Abstract
The anaerobic degradation of benzoate in bacteria involves the benzoyl-CoA central pathway. Azoarcus/Aromatoleum strains are a major group of anaerobic benzoate degraders, and the transcriptional regulation of the bzd genes was extensively studied in Azoarcus sp. CIB. In this work, we show that the bzdR regulatory gene and the PN promoter can also be identified upstream of the catabolic bzd operon in all benzoate-degrader Azoarcus/Aromatoleum strains whose genome sequences are currently available. All the PN promoters from Azoarcus/Aromatoleum strains described here show a conserved architecture including three operator regions (ORs), i.e., OR1 to OR3, for binding to the BzdR transcriptional repressor. Here, we demonstrate that, whereas OR1 is sufficient for the BzdR-mediated repression of the PN promoter, the presence of OR2 and OR3 is required for de-repression promoted by the benzoyl-CoA inducer molecule. Our results reveal that BzdR binds to the PN promoter in the form of four dimers, two of them binding to OR1. The BzdR/PN complex formed induces a DNA loop that wraps around the BzdR dimers and generates a superstructure that was observed by atomic force microscopy. This work provides further insights into the existence of a conserved BzdR-dependent mechanism to control the expression of the bzd genes in Azoarcus strains.
1. Introduction
Aromatic compounds are the most widespread organic compounds in nature after carbohydrates. Moreover, the release of aromatic compounds into the biosphere increased considerably over the last century as a consequence of industrial activity [1]. Many of these compounds are toxic and/or carcinogenic, thus representing major persistent environmental pollutants. Some specialized microorganisms, mainly bacteria and fungi, adapted to degrade a wide variety of aromatic compounds aerobically and/or anaerobically [2]. Whereas the aerobic degradation of aromatics is extensively studied [3,4,5], the anaerobic degradation pathways are much less well studied, especially regarding the genes involved in these anaerobic processes [6,7]. However, many environments are anoxic and, thus, the anaerobic degradation of aromatic compounds has great importance at the ecological level [6,7,8]. The anaerobic degradation of a wide variety of aromatic compounds converges into a few central pathways (catabolic funnel) that carry out the reductive de-aromatization and further conversion of the intermediates to the central metabolism. Most monocyclic aromatic compounds are channeled and activated to the central intermediate benzoyl-CoA [2,8,9]. Benzoate was used over the two last decades as a model compound to study the main pathway for anaerobic degradation of aromatic compounds, i.e., the central route of benzoyl-CoA [7,9]. The anaerobic degradation of benzoate takes place through a peripheral route consisting of a single reaction that activates benzoate to benzoyl-CoA. Benzoyl-CoA is then de-aromatized by the action of a reductase, the only oxygen-sensitive enzyme within the benzoyl-CoA pathway, which generates cyclohexa-1,5-diene-1-carbonyl-CoA, which is further converted into 3-hydroxypimelyl-CoA through a modified β-oxidation mechanism (Figure 1A) [2,7,8,9,10]. The genes encoding the enzymes involved in the anaerobic degradation of benzoate were reported in some phototrophic bacteria, e.g., Rhodopseudomonas palustris strains (bad genes), facultative anaerobes, e.g., Thauera, Azoarcus, Aromatoleum, Magnetospirillum, and Sedimenticola strains (bcr/bzd genes), and in strict anaerobes, e.g., Geobacter, Syntrophus, and Desulfobacula strains (bam genes) [8,9,10,11,12,13,14,15]. The expression of the catabolic genes is controlled by specific transcriptional regulators, but only a few such regulators are described so far, mainly the BadM protein in R. palustris [16,17], the BamVW and BgeR proteins in Geobacter strains [18,19], and the BzdR protein in Azoarcus sp. CIB [20,21].
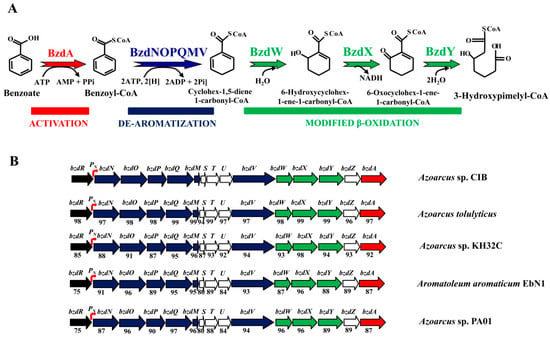
Figure 1.
Scheme of anaerobic degradation of benzoate and gene organization of the bzd cluster in different Azoarcus and Aromatoleum strains. (A) Scheme of the anaerobic degradation pathway of benzoate into benzoyl-CoA (red), the de-aromatization of benzoyl-CoA (blue), and the modified β-oxidation that produces 3-hydroxypimelyl-CoA. Enzyme abbreviations: BzdA, benzoate-CoA ligase; BzdNOPQMV, benzoyl-CoA reductase, ferredoxin, and putative reduced nicotinamide adenine dinucleotide phosphate (NADPH)::ferredoxin oxidoreductase; BzdW, cyclohex-1,5-diene-1-carbonyl-CoA hydratase; BzdX, 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase; BzdY, 6-oxocyclohexene-1-ene-carbonyl-CoA hydrolase. (B) Scheme of the bzd cluster in different Azoarcus/Aromatoleum strains. Genes are indicated in the same color code as the corresponding enzymes in panel (A), i.e., genes encoding the activation, de-aromatization, and modified β-oxidation are indicated in red, blue, and green color, respectively. The bzdR regulatory gene is shown in black, and the catabolic PN promoter is shown in red as a curved arrow. Genes of unknown function are colored in white. Below each gene, the percentage of the amino-acid sequence identity to the corresponding Azoarcus sp. CIB ortholog is indicated. The accession numbers of the corresponding genome sequences are as follows: Azoarcus sp. CIB (CP011072), Azoarcus sp. KH32C (AP012304), Aromatoleum aromaticum EbN1 (CR555306), Azoarcus tolulyticus strain ATCC51758 (NZ_FTMD00000000), and Azoarcus sp. PA01 (NZ_LARU00000000).
Among facultative anaerobes, bacteria of the Azoarcus genus, recently re-classified within the new Aromatoleum genus [22], represent one of the major groups of aromatic degraders. Azoarcus sp. CIB is a denitrifying β-Proteobacterium able to aerobically and/or anaerobically degrade a wide variety of aromatic compounds, most of which are channeled to the benzoyl-CoA central pathway encoded by the bzd genes. The bzd genes from strain CIB are clustered together in a large catabolic operon (bzdNOPQMSTUVWXYZA) driven by the PN promoter (Figure 1B). The specific transcriptional regulation of the bzd operon is carried out by the BzdR repressor encoded immediately upstream of the catabolic operon (Figure 1B) [20,23]. Benzoyl-CoA, the first intermediate during the anaerobic degradation of benzoate, is specifically recognized by BzdR and acts as the inducer molecule allowing the expression of the bzd catabolic genes [23]. BzdR binds to the PN promoter at three different regions that contain a direct repetition of the sequence TGCA [20]. In addition to BzdR, two additional regulatory proteins account for an overimposed regulation of the PN promoter. AcpR, a regulator belonging to the well-known fumarate and nitrate reductase (FNR)/cyclic AMP receptor protein (CRP) superfamily of transcriptional regulators, is essential for the activation of the PN promoter under oxygen deprivation conditions [24]. Furthermore, the AccR response regulator interacts with PN and inhibits its activity in response to several organic acids such as succinate, malate, or acetate, accounting for the carbon catabolite control of the bzd genes [23,25].
