Population Genetic Divergence and Environment Influence the Gut Microbiome in Oregon Threespine Stickleback
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collections
2.2. Gut Isolation and Soma Preservation and DNA Extraction for Genomic Analysis
2.3. Restriction Site-Associated DNA Sequencing (RAD) Library Construction and Single-Nucleotide Polymorphism Discovery and Genomic Analysis
2.4. Stickleback Population Structure
2.5. Microbiome DNA Extraction and 16S Sequencing
2.6. Microbiome DNA Sequence Processing Using DADA2 Pipeline
2.7. Microbiome Diversity
2.8. Linear Mixed Models
2.9. Microbiome Composition and Permutational Multivariate Analysis of Variance (PERMANOVA)
2.10. Amplicon Sequences Variants Abundance Enrichment Analysis
3. Results
3.1. Genetic Variation in Oregon Stickleback Is Partitioned Between Geographical Regions and Environment
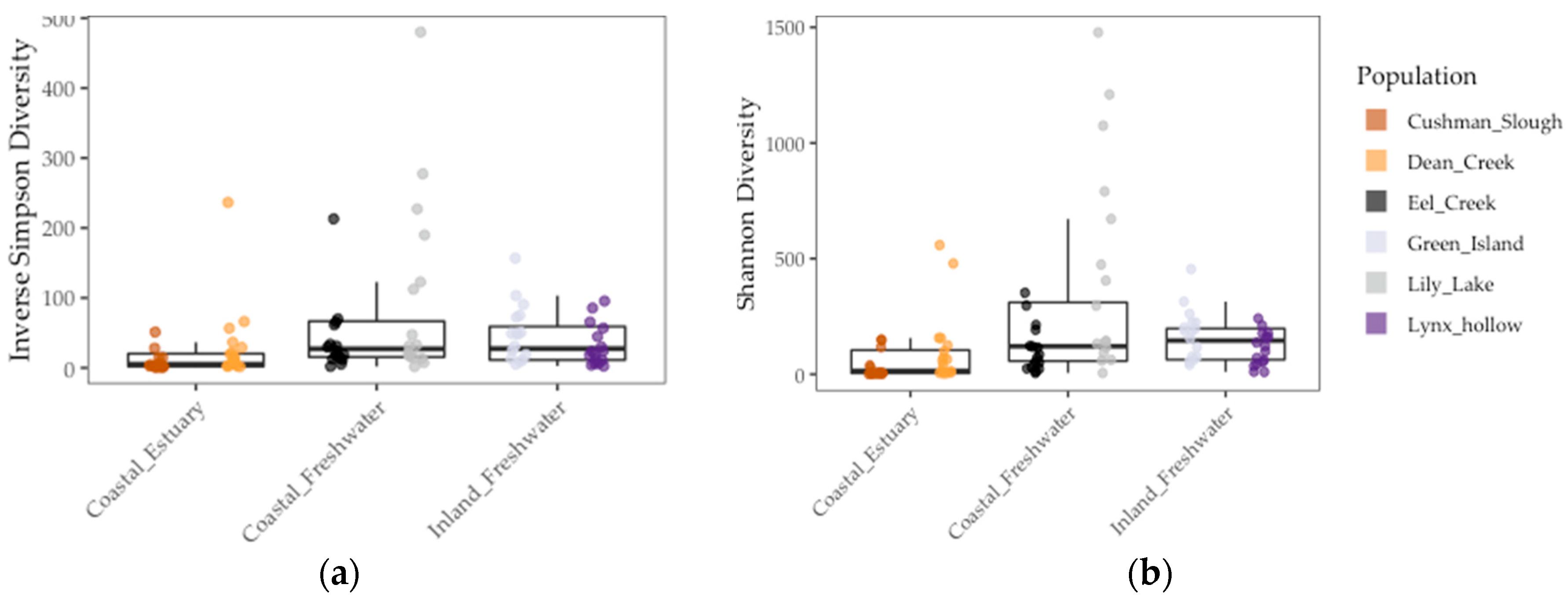
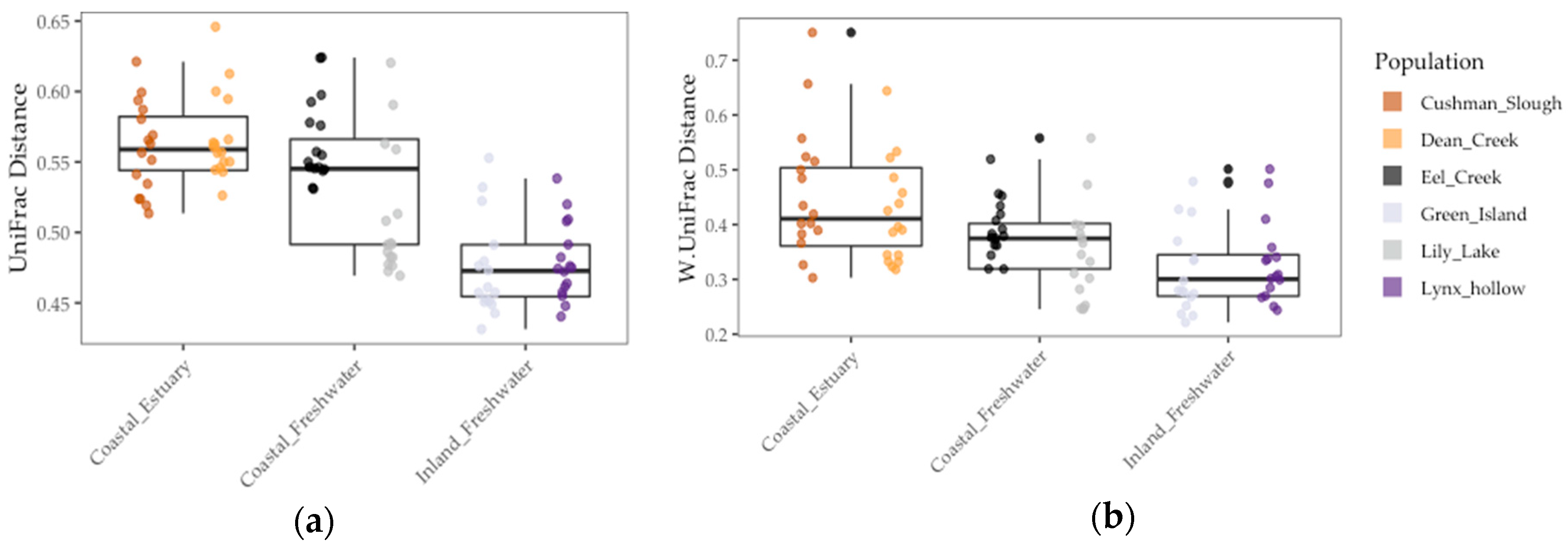
3.2. Stickleback Gut Microbiome Diversity and Composition Is Better Predicted by Population Genetic Divergence Than by Environmental and Geographic Differences
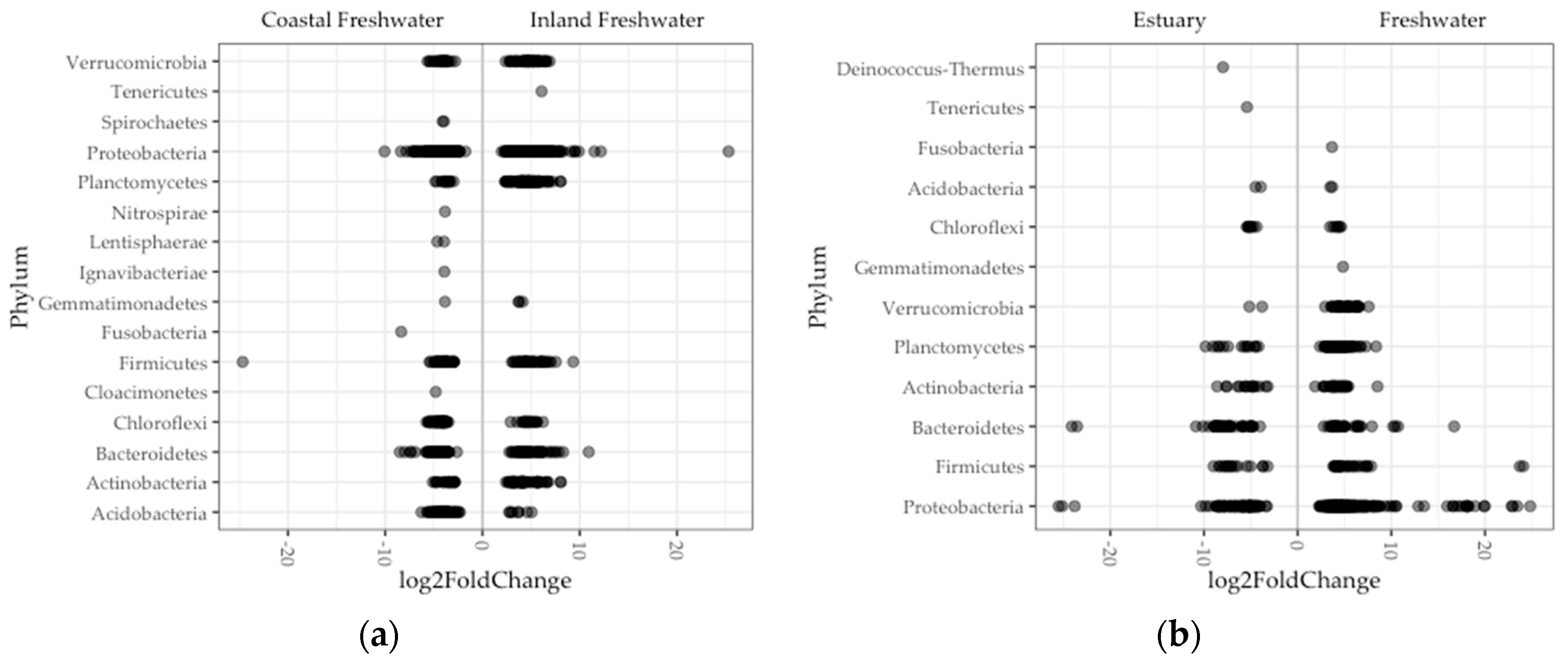
3.3. Differential Amplicon Sequences Variants Abundance Among Gut
4. Discussion
4.1. Host Population Genomic Divergence Correlated with Gut Microbiome Divergence
4.2. A Small Subset of Microbial Taxa Contributed to Microbiome Divergence Linked to Host Genetics and Environment
4.3. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of mammals and their gut microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Bäckhed, F. The gut microbiota—masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Kanther, M.; Rawls, J.F. Host-microbe interactions in the developing zebrafish. Curr. Opin. Immunol. 2010, 22, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Núñez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M. The gut microbiota and host health: A new clinical frontier. Gut 2015, 65, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Spector, T.D.; Clark, A.G.; Ley, R.E. Human genetics shape the gut microbiome. Cell 2014, 159, 789–799. [Google Scholar] [CrossRef]
