Genome-Wide Identification of Na+/H+ Antiporter (NHX) Genes in Sugar Beet (Beta vulgaris L.) and Their Regulated Expression under Salt Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Characterization of the NHX Genes in Sugar Beet
2.2. Phylogenetic Analysis
2.3. Chromosome Distributioof BvNHX Genes and Analysis of Ka/Ks Ratio
2.4. Analysis of Conserved Motifs, Gene Structures, and cis-Acting Elements
2.5. Three-Dimensional Structural Prediction of BvNHX Proteins
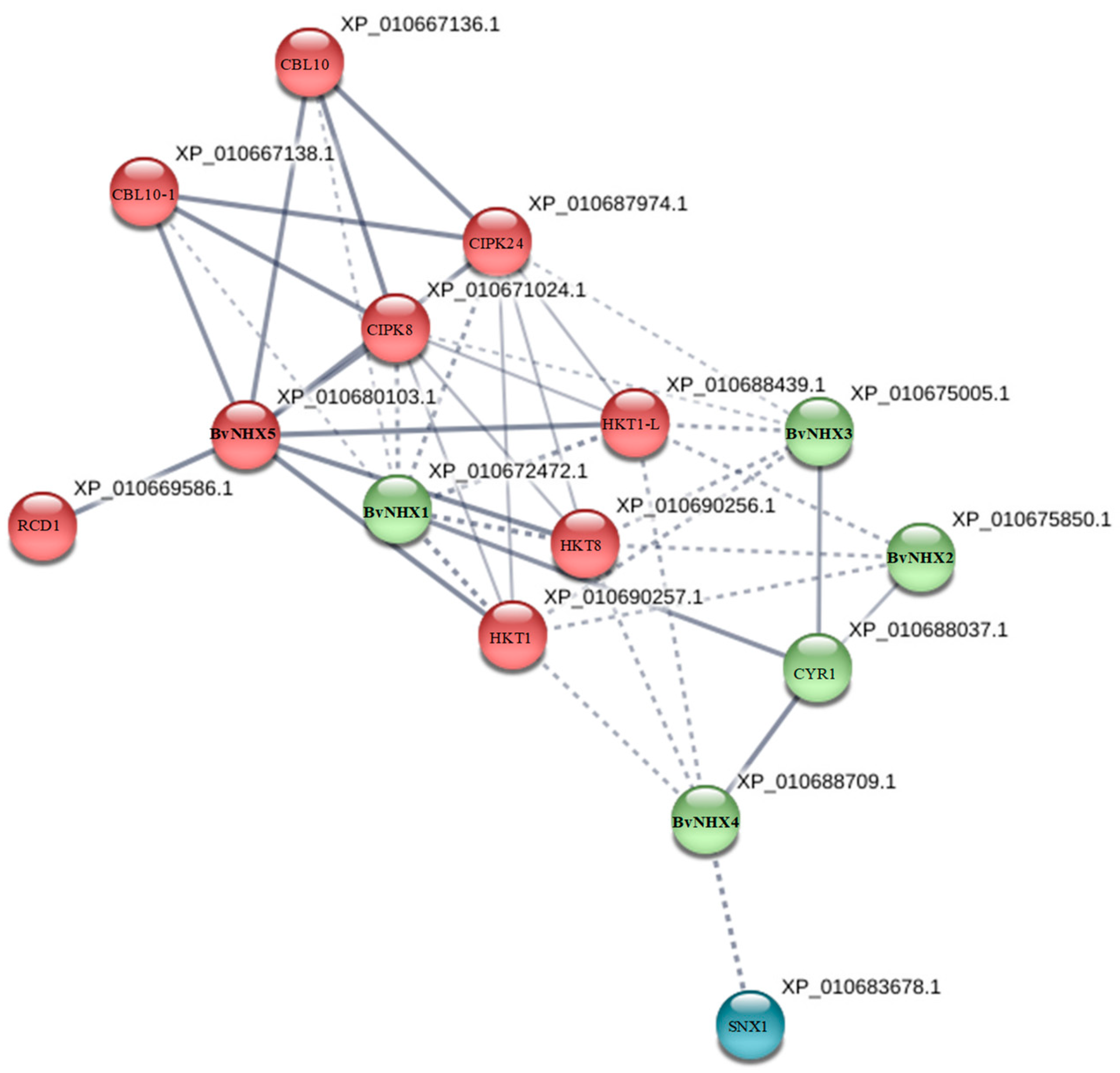
2.6. Protein-Protein Interaction Prediction of BvNHX Proteins
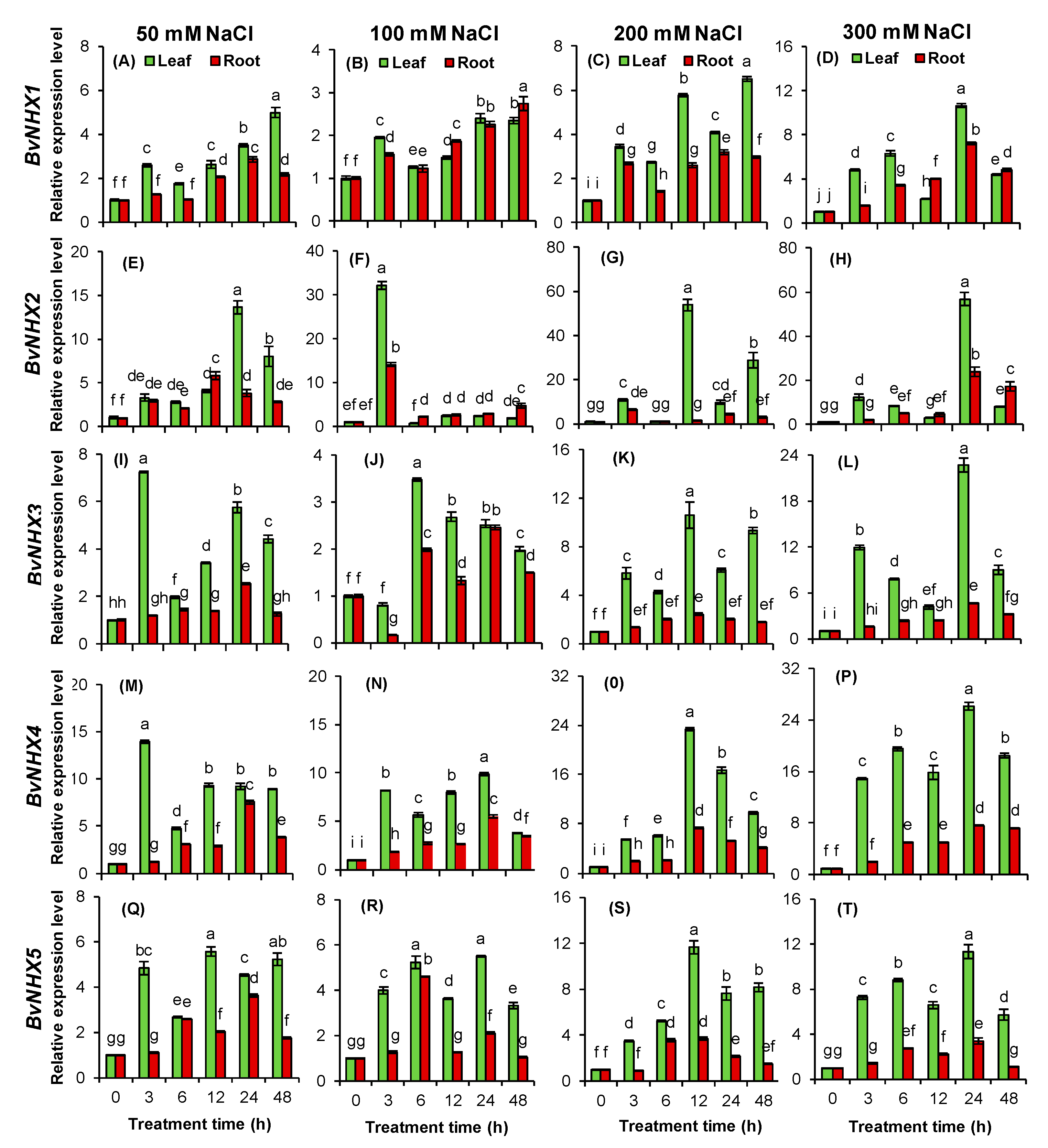
2.7. Plant Material, Treatment, and qRT-PCR Analysis
3. Results
3.1. Identification of BvNHX Genes
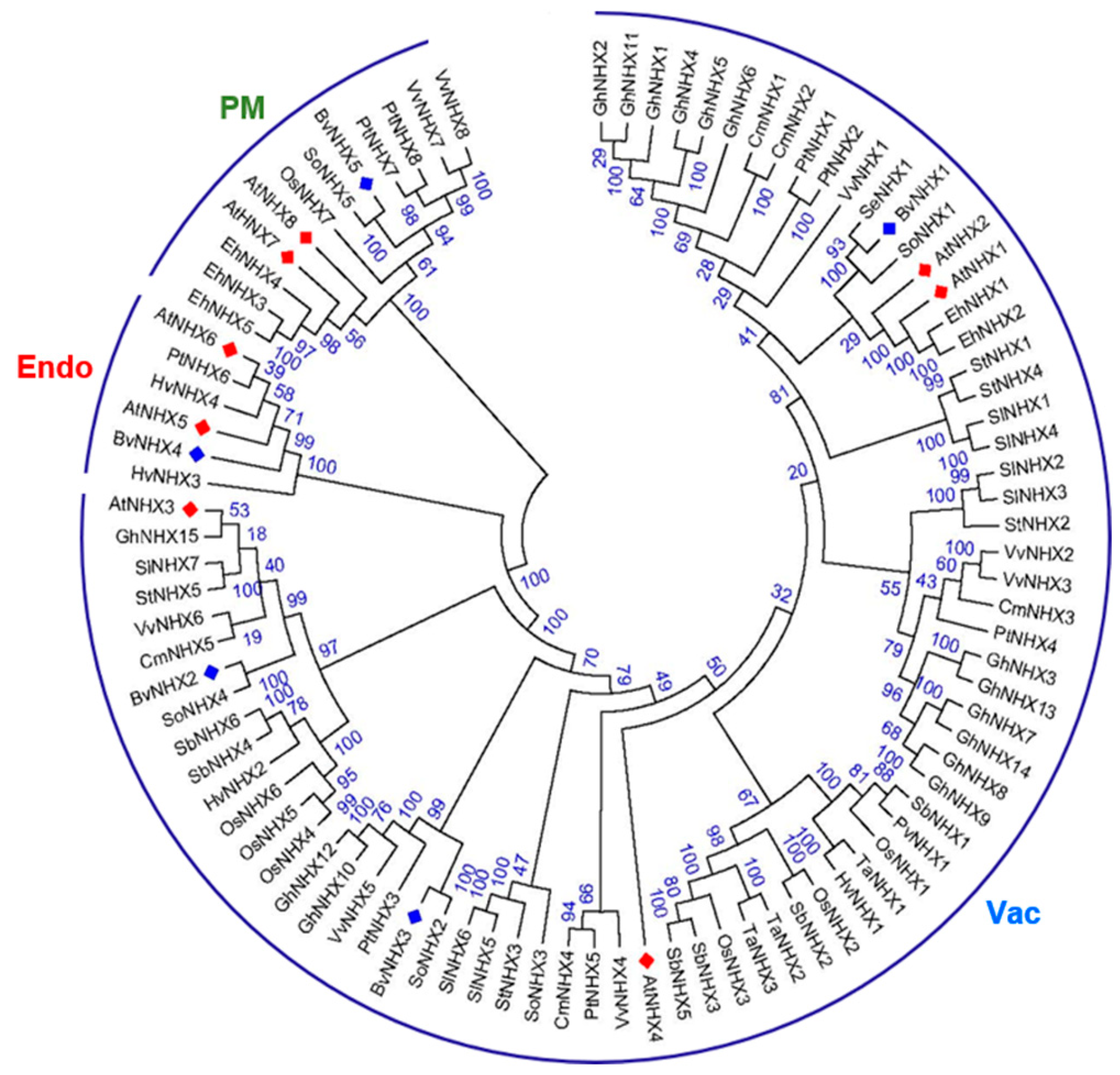
3.2. Phylogenetic Relationship of Sugar Beet and Other Plants in NHX Gene Families
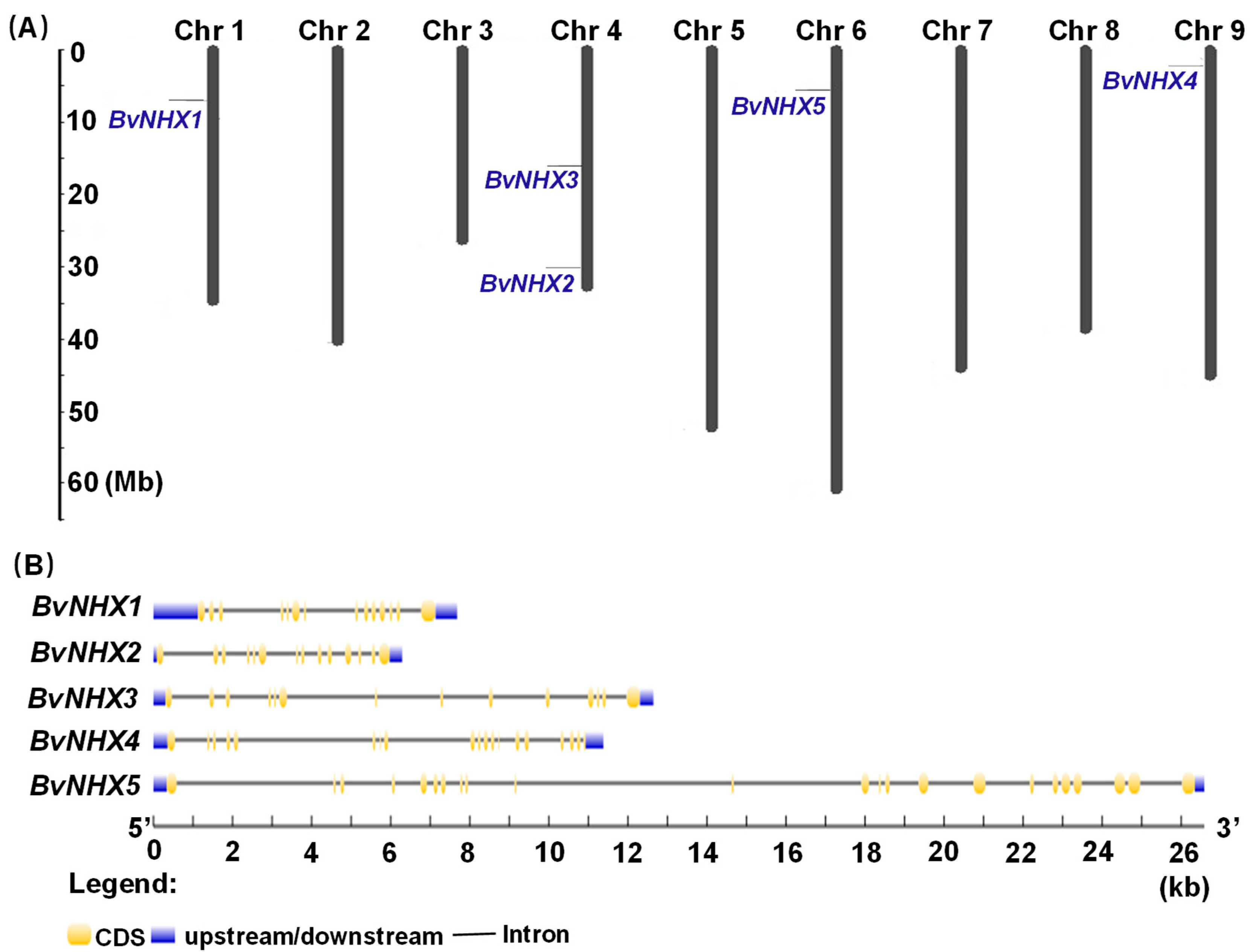
3.3. Chromosomal Location, Ka/Ks Ratio Calculation and Gene Structure Analysis of BvNHX Genes
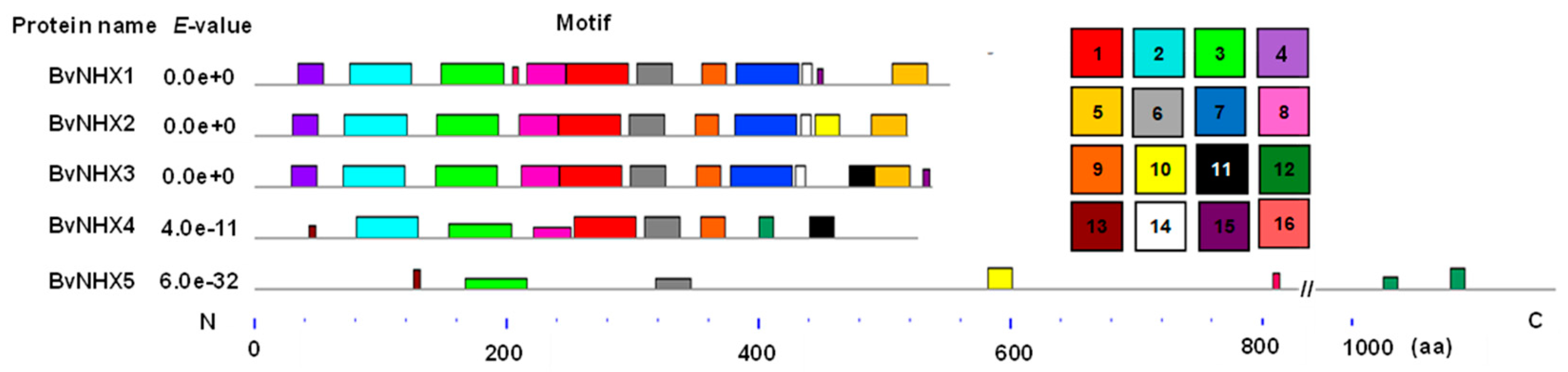
3.4. Analysis of the Conserved Motifs of BvNHX Proteins
3.5. Analysis of cis-Acting Elements in BvNHX Promoters
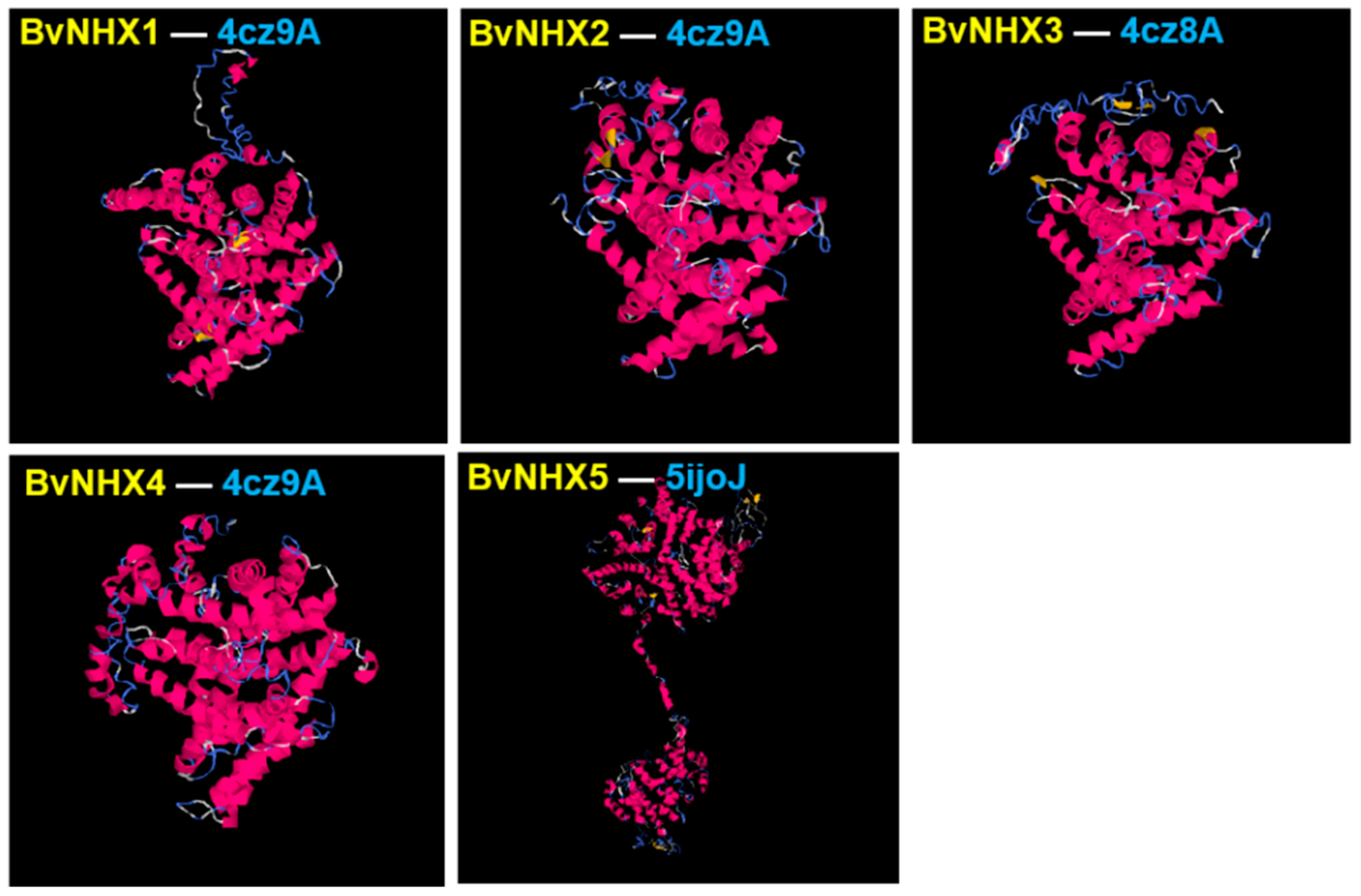
3.6. Analysis of BvNHX Proteins Structures
3.7. Protein-Protein Interaction Prediction of BvNHXs
3.8. Expression Patterns of BvNHX under Various Concentrations of NaCl
4. Discussion
4.1. Identification and Structure Analysis of BvNHX Genes
4.2. Phylogenetic, Conserved Motif, and Promoter Analysis of BvNHX Genes
4.3. Expression Analysis of BvNHX Genes under Salt Stress
4.4. The Protein-Protein Interaction Analysis Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| ABRE | ABA-responsive element |
| ARE | Anaerobic-responsive element |
| ATM | Ataxia telangiectasia mutated |
| CBL | Calcineurin B-like proteins |
| CDC2 | Cell division cycle protein 2 |
| CDS | Coding sequences |
| CIPK | CBL-interacting protein kinases |
| CMP | Calcium-binding protein |
| CYB5R1 | NADH-cytochrome b5 reductase 1 |
| DER | Drought-responsive element |
| ERE | Ethylene-responsive element |
| GSDS | Gene structure display serve |
| GSK3 | Glycogen synthase kinase 3 |
| HKT | High-affinity K+ transporter |
| IAA | Auxin |
| KEA | K+ efflux antiporter |
| LTR | Low-temperature responsiveness |
| ORF | Open reading frame |
| PDB | Protein data bank |
| pI | Isoelectric point |
| PKA | Protein kinase A |
| PKC | Protein kinase C |
| PM | Plasma membrane |
| PPI | Protein-protein interaction |
| SA | Salicylic acid |
| SOS1 | Salt overly sensitive 1 |
| SERE | Stress response element |
| 3-D | Three-dimension |
| TM | Transmembrane helical domain |
| VP | Vacuolar H+-PPase |
| MEME | Multiple expectation maximization for motif elicitation |
| MW | Molecular weight |
| NHX | Na+/H+ antiporter |
References
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Q.; Zhang, H.; Wang, T.; Chen, S.X.; Dai, S.J. Proteomics-based investigation of salt-responsive mechanisms in plant roots. J. Proteom. 2013, 82, 230–253. [Google Scholar] [CrossRef]
