Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. RNA-Seq
2.3. Reads Alignment, Transcript Assembly and Junction Prediction
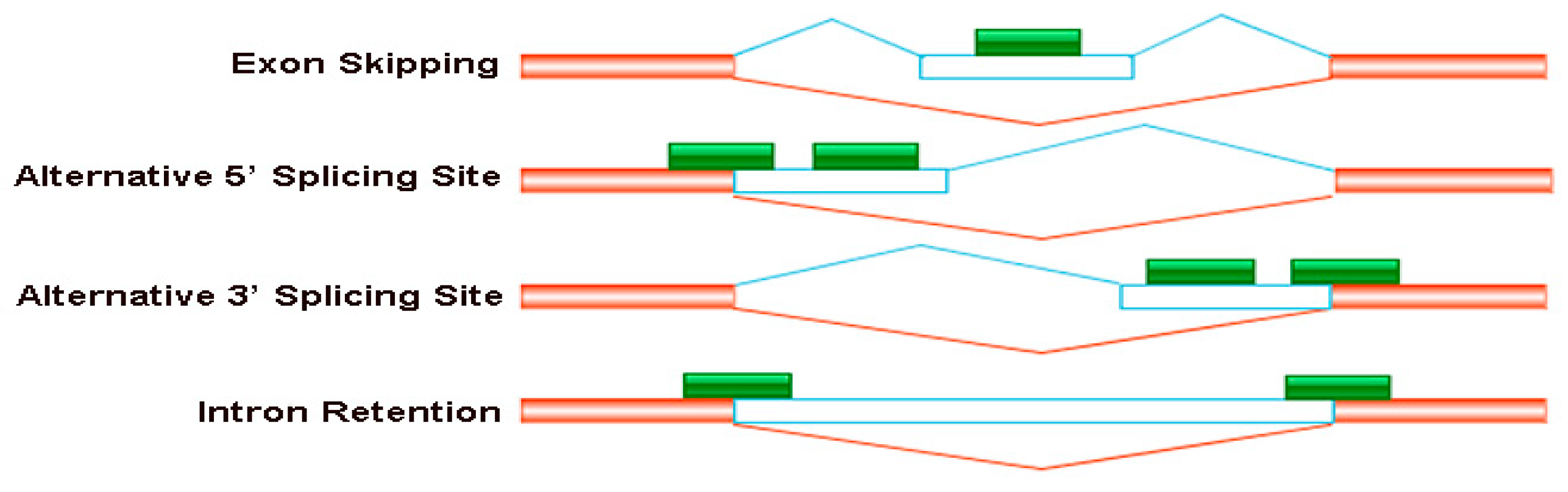
2.4. Detection of Alternative Splicing Events
2.5. Identification of Differential Alternative Splicing Events and Differentially Expressed Genes
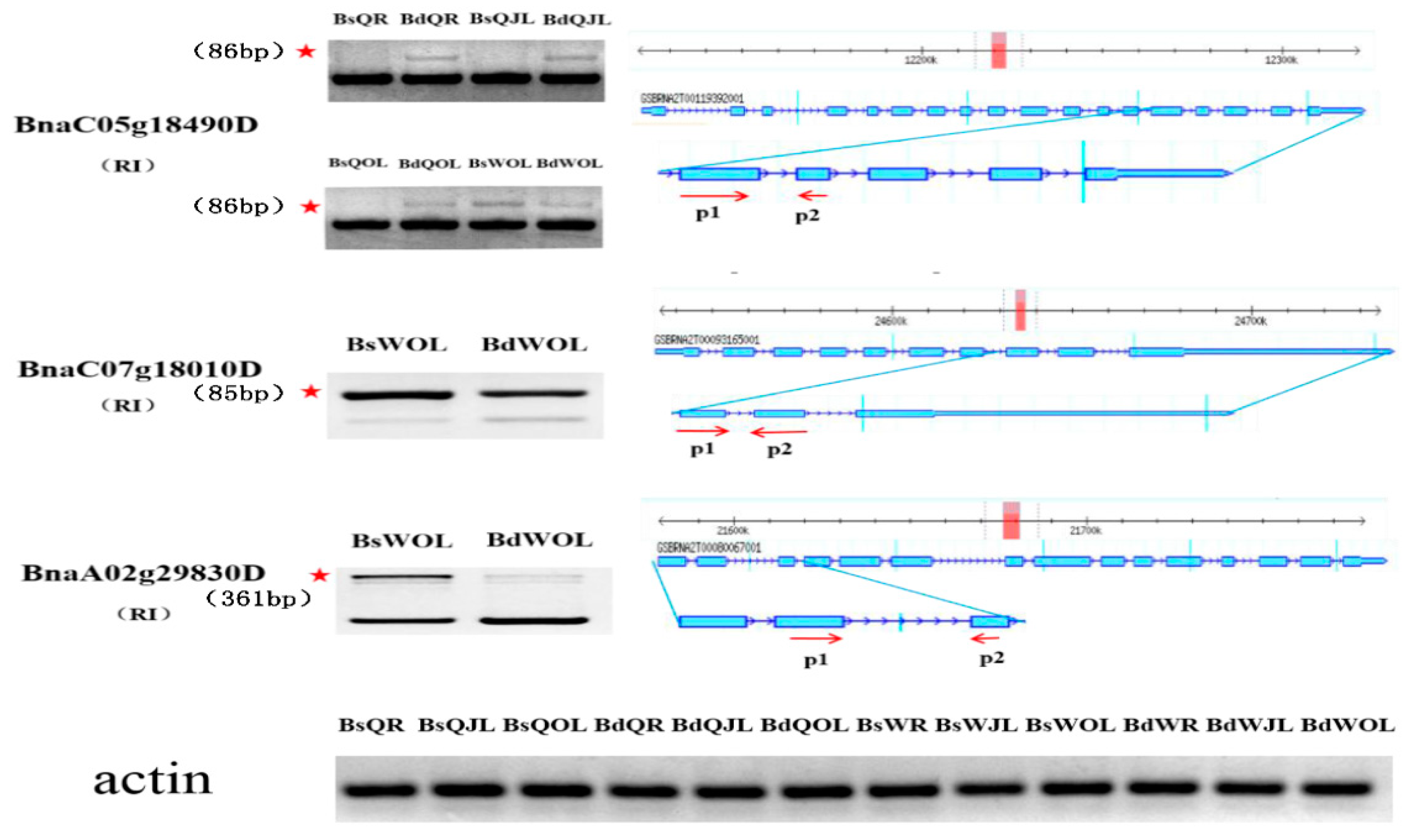
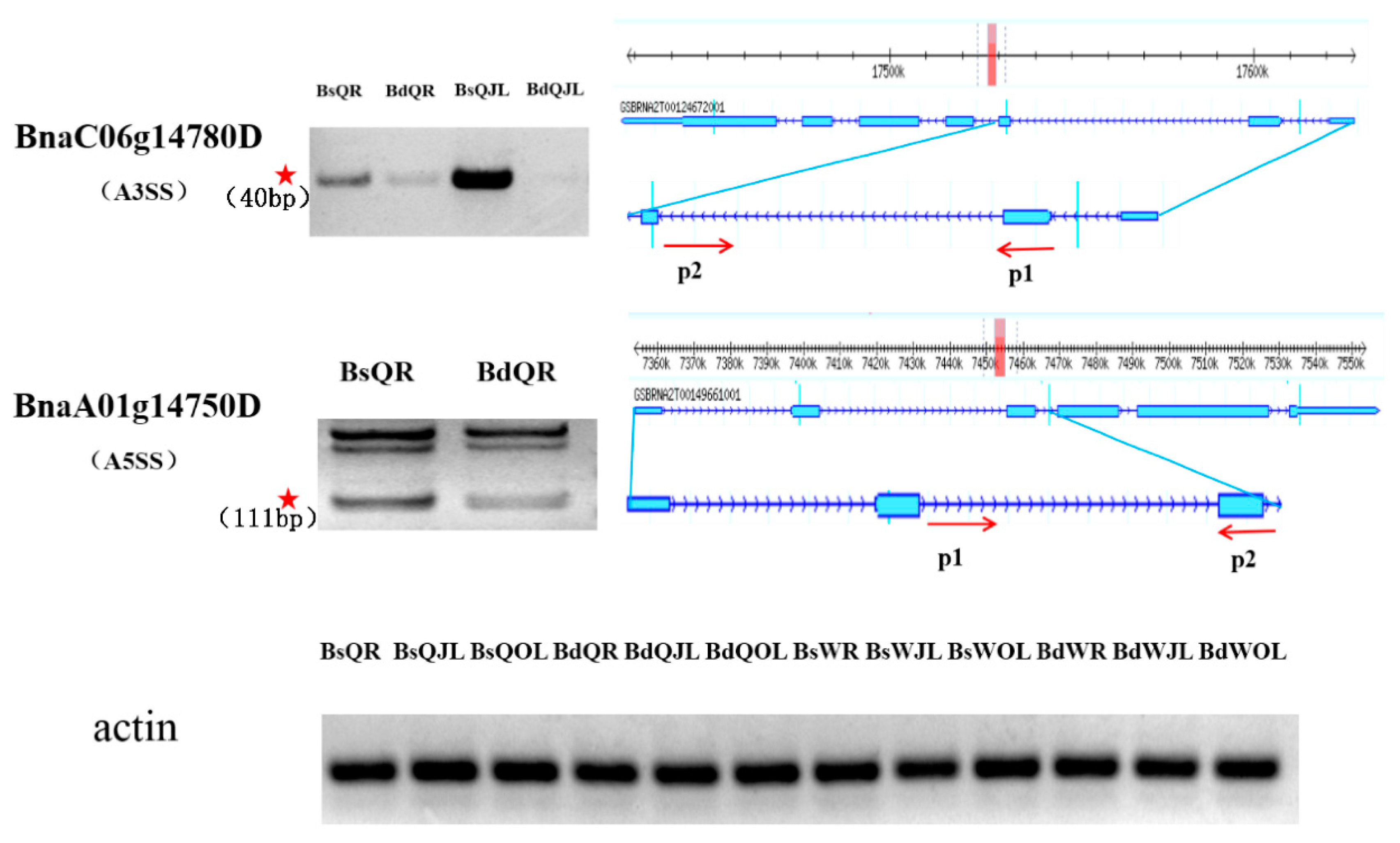
2.6. Semi-Quantitative RT-PCR Analysis
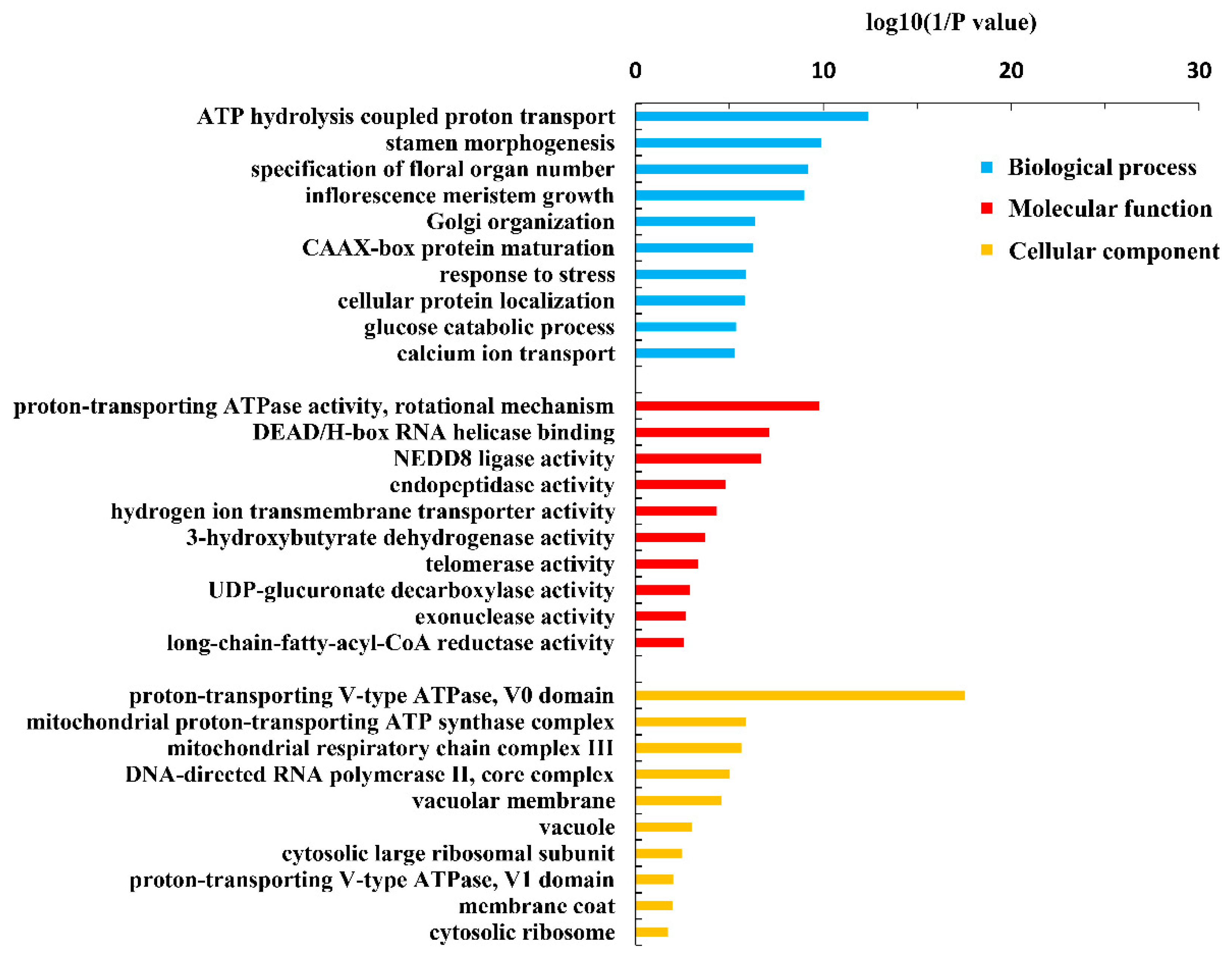
2.7. Gene Ontology (GO) Analysis and KEGG Analysis
3. Results
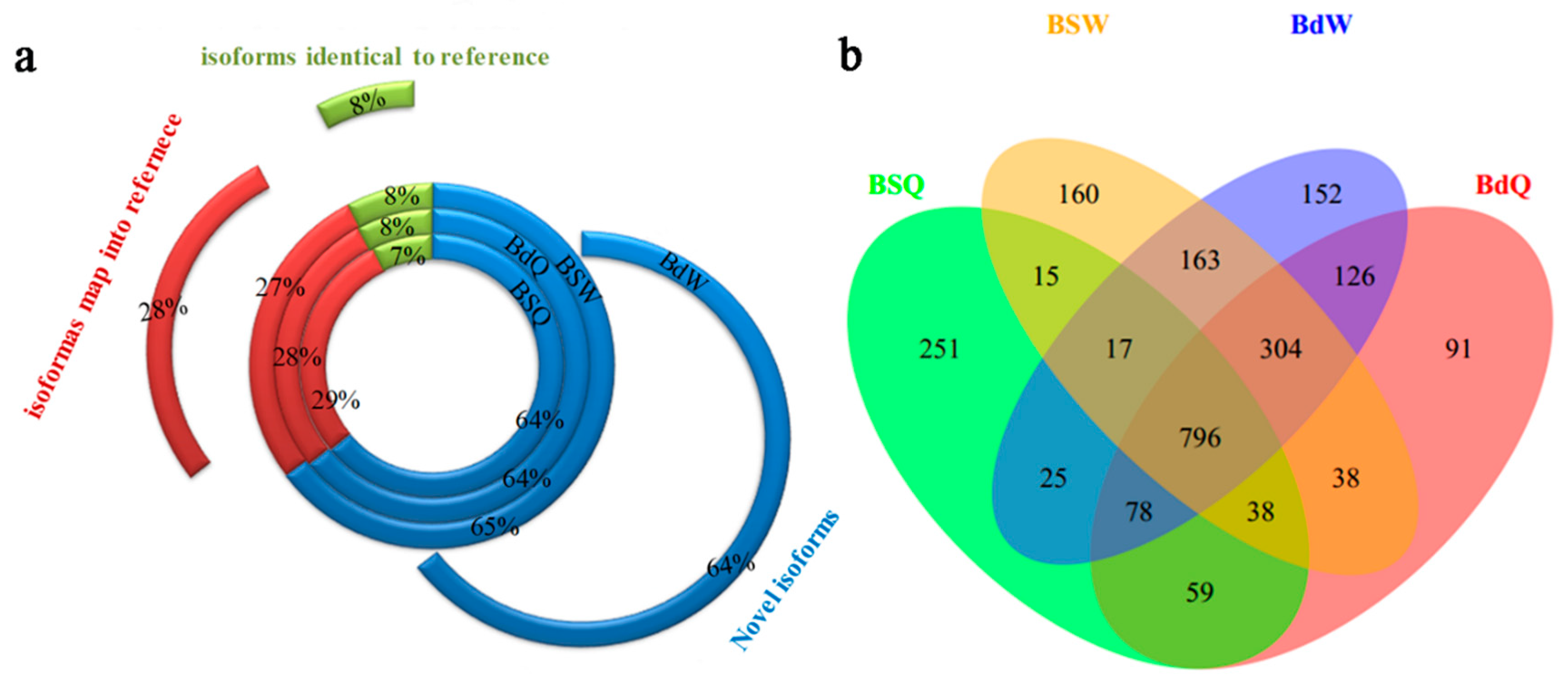
3.1. Novel Transcripts and Novel Genes in B. napus
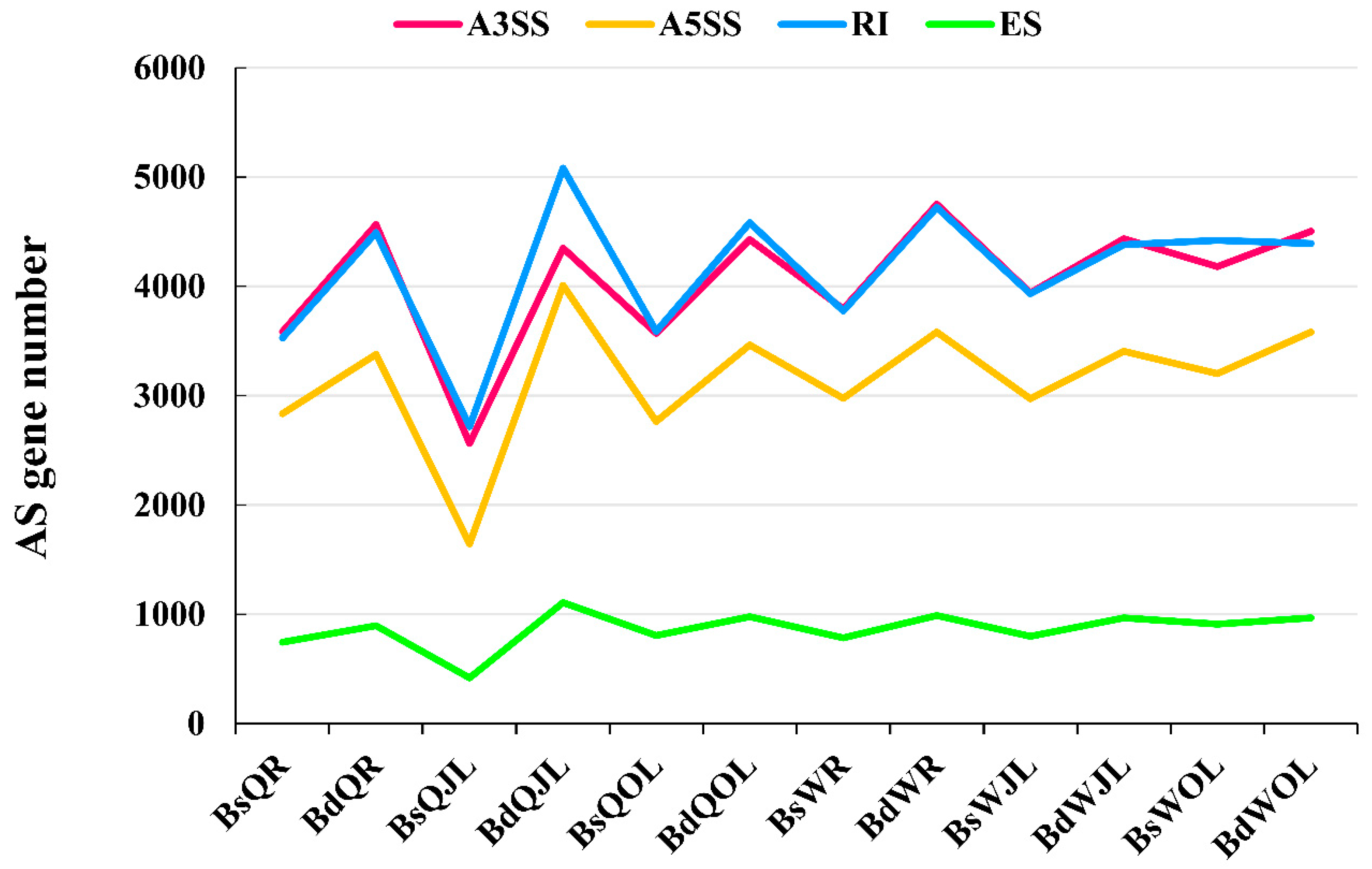
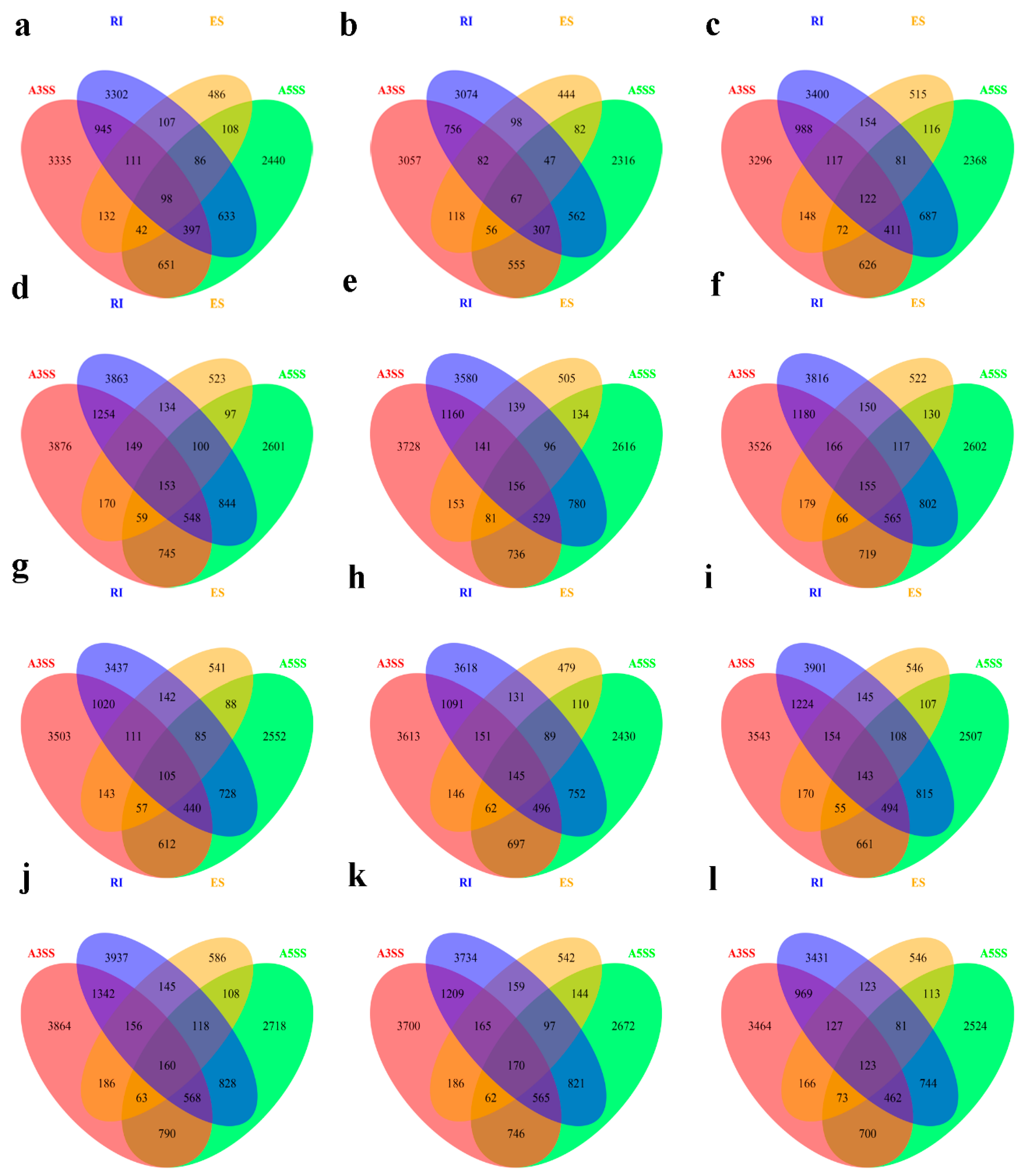
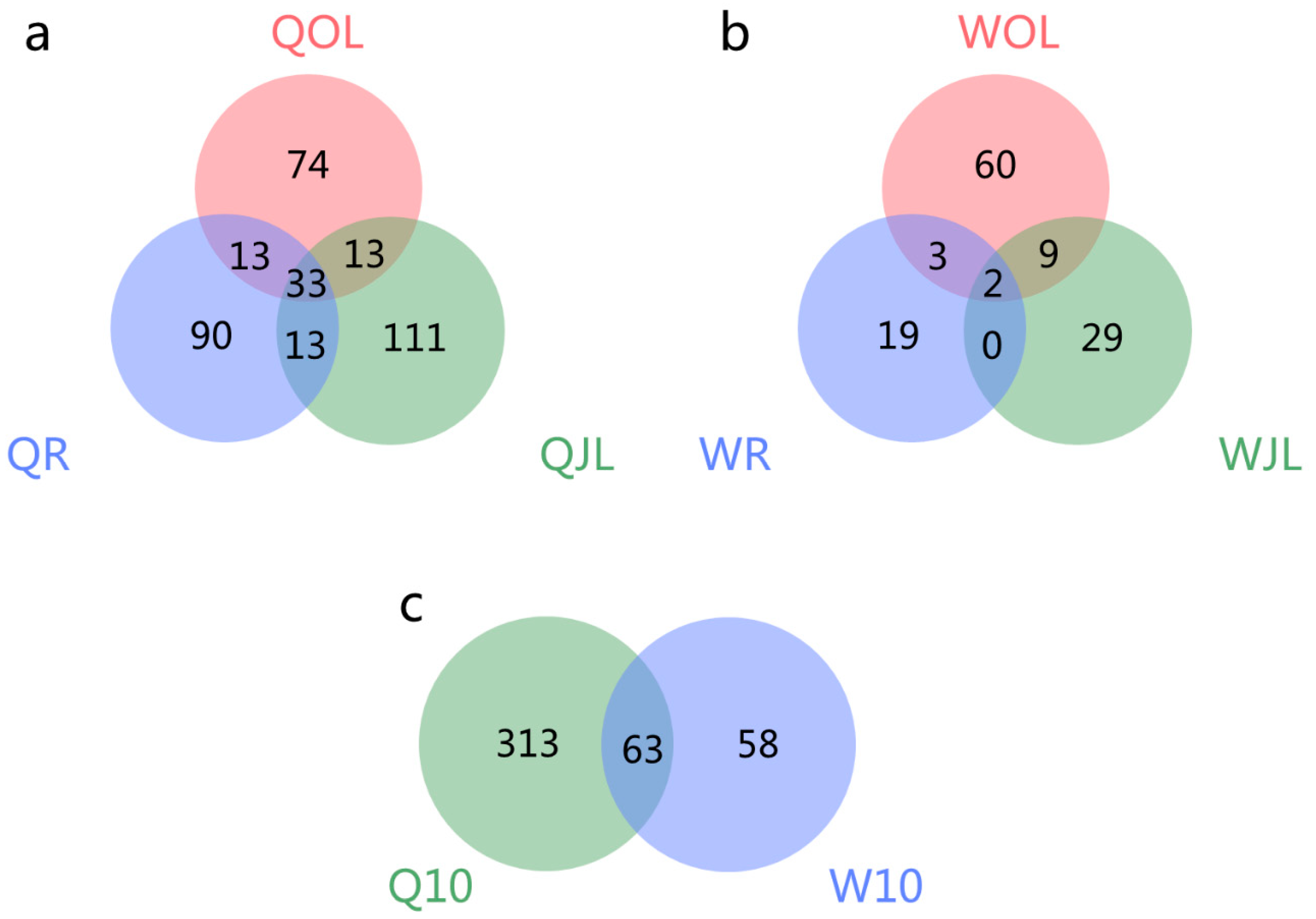
3.2. Identification of AS Events in QY10 and W10 under B Sufficient and Deficient Conditions
3.3. Differential Alternative Splicing in QY10 and W10 in B Deficient Conditions
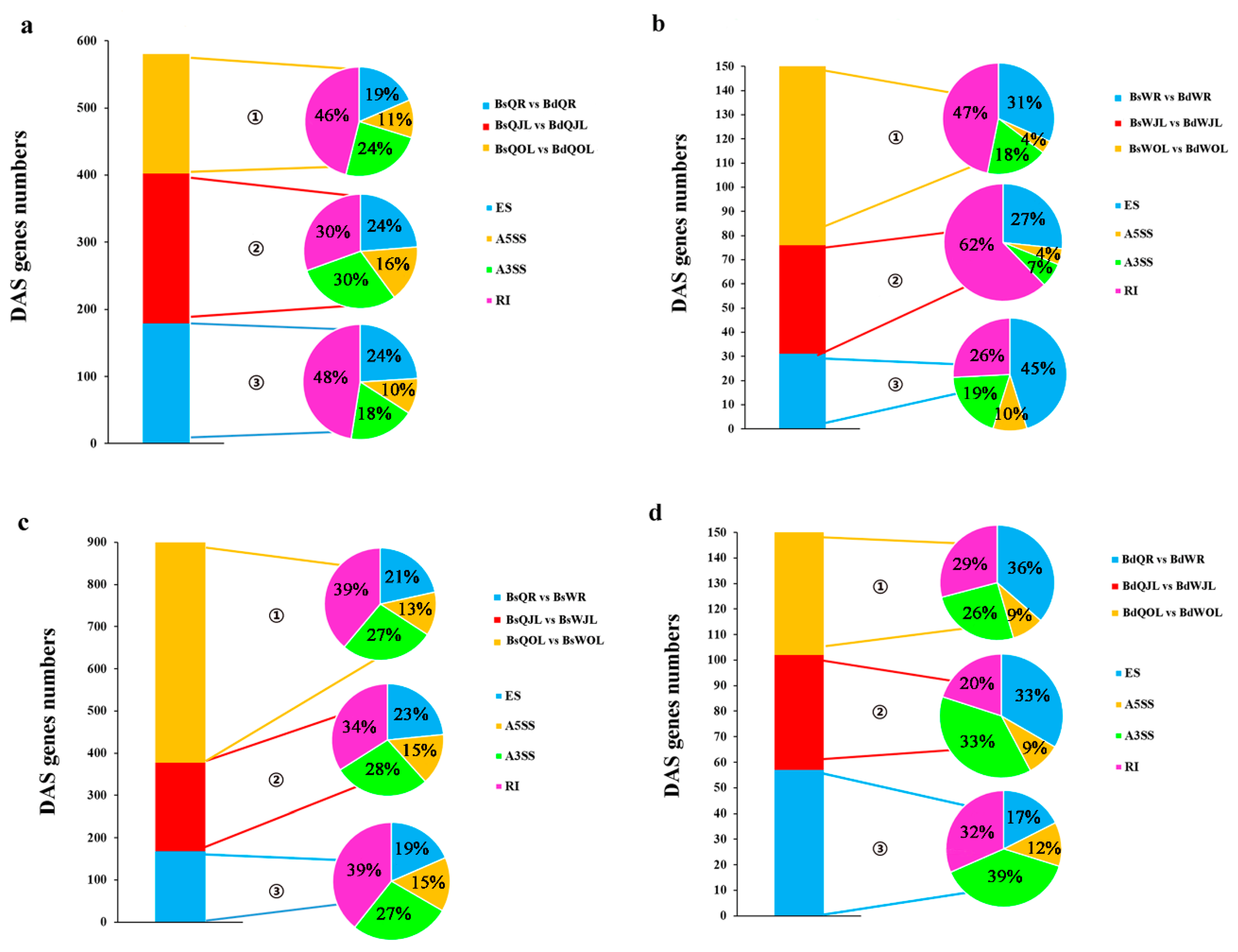
3.4. DAS Genes and DE Genes in QY10 and W10 in B Deficient Conditions
3.5. SR Splicing Factors in QY10 and W10 under B Deficient Condition
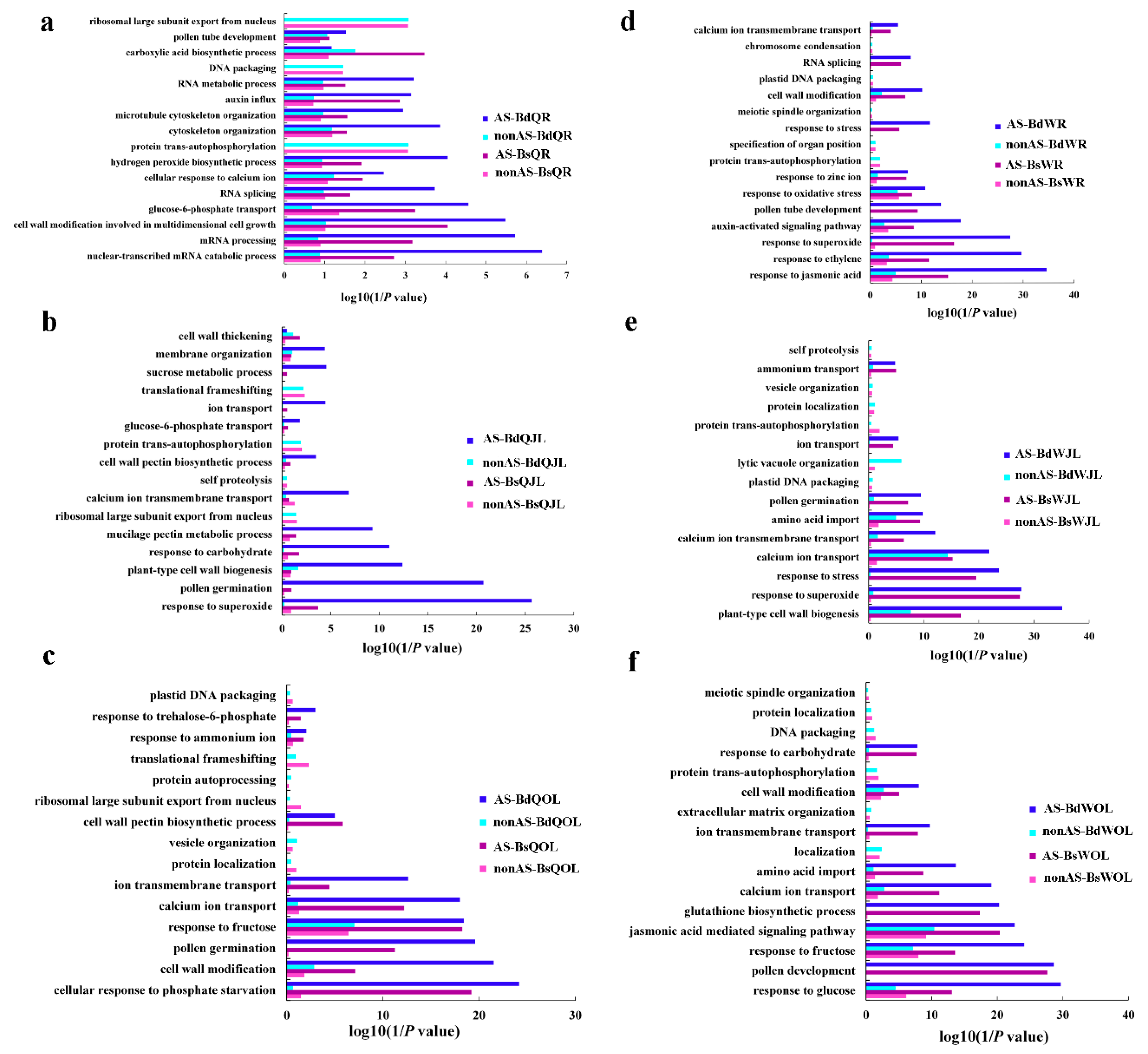
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Maniatis, T. Mechanisms of alternative pre-mRNA splicing. Science 1991, 251, 33–35. [Google Scholar] [CrossRef] [PubMed]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Huang, B.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xiao, X.; Van Nostrand, E.; Burge, C.B. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell 2006, 23, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W. Alternative splicing in plants—Coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lin, W.D.; Ray, P.; Lan, P.; Schmidt, W. Genome-wide detection of condition-sensitive alternative splicing in Arabidopsis roots. Plant Physiol. 2013, 162, 1750–1763. [Google Scholar] [CrossRef]
- Walters, B.; Lum, G.; Sablok, G.; Min, X.J. Genome-wide landscape of alternative splicing events in Brachypodium distachyon. DNA Res. 2013, 20, 163–171. [Google Scholar] [CrossRef]
- Lorković, Z.J.; Kirk, D.A.W.; Lambermon, M.H.; Filipowicz, W. Pre-mRNA splicing in higher plants. Trends Plant Sci. 2000, 5, 160–167. [Google Scholar] [CrossRef]