In this work, we expand the current knowledge on the organization of the bzd clusters in all Azoarcus/Aromatoleum strains sequenced so far that are able to degrade benzoate anaerobically. The evolutionary conservation of a common PN promoter architecture with three BzdR operator regions can be highlighted. The role of the three BzdR operator regions was studied, and the formation of a BzdR/PN superstructure was confirmed and visualized.
2. Material and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains and plasmids used in this study are listed in Table 1.

Table 1.
Bacterial strains and plasmids used in this study.
Escherichia coli cells were grown at 37 °C in lysogeny broth (LB) medium [34]. When required, E. coli cells were grown anaerobically at 30 °C in LB medium supplemented with 0.1% casamino acids [34] and 10 mM KNO3 as a terminal electron acceptor. Azoarcus strains were anaerobically grown at 30 °C in MC medium as described previously [23]. Where appropriate, antibiotics were added at the following concentrations: ampicillin, 100 µg∙mL−1; chloramphenicol, 30 µg∙mL−1; gentamicin, 7.5 µg∙mL−1; and kanamycin, 50 µg∙mL−1.
2.2. Molecular Biology Techniques
Recombinant DNA techniques were carried out according to published methods [26]. Plasmid DNA was prepared with a High Pure Plasmid Isolation Kit (Roche Applied Science, Penzberg, Germany). The DNA fragments were purified with Gene Clean Turbo (Q-BIOgene, Carlsbad, CA, USA). Oligonucleotides were supplied by Sigma (St. Louis, MO, USA). The sequence of inserts/DNA fragments was confirmed by DNA sequencing with an ABI Prism 377 automated DNA sequencer (Applied Biosystems, Foster City, CA, USA). Transformation of E. coli was carried out by using competent cells prepared by the RbCl method [34] or by electroporation (Gene Pulser, Bio-Rad, Cambridge, MA, USA) [34]. Plasmids were transferred from E. coli S17-1λpir (donor strain) to Azoarcus sp. CIB recipient strains via biparental filter mating as previously reported [23]. Proteins were analyzed by SDS-PAGE as described previously [26].
2.3. Sequence Data Analyses
For bioinformatic inspection of genes and regulatory regions of interest, we employed the BioEdit Sequence Alignment Editor [35]. The Basic Local Alignment Search Tool (BLAST) platform [36] was used for studying the similarity/identity of proteins encoded in the bzd cluster. The amino-acid sequences of the open reading frames of proteins encoded in the bzd operon were compared with those present in databases using the TBLASTN algorithm at the National Center for Biotechnology Information (NCBI) server [37] (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Nucleotide and protein alignments were done with ALIGN [38] and CLUSTALW [39], respectively, in the BioEdit editor.
2.4. Overproduction and Purification of His6-BzdR, His6-NBzdRL, and His6-FNR* Proteins
Plasmid pQE32-His6BzdR and pQE32-His6NBzdRL produce N-terminally His6-tagged BzdR and NBzdRL proteins (N-terminal domain plus the BzdR linker), respectively (Table 1). Plasmid pQE60-His6FNR* produces a C-terminally His6-tagged Fnr* (Table 1). All these plasmids express the cloned genes under control of the PT5 promoter and two lac operator boxes. The His6-tagged proteins were overproduced in E. coli M15 strain also harboring plasmid pREP4, and they were purified following protocols previously established [20].
2.5. Analytical Ultracentrifugation Assays
To perform experiments of sedimentation equilibrium of His6-NBzdRL protein bound to DNA, different concentrations of this protein (0.5 to 10 μM) were incubated with the DNA fragment (0.05 to 0.2 μM). The DNA fragments used were the PNI promoter (145 bp) and the complete PN promoter (PN, 253 bp) that were obtained by PCR using the oligonucleotide pairs BLINE (5’–CGTGCCTGACATTTGACTTAGATC–3’) and 3IVTPN (5’–CGGGAATTCCATCGAACTATCTCCTCTGATG–3’, EcoRI site underlined), and RET1 (5’–CCGAGCCTCGCGTTTTACTGC–3’) and 3IVTPN, respectively. The ultracentrifugation experiments were conducted in a buffer containing 50 mM NaH2PO4, 300 mM KCl, 100 mM imidazole, pH 8, as previously described [21]. Since the BzdR protein aggregates in experiments of sedimentation equilibrium at concentrations higher than 2 μM, we used the NBzdRL protein that is totally soluble at 10 μM. The presence of DNA was checked by measuring the absorbance at 260 nm. The distribution of sediment coefficients obtained in the sedimentation velocity experiments was determined using the SEDFIT program as reported previously [21].
2.6. Construction of PNI::lacZ and PNII::lacZ Translational Fusions and β-Galactosidase Assays
The PN::lacZ translational fusion was described previously [23]. To generate two truncated versions of the PN promoter, PNI and PNII, fused to the lacZ reporter gene, we firstly PCR-amplified the truncated promoters. The oligonucleotides used for amplification of the PNI promoter were BKIN (5’–GGGGTACCCGTGCCTGACATTTGACTTAGATC–3’; KpnI site underlined) and 5BZN (5’–GCTCTAGACCCATCGAACTATCTCCTCTGATG–3’; XbaI site underlined). For the amplification of the PNII promoter, the oligonucleotides used were NIU PN-II (5´–GGGGTACCCAAGAAAGATTGCAGTTTTCCATG; KpnI site underlined) and 5BZN. The template used for amplification of the promoter regions was the pECOR7 plasmid (Table 1). After PCR-amplification, both DNA fragments were digested with XbaI and KpnI restriction enzymes, and later cloned in plasmid pSJ3 in-frame with lacZ gene, thus generating the corresponding PNI::lacZ and PNII::lacZ translational fusions in plasmids pSJ3-PNI and pSJ3-PNII, respectively. These translational fusions were subsequently subcloned by EcoRI/HindIII double digestion into plasmid pBBR1MCS-5 generating plasmids, pBBR5-PNI and pBBR5-PNII, respectively. For determination of promoter activity, pSJ3-PNI and pSJ3-PNII were transformed in E. coli MC4100, while pBBR5-PNI and pBBR5-PNII were transferred to Azoarcus sp. CIB [23]. E. coli MC4100 carrying the PN::lacZ, PNI::lacZ, or PNII::lacZ fusions was grown anaerobically in LB-rich medium, whereas Azoarcus sp. CIB carrying the PN::lacZ, PNI::lacZ, or PNII::lacZ fusions was grown anaerobically in MC medium in the presence of 3 mM benzoate or 0.2% (w/v) succinate. Cultures were incubated at 30 °C until the mid-exponential phase. The β-galactosidase activity (in Miller Units) was determined in permeabilized cells (using 0.1% SDS and chloroform) according to the method described by Miller [34].