- Jha, U.C.; Bohra, A.; Jha, R.; Parida, S.K. Salinity stress response and ‘omics’ approaches for improving salinity stress tolerance in major grain legumes. Plant Cell Rep. 2019, 38, 255–277. [Google Scholar] [CrossRef] [PubMed]
- Hanin, M.; Ebel, C.; Ngom, M.; Laplaze, L.; Masmoudi, K. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front. Plant Sci. 2016, 7, 1787. [Google Scholar] [CrossRef]
- Liang, W.; Ma, X.; Peng, W.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rosales, M.P.; Gálvez, F.J.; Huertas, R.; Aranda, M.N.; Baghour, M.; Cagnac, O.; Venema, K. Plant NHX cation/proton antiporters. Plant Signal Behav. 2009, 4, 265–276. [Google Scholar] [CrossRef] [Green Version]
- Pehlivan, N.; Sun, L.; Jarrett, P.; Yang, X.; Mishra, N.; Chen, L.; Kadioglu, A.; Shen, G.; Zhang, H. Co-overexpressing a plasma membrane and a vacuolar membrane sodium/proton antiporter significantly improves salt tolerance in transgenic Arabidopsis plants. Plant Cell Physiol. 2016, 57, 1069–1084. [Google Scholar] [CrossRef]
- Gaxiola, R.A.; Palmgren, M.G.; Schumacher, K. Plant proton pumps. FEBS Lett. 2007, 581, 2204–2214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, A.K.; Wang, S.M.; Wu, G.Q.; Xi, J.J.; Zhang, J.L.; Wang, C.M. Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Sci. 2009, 176, 232–240. [Google Scholar] [CrossRef]
- Brett, C.L.; Donowitz, M.; Rao, R. Evolutionary origins of eukaryotic sodium/proton exchangers. Am. J. Physiol. 2005, 288, C223–C239. [Google Scholar] [CrossRef] [Green Version]
- Aharon, G.S.; Apse, M.P.; Duan, S.; Hua, X.; Blumwald, E. Characterization of a family of vacuolar Na+/H+ antiporters Arabidopsis thaliana. Plant Soil 2003, 253, 245–256. [Google Scholar] [CrossRef]
- Bassil, E.; Tajima, H.; Liang, Y.C.; Onto, M.A.; Ushijima, K.; Nakano, R.; Esumi, T.; Coku, A.; Belmonte, M.; Blumwald, E. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 2011, 23, 3482–3497. [Google Scholar] [CrossRef]
- Shi, H.Z.; Ishitani, M.; Kim, C.; Zhu, J.K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Apse, M.P.; Shi, H.; Blumwald, E. Topological analysis of a plant vacuolar Na+/H+ antiporter reveals a luminal C terminus that regulates antiporter cation selectivity. Proc. Natl. Acad. Sci. USA 2003, 100, 12510–12515. [Google Scholar] [CrossRef]
- Apse, M.P.; Sottosanto, J.B.; Blumwald, E. Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 2003, 36, 229–239. [Google Scholar] [CrossRef]
- Ohnishi, M.; Fukada-Tanaka, S.; Hoshino, A.; Takada, J.; Inagaki, Y.; Iida, S. Characterization of a novel Na+/H+ antiporter gene InNHX2 and comparison of InNHX2 with InNHX1, which is responsible for blue flower coloration by increasing the vacuolar pH in the Japanese morning glory. Plant Cell Physiol. 2005, 46, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Leidi, E.O.; Pardo, J.M. How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal Behav. 2010, 5, 792–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassil, E.; Coku, A.; Blumwald, E. Cellular ion homeostasis: Emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J. Exp. Bot. 2012, 63, 5727–5740. [Google Scholar] [CrossRef]
