Knockdown of Cytochrome P450 Genes Gh_D07G1197 and Gh_A13G2057 on Chromosomes D07 and A13 Reveals Their Putative Role in Enhancing Drought and Salt Stress Tolerance in Gossypium hirsutum
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Physiochemical Analysis of the Proteins Encoded by the Cotton Cytochrome P450 Genes
2.2. Chromosome Mapping and Subcellular Sublocalization Prediction of the Proteins Encoded by the G. hirsutum CYP450 Genes
2.3. RNA Sequencing Analysis of the Cotton Cytochrome (CYP450) Genes Profiled under Drought and Salt Stress Conditions in Various Tissues
2.4. Plant Materials and Abiotic Stress Treatment
2.5. RNA Isolation and RT-qPCR Analysis
2.6. Preparation of Inocula and Inoculation of Plants
2.7. Abiotic Stress Treatment of the VIG Plants, Wild Type and Infused with TRV:00 (Empty Vector)
2.8. Physiological and Biochemical Evaluations of the VIGS Plants and Wild-Type Cotton Seedlings under Drought and Salt Stress Conditions
2.9. Stress-Responsive Gene Profiling in VIGS Plants and Wild Types under Drought and Salt Stress Conditions
3. Results
3.1. Identification and Physiochemical Properties of the Cotton Cytochrome P450 (CYPs) Genes
3.2. Subcellular Localization Analysis of the Cotton Cytochrome P450
3.3. Cis-Regulatory Element Analysis of the Cotton Cytochrome P450
3.4. Chromosomal Mapping of the Upland G. hirsutum Cytochrome CYP450 Genes
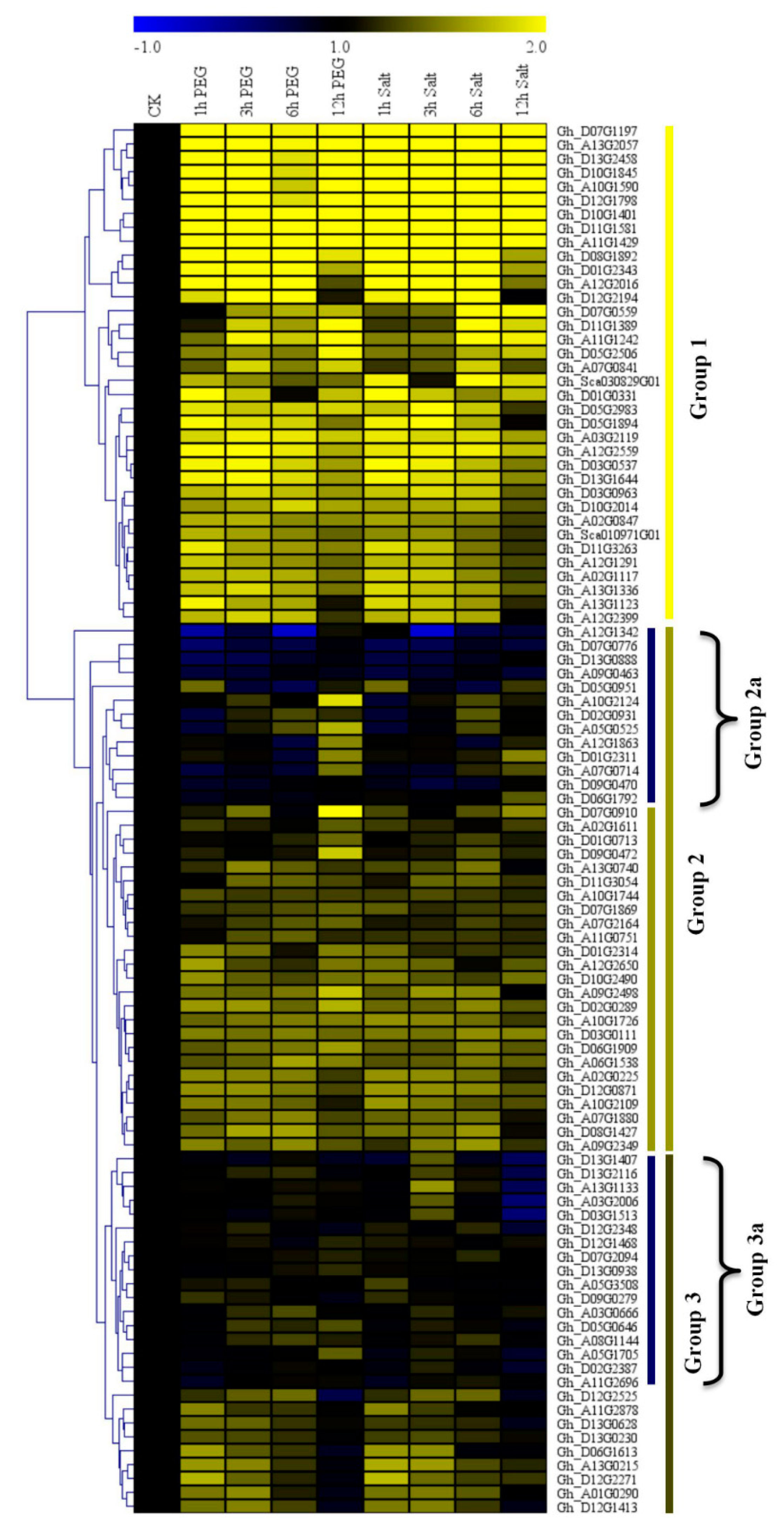
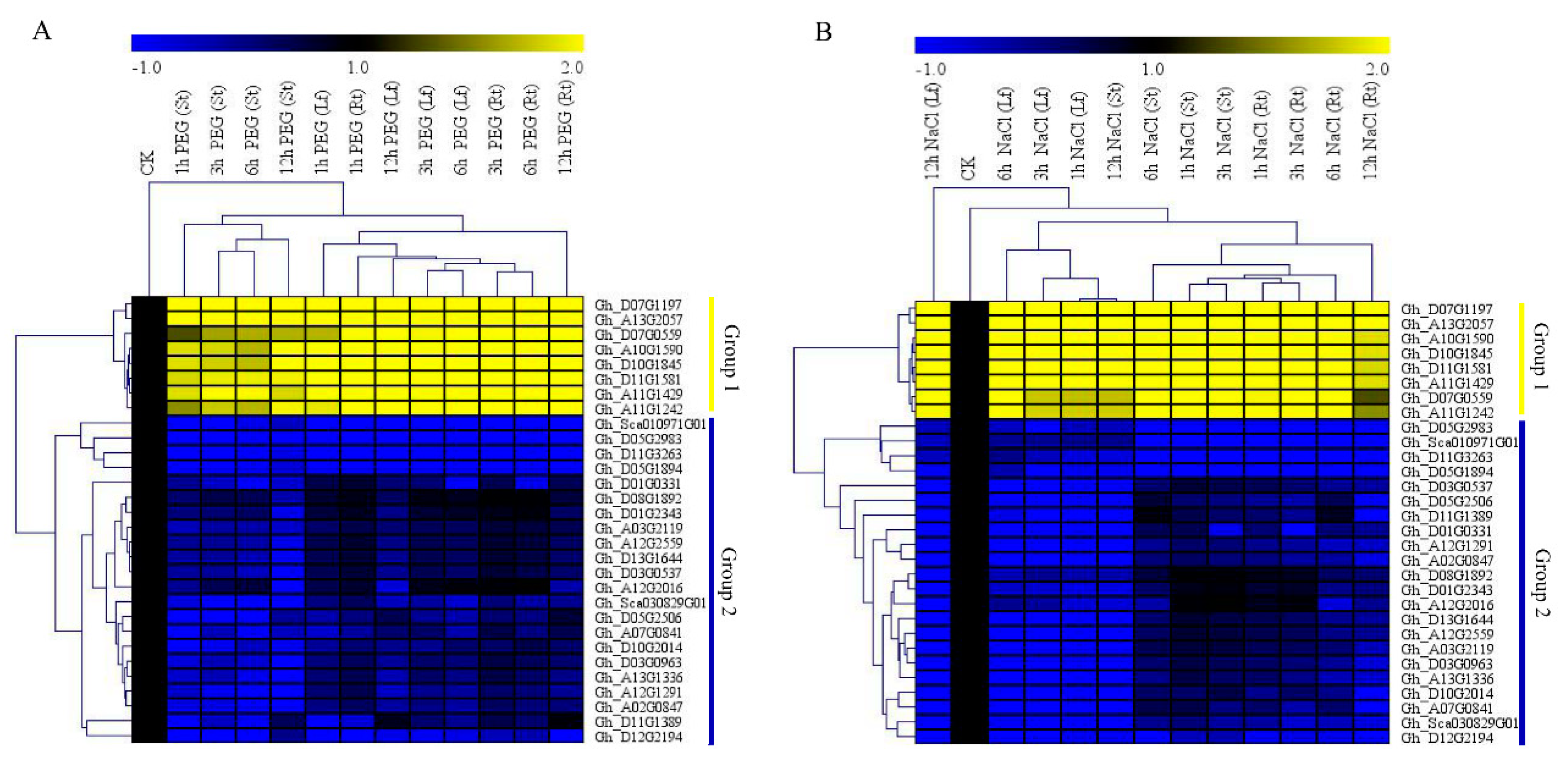
3.5. RNA Sequencing Analysis and RT-qPCR Validation of the Upland Cotton Cytochrome P450 Genes under Salt and Drought Stress Conditions
3.6. Expression Analysis of the Two Genes Gh_D07G1197 and Gh_A13G2057 in the Tissues of VIGS Seedlings and Wild-Type Cotton Seedlings
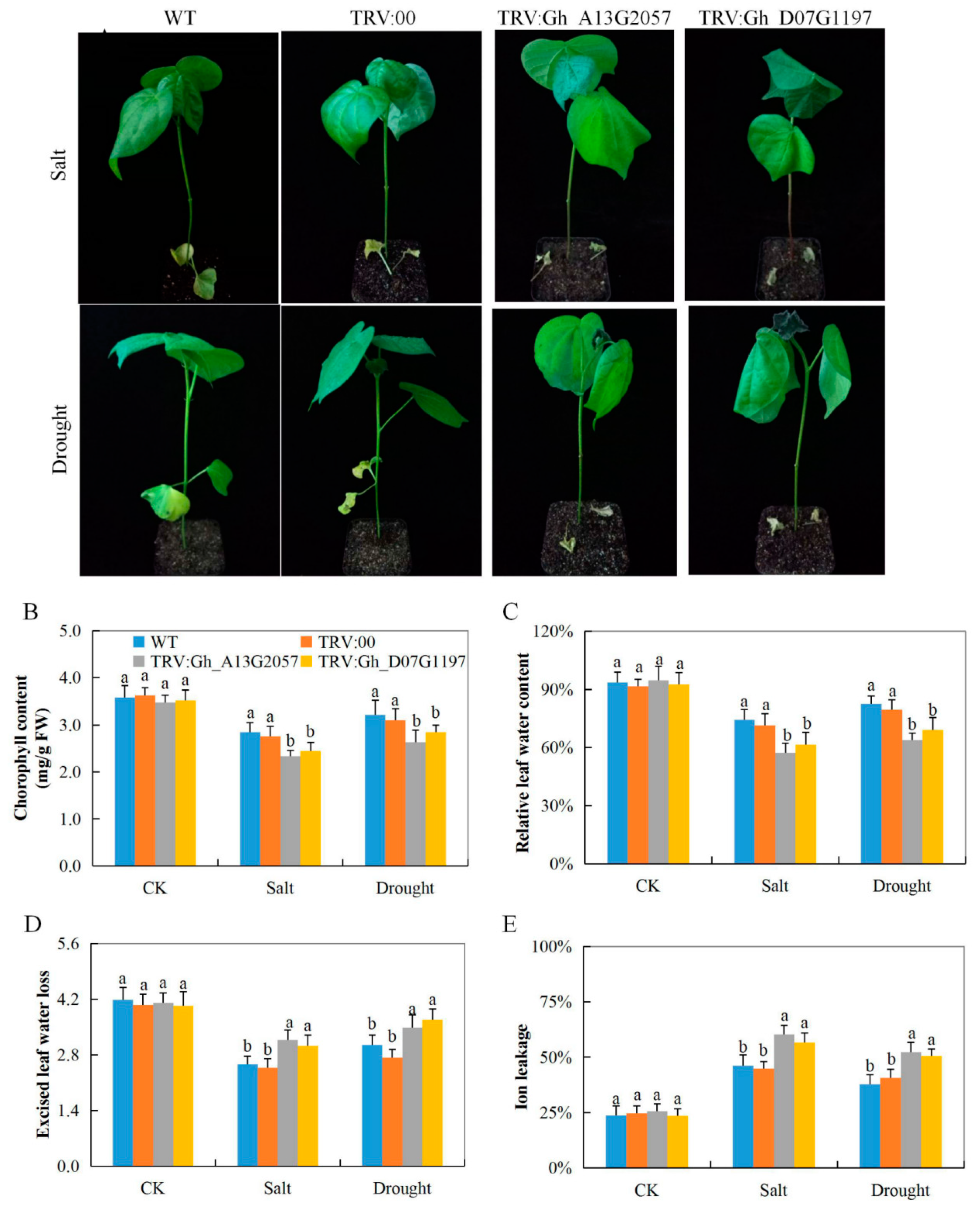
3.7. Evaluation of Performance of the VIGS Plants and Wild Cotton, G. hirsutum, under Drought and Salt Stress Condition
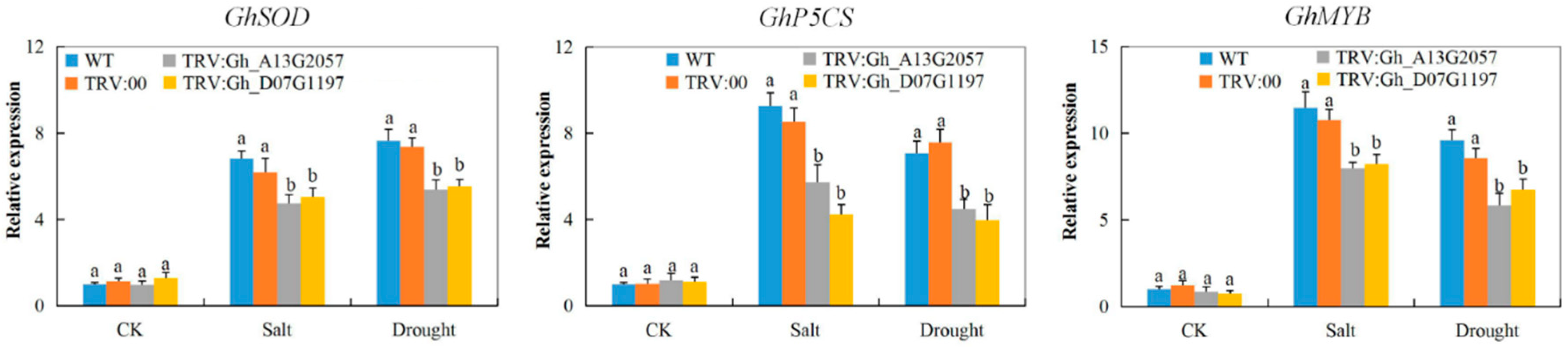
3.8. Stress-Responsive Gene Profiling on the Tissues of VIGS Plants and the Wild Types under Drought and Salt Stress Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shiota, N.; Kodama, S.; Inui, H.; Ohkawa, H. Expression of human cytochromes P450 1A1 and P450 1A2 as fused enzymes with yeast NADPH-cytochrome P450 oxidoreductase in transgenic tobacco plants. Biosci. Biotechnol. Biochem. 2000, 64, 2025–2033. [Google Scholar] [CrossRef]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: a success story. Genome Biol. 2000, 1, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, X.Y.; Guo, W.Z. The cytochrome P450 superfamily: Key players in plant development and defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar] [CrossRef]