2.7. In Vitro Transcription Assays
In vitro transcription assays were performed as previously published [40,41] using plasmids pJCD-PN, pJCD-PNI, and pJCD-PNII (0.5 nM) as supercoiled templates. To construct these plasmids, firstly, 585-bp, 139-bp, and 209-bp DNA fragments containing the PN, PNI, and PNII promoters, respectively, were PCR-amplified from the Azoarcus sp. strain CIB chromosome. The forward and reverse oligonucleotides 5IVTPN (5’–CGGAATTCCGTGCATCAATGATCCGGCAAG–3’; EcoRI site underlined) and 3IVTPN (5’–CGGAATTCCATCGAACTATCTCCTCTGATG–3’; EcoRI site underlined), BIN (5’–CGGAATTCCGTGCCTGACATTTGACTTAGATC–3’; EcoRI site underlined) and 3IVTPN, and BIIN (5’–CGGAATTCCAAGAAAGATTGCAGTTTTCCATG–3’; EcoRI site underlined) and 3IVTPN, respectively, were used for PCR-amplification. Then, the DNA fragments were EcoRI restricted, and cloned into the EcoRI-restricted pJCD01 cloning vector, giving rise to plasmids pJCD-PN, pJCD-PNI, and pJCD-PNII, respectively (Table 1). Reactions (50-µL mixtures) were performed in a buffer containing 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.1 mM bovine serum albumin, 10 mM dithiothreitol, and 1 mM ethylenediaminetetraacetic acid (EDTA). Unless otherwise indicated, each DNA template was premixed with 100 nM σ70-containing E. coli RNA polymerase (RNAP) holoenzyme (Amersham plc, Amersham, UK) and different amounts of purified His6-Fnr*. For multiple-round assays, transcription was then initiated by adding a mixture of 500 μM (each) adenine, cytosine, and guanine triphosphate (ATP, CTP, and GTP); 50 μM uridine triphosphate (UTP); and 2.5 μCi of [α32P]-UTP (3000 Ci∙mmol−1). After incubation for 15 min at 37 °C, the reactions were stopped with an equal volume of a solution containing 50 mM EDTA, 350 mM NaCl, and 0.5 mg of carrier transfer RNA (tRNA; yeast tRNA from Thermo Fisher Scientific, Whatham, MA, USA) per ml. The messenger RNA (mRNA) produced was then precipitated with ethanol, separated on a denaturing 7 M urea/4% polyacrylamide gel, and visualized by autoradiography. Transcript levels were quantified with a Bio-Rad Molecular Imager FX system and using the ImageJ software [42].
2.8. Gel Retardation Assays
The PNI DNA probe (145 bp) was obtained as described previously [21], digested with EcoRI, and labeled by filling in the overhanging EcoRI-digested end with [α-32P]dATP (6000 Ci∙mmol−1; Amersham Biosciences, Amersham, UK) and the Klenow fragment of E. coli DNA polymerase as described previously [20]. The DNA probe was mixed with the purified proteins at the concentration indicated in each assay. The retardation reaction mixtures contained 20 mM Tris-HCl pH 7.5, 10% glycerol, 2 mM β-mercaptoethanol, 50 mM KCl, 0.05 nM DNA probe, 250 μg∙mL−1 bovine serum albumin, and purified His6-BzdR protein in a 9-μL final volume. The samples were fractionated by electrophoresis in 5% polyacrylamide gels buffered with 0.5× TBE (45 mM Tris borate, 1 mM EDTA). The gels were dried onto Whatman 3MM paper and exposed to Hyperfilm MP (Amersham Biosciences, Little Chalfont, UK).
2.9. Analysis of Protein–DNA Interaction by Atomic Force Microscopy
To perform atomic force microscopy (AFM) experiments, a DNA fragment of 1225 bp, termed PNL, from position −505 to +720 and covering the intergenic region between bzdR and bzdN genes, as well as the 5´-end of the bzdN gene, was PCR-amplified using as template the plasmid pECOR7, and the oligonucleotides 3REG (5’–GGGGTACCCGTGCATCAATGATCCGGCAAG–3’; KpnI site underlined) and N3 (5’–TTCAGCATCTCGTTGTGCTC–3’). The purified His6-BzdR protein was obtained as detailed above. The binding reactions were performed for 15 min at room temperature using retardation buffer (20 mM Tris·HCl, pH 7.5, 10% glycerol, 2 mM β-mercaptoethanol, 50 mM KCl). The protein and DNA concentrations were optimized to ensure that the unbound protein (probably adsorbed to the surface) did not disturb the image. The incubation of the complexes was performed at a final concentration of 2 nM PNL fragment with [PNL]:[BzdR] ratios of 1:28 and 1:70. The samples of complexes were diluted 10 times in retardation buffer, in the presence of MgCl2 or NiCl2 at concentrations between 3 and 10 mM. Each reaction mixture was then deposited on a freshly exfoliated mica surface (which provides a clean and atomically flat substrate). After 1 min of incubation on the mica, the non-adsorbed material was removed by successive washes with the same adsorption buffer. Finally, the samples, imaged while immersed in buffer, were visualized with an atomic force microscope from Nanotec Electronica S.L (Tres Cantos, Spain) operated in jump mode [43]; the images were processed with the WSxM program [44].
3. Results and Discussion
3.1. The Genetic Organization of the bzd Cluster and the Architecture of the PN Promoter Are Conserved in the Azoarcus/Aromatoleum Genus
The anaerobic degradation of benzoate is a common feature among many strains of the Azoarcus/Aromatoleum genus [22]. By mining in the available genome sequences of Azoarcus aromatic degrader strains, i.e., those of A. tolulyticus, A. toluclasticus, Azoarcus sp. KH32C, and Azoarcus sp. PA01, we found the presence of genes homologous to the bzd genes responsible of the anaerobic degradation of benzoate in A. evansii, Azoarcus sp. CIB, and A. aromaticum EbN1 [8,45]. Interestingly, in all these Azoarcus strains, the bzd genes are arranged in a cluster that contains a bzdR regulatory gene located upstream of a putative bzd catabolic operon. The bzd operon is organized into at least three different functional modules: (i) the activation module (bzdA) that codes for the enzyme that generates benzoyl-CoA; (ii) the de-aromatization module (bzdNOPQMV) that encodes the benzoyl-CoA reductase and auxiliary enzymes; and (iii) the modified β-oxidation module (bzdWXY) that encodes the enzymes that generate 3-hydroxypimelyl-CoA (Figure 1) [8,10]. A close inspection of the promoter region of the predicted bzd operons from all Azoarcus strains revealed the conservation of the PN promoter described in Azoarcus sp. CIB [20] (Figure 2). Remarkably, it should be noted that this organization found in Azoarcus/Aromatoleum was not observed in closely related aromatic-compound-degrading denitrifying bacteria such as those of the genus Thauera.
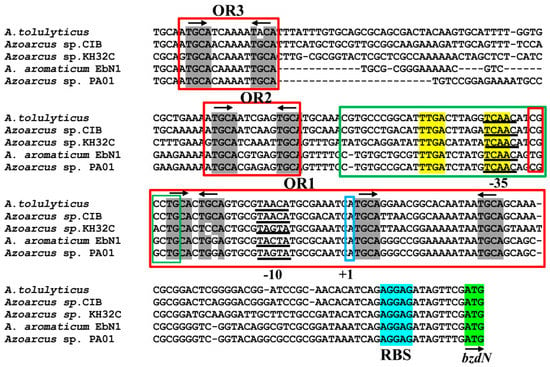
Figure 2.
