An Evolutionary Perspective of Dopachrome Tautomerase Enzymes in Metazoans
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieval and Processing of Sequence Datasets
2.2. Sequence Identification
2.3. Sequence Alignment and Phylogenetic Analysis
2.4. Re-Annotation of the C. gigas DCE/Yellow Gene Using RNA-Seq Data
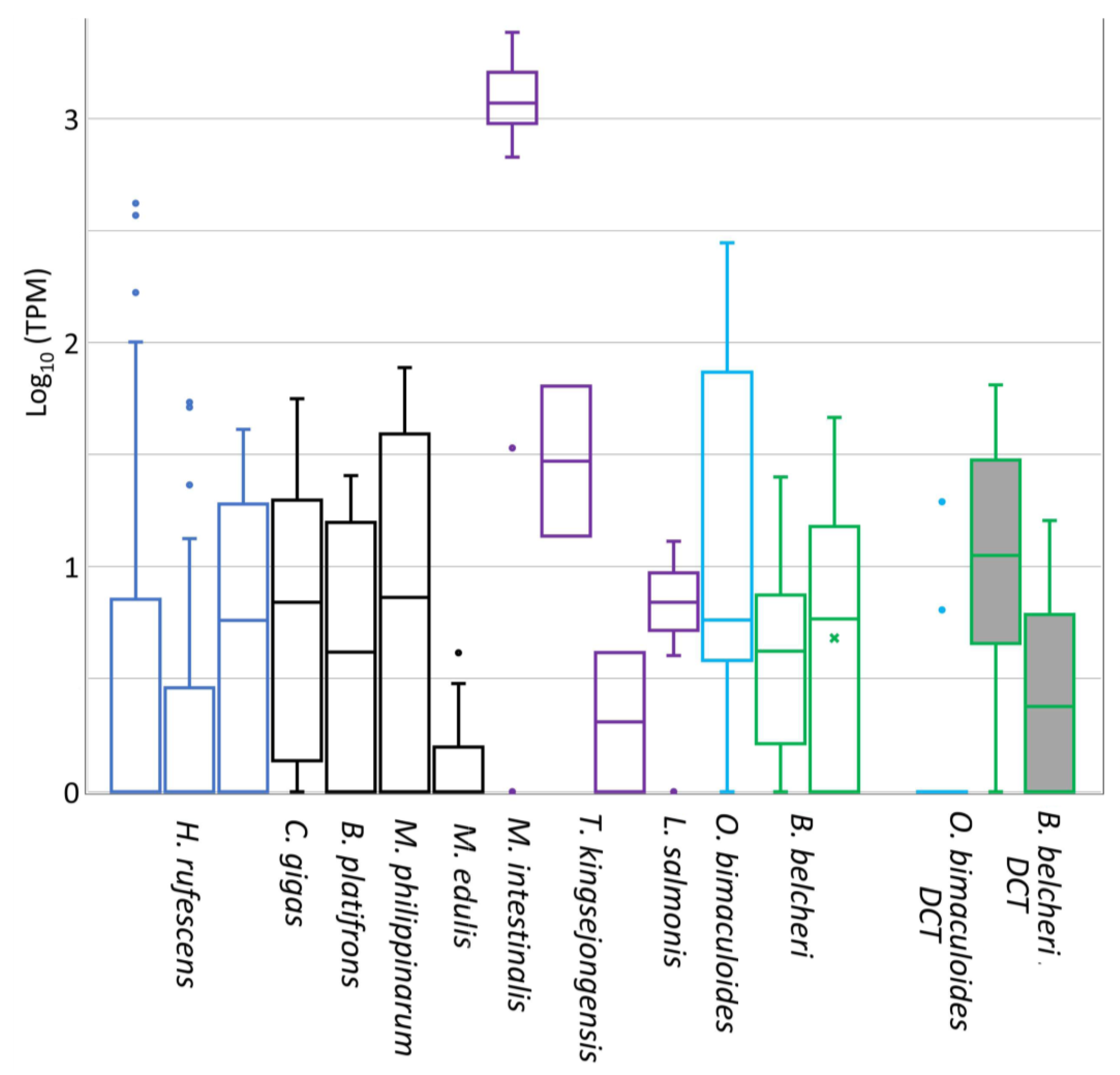
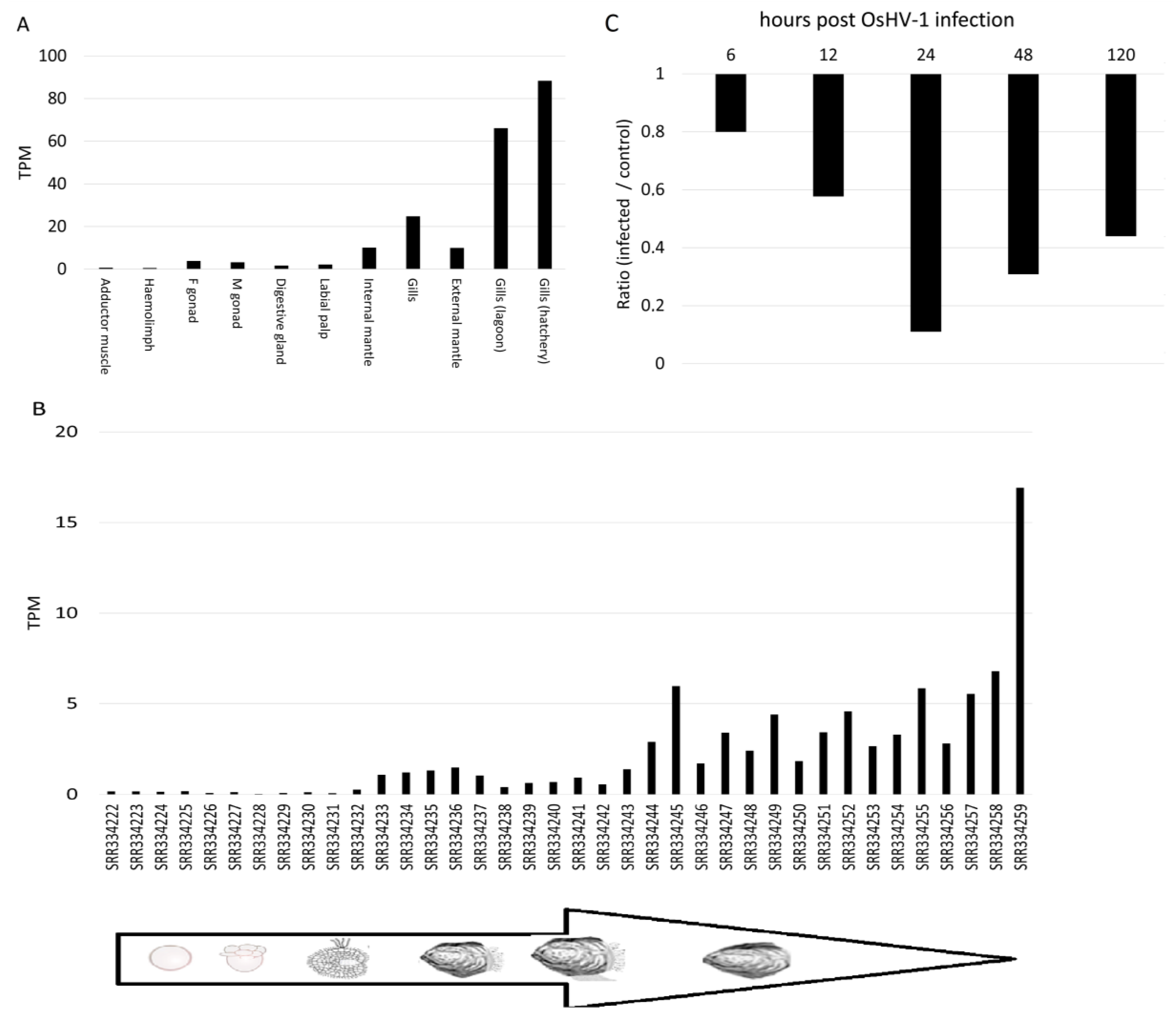
2.5. Gene Expression Analysis of DCE/Yellow Genes
2.6. Structural Analysis
3. Results
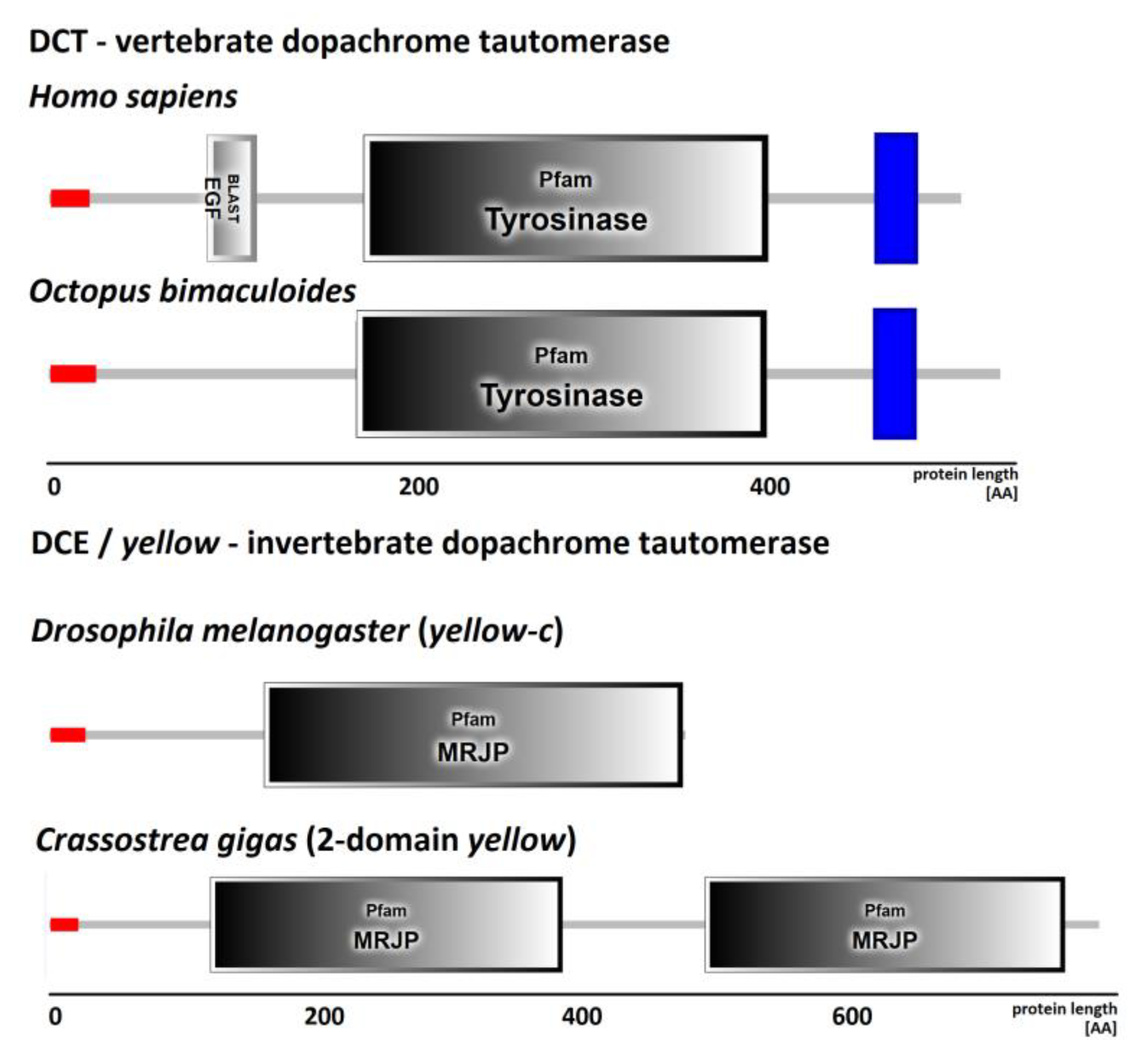
3.1. Taxonomic Distribution of Dopachrome Tautomerase Enzymes
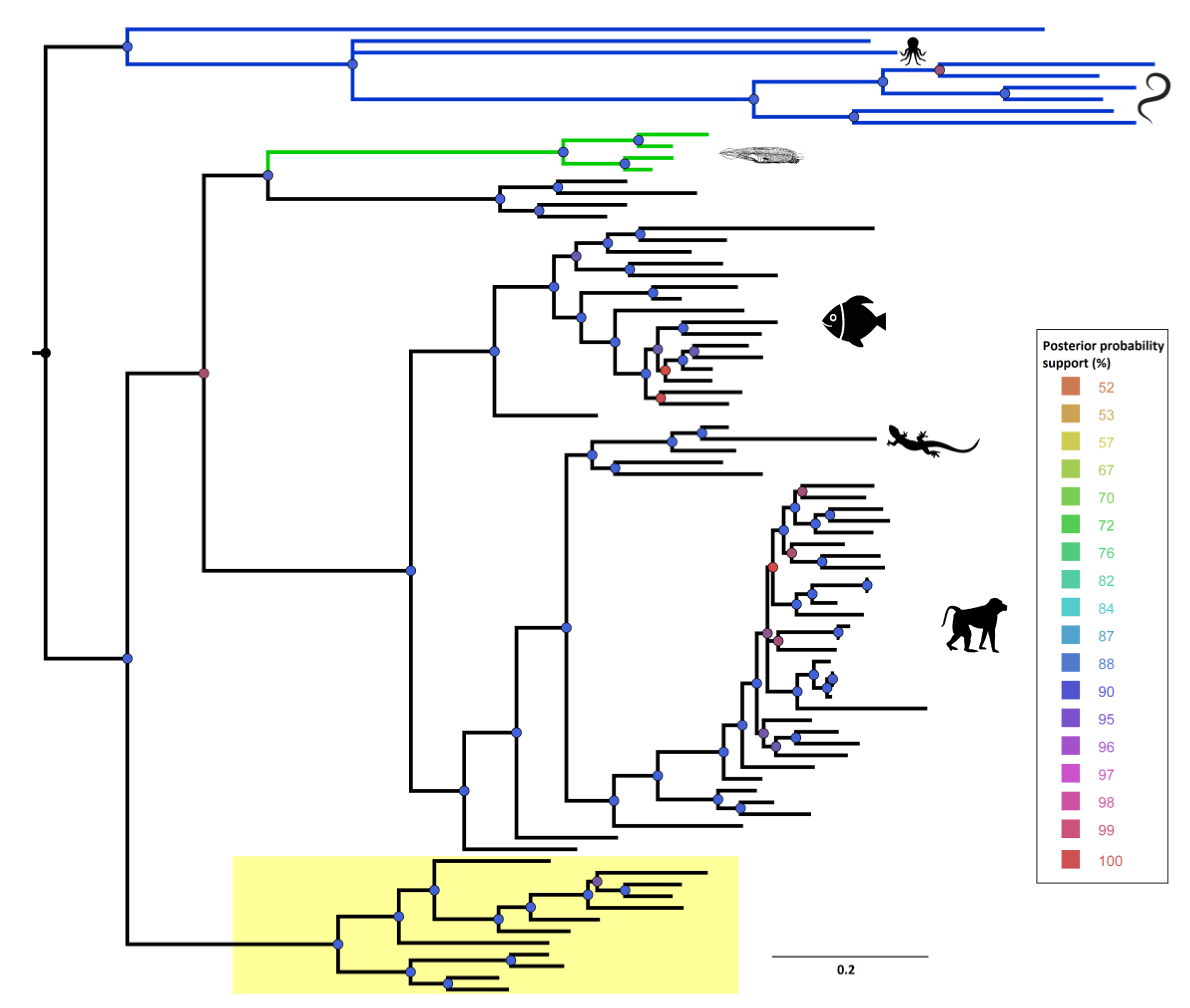
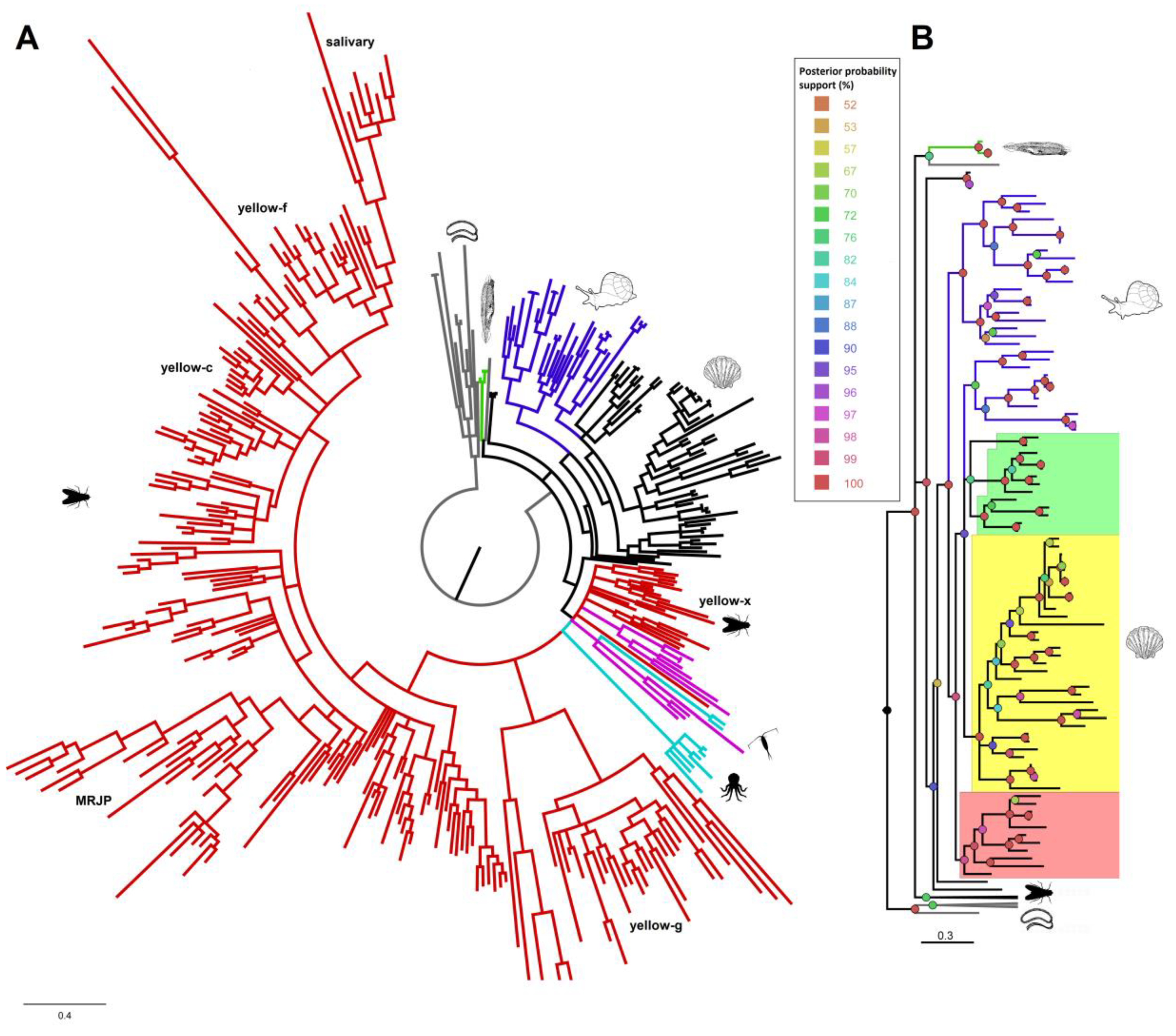
3.2. Phylogenetic Analysis of DCT and DCE/yellow Proteins
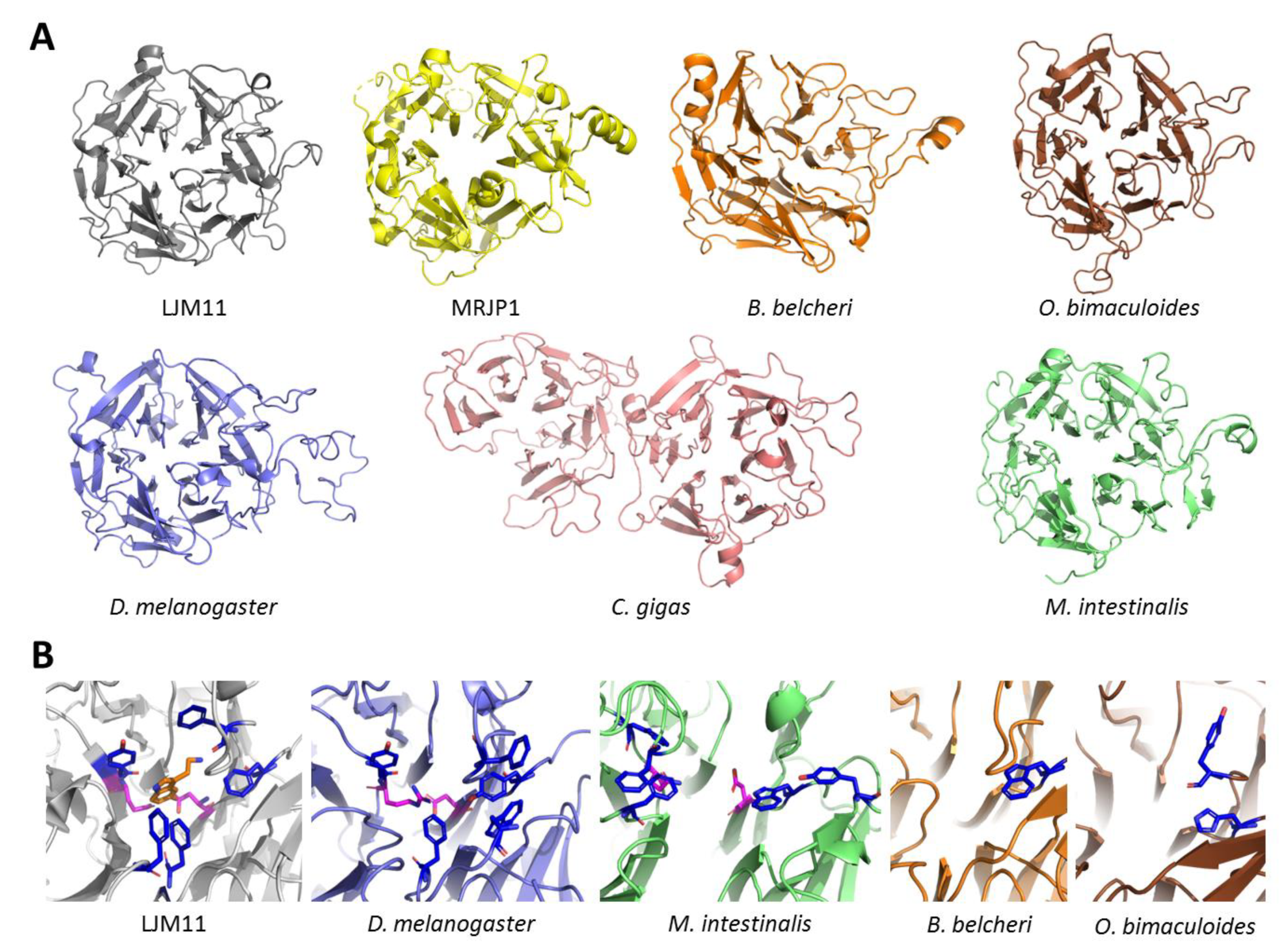
3.3. Structure of DCE/Yellow Proteins
3.4. Expression of DCE/Yellow Genes in Non-Insect Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Buchmann, K. Evolution of Innate Immunity: Clues from Invertebrates via Fish to Mammals. Front. Immunol. 2014, 5, 459. [Google Scholar] [CrossRef] [PubMed]
- Hillyer, J.F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 2016, 58, 102–118. [Google Scholar] [CrossRef] [PubMed]
- Dubovskiy, I.; Kryukova, N.; Glupov, V.; Ratcliffe, N. Encapsulation and nodulation in insects. Invertebr. Surviv. J. 2016, 13, 229–246. [Google Scholar]
- Cerenius, L.; Lee, B.L.; Söderhäll, K. The proPO-system: Pros and cons for its role in invertebrate immunity. Trends Immunol. 2008, 29, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Söderhäll, K.; Cerenius, L. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr. Opin. Immunol. 1998, 10, 23–28. [Google Scholar] [CrossRef]
- Solano, F. Melanins: Skin Pigments and Much More—Types, Structural Models, Biological Functions, and Formation Routes. Available online: https://www.hindawi.com/journals/njos/2014/498276/ (accessed on 19 March 2019).
- Cerenius, L.; Jiravanichpaisal, P.; Liu, H.-P.; Söderhill, I. Crustacean immunity. Adv. Exp. Med. Biol. 2010, 708, 239–259. [Google Scholar] [PubMed]
- Coaglio, A.L.; Ferreira, M.A.; Dos Santos Lima, W.; de Jesus Pereira, C.A. Identification of a phenoloxidase- and melanin-dependent defence mechanism in Achatina fulica infected with Angiostrongylus vasorum. Parasites Vectors 2018, 11, 113. [Google Scholar] [CrossRef]