- Huertas, R.; Rubio, L.; Cagnac, O.; García-Sánchez, M.J.; Alché Jde, D.; Venema, K.; Fernández, J.A. Rodríguez-Rosales, M.P. The K+/H+ antiporter LeNHX2 increases salt tolerance by improving K+ homeostasis in transgenic tomato. Plant Cell Environ. 2013, 36, 2135–2149. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–467. [Google Scholar] [CrossRef]
- Qiu, Q.S. Plant and yeast NHX antiporters: Roles in membrane trafficking. J. Integr. Plant Biol. 2012, 54, 66–72. [Google Scholar] [CrossRef]
- Li, N.; Wang, X.; Ma, B.; Du, C.; Zheng, L.; Wang, Y. Expression of a Na+/H+ antiporter RtNHX1 from recretohalophyte Reaumuria trigyna improved salt tolerance of transgenic Arabidopsis thaliana. J. Plant Physiol. 2017, 218, 109–120. [Google Scholar] [CrossRef]
- Bao, A.K.; Du, B.Q.; Touil, L.; Kang, P.; Wang, Q.L.; Wang, S.M. Co-expression of tonoplast Cation/H+ antiporter and H+-pyrophosphatase from xerophyte Zygophyllum xanthoxylum improves alfalfa plant growth under salinity, drought and field conditions. Plant Biotechnol. J. 2015, 14, 964–975. [Google Scholar] [CrossRef]
- Porcel, R.; Bustamante, A.; Ros, R.; Serrano, R.; Mulet Salort, J.M. BvCOLD1: A novel aquaporin from sugar beet (Beta vulgaris L.) involved in boron homeostasis and abiotic stress. Plant Cell Environ. 2018, 41, 2844–2857. [Google Scholar] [CrossRef]
- Kong, W.; Yang, S.; Wang, Y.; Bendahmane, M.; Fu, X. Genome-wide identification and characterization of aquaporin gene family in Beta vulgaris. Peer J. 2017, 5, e3747. [Google Scholar] [CrossRef]
- Wu, G.Q.; Feng, R.J.; Zhang, J.J. Evaluation of salinity tolerance in seedlings of sugar beet (Beta vulgaris L.) cultivars using proline, soluble sugars and cation accumulation criteria. Acta Physiol. Plant. 2013, 35, 2665–2674. [Google Scholar] [CrossRef]
- Bârsan, S.C.; Ivan, A.M.; Luca, L.C.; Luca, E. Sugar beet (Beta vulgaris L.) yields and potential for bioethanol production under irrigation regime. Notulae Botanicae Horti Agrobotanici Cluj-Napoca 2015, 43, 455–461. [Google Scholar] [CrossRef]
- Haankuku, C.; Epplin, F.M.; Kakani, V.G. Industrial sugar beets to biofuel: Field to fuel production system and cost estimates. Biomass Bioenergy 2015, 80, 267–277. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.Q.; Feng, R.J.; Liang, N.; Yuan, H.J.; Sun, W.B. Sodium chloride stimulates growth and alleviates sorbitol-induced osmotic stress in sugar beet seedlings. Plant Growth Regul. 2015, 75, 307–316. [Google Scholar] [CrossRef]
- Dohm, J.C.; Minoche, A.E.; Holtgräwe, D.; Capellagutiérrez, S.; Zakrzewski, F.; Tafer, H.; Rupp, O.; Sörensen, T.R.; Stracke, R.; Reinhardt, R. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 2014, 505, 546–549. [Google Scholar] [CrossRef]