Sequence comparison analysis between the Azoarcus sp. CIB PN promoter [20] and the PN promoters from other Azoarcus/Aromatoleum strains. The sequence of the PN promoter from strain CIB spanning from position −174 to +79, with respect to the transcription start site (+1, indicated) is shown and compared with that of PN promoters from other Azoarcus strains. The σ70-RNA polymerase (RNAP) recognition sequences −10 and −35 are underlined, the ribosome-binding sequence (RBS) is marked in cyan, and the ATG initiation codon of the bzdN gene is shown in green. The operator regions I, II, and III (OR1, OR2, and OR3), involved in the interaction with the BzdR protein, are boxed in red. The arrows indicate the palindromic TGCA sequences (in gray) present in each of the operator regions. The green box shows the AcpR-binding region (palindromic sequences are indicated in yellow).
As described before, the PN promoter of strain CIB shows the transcription start site located minus 75 nucleotides with respect to the ATG start codon of the bzdN gene, and the −10 and −35 boxes of interaction with the σ70 subunit of the RNAP were also identified [20] (Figure 2). Three BzdR operator regions were detected in the PN promoter by DNase I footprinting, i.e., operator region I (OR1), located between positions −32 and +31 (with respect to the transcription start site), operator region II (OR2) (−83 to −63), and operator region III (OR3) (−146 to −126) [20]. The operator region recognized by the AcpR activator protein overlaps the −35 box of RNAP [24] (green box, Figure 2). Interestingly, the PN promoter sequences of all bzd operons from Azoarcus strains exhibited the three BzdR operator regions (OR1, OR2, and OR3) and the operator region that binds to the AcpR transcriptional activator (Figure 2). The OR2 and OR3 regions of PN contain the conserved TGCA(N6)TGCA palindromic sequence where N6 is a six-nucleotide A-rich region in OR3. OR1 is longer and it contains two conserved palindromic sequences, TGCA(C)T(G/C)(C/G)A and TGCA(N15)TGCA, located between the −35 and −10 boxes and downstream of the transcription start site, respectively (Figure 2).
The TGCA palindromic structures were also described in other operator regions such as those recognized by the CopG repressor from Streptococcus strains, a 45 amino-acid homodimer encoded in the pMV158 plasmid family, at the Pcr promoter that drives the expression of the copG and repB genes involved in plasmid replication [46,47]. Another example of a regulatory protein that recognizes operator boxes that include the TGCA sequence is the XylS regulator, which controls the expression of the operon for m-xylene and toluene catabolism in Pseudomonas putida. XylS acts as an activator binding to a region of the Pm promoter that includes two imperfect palindromes of the TGCA(N6)GGNTA type [48]. The regulatory protein BenR, involved in the regulation of the aerobic catabolism of benzoate and other analogues in P. putida, also seems to recognize this type of imperfect palindrome [49].
The conservation of the three BzdR operator regions in all PN promoters identified so far suggests that they are required for the accurate regulation of this promoter. To further study the role of the three BzdR operator regions of PN, we performed in vivo and in vitro approaches with a set of truncated versions of this promoter.
3.2. In Vivo Studies on the Activity of Truncated PN Promoters
As described previously, the activity of the PN promoter requires the RNAP and the interaction with the AcpR, a transcriptional activator from the well-known FNR/CRP superfamily [50] that binds at a consensus sequence centered at position −41.5 from the transcription start site and overlapping the RNAP −35 box [24] (Figure 2). We demonstrated previously that the FNR protein from E. coli is able to bind and activate the PN promoter [24]. On the other hand, despite the PN promoter containing three BzdR operators, the OR1 region itself overlaps the RNAP −10 box and +1 sites (Figure 2), thus having the requested sequence for the interaction with RNAP. Therefore, it could be expected that a truncated promoter just containing OR1 and the AcpR operator could behave as a functional BzdR-controlled minimal promoter. To check this assumption, we engineered a set of truncated PN promoters (Figure 3A) and compared their behavior with that of the complete PN promoter by in vivo assays. To this end, the truncated PNI promoter (spanning from position −61 to +79; carries OR1) and truncated PNII promoter (spanning from position −112 to +79; carries OR1 and OR2) (Figure 3A) were cloned in plasmid pSJ3, rendering plasmids pSJ3-PNI (contains the PNI::lacZ translational fusion) and pSJ3-PNII (contains the PNII::lacZ translational fusion) (Table 1). These plasmid constructions were independently transformed into E. coli MC4100 (pCK01) and E. coli MC4100 (pCK01-BzdR), respectively (Table 1). The β-galactosidase activity assays revealed that promoters PNI and PNII showed activity levels similar to those obtained with the complete PN promoter cloned in plasmid pSJ3PN (Table 1) when present in the E. coli MC4100 (pCK01) strain (Figure 3B). Moreover, the BzdR protein was able to inhibit the activity of PNI and PNII as efficiently as in the case of the wild-type PN promoter in E. coli MC4100 (pCK01-BzdR) cells (Figure 3B). Therefore, these results show that PNI behaves as a functional minimal promoter that becomes fully active in the absence of BzdR and fully repressed in the presence of the BzdR protein.
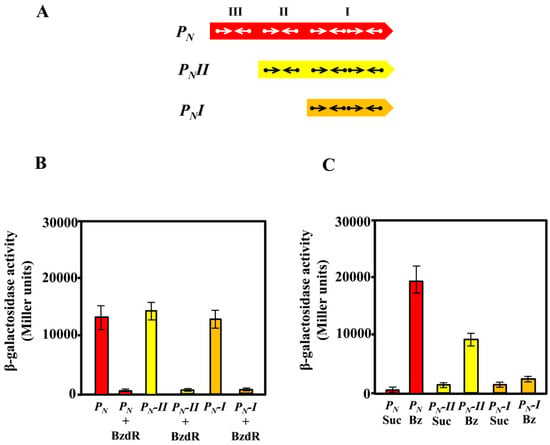
Figure 3.
Activity of the PN, PNI, and PNII promoters in Escherichia coli and Azoarcus sp. CIB cells. (A) Schematic representation of the PN, PNII, and PNI promoters. Arrows represent the inverted repeat TGCA sequences for BzdR binding at operator regions I, II, and III. (B) The activity of the promoters was analyzed in E. coli MC4100 cells harboring the control plasmid pCK01 or plasmid pCK01-BzdR that expresses the bzdR gene (BzdR), and the plasmids pSJ3-PN, pSJ3-PNI, or pSJ3-PNII that harbor the PN::lacZ (PN), PNI::lacZ (PN-I), or PNII::lacZ (PN-II) translational fusions, respectively. Growth was performed in lysogeny broth (LB) in anaerobic conditions for 16 h as described in Section 2.6. The β-galactosidase activity was measured as detailed in Section 2.6 [34]. Graphed values are the averages from three independent experiments ± SD (error bars). (C) Activity of the PN, PNII, and PNI promoters in Azoarcus sp. CIB cells. Azoarcus sp. CIB cells harboring plasmids pBBR5-PN, pBBR5-PNI, or pBBR5-PNII, which express the PN::lacZ (PN), PNI::lacZ (PN-I), or PNII::lacZ (PN-II) translational fusions, respectively, were anaerobically grown for 72 h in MC medium supplemented with 0.2% succinate (Suc) or 3 mM benzoate (Bz), and the β-galactosidase activity was measured as detailed in Section 2.6. Graphed values are the averages from three independent experiments ± SD (error bars).