- Allam, B.; Raftos, D. Immune responses to infectious diseases in bivalves. J. Invertebr. Pathol. 2015, 131, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xie, L.; Huang, J.; Chen, L.; Zhang, R. A novel putative tyrosinase involved in periostracum formation from the pearl oyster (Pinctada fucata). Biochem. Biophys. Res. Commun. 2006, 342, 632–639. [Google Scholar] [CrossRef]
- Allam, B.; Pales Espinosa, E.; Tanguy, A.; Jeffroy, F.; Le Bris, C.; Paillard, C. Transcriptional changes in Manila clam (Ruditapes philippinarum) in response to Brown Ring Disease. Fish Shellfish Immunol. 2014, 41, 2–11. [Google Scholar] [CrossRef]
- Bjärnmark, N.A.; Yarra, T.; Churcher, A.M.; Felix, R.C.; Clark, M.S.; Power, D.M. Transcriptomics provides insight into Mytilus galloprovincialis (Mollusca: Bivalvia) mantle function and its role in biomineralisation. Mar. Genom. 2016, 27, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Ivanina, A.V.; Falfushynska, H.I.; Beniash, E.; Piontkivska, H.; Sokolova, I.M. Biomineralization-related specialization of hemocytes and mantle tissues of the Pacific oyster Crassostrea gigas. J. Exp. Biol. 2017, 220, 3209–3221. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.B.; Fogel, N.S.; Lambert, J.D. Growth and morphogenesis of the gastropod shell. Proc. Natl. Acad. Sci. USA 2019, 116, 6878–6883. [Google Scholar] [CrossRef] [PubMed]
- Sugumaran, M. Reactivities of Quinone Methides versus o-Quinones in Catecholamine Metabolism and Eumelanin Biosynthesis. Int. J. Mol. Sci. 2016, 17, 1576. [Google Scholar] [CrossRef] [PubMed]
- D’Mello, S.A.N.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling Pathways in Melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef] [PubMed]
- Sugumaran, M.; Barek, H. Critical Analysis of the Melanogenic Pathway in Insects and Higher Animals. Int. J. Mol. Sci. 2016, 17, 1753. [Google Scholar] [CrossRef]
- Singh, S.; Malhotra, A.G.; Pandey, A.; Pandey, K.M. Computational model for pathway reconstruction to unravel the evolutionary significance of melanin synthesis. Bioinformation 2013, 9, 94–100. [Google Scholar] [CrossRef]
- Körner, A.M.; Pawelek, J. Dopachrome conversion: A possible control point in melanin biosynthesis. J. Investig. Dermatol. 1980, 75, 192–195. [Google Scholar] [CrossRef]
- Aksoy, P.; Meneses, P.I. The Role of DCT in HPV16 Infection of HaCaTs. PLoS ONE 2017, 12, e0170158. [Google Scholar] [CrossRef]