- Green, J.; Bohannan, B.J.M. Spatial scaling of microbial biodiversity. Trends Ecol. Evol. 2006, 21, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.R.; Stephens, W.Z.; Stagman, K.; Wong, S.; Rawls, J.F.; Guillemin, K.; Bohannan, B.J. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 2016, 10, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Stephens, W.Z.; Burns, A.R.; Stagman, K.; Wong, S.; Rawls, R.F.; Guillemin, K.; Bohannan, B.J. The composition of the zebrafish intestinal microbial community varies across development. ISME J. 2016, 10, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Dolan, J.R. An introduction to the biogeography of aquatic microbes. Aquat. Microb. Ecol. 2005, 41, 39–48. [Google Scholar] [CrossRef]
- Martiny, J.B.H.; Martiny, J.B.; Bohannan, B.J.; Brown, J.H.; Colwell, R.K.; Fuhrman, J.A.; Green, J.L.; Horner-Devine, M.C.; Kane, M.; Krumins, J.A.; et al. Microbial biogeography: Putting microorganisms on the map. Nat. Rev. Microbiol. 2006, 4, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Crump, B.C.; Hopkinson, C.S.; Sogin, M.L. Microbial biogeography along an estuarine salinity gradient: Combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 2004, 70, 1494–1505. [Google Scholar] [CrossRef]
- Fortunato, C.S.; Herfort, L.; Zuber, P.; Baptista, A.M. Spatial variability overwhelms seasonal patterns in bacterioplankton communities across a river to ocean gradient. ISME J. 2012, 6, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Sullam, K.E.; Essinger, S.D.; Lozupone, C.A.; O’Connor, M.P.; Rosen, G.L.; Knight, R.; Kilham, S.S.; Russell, J.A. Environmental and ecological factors that shape the gut bacterial communities of fish: A meta-analysis. Mol. Ecol. 2012, 21, 3363–3378. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, S.; Yeoman, C.J.; Sipos, M.; Torralba, M.; Wilson, B.A.; Goldberg, T.L.; Stumpf, R.M.; Leigh, S.R.; White, B.A.; Nelson, K.E. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Rawls, J.F.; Mahowald, M.A.; Ley, R.E.; Gordon, J.I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 2006, 127, 423–433. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Akkermans, A.D.L.; Akkermans-van Vliet, W.M.; de Visser, J.A.G.; de Vos, W.M. The host genotype affects the bacterial community in the human gastronintestinal tract. Microb. Ecol. Health Dis. 2001, 13, 129–134. [Google Scholar]
- Turnbaugh, P.J.; Hamady, H.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.; De Soyza, A.; Bourke, S.J.; Perry, J.D.; Cummings, S.P. Assessment of sample handling practices on microbial activity in sputum samples from patients with cystic fibrosis. Lett. Appl. Microbiol. 2010, 51, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Benson, A.K.; Kelly, S.A.; Legge, R.; Ma, F.; Low, S.J.; Kim, J.; Zhang, M.; Oh, P.L.; Nehrenberg, D.; Hua, K.; et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl. Acad. Sci. 2010, 107, 18933–18938. [Google Scholar] [CrossRef] [PubMed]
- Leamy, L.J.; Kelly, S.A.; Nietfeldt, J.; Legge, R.M.; Ma, F.; Hua, K.; Sinha, R.; Petereson, D.A.; Walter, J.; Benson, A.K.; et al. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. 2014, 15, 552. [Google Scholar] [CrossRef]
- Davenport, E.R.; Cusanovich, D.A.; Michelini, K.; Barreiro, L.B.; Ober, C.; Gilad, Y. Genome-wide association studies of the human gut microbiota. PLoS ONE 2015, 10, e0140301. [Google Scholar] [CrossRef] [PubMed]
- Davey, J.W.; Blaxter, M.L. RAD-seq: Next-generation population genetics. Brief. Funct. Genomics 2010, 9, 416–423. [Google Scholar] [CrossRef]
- Hohenlohe, P.A.; Bassham, S.; Etter, P.D.; Stiffler, N.; Johnson, E.A.; Cresko, W.A. Population genomics of parallel adaptation in threespine stickleback using sequenced rad tags. PLoS Genet. 2010, 6, e1000862. [Google Scholar] [CrossRef]
- Lescak, E.A.; Milligan-Myhre, K.C. Teleosts as model organisms to understand host-microbe interactions. J. Bacteriol. 2017, 199, e00868-16. [Google Scholar] [CrossRef]