- Adler, G.; Blumwald, E.; Bar-Zvi, D. The sugar beet gene encoding the sodium/proton exchanger 1 (BvNHX1) is regulated by a MYB transcription factor. Planta 2010, 232, 187–195. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucl. Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Tian, F.; Chang, E.; Li, Y.; Sun, P.; Hu, J.; Zhang, J. Expression and integrated network analyses revealed functional divergence of NHX-type Na+/H+ exchanger genes in poplar. Sci. Rep. 2017, 7, 2607. [Google Scholar] [CrossRef]
- Bjellqvist, B.; Hughes, G.J.; Pasquali, C.; Paquet, N.; Ravier, F.; Sanchez, J.C.; Frutiger, S.; Hochstrasser, D.F. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 1993, 14, 1023–1031. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Blom, N.; Sicheritz-Ponten, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Goldman, N.; Yang, Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 1994, 11, 725–736. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl. Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindvalov, I.N.; Bourne, P.E. The protein data bank. Nucl. Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucl. Acids Res. 2007, 35, 3375–3382. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucl. Acids Res. 2019, 47, 607–613. [Google Scholar] [CrossRef]
- Wu, G.Q.; Lin, L.Y.; Jiao, Q.; Li, S.J. Tetraploid exhibits more tolerant to salinity than diploid in sugar beet (Beta vulgaris L.). Acta Physiol. Plant. 2019, 41, 52. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Anton, N.; Makova, K.D.; Li, W.H. The Ka/Ks ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 2002, 12, 198–202. [Google Scholar]
- Chen, H.; Chen, X.; Wu, B.; Yuan, X.; Zhang, H.; Cui, X.; Liu, X. Whole-genome identification and expression analysis of K+ efflux antiporter (KEA) and Na+/H+ antiporter (NHX) families under abiotic stress in soybean. J. Integr. Agr. 2015, 14, 1171–1183. [Google Scholar] [CrossRef]
- Kumari, P.H.; Kumar, S.N.; Ramesh, K.; Reddy, P.S.; Nagaraju, M.; Prakash, A.B.; Shah, T.; Hederson, A.; Srivsastava, R.K.; Rajasheker, G.; et al. Genome-Wide Identification and analysis of Arabidopsis sodium proton antiporter (NHX) and human sodium proton exchanger (NHX) homologs in Sorghum bicolor. Genes 2018, 9, 236. [Google Scholar] [CrossRef]
- Wu, G.Q.; Wang, Q.; Bao, A.K.; Wang, S.M. Amiloride reduces sodium transport and accumulation in the succulent xerophyte Zygophyllum xanthoxylum under salt conditions. Biol. Trace Elem. Res. 2011, 139, 356–367. [Google Scholar] [CrossRef]
- Wang, B.; Zhai, H.; He, S.; Zhang, H.; Ren, Z.; Zhang, D.A. Vacuolar Na+/H+ antiporter gene, IbNHX2, enhances salt and drought tolerance in transgenic sweetpotato. Sci. Hortic. 2016, 201, 153–166. [Google Scholar] [CrossRef]