- Popa, I.L.; Milac, A.L.; Sima, L.E.; Alexandru, P.R.; Pastrama, F.; Munteanu, C.V.A.; Negroiu, G. Cross-talk between Dopachrome Tautomerase and Caveolin-1 Is Melanoma Cell Phenotype-specific and Potentially Involved in Tumor Progression. J. Biol. Chem. 2016, 291, 12481–12500. [Google Scholar] [CrossRef]
- Ainger, S.A.; Yong, X.L.; Wong, S.S.; Skalamera, D.; Gabrielli, B.; Leonard, J.H.; Sturm, R.A. DCT protects human melanocytic cells from UVR and ROS damage and increases cell viability. Exp. Dermatol. 2014, 23, 916–921. [Google Scholar] [CrossRef] [PubMed]
- Aso, Y.; Kramer, K.J.; Hopkins, T.L.; Whetzel, S.Z. Properties of tyrosinase and dopa quinone imine conversion factor from pharate pupal cuticle of Manduca sexta L. Insect Biochem. 1984, 14, 463–472. [Google Scholar] [CrossRef]
- Nash, W.G. Patterns of pigmentation color states regulated by the y locus in Drosophila melanogaster. Dev. Biol. 1976, 48, 336–343. [Google Scholar] [CrossRef]
- Geyer, P.K.; Spana, C.; Corces, V.G. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. EMBO J. 1986, 5, 2657–2662. [Google Scholar] [CrossRef] [PubMed]
- Albert, S.; Klaudiny, J. The MRJP/YELLOW protein family of Apis mellifera: Identification of new members in the EST library. J. Insect Physiol. 2004, 50, 51–59. [Google Scholar] [CrossRef]
- Han, Q.; Fang, J.; Ding, H.; Johnson, J.K.; Christensen, B.M.; Li, J. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem. J. 2002, 368, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, J.M.C.; Mans, B.J.; Arcà, B. An insight into the sialome of blood-feeding Nematocera. Insect Biochem. Mol. Biol. 2010, 40, 767–784. [Google Scholar] [CrossRef]
- Buttstedt, A.; Moritz, R.F.A.; Erler, S. Origin and function of the major royal jelly proteins of the honeybee (Apis mellifera) as members of the yellow gene family. Biol. Rev. Camb. Philos. Soc. 2014, 89, 255–269. [Google Scholar] [CrossRef]
- Vezeteu, T.V.; Bobiş, O.; Moritz, R.F.A.; Buttstedt, A. Food to some, poison to others—Honeybee royal jelly and its growth inhibiting effect on European Foulbrood bacteria. Microbiologyopen 2017, 6, e00397. [Google Scholar] [CrossRef]
- Buttstedt, A.; Moritz, R.F.; Erler, S. More than royal food—Major royal jelly protein genes in sexuals and workers of the honeybee Apis mellifera. Front. Zool. 2013, 10, 72. [Google Scholar] [CrossRef]