- Marquez, Y.; Brown, J.W.; Simpson, C.G.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef]
- Jiang, J.; Liu, X.; Liu, C.; Liu, G.; Li, S.; Wang, L. Integrating omics and alternative splicing reveals insights into grape response to high temperature. Plant Physiol. 2017, 173, 1502–1518. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Priest, H.D.; Givan, S.A.; Shen, R.; Bryant, D.W.; Fox, S.E.; Wong, W.K.; Mockler, T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010, 20, 45–58. [Google Scholar] [CrossRef]
- Lareau, L.F.; Brooks, A.N.; Soergel, D.A.; Meng, Q.; Brenner, S.E. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv. Exp. Med. Biol. 2007, 623, 190–211. [Google Scholar]
- Kalyna, M.; Simpson, C.G.; Syed, N.H.; Lewandowska, D.; Marquez, Y.; Kusenda, B.; Marshall, J.; Fuller, J.; Cardle, L.; McNicol, J. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2011, 40, 2454–2469. [Google Scholar] [CrossRef]
- Laloum, T.; Martín, G.; Duque, P. Alternative splicing control of abiotic stress responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Emrich, S.J.; Barbazuk, W.B.; Li, L.; Schnable, P.S. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007, 17, 69–73. [Google Scholar] [CrossRef]
- Brown, P.; Bellaloui, N.; Wimmer, M.; Bassil, E.; Ruiz, J.; Hu, H.; Pfeffer, H.; Dannel, F.; Römheld, V. Boron in plant biology. Plant Biol. 2002, 4, 205–223. [Google Scholar] [CrossRef]
- Broadley, M.; Brown, P.; Cakmak, I.; Rengel, Z.; Zhao, F. Function of nutrients: Micronutrients. In Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Marschner, P., Ed.; Academic Press: San Diego, CA, USA, 2012; pp. 191–248. [Google Scholar]
- Loomis, W.D.; Durst, R.W. Chemistry and biology of boron. Biofactors 1992, 3, 229–239. [Google Scholar]
- Wang, Y.; Lan, L. A study on the boron efficiency of rape (Brassica napus L.). J. Huazhong Agric. Univ. 1995, 21, 71–82. [Google Scholar]