- Guengerich, F.P.; Martin, M.V.; Sohl, C.D.; Cheng, Q. Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat. Protoc. 2009, 4, 1245–1251. [Google Scholar] [CrossRef]
- Dejong, C.A.; Wilson, J.Y. The cytochrome P450 superfamily complement (CYPome) in the Annelid Capitella teleta. PLoS ONE 2014, 9, e107728. [Google Scholar] [CrossRef]
- Bolwell, G.P.; Bozak, K.; Zimmerlin, A. Plant cytochrome p450. Phytochemistry 1994, 37, 1491–1506. [Google Scholar] [CrossRef]
- Nelson, D.; Werck-Reichhart, D. THE PLANT GENOME: AN EVOLUTIONARY VIEW ON STRUCTURE AND FUNCTION A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef] [PubMed]
- Balusamy, S.R.; Rahimi, S.; Yang, D.C. Characterization of squalene-induced PgCYP736B involved in salt tolerance by modulating key genes of abscisic acid biosynthesis. Int. J. Biol. Macromol. 2019, 121, 796–805. [Google Scholar] [CrossRef]
- Wang, C.; Yang, Y.; Wang, H.; Ran, X.; Li, B.; Zhang, J.; Zhang, H. Ectopic expression of a cytochrome P450 monooxygenase gene PtCYP714A3 from Populus trichocarpa reduces shoot growth and improves tolerance to salt stress in transgenic rice. Plant Biotechnol. J. 2016, 14, 1838–1851. [Google Scholar] [CrossRef]
- Nelson, D.R.; Ming, R.; Alam, M.; Schuler, M.A. Comparison of Cytochrome P450 Genes from Six Plant Genomes. Trop. Plant Biol. 2008, 1, 216–235. [Google Scholar] [CrossRef]
- Mao, G.; Seebeck, T.; Schrenker, D.; Yu, O. CYP709B3, a cytochrome P450 monooxygenase gene involved in salt tolerance in Arabidopsis thaliana. BMC Plant Biol. 2013, 13. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic Stress Responses and Microbe-Mediated Mitigation in Plants: The Omics Strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef]
- Udawat, P.; Jha, R.K.; Sinha, D.; Mishra, A.; Jha, B. Overexpression of a Cytosolic Abiotic Stress Responsive Universal Stress Protein (SbUSP) Mitigates Salt and Osmotic Stress in Transgenic Tobacco Plants. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Udawat, P.; Jha, R.K.; Mishra, A.; Jha, B. Overexpression of a Plasma Membrane-Localized SbSRP-Like Protein Enhances Salinity and Osmotic Stress Tolerance in Transgenic Tobacco. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Bartwal, A.; Mall, R.; Lohani, P.; Guru, S.K.; Arora, S. Role of secondary metabolites and brassinosteroids in plant defense against environmental stresses. J. Plant Growth Regul. 2013, 32, 216–232. [Google Scholar] [CrossRef]
- Schuler, M.A. Plant cytochrome P450 monooxygenases. Crit. Rev. Plant Sci. 2010, 15, 235–284. [Google Scholar] [CrossRef]