- Catchen, J.; Bassham, S.; Wilson, T.; Currey, M.; O’Brien, C.; Yeates, Q.; Cresko, W.A. The population structure and recent colonization history of Oregon threespine stickleback determined using RAD-seq. Mol. Ecol. 2013, 22, 2864–2883. [Google Scholar] [CrossRef]
- Small, C.M.; Milligan-Myhre, K.; Bassham, S.; Guillemin, K.; Cresko, W.A. Host genotype and microbiota contribute asymmetrically to transcriptional variation in the threespine stickleback gut. Genome Biol. Evol. 2017, 9, 504–520. [Google Scholar] [CrossRef]
- Milligan-Myhre, K.; Small, C.M.; Mittge, E.K.; Agarwal, M.; Currey, M.; Cresko, W.A.; Guillemin, K. Innate immune responses to gut microbiota differ between oceanic and freshwater threespine stickleback populations. Dis. Model. Mech. 2016, 9, 187–198. [Google Scholar] [CrossRef]
- Bolnick, D.I.; Snowberg, L.K.; Hirsch, P.E.; Lauber, C.L.; Knight, R.; Caporaso, J.G.; Svanbäck, R. Individuals’ diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch). Ecol. Lett. 2014, 17, 979–987. [Google Scholar] [CrossRef]
- Bolnick, D.I.; Snowberg, L.K.; Hirsch, P.E.; Lauber, C.L.; Org, E.; Parks, B.; Lusis, A.J.; Knight, R.; Caporaso, J.G.; Svanbäck, R. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat. Commun. 2014, 5, 4500. [Google Scholar] [CrossRef]
- Bolnick, D.I.; Snowberg, L.K.; Caporaso, J.G.; Lauber, C.; Knight, R.; Stutz, W.E. Major histocompatibility complex class IIb polymorphism influences gut microbiota composition and diversity. Mol. Ecol. 2014, 23, 4831–4845. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.; Snowberg, L.K.; Caporaso, J.; Knight, R.; Bolnick, D.I. Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 2015, 9, 2515–2526. [Google Scholar] [CrossRef]
- Currey, M.C.; Bassham, S.L.; Cresko, W.A. Genetic divergence outpaces phenotypic divergence among thresspine stickleback populations in old freshwater habitats. bioRxiv 2019. [Google Scholar] [CrossRef]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.D.; Reeder, J.; Lawrence, M.; Becker, G.; Brauer, M.J. GMAP and GSNAP for genomic sequence alignment: Enhancements to speed, accuracy, and functionality. Methods Mol. Biol. 2016, 1418, 283–334. [Google Scholar]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R: A language and environment for statistical computing. Available online: https://www.r-project.org/ (accessed on 1 February 2019).