- Xu, F.; Yang, Y.; Wang, Y.; Wu, L. Boron uptake and retranslocation in cultivars of Brassica napus differing in boron efficiency. In Boron in Plant and Animal Nutrition; Goldbach, H.E., Brown, P.H., Rerkasem, B., Thellier, M., Wimmer, M.A., Bell, R.W., Eds.; Springer: Boston, MA, USA, 2002; pp. 127–135. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Shi, L.; Wang, L.; Xu, F. Proteomic alterations of Brassica napus root in response to boron deficiency. Plant Mol. Biol. 2010, 74, 265–278. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Chen, S.; Shi, L.; Xu, F. Proteomics reveals the adaptability mechanism of Brassica napus to short-term boron deprivation. Plant Soil 2011, 347, 195–210. [Google Scholar] [CrossRef]
- Zhang, D.; Hua, Y.; Wang, X.; Zhao, H.; Shi, L.; Xu, F. A high-density genetic map identifies a novel major QTL for boron efficiency in oilseed rape (Brassica napus L.). PLoS ONE 2014, 9, e112089. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Zhang, D.; Zhou, T.; He, M.; Ding, G.; Shi, L.; Xu, F. Transcriptomics-assisted quantitative trait locus fine mapping for the rapid identification of a nodulin 26-like intrinsic protein gene regulating boron efficiency in allotetraploid rapeseed. Plant Cell Environ. 2016, 39, 1601–1618. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, H.; He, M.; Zhao, Z.; Cai, H.; Ding, G.; Shi, L.; Xu, F. The boron transporter BnaC4. BOR1; 1c is critical for inflorescence development and fertility under boron limitation in Brassica napus. Plant Cell Environ. 2017, 40, 1819–1833. [Google Scholar] [CrossRef]
- Chen, M.; Manley, J.L. Mechanisms of alternative splicing regulation: Insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009, 10, 741. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Lin, W.D.; Tu, S.L. Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol. 2014, 165, 826–840. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Cui, P.; Wang, Z.; Zhang, S.; Ali, S.; Xiong, L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genomics 2014, 15, 431. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.R.; Arnon, D.I. The water culture method for growing plant without soil. Calif. Agric. Exp. Stn. Circ. 1950, 347. [Google Scholar]