- Bancos, S. Diurnal Regulation of the Brassinosteroid-Biosynthetic CPD Gene in Arabidopsis. PLANT Physiol. 2006, 141, 299–309. [Google Scholar] [CrossRef]
- Ralston, L.; Kwon, S.T.; Schoenbeck, M.; Ralston, J.; Schenk, D.J.; Coates, R.M.; Chappell, J. Cloning, heterologous expression, and functional characterization of 5-epi-aristolochene-1,3-dihydroxylase from tobacco (Nicotiana tabacum). Arch. Biochem. Biophys. 2001, 393, 222–235. [Google Scholar] [CrossRef]
- Whitbred, J.M.; Schuler, M.A. Molecular characterization of CYP73A9 and CYP82A1 P450 genes involved in plant defense in pea. Plant Physiol. 2000, 124, 47–58. [Google Scholar] [CrossRef]
- Guttikonda, S.K.; Trupti, J.; Bisht, N.C.; Chen, H.; An, Y.Q.C.; Pandey, S.; Xu, D.; Yu, O. Whole genome co-expression analysis of soybean cytochrome P450 genes identifies nodulation-specific P450 monooxygenases. BMC Plant Biol. 2010, 10. [Google Scholar] [CrossRef]
- Nelson, D.R. Comparative Genomics of Rice and Arabidopsis. Analysis of 727 Cytochrome P450 Genes and Pseudogenes from a Monocot and a Dicot. PLANT Physiol. 2004, 135, 756–772. [Google Scholar] [CrossRef]
- Ma, B.; Luo, Y.; Jia, L.; Qi, X.; Zeng, Q.; Xiang, Z.; He, N. Genome-wide identification and expression analyses of cytochrome P450 genes in mulberry (Morus notabilis). J. Integr. Plant Biol. 2014, 56, 887–901. [Google Scholar] [CrossRef]
- Nelson, D.; Werck-Reichhart, D. A P450-centric view of plant evolution. Plant J. 2011, 66, 194–211. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Lu, H.; Wang, X.; Cai, X.; Zhou, Z.; Zhang, Z.; Salih, H.; Wang, K.; et al. Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 2018, 19. [Google Scholar] [CrossRef]
- Mmadi, M.; Dossa, K.; Wang, L.; Zhou, R.; Wang, Y.; Cisse, N.; Sy, M.; Zhang, X. Functional Characterization of the Versatile MYB Gene Family Uncovered Their Important Roles in Plant Development and Responses to Drought and Waterlogging in Sesame. Genes 2017, 8, 362. [Google Scholar] [CrossRef]
- Hamberger, B.; Bak, S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368. [Google Scholar] [CrossRef]
- Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Diouf, L.; Dong, Q.; Hu, Y.; Cai, X.; Xu, Y.; Hou, Y.; Zhou, Z.; et al. GBS mapping and analysis of genes conserved between Gossypium tomentosum and Gossypium hirsutum cotton cultivars that respond to drought stress at the seedling stage of the BC2F2 generation. Int. J. Mol. Sci. 2018, 19, 1614. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Z.; Li, F.; Ye, W.; Wang, J.; Song, G.; Yue, Z.; Cong, L.; Shang, H.; Zhu, S.; et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012, 44, 1098–1103. [Google Scholar] [CrossRef]
