The Challenge of the Sponge Suberites domuncula (Olivi, 1792) in the Presence of a Symbiotic Bacterium and a Pathogen Bacterium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Specimen Collection and Ethical Statement
2.2. Bacterial Cultures
2.3. Lipopolysaccharides Preparation
2.4. Sponge Cell Dissociation and Primmorphs Culture
2.5. Quantitative Reverse Transcription (qRT-PCR) Analyses
3. Results
3.1. Interactions between Sponge-Isolated Bacteria and Primmorphs Cultures: Impact at Macroscopic and Molecular Levels
3.2. Co-cultivation of Primmorphs and Alive Bacteria
3.3. Incubation of Primmorphs in the Presence of Bacterial Culture Supernatant
3.4. Effects of LPS on Immune and Apoptotic Genes
4. Discussion
4.1. TLR
4.2. BHP-2
4.3. Caspase
4.4. TRAF-6
4.5. MPEG
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gloeckner, V.; Wehrl, M.; Moitinho-Silva, L.; Gernert, C.; Schupp, P.; Pawlik, J.R.L.; Erpenbeck, D.; Wörheide, G.; Hentschel, U. The HMA-LMA dichotomy revisited an electron microscopical survey of 56 sponge species. Biol. Bull 2014, 227, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, U.; Hopke, J.; Horn, M.; Friedrich, A.B.; Wagner, M.; Hacker, J.; Moore, B.S. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 2002, 68, 4431–4440. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.W.; Hill, R.T.; Piel, J.; Thacker, R.W.; Hentschel, U. Soaking it up: The complex lives of marine sponges and their microbial associates. ISME J. 2007, 1, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Hentschel, U.; Usher, K.M.; Taylor, M.W. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 2006, 55, 167–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reveillaud, J.; Maignien, L.; Murat, E.A.; Huber, J.A.; Apprill, A.; Sogin, M.L.; Vanreusel, A. Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 2014, 8, 1198–1209. [Google Scholar] [CrossRef] [PubMed]
- Zilber-Rosenberg, I.; Rosenberg, E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol. Rev. 2008, 32, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Webster, N.S.; Thomas, T. The sponge hologenome. mBio 2016, 7, e00135–e00216. [Google Scholar] [CrossRef] [PubMed]
- Gardères, J.; Taupin, L.; Bin Saidin, J.; Dufour, A.; Le Pennec, G. N-acyl homoserine lactone production by bacteria within the sponge Suberites domuncula (Olivi, 1792) (Porifera, Demospongiae). Mar. Biol 2012. [Google Scholar] [CrossRef]
- Gardères, J.; Henry, J.; Bernay, B.; Ritter, A.; Zatylny-Gaudin, C.; Wiens, M.; Müller, W.E.; Le Pennec, G. Cellular effects of bacterial N-3-Oxo-dodecanoyl-L-Homoserine lactone on the sponge Suberites domuncula (Olivi, 1792): Insights into an intimate inter-kingdom dialogue. PLoS ONE 2014, 9, e97662. [Google Scholar]
- Wiens, M.; Korzhev, M.; Krasko, A.; Thakur, N.L.; Perović-Ottstadt, S.; Breter, H.J.; Ushijima, H.; Diehl-Seifert, B.; Müller, I.M.; Müller, W.E.G. Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway induction of a perforin-like molecule. J. Biol. Chem. 2005, 280, 27949. [Google Scholar] [CrossRef]