- Yuan, D.; Li, W.; Hua, Y.; King, G.J.; Xu, F.; Shi, L. Genome-wide identification and characterization of the aquaporin gene family and transcriptional responses to boron deficiency in Brassica napus. Front Plant Sci 2017, 8, 1336. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2018, 5, 621–628. [Google Scholar] [CrossRef]
- Foissac, S.; Sammeth, M. ASTALAVISTA: Dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic. Acids Res. 2007, 35, W297–W299. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Park, J.W.; Lu, Z.x.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Thatcher, S.R.; Danilevskaya, O.N.; Meng, X.; Beatty, M.; Zastrow-Hayes, G.; Harris, C.; Van Allen, B.; Habben, J.; Li, B. Genome-wide analysis of alternative splicing during development and drought stress in maize. Plant Physiol. 2016, 170, 586–599. [Google Scholar] [CrossRef]
- Li, P.; Ponnala, L.; Gandotra, N.; Wang, L.; Si, Y.; Tausta, S.L.; Kebrom, T.H.; Provart, N.; Patel, R.; Myers, C.R.; et al. The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 2010, 42, 1060. [Google Scholar] [CrossRef]
- Wu, H.P.; Su, Y.S.; Chen, H.C.; Chen, Y.R.; Wu, C.C.; Lin, W.D.; Tu, S.L. Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 2014, 15, R10. [Google Scholar] [CrossRef]
- Liu, Z.; Qin, J.; Tian, X.; Xu, S.; Wang, Y.; Li, H.; Wang, X.; Peng, H.; Yao, Y.; Hu, Z.; et al. Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2018, 16, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yang, S.S.; Liang, Z.; Feng, B.R.; Liu, L.; Huang, Y.B.; Tang, Y.X. Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 2012, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Barta, A.; Kalyna, M.; Reddy, A.S. Implementing a rational and consistent nomenclature for serine/arginine-rich protein splicing factors (SR proteins) in plants. Plant Cell 2010, 22, 2926–2929. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, M.; Tsumoto, A.; Shimamoto, K. The serine/arginine-rich protein family in rice plays important roles in constitutive and alternative splicing of pre-mRNA. Plant Cell 2006, 18, 146–158. [Google Scholar] [CrossRef]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007, 49, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- Duque, P. A role for SR proteins in plant stress responses. Plant Signal. Behav. 2011, 6, 49–54. [Google Scholar] [CrossRef]