- Ding, X.; Li, J.; Pan, Y.; Zhang, Y.; Ni, L.; Wang, Y.; Zhang, X. Genome-wide identification and expression analysis of the UGlcAE gene family in tomato. Int. J. Mol. Sci. 2018, 19, 1583. [Google Scholar] [CrossRef]
- Verma, D.; Lakhanpal, N.; Singh, K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. 2019, 20, 227. [Google Scholar]
- Mishra, S.; Shukla, A.; Upadhyay, S.; Sanchita; Sharma, P.; Singh, S.; Phukan, U.J.; Meena, A.; Khan, F.; Tripathi, V.; et al. Identification, occurrence, and validation of DRE and ABRE cis-regulatory motifs in the promoter regions of genes of Arabidopsis thaliana. J. Integr. Plant Biol. 2014, 56, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, R. Modulation of ethylene and ascorbic acid on reactive oxygen species scavenging in plant salt response. Front. Plant Sci. 2019, 10, 319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X. Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 2019, 50, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Liu, Z.; Xing, L.; Wei, Y.; Mao, J.; Meng, Y.; Bao, L.; Han, M.; Zhao, C.; Zhang, D. miRNAs associated with auxin signaling, stress response, and cellular activities mediate adventitious root formation in apple rootstocks. Plant Physiol. Biochem. 2019, 139, 66–81. [Google Scholar] [CrossRef]
- Xu, Z.; Raza, Q.; Xu, L.; He, X.; Huang, Y.; Yi, J.; Zhang, D.; Shao, H.B.; Ma, H.; Ali, Z. GmWRKY49, a salt-responsive nuclear protein, improved root length and governed better salinity tolerance in transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 809. [Google Scholar] [CrossRef]
- Wu, H. Plant salt tolerance and Na+ sensing and transport. Crop J. 2018, 6, 215–225. [Google Scholar] [CrossRef]
- Lu, W.; Guo, C.; Li, X.; Duan, W.; Ma, C.; Zhao, M.; Gu, J.; Du, X.; Liu, Z.; Xiao, K. Overexpression of TaNHX3, a vacuolar Na⁺/H⁺ antiporter gene in wheat, enhances salt stress tolerance in tobacco by improving related physiological processes. Plant Physiol. Biochem. 2014, 76, 17–28. [Google Scholar] [CrossRef]
- Jegadeeson, V.; Kumari, K.; Pulipati, S.; Parida, A.; Venkataraman, G. Expression of wild rice Porteresia coarctata PcNHX1 antiporter gene (PcNHX1) in tobacco controlled by PcNHX1 promoter (PcNHX1p) confers Na+ specific hypocotyl elongation and stem-specific Na+ accumulation in transgenic tobacco. Plant Physiol. Biochem. 2019, 139, 161–170. [Google Scholar] [CrossRef]
- Zhang, W.D.; Wang, P.; Bao, Z.; Ma, Q.; Duan, L.J.; Bao, A.K.; Zhang, J.L.; Wang, S.M. SOS1, HKT1;5, and NHX1 synergistically modulate Na+ homeostasis in the halophytic grass Puccinellia tenuiflora. Front. Plant Sci. 2017, 8, 576. [Google Scholar] [CrossRef]
- Ma, Q.; Li, Y.X.; Yuan, H.J.; Hu, J.; Wei, L.; Bao, A.K.; Zhang, J.L.; Wang, S.M. ZxSOS1 is essential for long-distance transport and spatial distribution of Na+ and K+ in the xerophyte Zygophyllum xanthoxylum. Plant Soil 2014, 374, 661–676. [Google Scholar] [CrossRef]