- Ferguson, L.C.; Green, J.; Surridge, A.; Jiggins, C.D. Evolution of the insect yellow gene family. Mol. Biol. Evol. 2011, 28, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Heavner, M.E.; Gueguen, G.; Rajwani, R.; Pagan, P.E.; Small, C.; Govind, S. Partial venom gland transcriptome of a Drosophila parasitoid wasp, Leptopilina heterotoma, reveals novel and shared bioactive profiles with stinging Hymenoptera. Gene 2013, 526, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Rohoušová, I.; Subrahmanyam, S.; Volfová, V.; Mu, J.; Volf, P.; Valenzuela, J.G.; Jochim, R.C. Salivary gland transcriptomes and proteomes of Phlebotomus tobbi and Phlebotomus sergenti, vectors of leishmaniasis. PLoS Negl. Trop. Dis. 2012, 6, e1660. [Google Scholar] [CrossRef] [PubMed]
- Vlkova, M.; Sima, M.; Rohousova, I.; Kostalova, T.; Sumova, P.; Volfova, V.; Jaske, E.L.; Barbian, K.D.; Gebre-Michael, T.; Hailu, A.; et al. Comparative analysis of salivary gland transcriptomes of Phlebotomus orientalis sand flies from endemic and non-endemic foci of visceral leishmaniasis. PLoS Negl. Trop. Dis. 2014, 8, e2709. [Google Scholar] [CrossRef] [PubMed]
- Gretscher, R.R.; Streicher, P.E.; Strauß, A.S.; Wielsch, N.; Stock, M.; Wang, D.; Boland, W.; Burse, A. A common theme in extracellular fluids of beetles: Extracellular superoxide dismutases crucial for balancing ROS in response to microbial challenge. Sci. Rep. 2016, 6, 24082. [Google Scholar] [CrossRef] [PubMed]
- Vicente, V.A.; Weiss, V.A.; Bombassaro, A.; Moreno, L.F.; Costa, F.F.; Raittz, R.T.; Leão, A.C.; Gomes, R.R.; Bocca, A.L.; Fornari, G.; et al. Comparative Genomics of Sibling Species of Fonsecaea Associated with Human Chromoblastomycosis. Front. Microbiol. 2017, 8, 1924. [Google Scholar] [CrossRef]
- Luna-Acosta, A.; Breitwieser, M.; Renault, T.; Thomas-Guyon, H. Recent findings on phenoloxidases in bivalves. Mar. Pollut. Bull. 2017, 122, 5–16. [Google Scholar] [CrossRef]
- Masonbrink, R.E.; Purcell, C.M.; Boles, S.E.; Whitehead, A.; Hyde, J.R.; Seetharam, A.S.; Severin, A.J. An Annotated Genome for Haliotis rufescens (Red Abalone) and Resequenced Green, Pink, Pinto, Black, and White Abalone Species. Genome Biol. Evol. 2019, 11, 431–438. [Google Scholar] [CrossRef]