- Small, M.C.; Currey, M.; Beck, E.A.; Bassham, S.; Cresko, W.A. Higly reproducable 16S sequencing facilitates measurement of the host genetic influences on the stickleback gut microbiome. bioRxiv 2018. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve bayesian classifier for rappid assignement of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagenr, H. Vegan: Community ecology package. Available online: https://cran.r-project.org/package=vegan (accessed on 1 February 2019).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 7, 1. [Google Scholar] [CrossRef]
- Kuznetsova, A.; Brockhoff, P.B.; Christensen, R.H.B. LmerTest: Tests for random and fixed effects for linear mixed effect models (lmer objects of lme4 package). J. Stat. Softw. 2017, 82, 13. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Holben, W.E.; Williams, P.; Saarinen, M.; Särkilahti, L.K.; Apajalahti, J.H.A. Phylogenetic analysis of intestinal microflora indicates a novel mycoplasma phylotype in farmed and wild salmon. Microb. Ecol. 2002, 44, 175–185. [Google Scholar] [CrossRef]
- Romero, J.; Ringø, E.; Merrifield, D.L. The gut microbiota of fish. In Aquaculture Nutrition, 1st ed.; Merrifield, D., Ringo, E., Eds.; John Wiley & Sons, Ltd: Chichester, UK, 2014; pp. 75–100. [Google Scholar]
- Llewellyn, M.S.; Boutin, S.; Hoseinifar, S.H.; Derome, N. Teleost microbiomes: the state of the art in their characterization, manipulation and importance in aquaculture and fisheries. Front. Microbiol. 2014, 5, 207. [Google Scholar] [CrossRef]
- Sasaki, Y.; Ishikawa, J.; Yamashita, A.; Oshima, K.; Kenri, T.; Furuya, K.; Yoshino, C.; Horino, A.; Shiba, T.; Sasaki, T.; et al. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 2002, 30, 5293–5300. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.R.; Watral, V.; Sichel, S.; Spagnoli, S.; Banse, A.V.; Mitthe, E.; Sharpton, T.J.; Guillemin, K.; Kent, M.L. Transmission of a common intestinal neoplasm in zebrafish by cohabitation. J. Fish Dis. 2018, 41, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.M.; Wiens, G.D.; Salinas, I. Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immuninol. 2019, 86, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Lyons, P.P.; Turnbull, J.F.; Dawson, K.A.; Crumlish, M. Phylogenetic and functional characterization of the distal intestinal microbiome of rainbow trout Oncorhynchus mykiss from both farm and aquarium settings. J. Appl. Microbiol. 2017, 122, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Gordon, J.I. The core gut microbiome, energy balance and obesity. J. Physiol. 2009, 587, 4153–4158. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Knight, R. Global patterns in bacterial diversity. Proc. Natl.Acad. Sci. 2007, 104, 11436–11440. [Google Scholar] [CrossRef] [PubMed]
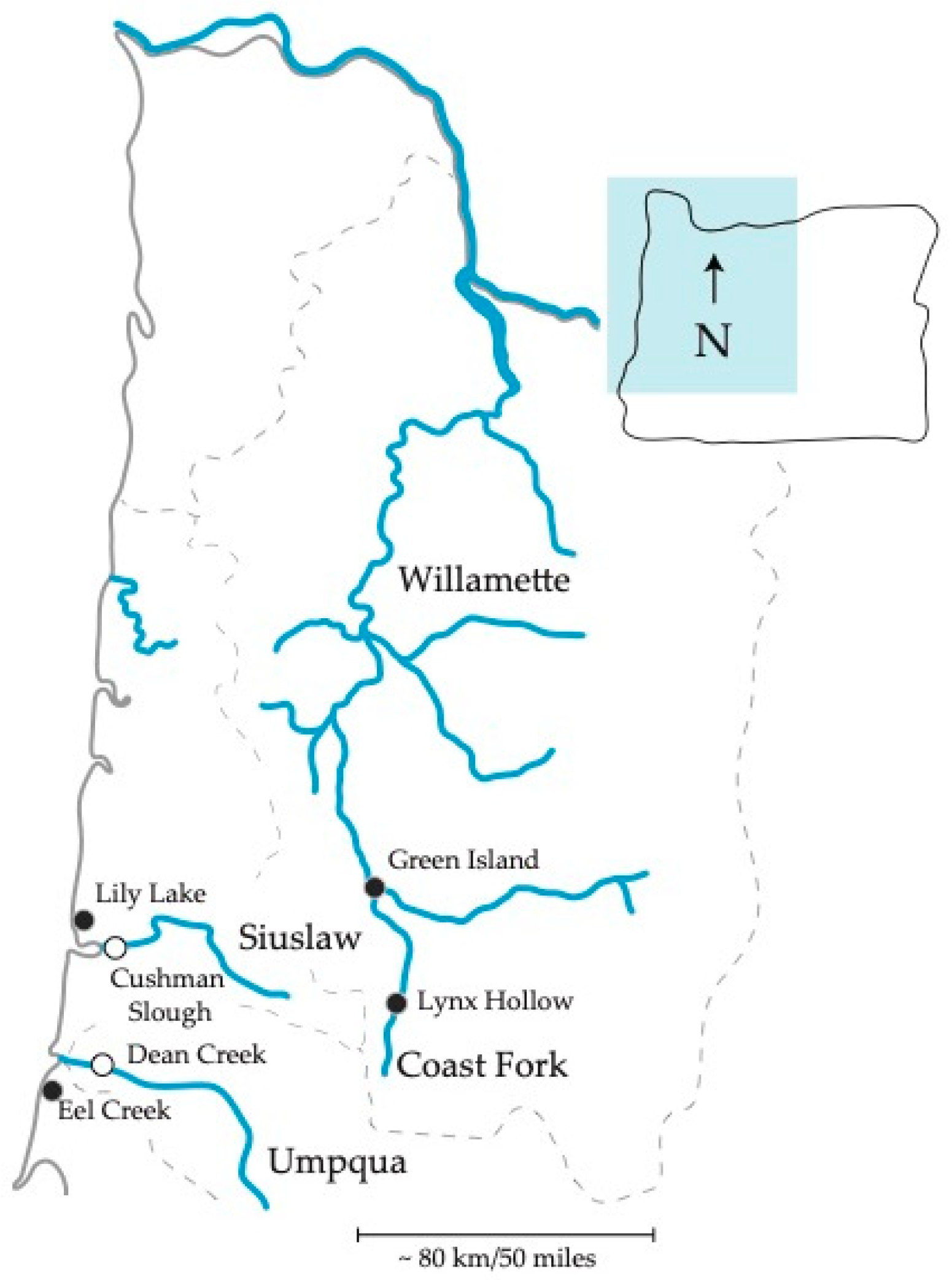

| Site | Region | Fin Clip Collected | Environment | Location | Gut Collected | Guts |
|---|---|---|---|---|---|---|
| Dean Creek | Coast | 2017 [37] | Estuary | N43.6960, W124.0170 | 06/13/17 | 20 |
| Eel Creek | Coast | 2017 [37] | Freshwater | N43.5869, W124.1861 | 06/13/17 | 20 |
| Cushman Slough | Coast | 2007,2009 [30] | Estuary | N43.9881, W124.0395 | 07/29/15 | 22 |
| Lily Lake | Coast | 2015 [37] | Freshwater | N44.0886, W124.1140 | 08/04/15 | 22 |
| Green Island | Inland | 2013 [37] | Freshwater | N44.1582, W123.1189 | 07/24/15 | 22 |
| Lynx Hollow | Inland | 2013 [37] | Freshwater | N43.8613, W123.0249 | 07/03/17 | 20 |
| Site | Input | Filtered | Merged | Nonchimeric |
|---|---|---|---|---|
| Cushman Slough | 9,256,421 | 4,888,393 | 4,298,407 | 4,264,096 |
| Dean Creek | 3,359,145 | 3,069,584 | 2,521,985 | 2,204,735 |
| Eel Creek | 2,287,323 | 2,055,175 | 1,905,651 | 1,663,629 |
| Lily Lake | 10,541,264 | 7,072,180 | 6,777,026 | 6,721,944 |
| Green Island | 8,616,378 | 4,260,381 | 3,699,754 | 3,681,706 |
| Lynx Hollow | 4,232,833 | 3,939,422 | 3,624,234 | 3,426,427 |