- Li, F.; Fan, G.; Wang, K.; Sun, F.; Yuan, Y.; Song, G.; Li, Q.; Ma, Z.; Lu, C.; Zou, C.; et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014, 46, 567–572. [Google Scholar] [CrossRef]
- Lu, X.; Fu, X.; Wang, D.; Wang, J.; Chen, X.; Hao, M.; Wang, J.; Gervers, K.A.; Guo, L.; Wang, S.; et al. Resequencing of cv CRI-12 family reveals haplotype block inheritance and recombination of agronomically important genes in artificial selection. Plant Biotechnol. J. 2018. [Google Scholar] [CrossRef]
- Lu, P.; Magwanga, R.O.; Guo, X.; Kirungu, J.N.; Lu, H.; Cai, X.; Zhou, Z.; Wei, Y.; Wang, X.; Zhang, Z.; et al. Genome-Wide Analysis of Multidrug and Toxic Compound Extrusion (MATE) Family in Diploid Cotton, Gossypium raimondii and Gossypium arboreum and Its Expression Analysis Under Salt, Cadmium and Drought Stress. G3 Genes|Genomes|Genetics 2018, 8, 2483–2500. [Google Scholar] [CrossRef]
- Megha, B.R. Evaluation of Hirsutum Cotton Genotypes for Water Stress using Peg-6000 by Slanting Glass Plate Technique. Int. J. Pure Appl. Biosci. 2017, 5, 740–750. [Google Scholar] [CrossRef]
- Lu, P.; Magwanga, R.O.; Lu, H.; Kirungu, J.N.; Wei, Y.; Dong, Q.; Wang, X.; Cai, X.; Zhou, Z.; Wang, K.; et al. A novel G-protein-coupled receptors gene from upland cotton enhances salt stress tolerance in transgenic Arabidopsis. Genes 2018, 9, 209. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.T.; McInturf, S.A.; Mendoza-Cózatl, D.G. Hydroponics: A Versatile System to Study Nutrient Allocation and Plant Responses to Nutrient Availability and Exposure to Toxic Elements. J. Vis. Exp. 2016. [Google Scholar] [CrossRef]
- Jordon-Thaden, I.E.; Chanderbali, A.S.; Gitzendanner, M.A.; Soltis, D.E. Modified CTAB and TRIzol Protocols Improve RNA Extraction from Chemically Complex Embryophyta. Appl. Plant Sci. 2015, 3, 1400105. [Google Scholar] [CrossRef]
- Lalitha, S. Primer Premier 5. Biotech Softw. Internet Rep. 2002, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.-Q.; Zhu, B.-Z.; Zhu, H.-L.; Jiang, W.-B.; Luo, Y.-B. Virus-induced gene silencing in tomato fruit. Plant J. 2005, 43, 299–308. [Google Scholar] [CrossRef]
- Gao, X.; Britt, R.C., Jr.; Shan, L.; He, P. Agrobacterium-Mediated Virus-Induced Gene Silencing Assay In Cotton. J. Vis. Exp. 2011. [Google Scholar] [CrossRef]
- Nxele, X.; Klein, A.; Ndimba, B.K. Drought and salinity stress alters ROS accumulation, water retention, and osmolyte content in sorghum plants. South African J. Bot. 2017, 108, 261–266. [Google Scholar] [CrossRef]