- Luo, Y.-J.; Takeuchi, T.; Koyanagi, R.; Yamada, L.; Kanda, M.; Khalturina, M.; Fujie, M.; Yamasaki, S.; Endo, K.; Satoh, N. The Lingula genome provides insights into brachiopod evolution and the origin of phosphate biomineralization. Nat. Commun. 2015, 6, 8301. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, Y.; Xu, T.; Zhang, Y.; Mu, H.; Zhang, Y.; Lan, Y.; Fields, C.J.; Hui, J.H.L.; Zhang, W.; et al. Adaptation to deep-sea chemosynthetic environments as revealed by mussel genomes. Nat. Ecol. Evol. 2017, 1, 0121. [Google Scholar] [CrossRef]
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Fan, G.; Jiao, Y.; Zhang, H.; Guo, X.; Huang, R.; Zheng, Z.; Bian, C.; Deng, Y.; Wang, Q.; et al. The pearl oyster Pinctada fucata martensii genome and multi-omic analyses provide insights into biomineralization. Gigascience 2017, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.; Kim, Y.-J.; Markkandan, K.; Shin, W.; Oh, S.; Woo, J.; Yoo, J.; An, H.; Han, K. The Whole-Genome and Transcriptome of the Manila Clam (Ruditapes philippinarum). Genome Biol. Evol. 2017, 9, 1487–1498. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.; Subramanian, S.; Suwansa-Ard, S.; Zhao, M.; O’Connor, W.; Raftos, D.; Elizur, A. The genome of the oyster Saccostrea offers insight into the environmental resilience of bivalves. DNA Res. 2018, 25, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, J.; Jiao, W.; Li, J.; Xun, X.; Sun, Y.; Guo, X.; Huan, P.; Dong, B.; Zhang, L.; et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 2017, 1, 0120. [Google Scholar] [CrossRef] [PubMed]
- Uliano-Silva, M.; Dondero, F.; Dan Otto, T.; Costa, I.; Lima, N.C.B.; Americo, J.A.; Mazzoni, C.J.; Prosdocimi, F.; Rebelo, M.D. A hybrid-hierarchical genome assembly strategy to sequence the invasive golden mussel Limnoperna fortunei. Gigascience 2017, 7, gix128. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.-M.; Lu-Sheng, X.; Umberto, R.; Biao, W.; Qing-Chen, W.; Xiao-Ke, D.; Zhi-Hong, L.; Chong-Ming, W. Supporting Data for “Chromosomal-Level Assembly of the Blood Clam, Scapharca (Anadara) Broughtonii, Using Long Sequence Reads and Hi-C”; GigaScience|Oxford Academic: Oxford, UK, 2019. [Google Scholar]
- Kang, S.; Ahn, D.-H.; Lee, J.H.; Lee, S.G.; Shin, S.C.; Lee, J.; Min, G.-S.; Lee, H.; Kim, H.-W.; Kim, S.; et al. The genome of the Antarctic-endemic copepod, Tigriopus kingsejongensis. Gigascience 2017, 6, giw010. [Google Scholar]