- Wiens, M.; Korzhev, M.; Perrovic-Ottstadt, S.; Luthringer, B.; Brandt, D.; Klein, S.; Müller, W.E.G. Toll-like receptors are part of the innate immune defense system of sponges (Demospongiae: Porifera). Mol. Biol. Evol. 2007, 24, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Thakur, A.N.; Thakur, N.L.; Indap, M.M.; Pandit, R.A.; Datar, V.V.; Müller, W.E. Antiangiogenic, antimicrobial, and cytotoxic potential of sponge-associated bacteria. Mar. Biotechnol. 2005, 7, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Rusch, D.; DeMaere, M.Z.; Yung, P.Y.; Lewis, M.; Halpern, A.; Heidelberg, K.B.; Egan, S.; Steinberg, P.D.; Kjelleberg, S. Functional genomic signatures of sponge bacteria reveal unique and shared features of symbiosis. ISME J. 2010, 4, 1557–1567. [Google Scholar] [CrossRef] [PubMed]
- Siegl, A.; Kamke, J.; Hochmuth, T.; Piel, J.; Richter, M.; Liang, C.; Dandekar, T.; Hentschel, U. Single-cell genomics reveals the lifestyle of Poribacteria, a candidate phylum symbiotically associated with marine sponges. ISME J. 2011, 5, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Müller, W.E.; Zahn, R.K.; Kurelec, B.; Lucu, C.; Müller, I.; Uhlenbrucg, G. Lectin, a possible basis for symbiosis between bacteria and sponges. J. Bacteriol. 1981, 145, 548–558. [Google Scholar]
- Gardères, J.; Bourgeut-Kondracki, M.L.; Hamer, B.; Batel, R.; Schröder, H.C.; Müller, W.E. Porifera lectins: Diversity, physiological roles and biotechnological potential. Mar. Drugs 2015, 13, 5059–5101. [Google Scholar] [CrossRef] [PubMed]
- Gardères, J.; Bedoux, G.; Koutsouveli, V.; Crequer, S.; Desriac, F.; Le Pennec, G. Lipopolysaccharides from Commensal and Opportunistic Bacteria: Characterization and Response of the Immune System of the Host Sponge Suberites domuncula. Mar. Drugs 2015, 13, 4985–5006. [Google Scholar] [CrossRef]
- Le Pennec, G.; Perovic, S.; Ammar, M.S.A.; Grebenjik, V.A.; Steffen, R.; Brümme, R.F.; Müller, W.E.G. Cultivation of primmorphs from the marine sponge Suberites domuncula: Morphogenetic potential of silicon and iron. Mar. Biotechnol. 2003, 100, 93–108. [Google Scholar] [CrossRef]
- Westphal, O.; Jann, K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 1965, 5, 83–91. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Morin, D.; Grasland, B.; Vallée-Réhel, K.; Dufau, C.; Haras, D. On-line high-performance liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum-sensing signal molecules, in the presence of biological matrices. J. Chromatogr. A. 2003, 1002, 79–91. [Google Scholar] [CrossRef]
- Takedad, K.; Kaishod, T.; Akira, S. Toll-like receptors. Annu. Rev. Immunol. 2003, 21, 335–376. [Google Scholar] [CrossRef] [PubMed]