To check the de-repression (induction) of the wild-type and truncated PN promoters by benzoyl-CoA, we expressed the PN::lacZ, PNI::lacZ, and PNII::lacZ fusions in Azoarcus sp. CIB. Azoarcus sp. CIB is able to produce benzoyl-CoA when benzoate is present in the culture medium. Benzoyl-CoA acts as inducer molecule allowing the de-repression of the system by inducing conformational changes in BzdR that lead to the release of the repressor from the promoter [21]. To perform the analysis in Azoarcus, the lacZ fusions were independently subcloned into the broad-host-range pBBR1MCS-5 vector generating plasmids pBBR5-PN, pBBR5-PNI, and pBBR5-PNII, respectively (Table 1). The plasmids were introduced by conjugation into Azoarcus sp. CIB, and the level of activity of each promoter was determined by β-galactosidase assays. As expected, the activity of the PN-derivative promoters expressed in Azoarcus cells grown in succinate (non-induction conditions) led to very low activity of all translational fusions (Figure 3C), revealing an efficient repression of the three PN promoters by the chromosomally encoded BzdR protein. When Azoarcus sp. CIB cells were grown in 3 mM benzoate (induction conditions), the complete PN promoter showed the expected activation (Figure 3C), thereby revealing the benzoyl-CoA-induced release of the BzdR repressor from the target promoter [20]. However, in contrast to the clear de-repression of the wild-type PN promoter in Azoarcus sp. CIB cells grown in benzoate, the PNII promoter showed a limited de-repression (Figure 3C), and almost no de-repression was observed with the truncated PNI promoter (Figure 3C).
In summary, the in vivo assays revealed that PNI is a fully active promoter in the absence of BzdR but a constitutively repressed promoter in the presence of BzdR. The OR2 and OR3 regions appear to be essential for the full de-repression (induction) of the PN promoter when benzoyl-CoA is produced in Azoarcus sp. CIB cells grown in benzoate. To further confirm the role of OR2 and OR3 in the de-repression of PN by benzoyl-CoA, several in vitro assays were performed.
3.3. Operator Regions II and III Are Needed for the Benzoyl-CoA-Dependent De-Repression of the PN Promoter
To further study the truncated PN promoters, we compared their activity levels to that of the wild-type PN promoter by performing in vitro transcription assays using the supercoiled plasmids pJCD-PN, pJCD-PNI, and pJCD-PNII (Table 1) as DNA templates, and the E. coli FNR* protein (a constitutively active AcpR ortholog [24]) as an activator. As shown in Figure 4, all three promoters generated the expected 184-nucleotide transcript in the absence of BzdR, but this transcript was lacking in the presence of the BzdR repressor. However, whereas a clear de-repression at the PN promoter was observed (about five times) when benzoyl-CoA was added to the BzdR-containing reaction, a slight de-repression (about two times) was observed with PNII, and benzoyl-CoA did not produce any de-repression effect at PNI (Figure 4). Thus, these results suggest that benzoyl-CoA was not able to alleviate the BzdR-mediated repression at PN when both OR2 and OR3 were missing. To check that the lack of de-repression at promoter PNI was a consequence of the promoter occupancy by BzdR even in the presence of the benzoyl-CoA inducer, gel shift assays were performed using the PNI probe in the presence of BzdR and increasing concentrations of benzoyl-CoA (Figure 5B). As expected, BzdR produced retardation of the PNI promoter probe, but increasing concentrations of benzoyl-CoA up to 2 mM, which released the BzdR repressor from the wild-type PN promoter (Figure 5A), were unable to induce the dissociation of the BzdR/PNI complex (Figure 5B). Since benzoyl-CoA promotes the release of BzdR from the PN promoter (Figure 5A) [20], this result suggests that the constitutive repression of PNI by BzdR is due to the irreversible binding of the repressor to the target promoter.
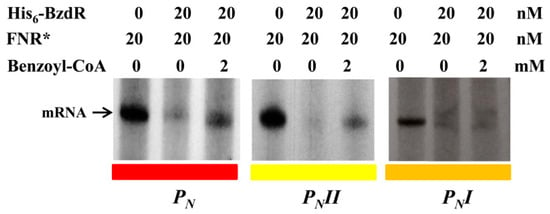
Figure 4.
In vitro activity of the PN, PNII, and PNI promoters. Multiple-round transcription reactions were carried out as detailed in Section 2.7 by using pJCD-PN, pJCD-PNI, and pJCD-PNII plasmids harboring PN, PNI, and PNII promoter templates, respectively, which produced the corresponding messenger RNA (mRNA). All the in vitro transcription reactions were performed with 100 nM E. coli σ70-RNAP holoenzyme. Purified FNR* was used at 20 nM. When required, His6-BzdR protein was used at 20 nM, and benzoyl-CoA was added at 2 mM.
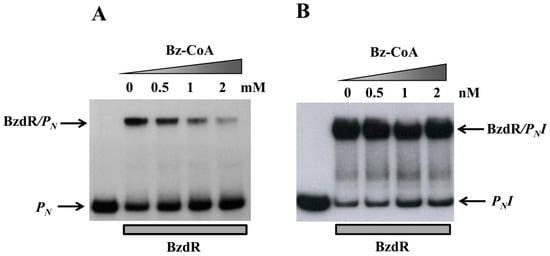
Figure 5.
Gel retardation analysis of BzdR binding to the PN (A) or the PNI promoter (B). Gel retardation analysis was performed as described in Section 2.8 by using 10 nM purified His6-BzdR in the presence of increasing concentrations (from 0 to 2 mM) of benzoyl-CoA (Bz-CoA). The free PN probe and the BzdR/PN complex (A) or PNI probe and the BzdR/PNI complex (B) are indicated by arrows.
We demonstrated previously that the binding of BzdR to its cognate promoter is cooperative [21]. Sedimentation velocity analyses showed that 8–10 molecules of BzdR bind to the PN promoter, and it was postulated that each of the four TGCA palindromic regions bind a BzdR dimer, i.e., four BzdR molecules bound at OR1, two at OR2, and another two at OR3 [21]. Thus, it could be expected that the lack of de-repression at the PNI promoter in the presence of benzoyl-CoA inducer could be due to an excess of BzdR molecules bound to OR1 when both OR2 and OR3 are missing in the target promoter.
To check this hypothesis, we performed analytical ultracentrifugation experiments with the N-terminal domain of the BzdR protein (NBzdRL protein; residues 1–90) that is able to bind to PN [30]. Previous sedimentation velocity experiments revealed that NBzdRL was a dimer with S-values of 2.3 S, demonstrating that BzdR dimerization is an intrinsic property of the N-terminal domain [30]. The results presented here revealed that eight molecules of NBzdRL were bound to the wild-type PN promoter (Figure 6A), confirming the previous results obtained with the complete BzdR protein [30]. More relevant, the ultracentrifugation analyses showed that four molecules of NBzdRL were bound to the PNI fragment (Figure 6B). Hence, these results provide the first experimental evidence that two BzdR dimers bind to OR1 and support the previous assumption of the distribution of the eight BzdR molecules along the PN promoter [21].
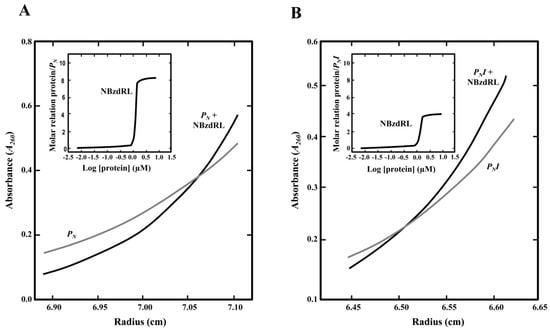
Figure 6.