- Robb, S.M.C.; Gotting, K.; Ross, E.; Sánchez Alvarado, A. SmedGD 2.0: The Schmidtea mediterranea genome database. Genesis 2015, 53, 535–546. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Penn, O.; Privman, E.; Landan, G.; Graur, D.; Pupko, T. An alignment confidence score capturing robustness to guide tree uncertainty. Mol. Biol. Evol. 2010, 27, 1759–1767. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Jouaux, A.; Ford, S.E.; Lelong, C.; Sourdaine, P.; Mathieu, M.; Guo, X. Transcriptome analysis reveals strong and complex antiviral response in a mollusc. Fish Shellfish Immunol. 2015, 46, 131–144. [Google Scholar] [CrossRef]
- Feis, M.E.; John, U.; Lokmer, A.; Luttikhuizen, P.C.; Wegner, K.M. Dual transcriptomics reveals co-evolutionary mechanisms of intestinal parasite infections in blue mussels Mytilus edulis. Mol. Ecol. 2018, 27, 1505–1519. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. A model based criterion for gene expression calls using RNA-seq data. Theory Biosci. 2013, 132, 159–164. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Chen, X.; Cao, Y.; Zhan, S.; Zhang, Y.; Tan, A.; Huang, Y. Identification of yellow gene family in Agrotis ipsilon and functional analysis of Aiyellow-y by CRISPR/Cas9. Insect Biochem. Mol. Biol. 2018, 94, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Oliveira, F.; Chang, B.W.; Collin, N.; Gomes, R.; Teixeira, C.; Reynoso, D.; my Pham, V.; Elnaiem, D.E.; Kamhawi, S.; et al. Structure and Function of a “Yellow” Protein from Saliva of the Sand Fly Lutzomyia longipalpis That Confers Protective Immunity against Leishmania major Infection. J. Biol. Chem. 2011, 286, 32383–32393. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Li, M.; Guo, H.; Peng, W.; Xue, X.; Hu, Y.; Liu, Y.; Zhao, Y.; Fang, X.; Wang, K.; et al. Architecture of the native major royal jelly protein 1 oligomer. Nat. Commun. 2018, 9, 3373. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, J.; Bredeston, L.; Malanga, G.; Mordoh, J. Role of melanin as a scavenger of active oxygen species. Pigment Cell Res. 1993, 6, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Lindsay-Mosher, N.; Pearson, B.J. The true colours of the flatworm: Mechanisms of pigment biosynthesis and pigment cell lineage development in planarians. Semin. Cell Dev. Biol. 2019, 87, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Waite, J.H.; Wilbur, K.M. Phenoloxidase in the periostracum of the marine bivalve Modiolus demissus dillwyn. J. Exp. Zool. 1976, 195, 359–367. [Google Scholar] [CrossRef]