- Kirsching, C.J.; Bauer, S. Toll-like receptors: cellular signal transducers for exogenous molecular patterns causing immune response. Int. J. Med. Microbiol. 2001, 291, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R. Recognition of microorganisms and activation of the immune response. Nature 2007, 449, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Cai, S.Y.; Shai, J.Z.; Chen, J. Toll-like receptors, associated biological roles, and signaling networks in non-mammals. Front. Immun. 2018, 1523. [Google Scholar] [CrossRef]
- Gauthier, M.E.A.; Pasquier, L.D.; Degnan, B.M. The genome of the sponge Amphimedon queenslandica provides new perspectives into the origin of toll-like and interleukin 1 receptor pathways. Evol. Dev. 2010, 12, 519. [Google Scholar] [CrossRef]
- D’Hennezel, E.; Abubucker, S.; Murhy, L.O.; Cullen, T.W. Total lipopolysaccharide from the human gut microbiome silences Toll-liker receptor signaling. mSystems 2017, 2. [Google Scholar] [CrossRef]
- Patterson, A.M.; Mulder, I.E.; Travis, A.J.; Lan, A.; Cerf-Bensussan, N.; Gaboriau-Routhiau, V.; Garden, K.; Logan, E.; Delday, M.I. Human gut symbiont Roseburia hominis promotes and regulates innate immunity. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef]
- Rasmont, R. Une technique de culture des éponges d’eau douce en milieu contrôlé. Idib 1963, 91, 147–156. [Google Scholar]
- Killian, E.F. Zur biologie der einheimischen spogilliden. Ergebnisse und problem. Zool. Beitr. 1964, 10, 85–159. [Google Scholar]
- Schmidt, I. Phagocytose et pinocytose chez les spongillidae. Etude in vivo de l’ingestion de bactéries et de protéines marquées à l’aide d’un colorant fluorescent en lumière ultraviolette. Z. Vergl. Physiol. 1969, 66, 398–420. [Google Scholar] [CrossRef]
- Kale, J.; Osterlund, E.J.; Andrews, D.D.W. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Diff. 2018, 25, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Han, C.; Liu, J. The role of Toll-like receptors in oncotherapy. Oncol. Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Wiens, M.; Perovic-Ottstadt, S.; Müller, I.M.; Müller, W.E.G. Allograft rejection in the mixed cell reaction system of the demosponge Suberites domuncula is controlled by differential expression of apoptotic genes. Immunogenetics 2004, 56, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Wiens, M.; Belikov, S.I.; Kaluzhnaya, O.V.; Schröder, H.C.; Hamer, B.; Perovic-Ottstadt, S.; Borejko, A.; Luthringer, B.; Müller, I.M.; Müller, W.E. Axial (apical-basal) expression of pro-apoptotic and pro-survival genes in the lake baikal demosponge Lubomirskia baicalensis. DNA Cell Biol. 2006, 25, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Longo, C.; Cardone, F.; Corriero, G.; Licciano, M.; Pierri, C.; Stabili, L. The co-occurrence of the demosponge Hymeniacidon perlevis and the edible mussel Mytilus galloprovincialis as a new toll for bacterial load mitigation in aquaculture. Environ. Sci. Pollut. Res. Int. 2016, 23, 3736–3746. [Google Scholar] [CrossRef]