| Eel Creek | Cushman Slough | Lily Lake | Green Island | Lynx Hollow | |
|---|---|---|---|---|---|
| Dean Creek | 0.1293 | 0.0212 | 0.0698 | 0.3117 | 0.3942 |
| Eel Creek | 0.0919 | 0.0793 | 0.4709 | 0.5748 | |
| Cushman Slough | 0.0653 | 0.2392 | 0.2789 | ||
| Lily Lake | 0.3028 | 0.3661 | |||
| Green Island | 0.1488 |
| LMM | Inverse Simpson | Shannon | Unweighted UniFrac | Weighted UniFrac | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fixed | F | df | p | F | df | p | F | df | p | F | df | p |
| Environment | 2.03 | 1 | 0.23 | 2.02 | 1 | 0.22 | 3.36 | 1 | 0.14 | 6.94 | 1 | <0.08 |
| Random | χ2 | χ2 | χ2 | χ2 | ||||||||
| Pop. FST | 5.53 | 1 | <0.02 | 13.77 | 1 | <0.001 | 29.32 | 1 | <0.001 | 1.07 | 1 | 0.30 |
| Sample Period | 0.0 | 1 | 1.0 | 0.00 | 1 | 1.0 | 0.0 | 1 | 1.0 | 0.22 | 1 | 0.64 |
| PERMANOVA | R2 | df | p-value |
|---|---|---|---|
| Population FST | 0.080 | 1 | 0.001 |
| Environment | 0.058 | 1 | 0.001 |
| Sample Period | 0.053 | 1 | 0.001 |
| River Miles | 0.053 | 1 | 0.001 |
| Population Heterozygosity | 0.050 | 1 | 0.001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Steury, R.A.; Currey, M.C.; Cresko, W.A.; Bohannan, B.J.M. Population Genetic Divergence and Environment Influence the Gut Microbiome in Oregon Threespine Stickleback. Genes 2019, 10, 484. https://doi.org/10.3390/genes10070484
Steury RA, Currey MC, Cresko WA, Bohannan BJM. Population Genetic Divergence and Environment Influence the Gut Microbiome in Oregon Threespine Stickleback. Genes. 2019; 10(7):484. https://doi.org/10.3390/genes10070484
Chicago/Turabian StyleSteury, Robert A., Mark C. Currey, William A. Cresko, and Brendan J. M. Bohannan. 2019. "Population Genetic Divergence and Environment Influence the Gut Microbiome in Oregon Threespine Stickleback" Genes 10, no. 7: 484. https://doi.org/10.3390/genes10070484
APA StyleSteury, R. A., Currey, M. C., Cresko, W. A., & Bohannan, B. J. M. (2019). Population Genetic Divergence and Environment Influence the Gut Microbiome in Oregon Threespine Stickleback. Genes, 10(7), 484. https://doi.org/10.3390/genes10070484