- Gao, J.; Sun, J.; Cao, P.; Ren, L.; Liu, C.; Chen, S.; Chen, F.; Jiang, J. Variation in tissue Na+ content and the activity of SOS1 genes among two species and two related genera of Chrysanthemum. BMC Plant Biol. 2016, 16, 98. [Google Scholar] [CrossRef]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J.M. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef]
- Yadav, N.S.; Shukla, P.S.; Jha, A.; Agarwal, P.K.; Jha, B. The SbSOS1 gene from the extreme halophyte Salicornia brachiata enhances Na+ loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biol. 2012, 12, 188. [Google Scholar] [CrossRef]
- Wayne, L.L.; Wallis, J.G.; Kumar, R.; Markham, J.E.; Browse, J. Cytochrome b5 reductase encoded by CBR1 is essential for a functional male gametophyte in Arabidopsis. Plant Cell 2013, 25, 3052–3066. [Google Scholar] [CrossRef]
- Oh, Y.J.; Kim, H.; Seo, S.H.; Hwang, B.G.; Chang, Y.S.; Lee, J.; Lee, D.W.; Sohn, E.J.; Lee, S.J.; Lee, Y.; et al. Cytochrome b5 reductase 1 triggers serial reactions that lead to iron uptake in plants. Mol. Plant 2016, 9, 501–513. [Google Scholar] [CrossRef]
- Wu, G.Q.; Xi, J.J.; Wang, Q.; Ma, Q.; Bao, A.K.; Zhang, J.L.; Wang, S.M. The ZxNHX gene encoding tonoplast Na+/H+ antiporter in the xerophyte Zygophyllum xanthoxylum plays important roles in response to salt and drought. J. Plant Physiol. 2011, 168, 758–767. [Google Scholar] [CrossRef]
- Bao, A.K.; Wang, Y.W.; Xi, J.J.; Liu, C.; Zhang, J.L.; Wang, S.M. Co-expression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1-1 enhances salt and drought tolerance in transgenic Lotus corniculatus by increasing cations accumulation. Funct. Plant Biol. 2014, 41, 203–214. [Google Scholar] [CrossRef]
- Wu, G.Q.; Feng, R.J.; Wang, S.M.; Wang, C.M.; Bao, A.K.; Wei, L.; Yuan, H.J. Co-expression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1-1 confers enhanced salinity tolerance in chimeric sugar beet (Beta vulgaris L.). Front. Plant Sci. 2015, 6, 581. [Google Scholar] [CrossRef]
- Yu, Q.; An, L.; Li, W. The CBL-CIPK network mediates different signaling pathways in plants. Plant Cell Rep. 2014, 33, 203–214. [Google Scholar] [CrossRef]
- Miranda, R.S.; Alvarez-Pizarro, J.C.; Costa, J.H.; Paula, S.O.; Prisco, J.T.; Gomes-Filho, E. Putative role of glutamine in the activation of CBL/CIPK signaling pathways during salt stress in sorghum. Plant Signal Behav. 2017, 12, e1361075. [Google Scholar] [CrossRef]
- Quintero, F.J.; Martinez-Atienza, J.; Villalta, I.; Jiang, X.Y.; Kim, W.Y.; Ali, Z.; Fujii, H.; Mendoza, I.; Yun, D.J.; Zhu, J.K.; et al. Activation of the plasma membrane Na/H antiporter Salt-Overly-Sensitive 1 (SOS1) by phosphorylation of an auto-inhibitory C-terminal domain. Proc. Natl. Acad. Sci. USA 2011, 108, 2611–2616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, Y.; Zhang, M.; Zhang, J.; Duan, L.; Li, Z. SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. J. Plant Physiol. 2012, 169, 255–261. [Google Scholar] [CrossRef] [PubMed]