- Müller, W.E.G. Molecular phylogeny of Eumetazoa: Genes in sponges (Porifera) give evidence for monophyly of animals. Prog. Mol. Subcell Biol. 1998, 19, 89–132. [Google Scholar] [PubMed]
- Kumar, S. Caspase function in programmed cell death. Death Differ. 2007, 14, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Tsatsanoris, J.A.; Franch-Arroyo, S.; Resch, U.; Charpentier, E. Extracellular vesicle RNA: a universal mediator of microbial communication. Opinion. 2018, 26, 401–410. [Google Scholar]
- Kell, D.B.; Pretorius, E. On the translocation of bacteria and their lipopolysaccharides between blood and peripheral locations in chronic, inflammatory diseases: the central roles of LPS and LPS-induced cell death. Integr Biol. 2015, 7, 1339–1377. [Google Scholar] [CrossRef] [Green Version]
- Freche, B.; Reig, N.; Vand der Goot, F.G. The role of the inflammasome in cellular response to toxins and bacterial effectors. Semin. Immunopathol. 2007, 29, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Simakov, O.; Chapman, J.; Fahey, B.; Gauthier, M.E.; Mitros, T.; Richards, G.S.; Conaco, C.; Dacre, M.; Hellsten, U.; et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 2010, 466, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.C.; Lee, J.; Choi, Y. Tumor necrosis factor receptor-associated factor 6 (TRAF-6) regulation of development, function, and homeostasis of the immune system. Immunol. Rev. 2015, 266, 72–92. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H.; Ishino, T.; Shimamoto, Y.; Okabe, J.; Miyamoto, T.; Takahashi, E.; Miyoshi, I. Ubiquitin-specific protease 2 modulates the lipopolysaccharide-elicited expression of proinflamattory cytokines in macrophages-like HL-60 cells. Mediat. Inflamm. 2017, 2017, 6909415. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; McBride, D.W.; Zhang, J.H. Axl activation attenuates neuroinflammation by inhibiting the TLR/TRAF/NF-ΚB pathway after MCAO in rats. Neurobiol. Dis. 2018, 110, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Deepika, A.; Sreedharan, K.; Paria, A.; Makesh, M.; Rajendran, K.V. Toll-pathway in tiger shrimp (Penaeus monodon) responds to white spot syndrome virus infection: evidence through molecular characterization and expression profiles of MyD88, TRAF-6 and TLR genes. Fish Shellf. Immunol. 2014, 41, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Huang, Y.; Wang, B.; Jian, J.; Xu, Y. Tumor necrosis factor receptor-associated factor 6 (TRAF-6) participates in peroxinectin gene expression in Fenneropenaeus penicillatus. Fish Shellf. Immunol. 2017, 64, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Qu, F.; Xiang, Z.; Zhou, Y.; Qin, Y.; Yu, Z. Tumor necrosis factor receptor-associated factor 3 from Anodonta woodiana is an important factor in bivalve immune response to pathogen infection. Fish Shellf. Immunol. 2017, 71, 151–159. [Google Scholar] [CrossRef]