| No. of Reads (Millions) | Reads Percentage (%) | |
|---|---|---|
| Starting reads | 2078.8 | - |
| Clean reads | 1921.9 | 92.45 |
| Mapped reads | 1310.9 | 68.2 |
| Sample Name | Novel Transcripts | Reference Like Transcripts | Novel Genes | Reference Like Genes |
|---|---|---|---|---|
| BsQR1 | 32,762 | 16,656 | 1019 | 5877 |
| BsQR2 | 33,451 | 16,384 | 1136 | 5766 |
| BsQR3 | 33,662 | 16,661 | 1096 | 5841 |
| BsQJL1 | 33,763 | 15,740 | 1147 | 5539 |
| BsQJL2 | 35,091 | 15,637 | 1248 | 5742 |
| BsQJL3 | 33,274 | 15,608 | 1221 | 5400 |
| BsQOL1 | 34,228 | 14,921 | 1288 | 5033 |
| BsQOL2 | 34,501 | 14,959 | 1299 | 5074 |
| BsQOL3 | 33,610 | 14,592 | 1330 | 4927 |
| BdQR1 | 35,565 | 16,441 | 1415 | 5648 |
| BdQR2 | 35,780 | 16,570 | 1549 | 5643 |
| BdQR3 | 36,428 | 16,731 | 1545 | 5658 |
| BdQJL1 | 37,831 | 16,365 | 1601 | 5455 |
| BdQJL2 | 36,892 | 16,614 | 1518 | 5625 |
| BdQJL3 | 37,677 | 16,124 | 1500 | 5391 |
| BdQOL1 | 37,624 | 16,125 | 1685 | 5291 |
| BdQOL2 | 35,766 | 16,028 | 1473 | 5339 |
| BdQOL3 | 36,421 | 16,189 | 1513 | 5383 |
| BsWR1 | 34,561 | 16,128 | 1357 | 5563 |
| BsWR2 | 35,245 | 16,801 | 1433 | 5777 |
| BsWR3 | 34,940 | 17,124 | 1417 | 5895 |
| BsWJL1 | 36,579 | 16,029 | 1513 | 5412 |
| BsWJL2 | 38,206 | 16,252 | 1470 | 5382 |
| BsWJL3 | 37,242 | 16,214 | 1529 | 5468 |
| BsWOL1 | 38,490 | 15,781 | 1812 | 5114 |
| BsWOL2 | 37,855 | 15,850 | 1767 | 5161 |
| BsWOL3 | 37,224 | 15,411 | 1713 | 5025 |
| BdWR1 | 35,174 | 16,653 | 1407 | 5738 |
| BdWR2 | 35,790 | 17,013 | 1483 | 5830 |
| BdWR3 | 36,108 | 16,838 | 1494 | 5766 |
| BdWJL1 | 36,001 | 15,875 | 1498 | 5328 |
| BdWJL2 | 37,751 | 16,557 | 1588 | 5538 |
| BdWJL3 | 36,854 | 15,976 | 1523 | 5360 |
| BdWOL1 | 36,841 | 16,041 | 1559 | 5325 |
| BdWOL2 | 36,942 | 15,973 | 1485 | 5335 |
| BdWOL3 | 35,714 | 15,746 | 1517 | 5284 |
| Cultivars | GeneID | Gene Description | DAS-Type |
|---|---|---|---|
| QY10 | BnaC09g39180D | alpha/beta-Hydrolases superfamily protein | A5SS |
| BnaA06g17710D | alpha-glucan phosphorylase 2 (PHS2) | RI | |
| BnaC01g05800D | AME3 | RI | |
