopic Intrasplenic Xenograft Mouse Model of Canine Histiocytic Sarcoma and Its Use in Evaluating the Efficacy of Treatment with Dasatinib. Comp. Med. 2019. [Google Scholar] [CrossRef] [PubMed]
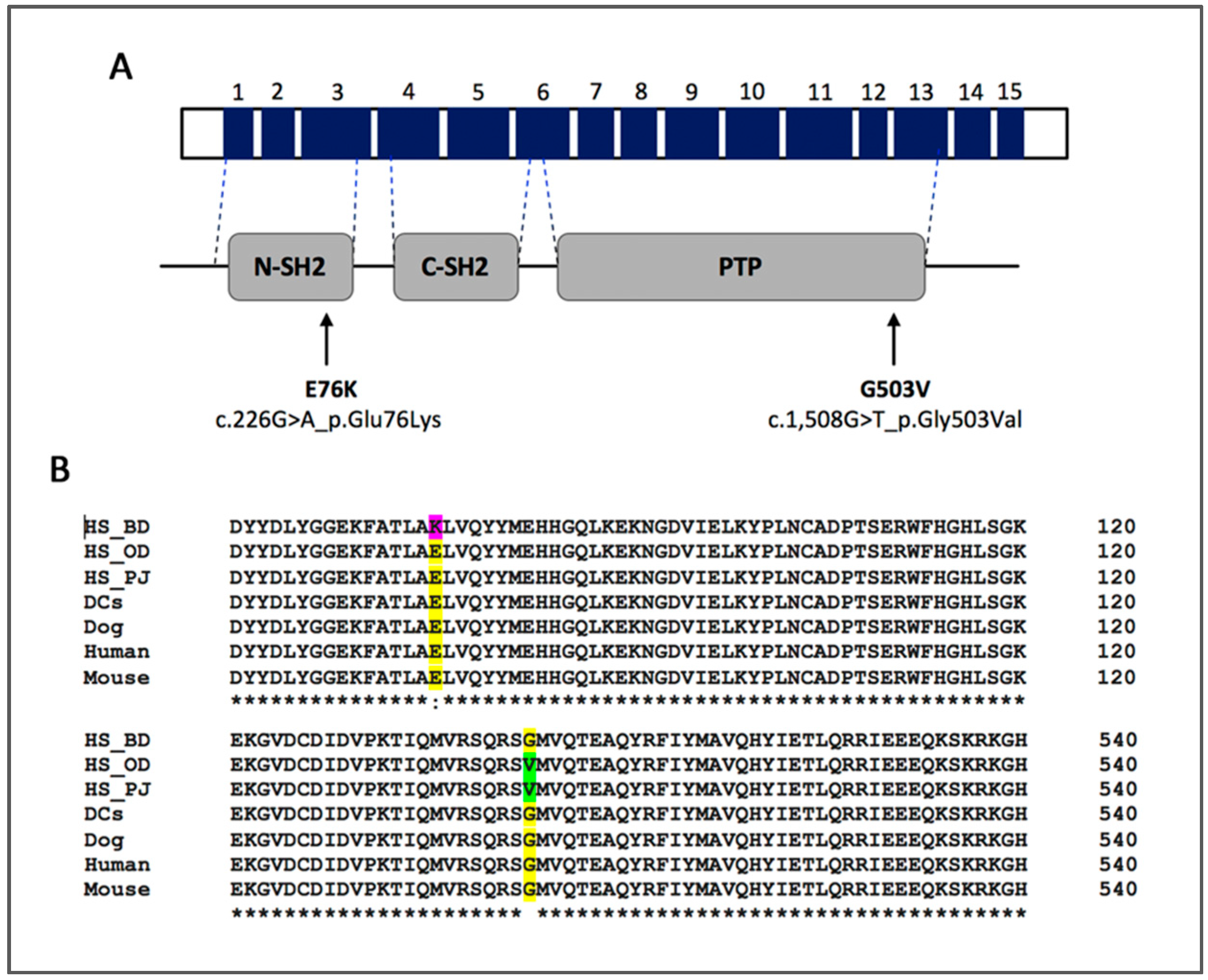
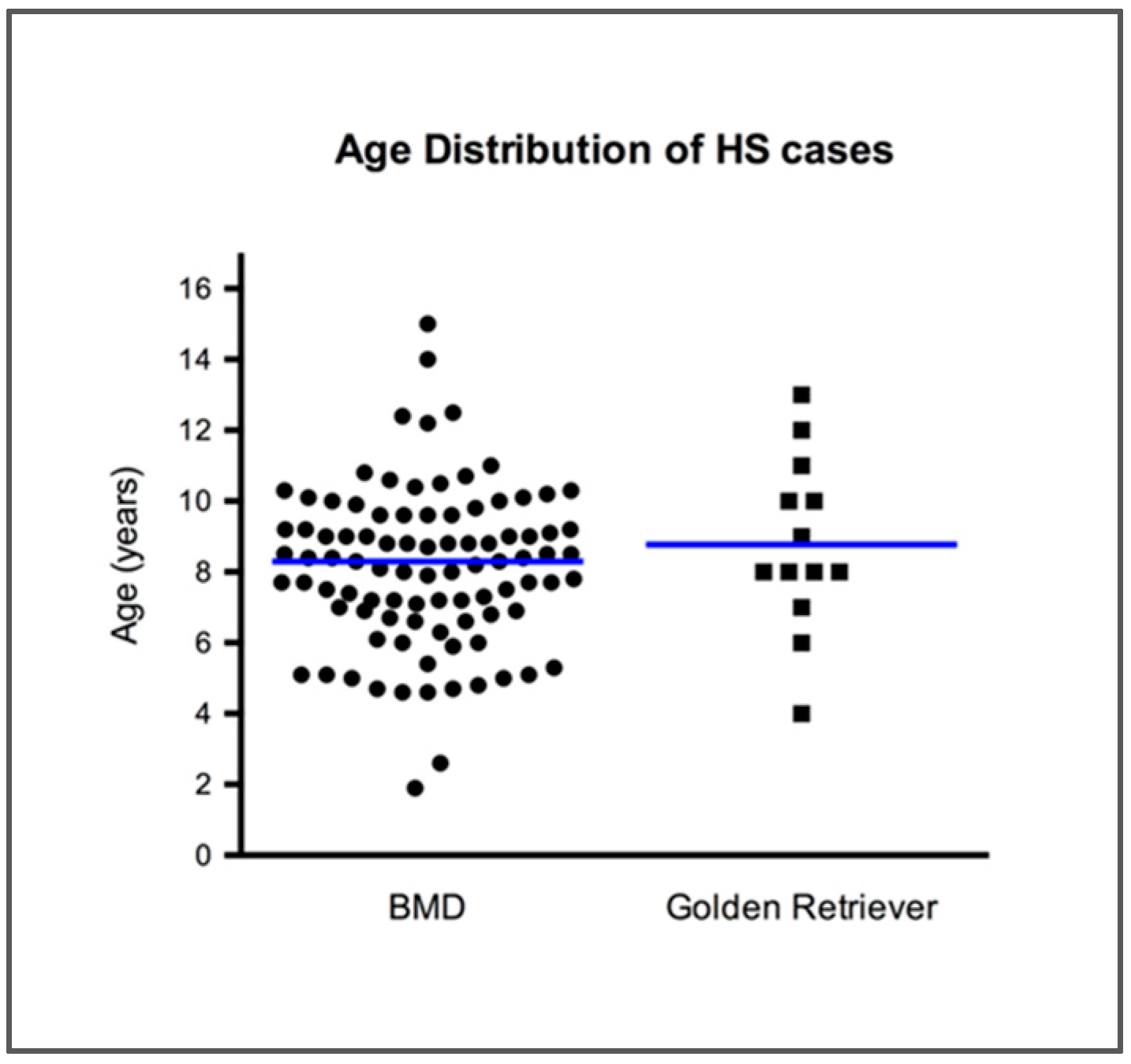
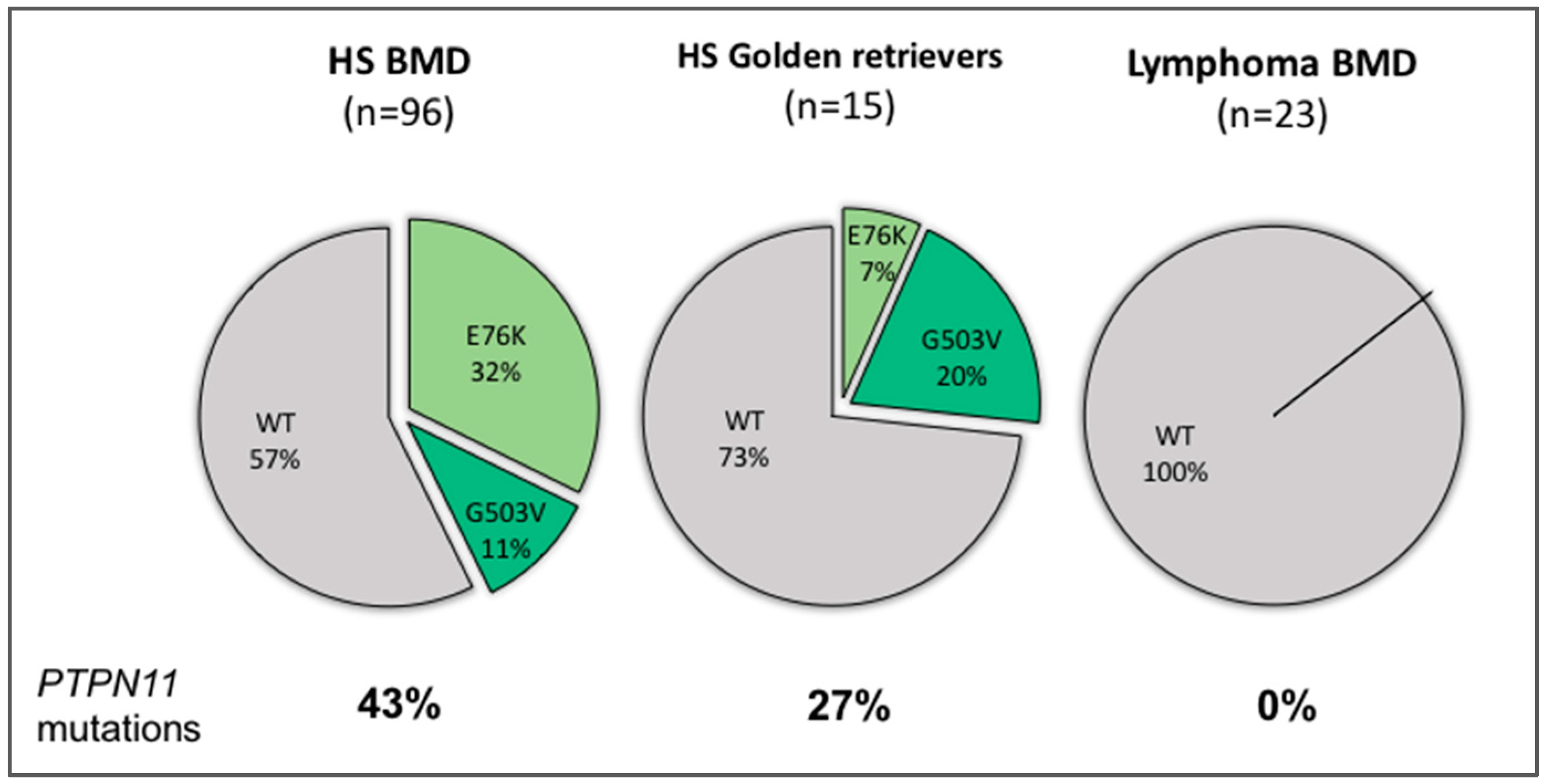
| Characteristics | N | PTPN11 Mutant | PTPN11 Wild-type | P | E76K | P | G503V | P |
|---|---|---|---|---|---|---|---|---|
| HS Bernese mountain dogs | 96 | 41 (43%) | 55 (57%) | 31 (32%) | 10 (10%) | |||
| Age | ||||||||
| <5.5 years | 14 | 6 (43%) | 8 (57%) | NS | 3 (21%) | NS | 3 (21%) | NS |
| 5.5–10 years | 60 | 19 (32%) | 41 (68%) | 16 (27%) | 3 (5%) | |||
| >10 years | 18 | 11 (61%) | 7 (39%) | 0.03 | 8 (44%) | 3 (17%) | ||