- Sun, W.W.; Zhang, X.X.; Wan, W.S.; Wang, S.Q.; Wen, X.B.; Zheng, H.P.; Zhang, Y.L.; Li, S.K. Tumor necrosis factor receptor-associated factor 6 (TNFR6) participates in anti-lipopolysaccharide factors (ALFs) gene expression in mud crab. Dev. Comp. Immunol. 2017, 67, 361–376. [Google Scholar] [CrossRef]
- Reiter, Y.; Ciobatariu, A.; Jones, J.; Morgan, B.P.; Fishelson, Z. Complement membrane attack complex, perforin, and bacterial exotoxins induce in K562 cells calcium-dependent cross-protection from lysis. J. Immunol. 1995, 155, 2203–2210. [Google Scholar]
- Xiong, P.; Shiratsuchi, M.; Matsushima, T.; Liao, J.; Tanaka, E.; Nakashima, Y.; Takayanagi, R.; Ogawa, Y. Regulation of expression and trafficking of perforin-2 by LPS and TNF-α. Cell Immunol. 2017, 320, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Krause, K.; Amer, A.O. Caspase exploitation by Legionella pneumophila. Front Microbiol. 2016, 7, 515. [Google Scholar] [CrossRef] [PubMed]
- Hochstrasser, H.; Hilbi, H. Intra-species and inter-kingdom signaling of Legionella pneumophila. Front Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Feuda, R.; Dohrmann, M.; Pett, W.; Philippe, H.; Rota-Stabelli, O.; Lartillot, N.; Wörheide, G.; Pisani, D. Improved modelling of compositional heterogeneity supports sponges as sister to all other animals. Curr. Bio. 2017, 27, 3864–3870. [Google Scholar] [CrossRef] [PubMed]
 OD600nm after inoculation of bacteria at T0;
OD600nm after inoculation of bacteria at T0;  OD600nm after 16h of co-culture.
OD600nm after 16h of co-culture.
 OD600nm after inoculation of bacteria at T0;
OD600nm after inoculation of bacteria at T0;  OD600nm after 16h of co-culture.
OD600nm after 16h of co-culture.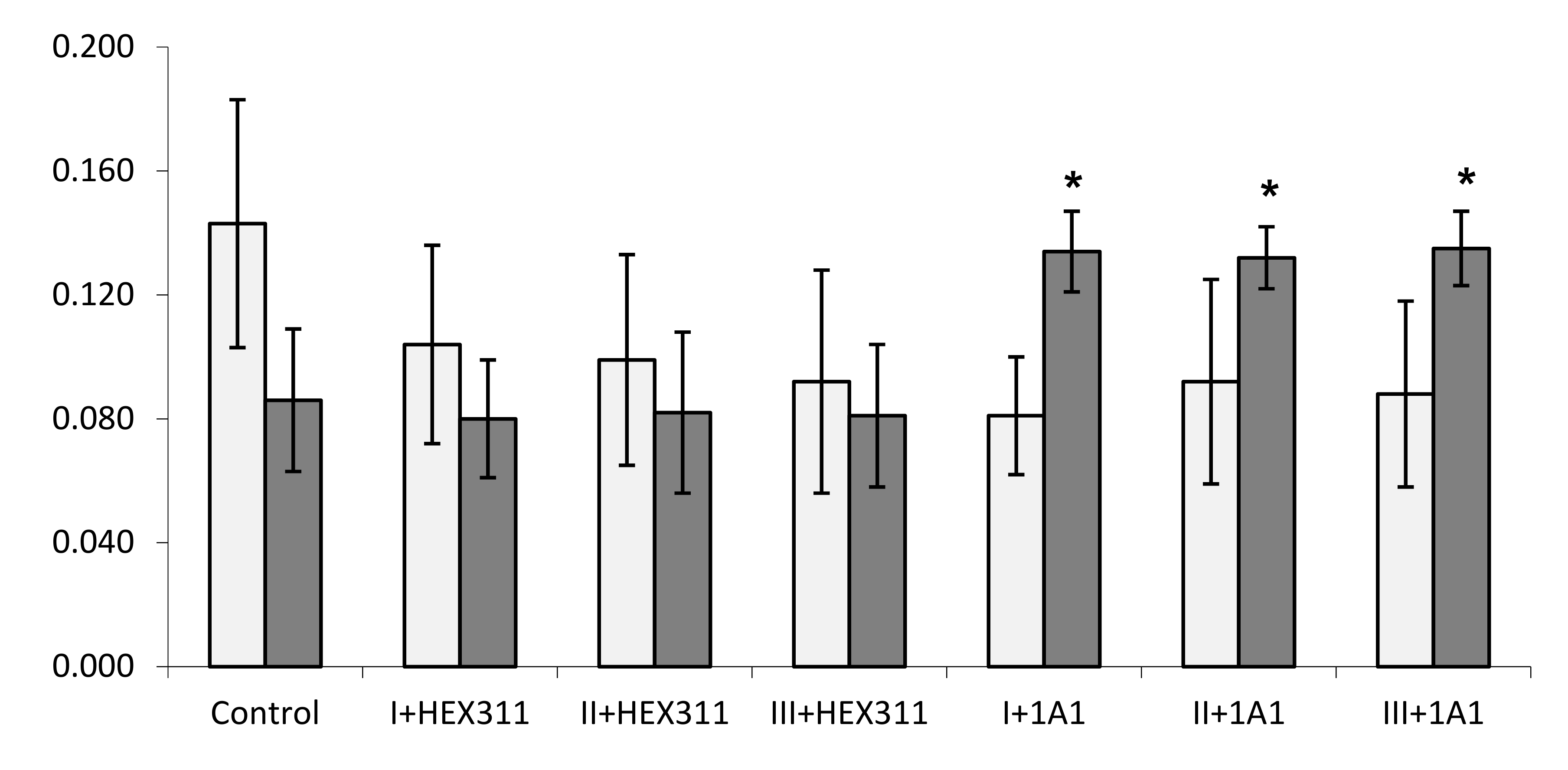
 ) or Pseudolateromonas sp. 1A1 (
) or Pseudolateromonas sp. 1A1 (  ). (*) Statistical differences between the controlled and the stimulated condition are materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.
). (*) Statistical differences between the controlled and the stimulated condition are materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.
 ) or Pseudolateromonas sp. 1A1 (
) or Pseudolateromonas sp. 1A1 (  ). (*) Statistical differences between the controlled and the stimulated condition are materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.
). (*) Statistical differences between the controlled and the stimulated condition are materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.
 ) or Pseudolateromonas sp. 1A1 (
) or Pseudolateromonas sp. 1A1 (  ), respectively. (*) Statistical differences between the controlled and the stimulated condition are materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.
), respectively. (*) Statistical differences between the controlled and the stimulated condition are materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.
 ) or Pseudolateromonas sp. 1A1 (
) or Pseudolateromonas sp. 1A1 (  ), respectively. (*) Statistical differences between the controlled and the stimulated condition are materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.