| BnaC03g31650D | ATOZI1 | A3SS | |
| BnaC09g20000D | calcineurin B-like protein 9 (CBL9) | ES | |
| BnaC06g30700D | cAMP-regulated phosphoprotein 19-related protein | A3SS | |
| BnaC05g02560D | casein kinase like 13 (CKL13) | A5SS | |
| BnaC08g08360D | CONSTITUTIVE PHOTOMORPHOGENIC 9 (COP9) | RI | |
| BnaA03g27430D | DEAD box RNA helicase family protein | RI | |
| BnaA09g27350D | dormancy-associated protein-like 1 (DYL1) | RI | |
| BnaA03g39560D | enhancer of ag-4 2 (hua2) | RI | |
| BnaC03g57920D | FK506 binding protein 53 (fkbp53) | A3SS | |
| BnaAnng37730D | glutathione S-transferase phi 8 (GSTF8) | ES | |
| BnaA07g00230D | actin 7 (ACT7) | RI | |
| BnaA04g12350D | glycine rich protein 7 (atgrp7) | A5SS | |
| BnaC04g04470D | homeodomain GLABROUS 1 (HDG1) | RI | |
| BnaA06g30540D | NADH-ubiquinone oxidoreductase B8 subunit, putative | ES | |
| BnaA06g38980D | nitrilase 2 (NIT2) | ES | |
| BnaC09g20910D | peptide transporter 2 (PTR2) | RI | |
| BnaC05g18490D | Phosphoglucomutase/phosphomannomutase family protein | RI | |
| BnaA07g16600D | PUR5 | A3SS | |
| BnaC05g24350D | radical-induced cell death1 (rcd1) | RI | |
| BnaA03g30650D | Ribosomal L29 family protein | ES | |
| BnaC09g54460D | Ribosomal protein S13/S18 family | A3SS | |
| BnaA07g25750D | RNA-binding (RRM/RBD/RNP motifs) family protein | RI | |
| BnaC06g14780D | RSZ32 | A3SS | |
| BnaA07g16660D | sedoheptulose-bisphosphatase (SBPASE) | ES | |
| BnaC05g08610D | sugar transporter 1 (STP1) | A3SS | |
| BnaC04g31660D | TLD-domain containing nucleolar protein | RI | |
| BnaA03g07610D | Translation elongation factor EF1B | A3SS | |
| BnaC04g56630D | unknown protein | RI | |
| BnaCnng63660D | unknown protein | RI | |
| BnaC03g03780D | VND-interacting 1 (VNI1) | A3SS | |
| W10 | BnaCnng40950D | Cystatin/monellin superfamily protein | A3SS |
| BnaC07g51220D | nicotinate phosphoribosyltransferase 1 (NAPRT1) | A5SS |
| Tissues | KEGG Pathways | Pathway ID | No. of DAS Gene | p-Value |
|---|---|---|---|---|
| QR | Spliceosome | ko03040 | 13 | 3.08E-06 |
| Carbon metabolism | ko01200 | 7 | 0.034913379 | |
| Carbon fixation in photosynthetic organisms | ko00710 | 3 | 0.034913379 | |