- Clarke, J.M.; McCaig, T.N. Excised-leaf water retention capability as an indicator of drought resistance of Triticum genotypes. Can. J. Plant Sci. 1982, 62, 571–578. [Google Scholar] [CrossRef]
- Fokar, M.; Nguyen, H.T.; Blum, A. Heat tolerance in spring wheat. I. Estimating cellular thermotolerance and its heritability. Euphytica 1998, 104, 1–8. [Google Scholar] [CrossRef]
- Majidi, M.M.; Rashidi, F.; Sharafi, Y. Physiological traits related to drought tolerance in Brassica. GUASNR Int. J. Plant Prod. 2015, 9, 1735–6814. [Google Scholar]
- Bartosz, G. Superoxide Dismutases and Catalase. Oxid. Antioxid. Def. Syst. 2005, 2, 109–149. [Google Scholar] [CrossRef]
- Li, L.; Cheng, H.; Gai, J.; Yu, D. Genome-wide identification and characterization of putative cytochrome P450 genes in the model legume Medicago truncatula. Planta 2007, 226, 109–123. [Google Scholar] [CrossRef]
- Rai, A.; Singh, R.; Shirke, P.A.; Tripathi, R.D.; Trivedi, P.K.; Chakrabarty, D. Expression of rice CYP450-like gene (Os08g01480) in Arabidopsis modulates regulatory network leading to heavy metal and other abiotic stress tolerance. PLoS ONE 2015, 10, e0138574. [Google Scholar] [CrossRef]
- Amara, I.; Zaidi, I.; Masmoudi, K.; Dolors Ludevid, M.; Pagès, M.; Goday, A.; Brini, F. Insights into Late Embryogenesis Abundant (LEA) Proteins in Plants: From Structure to the Functions. Am. J. Plant Sci. 2014, 5, 3440–3455. [Google Scholar] [CrossRef]
- Usui, E.; Noshiro, M.; Ohyama, Y.; Okuda, K. Unique property of liver mitochondrial P450 to catalyze the two physiologically important reactions involved in both cholesterol catabolism and vitamin D activation. FEBS Lett. 1990, 274, 175–177. [Google Scholar] [CrossRef]
- Ro, D.K.; Mah, N.; Ellis, B.E.; Douglas, C.J. Functional Characterization and Subcellular Localization of Poplar (Populus trichocarpa × Populus deltoides) Cinnamate 4-Hydroxylase1. Plant Physiol. 2001, 126, 317–329. [Google Scholar] [CrossRef]
- Watson, C.J.W.; Froehlich, J.E.; Josefsson, C.A.; Chapple, C.; Durst, F.; Benveniste, I.; Coolbaugh, R.C. Localization of CYP86B1 in the outer envelope of chloroplasts. Plant Cell Physiol. 2001, 42, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Pinot, F.; Schaller, H.; Franke, R.; Benveniste, I.; Meyer, D.; Compagnon, V.; Schreiber, L.; Diehl, P. CYP86B1 Is Required for Very Long Chain -Hydroxyacid and -Dicarboxylic Acid Synthesis in Root and Seed Suberin Polyester. Plant Physiol. 2009, 150, 1831–1843. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Saito, S.; Hirai, N.; Matsumoto, C.; Ohigashi, H.; Ohta, D. Arabidopsis CYP707As Encode (1) -Abscisic Acid 89-Hydroxylase, a Key Enzyme in the Oxidative Catabolism of Abscisic Acid 1. Society 2004, 134, 1439–1449. [Google Scholar] [CrossRef]