Sedimentation equilibrium analysis to study the interaction of purified NBzdRL protein with PN and PNI DNA fragments. (A) Distribution of the gradient in sedimentation equilibrium of the PN fragment (PN, gray line) or PN fragment with His6-NBzdRL protein (black line). The inset graph shows the protein/DNA molar ratio as the concentration of His6-NBzdRL increases. (B) Distribution of the gradient in sedimentation equilibrium of the PNI fragment (gray line) and PNI + His6-NBzdRL complex (black line). The inset graph shows the protein/DNA molar ratio as the concentration of NBzdRL increases.
3.4. Visualization of the BzdR/PN Complex
The results presented above indicate that three BzdR operator regions are conserved in all PN promoters identified so far, and they suggest that binding of four BzdR dimers to the wild-type PN promoter should lead to the formation of a big protein–DNA complex. Atomic force microscopy (AFM) permits direct visualization of the tridimensional structure of protein/DNA complexes arranged on an atomically flat surface and supplies data about the topology and stoichiometry of the complexes [51,52,53,54]. A 1225-bp DNA sequence (414 nm in length) spanning from position −505 to +720 with respect to the transcription start site was used as template (PNL fragment) to visualize the structure of PN interacting with BzdR when using a 50:1 [BzdR]:[DNA] ratio (Figure 7A). Images obtained when the three operator boxes were occupied by BzdR revealed a compact structure (Figure 7B). The lengths of the free (unoccupied) DNA that emerged at both sides of the compact structure were 108.9 ± 20 nm and 199.9 ± 44 nm (n = 16), respectively (Figure 7C, shown for one of the 16 samples). These sizes are slightly lower than those theoretically expected, i.e., 121 nm and 233 nm, respectively (Figure 7A). The total extension of the BzdR–DNA complex was reduced by 14 ± 4% with respect to the length of the naked PNL DNA fragment (Figure 7C).
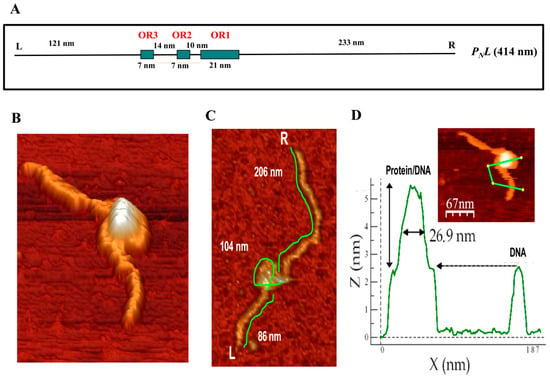
Figure 7.
Analysis of the binding of BzdR to the PN promoter by atomic force microscopy (AFM). (A) Scheme of the PNL DNA fragment used as a template. The DNA fragment extends from position −505 to +720 with respect to the transcription start site of the PN promoter. The operator regions recognized by BzdR are represented by green boxes with their corresponding numbers (OR1, OR2, and OR3). The distances separating each of the indicated regions, the extension of each operator region in nm, and the left (L) and right (R) ends of the PNL fragment are also indicated. (B) Image of a superstructure of the BzdR/PNL complex. (C) Analysis of distances in a BzdR/PNL superstructure (one image, n = 16). The distances of both arms and the loop of the superstructure are detailed. L and R indicate the left and right ends of the PNL DNA fragment, respectively. (D) Image of a BzdR/PNL superstructure and its corresponding height profile. The green line of the image corresponds to the profile outlined in the lower graph. In the graph, the length (in nm) of the line is represented on the x-axis and the height on the y-axis. The first peak corresponds to the BzdR/PNL superstructure and the second peak corresponds to the naked DNA.
These results are compatible with the formation of one loop of DNA around the BzdR proteins in the BzdR–DNA complex. The length of the DNA, including the DNA that is forming the loop, was estimated to be 398.6 ± 28.9 nm, which is compatible with the total length of the PNL DNA fragment (414 nm) and with the previous length calculated for the PNL naked template (400 ± 13 nm; [55]). Furthermore, the width of the BzdR–DNA “superstructure” had a value of 26.9 ± 6 nm (n = 10) (Figure 7D). Since a BzdR dimer resolved by electron microscopy is like a cylinder of 9 nm × 7 nm [21], the width of the superstructure observed by AFM is compatible with 4–5 dimers (8–10 molecules) of BzdR protein interacting with OR regions I, II, and III. These results are in good agreement with those obtained by analytical ultracentrifugation experiments (Figure 6A) [21]. In summary, the AFM results strongly suggest that the interaction of BzdR with the three operator regions of PN leads to the formation of a superstructure compatible with a DNA loop embracing 4–5 BzdR dimers by turning on itself (Figure 7). This type of superstructure was also described for other DNA–protein complexes such as that formed by the DNA/MutS protein [56], although other transcriptional regulators, such as the TodT protein that controls the aerobic toluene degradation pathway in P. putida, interact with the target DNA following a fork-type model [57]. The existence of a DNA loop wrapping the BzdR dimers as observed by AFM is also consistent with the DNase I footprinting studies that showed hyper-reactive nucleotides between OR1 and OR2 and between OR2 and OR3 [18], which might reflect BzdR-induced DNA bends leading to the loop formation.
4. Conclusions
In this work, we showed that the genes responsible for the anaerobic degradation of benzoate in all aromatic-degrader Azoarcus/Aromatoleum strains whose genome sequences are currently available are organized in a well-conserved bzd cluster. Thus, a bzdR regulatory gene and a PN promoter could be identified upstream of the catabolic bzd operon in all bzd clusters analyzed. Interestingly, all PN promoters show a conserved architecture that includes three BzdR operator regions (OR1–3). Whereas OR1 is sufficient for the BzdR-mediated repression of the PN promoter, the presence of OR2 and OR3 is required for the de-repression promoted by the benzoyl-CoA inducer molecule. Our results suggest that the dimeric protein BzdR binds to the PN promoter in the form of four dimers; two of the dimers are initially bound to OR1, and the complex formed may favor the interaction of two additional dimers at OR2 and OR3. The BzdR/PN complex formed induces a DNA loop that wraps around the BzdR dimers and generates a superstructure that was observed by AFM. This three-dimensional (3D) configuration may keep the −10/−35 sequences of PN inaccessible to the RNAP for achieving an efficient repression and, at the same time, could facilitate the PN de-repression when benzoyl-CoA is generated. However, the elucidation of the molecular mechanism underlying the participation of OR2 and OR3 in the de-repression of PN requires further research. In this sense, it should be taken into account that regulatory proteins other than BzdR, e.g., AcpR and AccR, are also involved in the control of the PN promoter [24,25] and, hence, more complex structure–function relationships at the PN promoter should be expected. In any case, the work presented here strongly suggests the existence of a common BzdR-dependent mechanism to control the expression of the bzd genes in Azoarcus/Aromatoleum strains.
Author Contributions
G.D.-R. carried out the genetic manipulations, physiology experiments, and in vitro enzyme assays. P.G.-d.-A. and M.V. designed and performed the AFM experiments and interpreted all the data obtained from this technique. G.D.-R., E.D., and M.C. conceived the whole study, designed the experiments, and contributed to the discussion of the research and interpretation of the data. G.D.-R., E.D., and M.C. wrote the article.