| Biosynthesis of amino acids | ko01230 | 6 | 0.034913379 | |
| mRNA surveillance pathway | ko03015 | 4 | 0.034913379 | |
| Glycolysis / Gluconeogenesis | ko00010 | 3 | 0.034913379 | |
| Citrate cycle (TCA cycle) | ko00020 | 2 | 0.034913379 | |
| Glyoxylate and dicarboxylate metabolism | ko00630 | 2 | 0.034913379 | |
| Pyruvate metabolism | ko00620 | 2 | 0.034913379 | |
| Sulfur metabolism | ko00920 | 1 | 0.034913379 | |
| Nitrogen metabolism | ko00910 | 1 | 0.034913379 | |
| Calcium signaling pathway | ko04020 | 1 | 0.034913379 | |
| Galactose metabolism | ko00052 | 1 | 0.035225944 | |
| MAPK signaling pathway | ko04010 | 1 | 0.035225944 | |
| RNA transport | ko03013 | 2 | 0.037548325 | |
| Arginine and proline metabolism | ko00330 | 1 | 0.038190214 | |
| Starch and sucrose metabolism | ko00500 | 2 | 0.039194028 | |
| Amino sugar and nucleotide sugar metabolism | ko00520 | 1 | 0.043604665 | |
| Oxidative phosphorylation | ko00190 | 1 | 0.046236768 | |
| Plant hormone signal transduction | ko04075 | 2 | 0.047198466 | |
| QJL | Carbon fixation in photosynthetic organisms | ko00710 | 4 | 0.011345749 |
| Tryptophan metabolism | ko00380 | 3 | 0.011345749 | |
| Pentose phosphate pathway | ko00030 | 3 | 0.011345749 | |
| Glycolysis / Gluconeogenesis | ko00010 | 3 | 0.019619536 | |
| Spliceosome | ko03040 | 4 | 0.019619536 | |
| Glyoxylate and dicarboxylate metabolism | ko00630 | 2 | 0.019619536 | |
| Pyruvate metabolism | ko00620 | 2 | 0.019619536 | |
| ABC transporters | ko02010 | 1 | 0.019619536 | |
| Carbon metabolism | ko01200 | 4 | 0.019619536 | |
| Fructose and mannose metabolism | ko00051 | 1 | 0.019619536 | |
| MAPK signaling pathway | ko04010 | 1 | 0.019619536 | |
| Glycine, serine and threonine metabolism | ko00260 | 1 | 0.020680045 | |
| Arginine and proline metabolism | ko00330 | 1 | 0.021716355 | |
| Starch and sucrose metabolism | ko00500 | 2 | 0.021716355 | |
| Peroxisome | ko04146 | 1 | 0.021716355 | |
| Pentose and glucuronate interconversions | ko00040 | 1 | 0.021716355 | |
| Glutathione metabolism | ko00480 | 1 | 0.021865611 | |
| Amino sugar and nucleotide sugar metabolism | ko00520 | 1 | 0.024431115 | |