), respectively. (*) Statistical differences between the controlled and the stimulated condition are materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.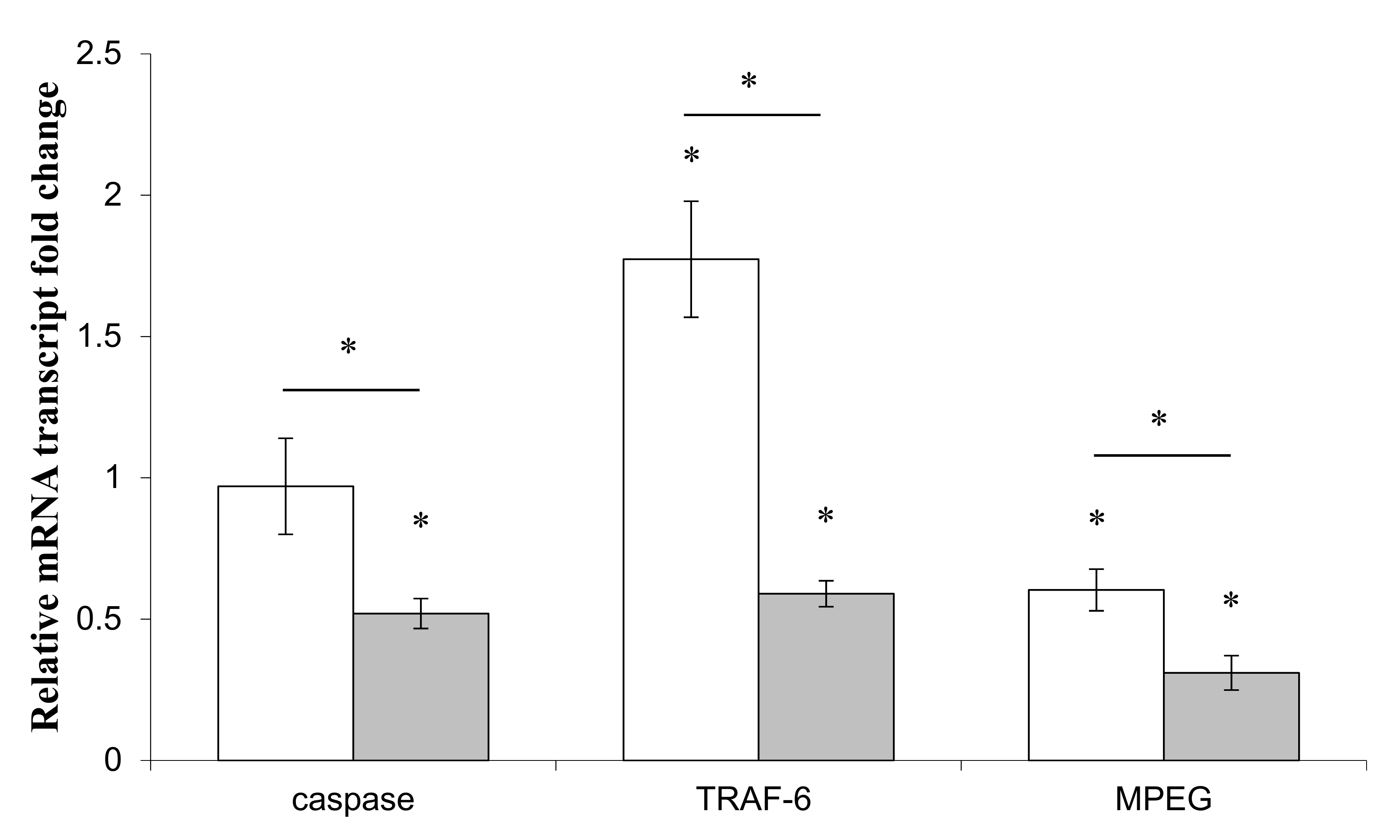
 ) or Pseudoalteromonas sp. 1A1 (
) or Pseudoalteromonas sp. 1A1 (  ) and the control bacterium Escherichia coli (
) and the control bacterium Escherichia coli (  ). (*) Statistical differences between the standard and the stimulated condition a materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.
). (*) Statistical differences between the standard and the stimulated condition a materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.
 ) or Pseudoalteromonas sp. 1A1 (
) or Pseudoalteromonas sp. 1A1 (  ) and the control bacterium Escherichia coli (
) and the control bacterium Escherichia coli (  ). (*) Statistical differences between the standard and the stimulated condition a materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.