Funding
This work was supported by grant BIO2016-79736-R from the Ministry of Economy and Competitiveness of Spain, by grant CSIC 2016 20E093 from the CSIC, and by European Union H2020 grant 760994.
Acknowledgments
The author thanks A. Valencia for the technical assistance and S.L. Secugen for DNA sequencing. We are indebted to C. Alfonso and G. Rivas for their help in the analytical ultracentrifugation experiments.
Conflicts of Interest
The authors declare that there are no competing interests.
References
- Pieper, D.H.; Reineke, W. Engineering bacteria for bioremediation. Curr. Opin. Biotechnol. 2000, 11, 262–270. [Google Scholar] [CrossRef]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds—From one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Brzeszcz, J.; Kaszycki, P. Aerobic bacteria degrading both n-alkanes and aromatic hydrocarbons: And undervalued strategy for metabolic diversity and flexibility. Biodegradation 2018, 29, 359–407. [Google Scholar] [CrossRef] [PubMed]
- Díaz, E.; Jiménez, J.I.; Nogales, J. Aerobic degradation of aromatic compounds. Curr. Opin. Biotechnol. 2013, 24, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.; Jauregui, R.; Vilchez-Vargas, R.; Junca, H.; Pieper, D.H. AromaDeg, a novel database for phylogenomics of aerobic bacterial degradation of aromatics. Database 2014, 2014, bau118. [Google Scholar] [CrossRef] [PubMed]
- Rabus, R.; Boll, M.; Heider, J.; Meckenstock, R.U.; Buckel, W.; Einsle, O.; Ermler, U.; Golding, B.T.; Gunsalus, R.P.; Kroneck, P.M.H.; et al. Anaerobic microbial degradation of hydrocarbons: From enzymatic reactions to the environment. J. Mol. Microbiol. Biotechnol. 2016, 26, 5–28. [Google Scholar] [CrossRef]
- Lovley, D.R. Biorremediation. Anaerobes to the rescue. Science 2001, 293, 1444–1446. [Google Scholar] [CrossRef]
- Carmona, M.; Zamarro, M.T.; Blázquez, B.; Durante-Rodríguez, G.; Juárez, J.F.; Valderrama, J.A.; López-Barragán, M.J.; García, J.L.; Díaz, E. Anaerobic catabolism of aromatic compounds: A genetic and genomic view. Microbiol. Mol. Biol. Rev. 2009, 73, 71–133. [Google Scholar] [CrossRef]
- Boll, M.; Löffler, C.; Morris, B.E.; Kung, J.W. Anaerobic degradation of homocyclic aromatic compounds via arylcarboxyl-coenzyme A esters: Organisms, strategies and key enzymes. Environ. Microbiol. 2014, 16, 612–627. [Google Scholar] [CrossRef]
- Durante-Rodríguez, G.; Gómez-Álvarez, H.; Blázquez, B.; Fernández-Llamosas, H.; Martín-Moldes, Z.; Sanz, D.; Nogales, J.; Carmona, M.; Díaz, E. Anaerobic pathways for the catabolism of aromatic compounds. In Lignin Valorization: Emerging Approaches; Beckham, T., Ed.; The Royal Society of Chemistry Publishing: Cambridge, UK, 2018; pp. 333–390. [Google Scholar]
- Egland, P.G.; Pelletier, D.A.; Dispensa, M.; Gibson, J.; Harwood, C.S. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. USA 1997, 94, 6484–6489. [Google Scholar] [CrossRef]
- Wischgoll, S.; Heintz, D.; Peters, F.; Erxleben, A.; Sarnighausen, E.; Reski, R.; van Dorsselaer, A.; Boll, M. Gene clusters involved in anaerobic benzoate degradation in Geobacter metallireducens. Mol. Microbiol. 2005, 58, 1238–1252. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.E.; He, Q.; Nevin, K.P.; He, Z.; Zhou, J.; Lovley, D.R. Genomic and microarray analysis of aromatics degradation in Geobacter metallireducens and comparison to a Geobacter isolate from a contaminated field site. BMC Genom. 2007, 8, 180. [Google Scholar] [CrossRef] [PubMed]
- McInerney, M.J.; Rohlin, L.; Mouttaki, H.; Kim, U.; Krupp, R.S.; Rios-Hernandez, L.; Sieber, J.; Struchtemeyer, C.G.; Bhattacharyya, A.; Campbell, J.W.; et al. The genome of Syntrophus aciditrophicus: Life at the thermodynamic limit of microbial growth. Proc. Natl. Acad. Sci. USA 2007, 104, 7600–7605. [Google Scholar] [CrossRef] [PubMed]
- Wöhlbrand, L.; Jacob, J.H.; Kube, M.; Mussmann, M.; Jarling, R.; Beck, A.; Amann, R.; Wilkes, H.; Reinhardt, R.; Rabus, R. Complete genome, catabolic sub-proteomes and key-metabolites of Desulfobacula toluolica Tol2, a marine, aromatic compound-degrading, sulfate-reducing bacterium. Environ. Microbiol. 2013, 15, 1334–1355. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, H.; Hirakawa, Y.; Greenberg, E.P.; Harwood, C.S. BadR and BadM proteins transcriptionally regulate two operons needed for anaerobic benzoate degradation by Rhodopseudomonas palustris. Appl. Environ. Microbiol. 2015, 81, 4253–4262. [Google Scholar] [CrossRef] [PubMed]
- Van Drisse, C.M.; Escalante-Semerena, J.C. Small-molecule acetylation controls the degradation of benzoate and photosynthesis in Rhodopseudomonas palustris. MBio 2018, 16, e01895-18. [Google Scholar]
- Ueki, T. Identification of a transcriptional repressor involved in benzoate metabolism in Geobacter bemidjiensis. Appl. Environ. Microbiol. 2011, 77, 7058–7062. [Google Scholar] [CrossRef] [PubMed]
- Juárez, J.F.; Zamarro, M.T.; Barragán, M.J.; Blázquez, B.; Boll, M.; Kuntze, K.; García, J.L.; Díaz, E.; Carmona, M. Identification of the Geobacter metallireducens bamVW two-component system, involved in transcriptional regulation of aromatic degradation. Appl. Environ. Microbiol. 2010, 76, 383–385. [Google Scholar] [CrossRef]
- Barragán, M.J.; Blázquez, B.; Zamarro, M.T.; Mancheño, J.M.; García, J.L.; Díaz, E.; Carmona, M. BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 2005, 280, 10683–10694. [Google Scholar] [CrossRef]
- Durante-Rodríguez, G.; Valderrama, J.A.; Mancheño, J.M.; Rivas, G.; Alfonso, C.; Arias-Palomo, E.; Llorca, O.; García, J.L.; Díaz, E.; Carmona, M. Biochemical characterization of the transcriptional regulator BzdR from Azoarcus sp. CIB. J. Biol. Chem. 2010, 285, 35694–35705. [Google Scholar] [CrossRef]
- Rabus, R.; Wöhlbrand, L.; Thies, D.; Meyer, M.; Reinhold-Hurek, B.; Kämpfer, P. Aromatoleum gen. nov., a novel genus accommodating the phylogenetic lineage including Azoarcus evansii and related species, and proposal of Aromatoleum aromaticum sp. nov., Aromatoleum petrolei sp. nov., Aromatoleum bremense sp. nov., Aromatoleum toluolicum sp. nov. and Aromatoleum diolicum sp. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 982–997. [Google Scholar] [PubMed]
- López-Barragán, M.J.; Carmona, M.; Zamarro, M.T.; Thiele, B.; Boll, M.; Fuchs, G.; García, J.L.; Díaz, E. The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J. Bacteriol. 2004, 186, 5762–5774. [Google Scholar] [CrossRef] [PubMed]