- Umezawa, T.; Okamoto, M.; Kushiro, T.; Nambara, E.; Oono, Y.; Seki, M.; Kobayashi, M.; Koshiba, T.; Kamiya, Y.; Shinozaki, K. CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 2006, 46, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P.; Guerdoux-Jamet, P.; Moszer, I.; Viari, A.; Danchin, A. Implication of gene distribution in the bacterial chromosome for the bacterial cell factory. J. Biotechnol. 2000, 78, 209–219. [Google Scholar] [CrossRef]
- Lockyer, A.E.; Noble, L.R.; Rollinson, D.; Jones, C.S. Isolation and characterization of the full-length cDNA encoding a member of a novel cytochrome p450 family (CYP320A1) from the tropical freshwater snail, Biomphalaria glabrata, intermediate host for Schistosoma mansoni. Mem. Inst. Oswaldo Cruz 2005, 100, 259–262. [Google Scholar] [CrossRef]
- Tavares, K.C.S.; de Mello e Pinho, R.; de Sá Carneiro, I.; de Aguiar, L.H.; Calderón, C.E.M.; Martins, L.T.; Ambrósio, C.E.; Maga, E.A.; Bertolini, M.; Murray, J.D.; et al. Efficient RNAi-induced protein knockdown in somatic cells using diced or chemically produced small interfering RNAs (siRNA). Acta Sci. Vet. 2012, 40, 1048. [Google Scholar]
- Miller, G.; Honig, A.; Stein, H.; Suzuki, N.; Mittler, R.; Zilberstein, A. Unraveling Δ1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J. Biol. Chem. 2009, 284, 26482–26492. [Google Scholar] [CrossRef]
- Pavei, D.; Gonçalves-Vidigal, M.C.; Schuelter, A.R.; Schuster, I.; Vieira, E.S.N.; Vendruscolo, E.C.G.; Poletine, J.P. Response to water stress in transgenic (p5cs gene) wheat plants (Triticum aestivum L.). Aust. J. Crop Sci. 2016, 10, 776–783. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, H.; Liu, Z.; Wu, S.; Yang, Y.; Feyereisen, R.; Heckel, D.G.; Wu, Y. Phylogenetic and functional characterization of ten P450 genes from the CYP6AE subfamily of Helicoverpa armigera involved in xenobiotic metabolism. Insect Biochem. Mol. Biol. 2018, 93, 79–91. [Google Scholar] [CrossRef]
- Gao, J.; Lan, T. Functional characterization of the late embryogenesis abundant (LEA) protein gene family from Pinus tabuliformis (Pinaceae) in Escherichia coli. Sci. Rep. 2016, 6, 19467. [Google Scholar] [CrossRef]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 791–812. [Google Scholar] [CrossRef]
- Hedayati, P.; Monfard, H.H.; Isa, N.M.; Hwang, D.J.; Zain, C.R.C.M.; Uddin, M.I.; Zuraida, A.R.; Ismail, I.; Zainal, Z. Overexpression of the stress-associated protein (SAP) SAPMR219 in Oryza sativa (var. MR219) increases salt tolerance in transformed Arabidopsis thaliana plants. Plant Omics 2015, 8, 292–299. [Google Scholar]