). (*) Statistical differences between the standard and the stimulated condition a materialized by an asterisk and statistical differences between primmorphs stimulated by the two bacteria are materialized by a horizontal bar.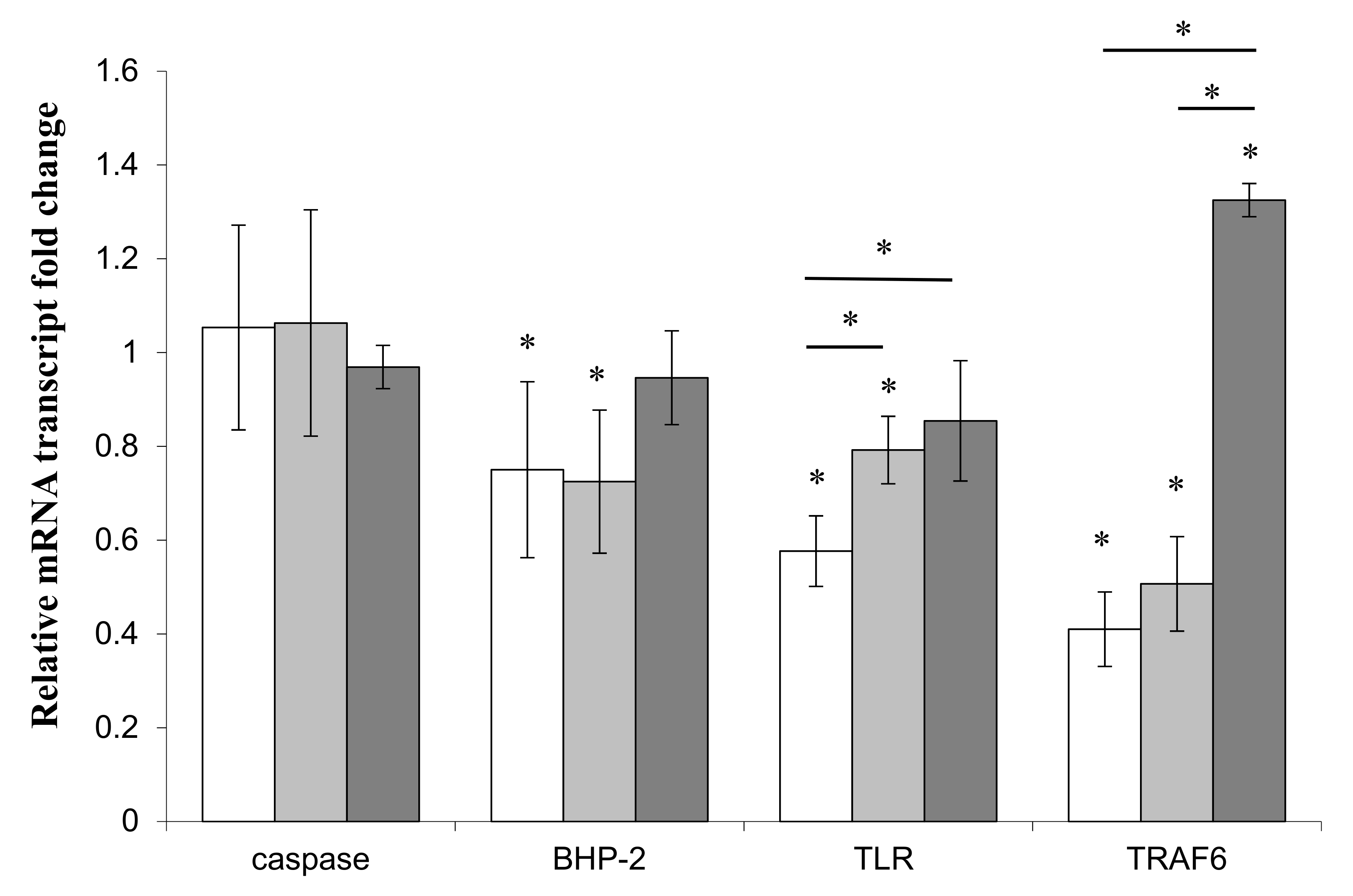
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Pennec, G.; Gardères, J. The Challenge of the Sponge Suberites domuncula (Olivi, 1792) in the Presence of a Symbiotic Bacterium and a Pathogen Bacterium. Genes 2019, 10, 485. https://doi.org/10.3390/genes10070485
Le Pennec G, Gardères J. The Challenge of the Sponge Suberites domuncula (Olivi, 1792) in the Presence of a Symbiotic Bacterium and a Pathogen Bacterium. Genes. 2019; 10(7):485. https://doi.org/10.3390/genes10070485
Chicago/Turabian StyleLe Pennec, Gaël, and Johan Gardères. 2019. "The Challenge of the Sponge Suberites domuncula (Olivi, 1792) in the Presence of a Symbiotic Bacterium and a Pathogen Bacterium" Genes 10, no. 7: 485. https://doi.org/10.3390/genes10070485
APA StyleLe Pennec, G., & Gardères, J. (2019). The Challenge of the Sponge Suberites domuncula (Olivi, 1792) in the Presence of a Symbiotic Bacterium and a Pathogen Bacterium. Genes, 10(7), 485. https://doi.org/10.3390/genes10070485