- Husnik, F.; McCutcheon, J.P. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 2018, 16, 67–79. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Glöckner, G.; Hülsmann, N.; Schleicher, M.; Noegel, A.A.; Eichinger, L.; Gallinger, C.; Pawlowski, J.; Sierra, R.; Euteneuer, U.; Pillet, L.; et al. The genome of the foraminiferan Reticulomyxa filosa. Curr. Biol. 2014, 24, 11–18. [Google Scholar] [CrossRef]
- Merk, M.; Mitchell, R.A.; Endres, S.; Bucala, R. D-dopachrome tautomerase (D-DT or MIF-2): Doubling the MIF cytokine family. Cytokine 2012, 59, 10–17. [Google Scholar] [CrossRef]
- Rosengren, E.; Bucala, R.; Aman, P.; Jacobsson, L.; Odh, G.; Metz, C.N.; Rorsman, H. The immunoregulatory mediator macrophage migration inhibitory factor (MIF) catalyzes a tautomerization reaction. Mol. Med. 1996, 2, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Han, B.; Feng, M.; Fang, Y.; Zhang, L.; Hu, H.; Hao, Y.; Qi, Y.; Zhang, X.; Li, J. Functional and Proteomic Investigations Reveal Major Royal Jelly Protein 1 Associated with Anti-hypertension Activity in Mouse Vascular Smooth Muscle Cells. Sci. Rep. 2016, 6, 30230. [Google Scholar] [CrossRef] [PubMed]
- Rosani, U.; Domeneghetti, S.; Gerdol, M.; Pallavicini, A.; Venier, P. Expansion and loss events characterized the occurrence of MIF-like genes in bivalves. FSIM 2019. submitted for publication. [Google Scholar]
- Sparkes, A.; De Baetselier, P.; Roelants, K.; De Trez, C.; Magez, S.; Van Ginderachter, J.A.; Raes, G.; Bucala, R.; Stijlemans, B. Reprint of: The non-mammalian MIF superfamily. Immunobiology 2017, 222, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, B.; Del Marmol, V.; Jackson, I.J.; Cherif, D.; Dubertret, L. Molecular characterization of a human tyrosinase-related-protein-2 cDNA. Patterns of expression in melanocytic cells. Eur. J. Biochem. 1994, 219, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Lupetti, R.; Pisarra, P.; Verrecchia, A.; Farina, C.; Nicolini, G.; Anichini, A.; Bordignon, C.; Sensi, M.; Parmiani, G.; Traversari, C. Translation of a retained intron in tyrosinase-related protein (TRP) 2 mRNA generates a new cytotoxic T lymphocyte (CTL)-defined and shared human melanoma antigen not expressed in normal cells of the melanocytic lineage. J. Exp. Med. 1998, 188, 1005–1016. [Google Scholar] [CrossRef]