- Chen, Z.; Brown, R.L.; Cleveland, T.E. Evidence for an association in corn between stress tolerance and resistance to Aspergillus flavus infection and aflatoxin contamination. J. Biotechnol. 2004, 3, 693–699. [Google Scholar]
- Zhou, M.-L.; Ma, J.-T.; Pang, J.-F.; Zhang, Z.-L.; Tang, Y.-X.; Wu, Y.-M. Regulation of plant stress response by dehydration responsive element binding (DREB) transcription factors. African J. Biotechnol. 2010, 9, 9255–9279. [Google Scholar] [CrossRef]
- An, D.; Ma, Q.; Wang, H.; Yang, J.; Zhou, W.; Zhang, P. Cassava C-repeat binding factor 1 gene responds to low temperature and enhances cold tolerance when overexpressed in Arabidopsis and cassava. Plant Mol. Biol. 2017, 94, 109–124. [Google Scholar] [CrossRef]
- Sun, H.; Hu, M.; Li, J.; Chen, L.; Li, M.; Zhang, S.; Zhang, X.; Yang, X. Comprehensive analysis of NAC transcription factors uncovers their roles during fiber development and stress response in cotton. BMC Plant Biol. 2018, 18. [Google Scholar] [CrossRef]
- Saastamoinen, M.; Brakefield, P.M.; Ovaskainen, O. Environmentally induced dispersal-related life-history syndrome in the tropical butterfly, Bicyclus anynana. J. Evol. Biol. 2012, 25, 2264–2275. [Google Scholar] [CrossRef]
- Nazari, M.; Maali Amiri, R.; Mehraban, F.H.; Khaneghah, H.Z. Change in antioxidant responses against oxidative damage in black chickpea following cold acclimation. Russ. J. Plant Physiol. 2012, 59, 183–189. [Google Scholar] [CrossRef]
- Bonnes-Taourel, D.; Guérin, M.C.; Torreilles, J. Is malonaldehyde a valuable indicator of lipid peroxidation? Biochem. Pharmacol. 1992, 44, 985–988. [Google Scholar] [CrossRef]
- Mayne, S.T. Oxidative stress, dietary antioxidant supplements, and health: Is the glass half full or half empty? Cancer Epidemiol. Biomarkers Prev. 2013, 22, 2145–2147. [Google Scholar] [CrossRef]
- Stein, H.; Honig, A.; Miller, G.; Erster, O.; Eilenberg, H.; Csonka, L.N.; Szabados, L.; Koncz, C.; Zilberstein, A. Elevation of free proline and proline-rich protein levels by simultaneous manipulations of proline biosynthesis and degradation in plants. Plant Sci. 2011, 181, 140–150. [Google Scholar] [CrossRef]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef]
- Kubala, S.; Wojtyla, L.; Quinet, M.; Lechowska, K.; Lutts, S.; Garnczarska, M. Enhanced expression of the proline synthesis gene P5CSA in relation to seed osmopriming improvement of Brassica napus germination under salinity stress. J. Plant Physiol. 2015, 183, 1–12. [Google Scholar] [CrossRef]