| No. | Gene Name | Forward Primer Sequence (5’-3’) | Reverse Primer Sequence (5’-3’) |
|---|---|---|---|
| 1 | BvACTIN | ACTGGTATTGTGCTTGACTC | ATGAGATAATCAGTGAGATC |
| 2 | BvNHX1 | TCGATGATTCTTTCATGAGG | GCCAACTGCCTCATACTCTG |
| 3 | BvNHX2 | GTGTTAGATTTGGGTGTGCAAATAGC | GCAGTGATGGATTCATTGACCCAACG |
| 4 | BvNHX3 | GTTTGGTGTACCAAATATAGTGCATG | TGATAGATTCATTAGCCCAACGATTC |
| 5 | BvNHX4 | TCCTACAAATGATCCGCTGATATCTC | TGACGACGTAAAACATGACCAAGTAC |
| 6 | BvNHX5 | GCCAGCTATGGCAGCTTATC | ATCCAAGGCCAATGCCGATG |
| Gene Name | Gene ID | Gene Code | Chr | Exons Count | CDS (bp) | ORF (aa) | pI | MW (kDa) | TM | Plant-mPLoc |
|---|---|---|---|---|---|---|---|---|---|---|
| BvNHX1 | Xkmw | Bv1g006450_xkmw.t1 | 1 | 15 | 1659 | 552 | 6.31 | 61.3 | 12 | Vac |
| BvNHX2 | Zfce | Bv4g089680_zfce.t1 | 4 | 14 | 1560 | 519 | 8.45 | 58.2 | 11 | Vac |
| BvNHX3 | Jkji | Bv4g083400_jkji.t1 | 4 | 14 | 1617 | 538 | 6.33 | 59.3 | 12 | Vac |
| BvNHX4 | Pswr | Bv9g203100_pswr.t1 | 9 | 26 | 1584 | 527 | 5.5 | 58.2 | 12 | Endo |
| BvNHX5 | Sjdh | Bv6g131830_sjdh.t1 | 6 | 23 | 3489 | 1162 | 6.34 | 128.5 | 12 | PM |
| Similarity (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein names | BvNHX1 | BvNHX2 | BvNHX3 | BvNHX4 | BvNHX5 | AtNHX1 | AtNHX2 | AtNHX3 | AtNHX4 | AtNHX6 | AtHNX7 | AtNHX8 | |
| Divergence | BvNHX1 | 78.8 | 79.6 | 64.4 | 8.6 | 89.2 | 89.5 | 78.5 | 84.0 | 65.0 | 10.1 | 43.1 | |
| BvNHX2 | 57.7 | 77.8 | 65.5 | 9.1 | 79.5 | 79.5 | 86.5 | 79.1 | 65.8 | 11.1 | 43.3 | ||
| BvNHX3 | 61.3 | 68.9 | 65.0 | 9.4 | 79.9 | 79.5 | 79.1 | 78.2 | 64.9 | 11.1 | 43.0 | ||
| BvNHX4 | 183.2 | 166.1 | 184.7 | 10.1 | 65.2 | 64.4 | 64.9 | 64.7 | 86.8 | 12.0 | 43.6 | ||
| BvNHX5 | 299.0 | 257.0 | 265.0 | 237.0 | 9.4 | 9.1 | 9.5 | 8.9 | 9.8 | 61.9 | 43.8 | ||
| AtNHX1 | 25.2 | 57.5 | 62.6 | 177.6 | 265.0 | 94.2 | 79.5 | 83.7 | 65.4 | 10.6 | 43.3 | ||
| AtNHX2 | 25.3 | 56.5 | 63.5 | 186.7 | 281.0 | 12.3 | 79.4 | 84.1 | 65.5 | 10.6 | 43.2 | ||
| AtNHX3 | 62.4 | 35.2 | 62.9 | 178.0 | 252.0 | 59.3 | 60.3 | 78.7 | 65.6 | 11.1 | 43.8 | ||
| AtNHX4 | 35.4 | 59.9 | 61.9 | 170.1 | 264.0 | 37.6 | 36.5 | 62.5 | 66.4 | 10.3 | 42.7 | ||
| AtNHX6 | 175.9 | 175.9 | 191.7 | 30.9 | 244.0 | 174.1 | 172.2 | 177.3 | 168.4 | 11.8 | 43.3 | ||
| AtHNX7 | 274.0 | 229.0 | 243.0 | 221.0 | 50.8 | 252.0 | 255.0 | 236.0 | 248.0 | 226.0 | 51.0 | ||
| AtNHX8 | 267.0 | 245.0 | 273.0 | 254.0 | 46.8 | 254.0 | 261.0 | 241.0 | 255.0 | 261.0 | 31.1 | ||
| Gene 1 | Gene 2 | Ks | Ka | Ka/Ks |
|---|---|---|---|---|
| BvNHX1 | BvNHX5 | 0.1122 | 11.1040 | 99.0000 |
| BvNHX2 | BvNHX5 | 3.7373 | 7.9949 | 2.1392 |
| BvNHX3 | BvNHX5 | 18.8076 | 3.4894 | 0.1855 |
| BvNHX4 | BvNHX5 | 2.9900 | 5.5013 | 1.8399 |
| Functional Class | Elements | Function | Sequence | Genes | ||||
|---|---|---|---|---|---|---|---|---|
| BvNHX1 | BvNHX2 | BvNHX3 | BvNHX4 | BvNHX5 | ||||
| Hormone | ABRE | Abscisic acid-responsive element | ACGTG | 0 | 0 | 0 | 0 | 1 |