- Suzuki, T.K.; Koshikawa, S.; Kobayashi, I.; Uchino, K.; Sezutsu, H. Modular cis-regulatory logic of yellow gene expression in silkmoth larvae. Insect Mol. Biol. 2019. [Google Scholar] [CrossRef]
| Group | Phylum | Class | DCT | DCE (Gene) | DCE (Transcript) |
|---|---|---|---|---|---|
| Lophotrochozoa | Mollusca | Bivalvia | no | X | X |
| Gastropoda | no | X | X | ||
| Cephalopoda | X | X | X | ||
| Annelida | no | no | nt | ||
| Nemertea | Pilidiophora | no | no | no | |
| Brachiopoda | Lingulata | no | no | nt | |
| Phoronida | no | no | no | ||
| Rotifera | Bdelloidea | no | no | nt | |
| Ecdysozoa | Nematoda | \ | X * | no | nt |
| Arthropoda | Malacostraca | no | no | nt | |
| Hexanauplia | no | X | X | ||
| Collembola | no | X | nt | ||
| Insecta | no | X | nt | ||
| Platyhelminthes | X | no | nt | ||
| Deuterostomia | Chordata | Ascidiacea | X | no | nt |
| Branchiostomidae | X | X | nt | ||
| Craniata | X | no | nt |
| L. longipalpis | D. melanogaster | M. intestinalis | O. bimaculoides | B. belcheri | A. variabilis |
|---|---|---|---|---|---|
| Tyr-90 | Tyr-143 | Phe-127 | --- | --- | --- |
| Phe-178 | Trp-265 | Phe-151 | --- | --- | --- |
| Phe-223 | Phe-270 | Phe-193 | --- | --- | --- |
| Phe-325 | Tyr-378 | Trp-366 | His-333 | Trp-344 | Trp-291 |
| Phe-344 | Phe-400 | Tyr-245 | Tyr-352 | --- | --- |
| Gln-91 | --- | Gln-129 | --- | --- | --- |
| Asp-328 | Asp-381 | Asp-368 | --- | --- | --- |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosani, U.; Domeneghetti, S.; Maso, L.; Wegner, K.M.; Venier, P. An Evolutionary Perspective of Dopachrome Tautomerase Enzymes in Metazoans. Genes 2019, 10, 495. https://doi.org/10.3390/genes10070495
Rosani U, Domeneghetti S, Maso L, Wegner KM, Venier P. An Evolutionary Perspective of Dopachrome Tautomerase Enzymes in Metazoans. Genes. 2019; 10(7):495. https://doi.org/10.3390/genes10070495
Chicago/Turabian StyleRosani, Umberto, Stefania Domeneghetti, Lorenzo Maso, K. Mathias Wegner, and Paola Venier. 2019. "An Evolutionary Perspective of Dopachrome Tautomerase Enzymes in Metazoans" Genes 10, no. 7: 495. https://doi.org/10.3390/genes10070495
APA StyleRosani, U., Domeneghetti, S., Maso, L., Wegner, K. M., & Venier, P. (2019). An Evolutionary Perspective of Dopachrome Tautomerase Enzymes in Metazoans. Genes, 10(7), 495. https://doi.org/10.3390/genes10070495