- Syvertsen, J.P.; Garcia-Sanchez, F. Multiple abiotic stresses occurring with salinity stress in citrus. Environ. Exp. Bot. 2014, 103, 128–137. [Google Scholar] [CrossRef]
- Bhutta, M.A.; Qureshi, M.K.; Shabaz, M.K.; Ahmad, M.; Ul-Malook, S.; Ali, Q. Oxidative damage caused by reactive oxygen species under drought stress in Gossypium hirsutum. Life Sci. J. 2015, 12. [Google Scholar] [CrossRef]
- Aydin, S.; Büyük, I.; Aras, E.S. Expression of SOD gene and evaluating its role in stress tolerance in NaCl and PEG stressed Lycopersicum esculentum. Turk. J. Botany 2014, 38, 89–98. [Google Scholar] [CrossRef]
- Erpen, L.; Devi, H.S.; Grosser, J.W.; Dutt, M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell. Tissue Organ Cult. 2018, 132, 1–25. [Google Scholar] [CrossRef]
- Buck, T.M.; Wright, C.M.; Brodsky, J.L. The activities and function of molecular chaperones in the endoplasmic reticulum. Semin. Cell Dev. Biol. 2007, 18, 751–761. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Magwanga, R.O.; Lu, P.; Kirungu, J.N.; Dong, Q.; Cai, X.; Zhou, Z.; Wang, X.; Hou, Y.; Xu, Y.; Peng, R.; et al. Knockdown of Cytochrome P450 Genes Gh_D07G1197 and Gh_A13G2057 on Chromosomes D07 and A13 Reveals Their Putative Role in Enhancing Drought and Salt Stress Tolerance in Gossypium hirsutum. Genes 2019, 10, 226. https://doi.org/10.3390/genes10030226
Magwanga RO, Lu P, Kirungu JN, Dong Q, Cai X, Zhou Z, Wang X, Hou Y, Xu Y, Peng R, et al. Knockdown of Cytochrome P450 Genes Gh_D07G1197 and Gh_A13G2057 on Chromosomes D07 and A13 Reveals Their Putative Role in Enhancing Drought and Salt Stress Tolerance in Gossypium hirsutum. Genes. 2019; 10(3):226. https://doi.org/10.3390/genes10030226
Chicago/Turabian StyleMagwanga, Richard Odongo, Pu Lu, Joy Nyangasi Kirungu, Qi Dong, Xiaoyan Cai, Zhongli Zhou, Xingxing Wang, Yuqing Hou, Yanchao Xu, Renhai Peng, and et al. 2019. "Knockdown of Cytochrome P450 Genes Gh_D07G1197 and Gh_A13G2057 on Chromosomes D07 and A13 Reveals Their Putative Role in Enhancing Drought and Salt Stress Tolerance in Gossypium hirsutum" Genes 10, no. 3: 226. https://doi.org/10.3390/genes10030226
APA StyleMagwanga, R. O., Lu, P., Kirungu, J. N., Dong, Q., Cai, X., Zhou, Z., Wang, X., Hou, Y., Xu, Y., Peng, R., Agong, S. G., Wang, K., & Fang, L. (2019). Knockdown of Cytochrome P450 Genes Gh_D07G1197 and Gh_A13G2057 on Chromosomes D07 and A13 Reveals Their Putative Role in Enhancing Drought and Salt Stress Tolerance in Gossypium hirsutum. Genes, 10(3), 226. https://doi.org/10.3390/genes10030226