| Unknown age | ||||||||
| Sex | ||||||||
| Female | 43 | 17 (39%) | 26 (60%) | NS | 13 (30%) | NS | 4 (9%) | NS |
| Male | 50 | 21 (42%) | 29 (58%) | 15 (30%) | 6 (12%) | |||
| HS golden retrievers | 13 | 3 (23%) | 10 (77%) | NS | 2 (15%) | NS | 1 (8%) | NS |
| Lymphoma BMDs | 23 | 0 | 23 (100%) | 0.0001 | - | - | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takada, M.; Smyth, L.A.; Thaiwong, T.; Richter, M.; Corner, S.M.; Schall, P.Z.; Kiupel, M.; Yuzbasiyan-Gurkan, V. Activating Mutations in PTPN11 and KRAS in Canine Histiocytic Sarcomas. Genes 2019, 10, 505. https://doi.org/10.3390/genes10070505
Takada M, Smyth LA, Thaiwong T, Richter M, Corner SM, Schall PZ, Kiupel M, Yuzbasiyan-Gurkan V. Activating Mutations in PTPN11 and KRAS in Canine Histiocytic Sarcomas. Genes. 2019; 10(7):505. https://doi.org/10.3390/genes10070505
Chicago/Turabian StyleTakada, Marilia, Lauren A. Smyth, Tuddow Thaiwong, Marlee Richter, Sarah M. Corner, Peter Z. Schall, Matti Kiupel, and Vilma Yuzbasiyan-Gurkan. 2019. "Activating Mutations in PTPN11 and KRAS in Canine Histiocytic Sarcomas" Genes 10, no. 7: 505. https://doi.org/10.3390/genes10070505
APA StyleTakada, M., Smyth, L. A., Thaiwong, T., Richter, M., Corner, S. M., Schall, P. Z., Kiupel, M., & Yuzbasiyan-Gurkan, V. (2019). Activating Mutations in PTPN11 and KRAS in Canine Histiocytic Sarcomas. Genes, 10(7), 505. https://doi.org/10.3390/genes10070505