| ERE | Ethylene-responsive element | ATTTCATA | 1 | 0 | 1 | 1 | 0 | |
| TCA-element | Involved in salicylic acid responsiveness | CCATCTTTTT | 2 | 0 | 0 | 0 | 1 | |
| TGA-element | Auxin-responsive element | AACGAC | 0 | 0 | 1 | 0 | 1 | |
| Stress | ARE | Anaerobic-responsive element | AAACCA | 1 | 0 | 0 | 2 | 0 |
| DRE | Drought-responsive element | GCCGAC | 1 | 0 | 0 | 0 | 0 | |
| LTR | Low-temperature responsiveness | CCGAAA | 0 | 0 | 0 | 1 | 0 | |
| WUN-motif | Wound-responsive element | AAATTACTA | 0 | 0 | 1 | 0 | 0 | |
| MYB | Drought-responsive element | CAACCA | 1 | 0 | 0 | 0 | 0 | |
| W box | Salt-responsive element | TTGACC | 0 | 0 | 0 | 1 | 0 | |
| STRE | Stress response element | AGGGG | 0 | 0 | 0 | 0 | 1 | |
| Others | Box 4 | Involved in light responsiveness | ATTAAT | 1 | 0 | 1 | 0 | 0 |
| GT1-motif | Involved in light responsiveness | GGTTAAT | 1 | 0 | 0 | 0 | 0 | |
| TCT-motif | Involved in light responsiveness | TCTTAC | 1 | 0 | 0 | 1 | 1 | |
| Gap-box | Involved in light responsiveness | CAAATGAA | 0 | 0 | 1 | 0 | 0 | |
| AE-box | Modul for light response | AGAAACTT | 0 | 0 | 0 | 1 | 0 | |
| O2-site | Zein metabolism regulation | GATGACATGA | 0 | 0 | 0 | 1 | 2 | |
| G-box | Involved in light responsiveness | TACGTG | 0 | 0 | 0 | 0 | 1 | |
| Protein | C-Score | TM-Score | RMSD (Å) | Best Identified Structural Analogs in PDB | ||||
|---|---|---|---|---|---|---|---|---|
| PDB Hit | TM-Score a | RMSD a | IDEN a | Cov | ||||
| BvNHX1 | −1.03 | 0.58 ± 0.14 | 9.9 ± 4.6 | 4cz9A | 0.701 | 1.08 | 0.221 | 0.712 |
| BvNHX2 | −1.83 | 0.49 ± 0.15 | 11.8 ± 4.5 | 4cz9A | 0.739 | 1.19 | 0.21 | 0.753 |
| BvNHX3 | −1.64 | 0.51 ± 0.15 | 11.4 ± 4.5 | 4cz8A | 0.711 | 1.44 | 0.202 | 0.729 |
| BvNHX4 | −0.61 | 0.52 ± 0.15 | 11.3 ± 4.6 | 4cz9A | 0.724 | 1.28 | 0.195 | 0.74 |
| BvNHX5 | −1.26 | 0.56 ± 0.15 | 12.4 ± 4.3 | 5ijoJ | 0.902 | 1.59 | 0.087 | 0.915 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, G.-Q.; Wang, J.-L.; Li, S.-J. Genome-Wide Identification of Na+/H+ Antiporter (NHX) Genes in Sugar Beet (Beta vulgaris L.) and Their Regulated Expression under Salt Stress. Genes 2019, 10, 401. https://doi.org/10.3390/genes10050401
Wu G-Q, Wang J-L, Li S-J. Genome-Wide Identification of Na+/H+ Antiporter (NHX) Genes in Sugar Beet (Beta vulgaris L.) and Their Regulated Expression under Salt Stress. Genes. 2019; 10(5):401. https://doi.org/10.3390/genes10050401
Chicago/Turabian StyleWu, Guo-Qiang, Jin-Long Wang, and Shan-Jia Li. 2019. "Genome-Wide Identification of Na+/H+ Antiporter (NHX) Genes in Sugar Beet (Beta vulgaris L.) and Their Regulated Expression under Salt Stress" Genes 10, no. 5: 401. https://doi.org/10.3390/genes10050401
APA StyleWu, G.-Q., Wang, J.-L., & Li, S.-J. (2019). Genome-Wide Identification of Na+/H+ Antiporter (NHX) Genes in Sugar Beet (Beta vulgaris L.) and Their Regulated Expression under Salt Stress. Genes, 10(5), 401. https://doi.org/10.3390/genes10050401