- Durante-Rodríguez, G.; Zamarro, M.T.; García, J.L.; Díaz, E.; Carmona, M. Oxygen-dependent regulation of the central pathway for the anaerobic catabolism of aromatic compounds in Azoarcus sp. strain CIB. J. Bacteriol. 2006, 188, 2343–2354. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, J.A.; Shingler, V.; Carmona, M.; Díaz, E. AccR is a master regulator involved in carbon catabolite repression of the anaerobic catabolism of aromatic compounds in Azoarcus sp. CIB. J. Biol. Chem. 2014, 289, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Rusell, D. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- De Lorenzo, V.; Timmis, K.N. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994, 235, 386–405. [Google Scholar] [PubMed]
- Casadaban, M.J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 1976, 104, 541–555. [Google Scholar] [CrossRef]
- Wing, H.J.; Green, J.; Guest, J.R.; Busby, S.J. Role of activating region 1 of Escherichia coli FNR protein in transcription activation at class II promoter. J. Biol. Chem. 2000, 275, 29061–29065. [Google Scholar] [CrossRef]
- Durante-Rodríguez, G.; Mancheño, J.M.; Rivas, G.; Alfonso, C.; García, J.L.; Díaz, E.; Carmona, M. Identification of a missing link in the evolution of an enzyme into a transcriptional regulator. PLoS ONE 2013, 8, e57518. [Google Scholar] [CrossRef]
- Kovach, M.; Elzer, P.H.; Hill, D.S.; Robertson, G.T.; Farris, M.A.; Roop, R.M.I.; Peterson, K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Marshall, C.; Labrousse, V.; Kreimer, M.; Weichart, D.; Kolb, A.; Hengge-Aronis, R. Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on σs and requires activation by cAMP-CRP. J. Mol. Biol. 1998, 276, 339–353. [Google Scholar] [CrossRef]
- Ferrández, A.; Miñambres, B.; García, B.; Olivera, E.R.; Luengo, J.M.; García, J.L.; Díaz, E. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 1998, 273, 25974–26986. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Press: New York, NY, USA, 1972; pp. 352–355. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (NCBI) Server. Available online: http://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 26 June 2019).
- Wilbur, W.J.; Lipman, D.J. Rapid similarity searches of nucleic acid and protein data banks. Proc. Natl. Acad. Sci. USA 1983, 80, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Claverie, F.; Magasanik, B. Positive and negative effects of DNA bending on activation of transcription from a distant site. J. Mol. Biol. 1992, 227, 996–1008. [Google Scholar] [CrossRef]
- Carmona, M.; Magasanik, B. Activation of transcription at σ54-dependent promoters on linear templates requires intrinsic or induced bending of the DNA. J. Mol. Biol. 1996, 261, 348–356. [Google Scholar] [CrossRef]
- Collins, T.J. ImageJ for microscopy. Biotechniques 2007, 43, 25–30. [Google Scholar] [CrossRef]
- Moreno-Herrero, F.; De Pablo, P.J.; Fernández-Sánchez, R.; Colchero, J.; Gómez-Herrero, J.; Baró, A.M. Scanning force microscopy jumping and tapping modes in liquids. Appl. Phys. Lett. 2002, 81, 2620–2622. [Google Scholar] [CrossRef]
- Horcas, I.; Fernández, R.; Gómez-Rodríguez, J.M.; Colchero, J.; Gómez-Herrero, J.; Baro, A.M. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007, 78, 013705. [Google Scholar] [CrossRef]
- Rabus, R.; Trautwein, K.; Wöhlbrand, L. Towards habitat-oriented systems biology of “Aromatoleum aromaticum” EbN1: Chemical sensing, catabolic network modulation and growth control in anaerobic aromatic compound degradation. Appl. Microbiol. Biotechnol. 2014, 98, 3371–3388. [Google Scholar] [CrossRef] [PubMed]
- Del Solar, G.H.; Pérez-Martín, J.; Espinosa, M. Plasmid pLS1-enconded RepA protein regulates transcription from repAB promoter by binding to a DNA sequence containing a 13-base pair symmetric element. J. Biol. Chem. 1990, 265, 12569–12575. [Google Scholar] [PubMed]
- Gomis-Ruth, F.X.; Sola, M.; Acebo, P.; Parraga, A.; Guasch, A.; Eritja, R.; González, A.; Espinosa, M.; del Solar, G.; Coll, M. The structure of plasmid-encoded transcriptional repressor CopG unligated and bound to its operator. EMBO J. 1998, 17, 7404–7415. [Google Scholar] [CrossRef] [PubMed]
- González-Pérez, M.M.; Ramos, J.L.; Gallegos, M.T.; Marqués, S. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J. Biol. Chem. 1999, 274, 2286–2290. [Google Scholar] [CrossRef] [PubMed]
- Cowles, C.E.; Nichols, N.N.; Harwood, C.S. BenR, a XylS homologue regulates three different pathways of aromatic acid degradation in Pseudomonas putida. J. Bacteriol. 2000, 182, 6339–6346. [Google Scholar] [CrossRef] [PubMed]
- Körner, H.; Sofia, H.J.; Zumft, W.G. Phylogeny of the bacterial superfamily of Crp-Fnr transcriptional regulators: Exploiting the metabolic spectrum by controlling alterative gene programas. FEMS Microbiol. Rev. 2003, 27, 559–592. [Google Scholar] [CrossRef]
- Hansma, H. Surface biology of DNA by atomic force microscopy. Annu. Rev. Phys. Chem. 2001, 52, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Rivetti, C.; Guthold, M. Single DNA molecule analysis of transcription complexes. Methods Enzymol. 2003, 371, 34–50. [Google Scholar]
- Yang, Y.; Sass, L.; Du, C.; Hsieh, P.; Erie, D. Determination of protein-DNA binding constants and specificities from statistical analyses of single molecule: MutS-DNA interactions. Nucleic Acids Res. 2005, 33, 4322–4334. [Google Scholar] [CrossRef]
- Zhang, J.; Zeuner, Y.; Kleefeld, A.; Unden, G.; Janshoff, A. Multiple site-specific binding of Fis protein to Escherichia coli nuoA-N promoter DNA and its impact on DNA topology visualized by means of scanning force microscopy. ChemBioChem 2004, 5, 1286–1289. [Google Scholar] [CrossRef]
- Gutiérrez-del-Arroyo, P. Caracterización Mediante Microscopía de Fuerzas Atómicas de Complejos de Regulación Transcripcional. Ph.D. Thesis, Universidad Autónoma de Madrid, Madrid, Spain, 2009. [Google Scholar]
- Jia, Y.; Bi, L.; Li, F.; Chen, Y.; Zhang, C.; Zhang, X. α-shaped DNA loops induced by MutS. Biochem. Biophys. Res. Commun. 2008, 372, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Lacal, J.; Guazzaroni, M.E.; Gutiérrez del Arroyo, P.; Busch, A.; Vélez, M.; Krell, T.; Ramos, J.L. Two level of comparativeness in the binding of TodT to the tod operon promoter. J. Mol. Biol. 2008, 384, 1037–1047. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).