| Plant hormone signal transduction | ko04075 | 2 | 0.026222312 | |
| RNA transport | ko03013 | 1 | 0.02673548 | |
| Biosynthesis of amino acids | ko01230 | 1 | 0.029409127 | |
| QOL | Spliceosome | ko03040 | 7 | 0.030261395 |
| AMPK signaling pathway | ko04152 | 5 | 0.030261395 | |
| Pyruvate metabolism | ko00620 | 3 | 0.047340426 | |
| Tryptophan metabolism | ko00380 | 2 | 0.049084834 | |
| WR | Ribosome | ko03010 | 2 | 0.000561484 |
| WJL | Peroxisome | ko04146 | 2 | 0.027237847 |
| AMPK signaling pathway | ko04152 | 2 | 0.042503393 | |
| Glyoxylate and dicarboxylate metabolism | ko00630 | 1 | 0.190757264 | |
| Glycolysis / Gluconeogenesis | ko00010 | 1 | 0.278858201 | |
| WOL | Starch and sucrose metabolism | ko00500 | 3 | 0.034551184 |
| Amino sugar and nucleotide sugar metabolism | ko00520 | 2 | 0.076377304 | |
| Pentose phosphate pathway | ko00030 | 1 | 0.165990643 | |
| Galactose metabolism | ko00052 | 1 | 0.173014536 |
| Sample Names | DAS Genes | DE Genes | Overlap |
|---|---|---|---|
| BsQR vs. BdQR | 179 | 3404 | 32 |
| BsQJL vs. BdQJL | 223 | 1482 | 30 |
| BsQOL vs. BdQOL | 178 | 1364 | 8 |
| BsWR vs. BdWR | 32 | 2053 | 7 |
| BsWJL vs. BdWJL | 47 | 1054 | 1 |
| BsWOL vs. BdWOL | 85 | 1181 | 13 |
| BsQR vs. BsWR | 174 | 3253 | 10 |
| BsQJL vs. BsWJL | 217 | 744 | 4 |
| BsQOL vs. BsWOL | 589 | 2769 | 31 |
| BdQR vs. BdWR | 37 | 102 | 3 |
| BdQJL vs. BdWJL | 49 | 51 | 0 |
| BdQOL vs. BdWOL | 62 | 196 | 0 |
| Gene Name | Target Genes | Z-Score | Gene Description | DAS Type-Regulation | |||||
|---|---|---|---|---|---|---|---|---|---|
| Boron Deficient/Boron Sufficient | |||||||||
| QR | QJL | QOL | WR | WJL | WOL | ||||
| BnaC06g14780D | BnaC04g52770D | 0.92 | Basic-leucine zipper (bZIP) transcription factor | RI | - | - | - | - | - |
| BnaC01g37580D | 0.91 | Protein kinase domain | ES | - | ES | ES | - | - | |
| BnaA09g52970D | 0.88 | Expansin | RI | - | - | - | - | - | |
| BnaA01g14590D | 0.87 | Zinc finger, RING-type | - | - | RI | - | - | - | |
| BnaC04g12670D | 0.86 | Folate-biopterin transporter | - | A3SS | - | - | - | - | |
| BnaA05g30860D | 0.83 | Glycosyl hydrolase family 100 | - | - | ES | - | - | - | |
| BnaC02g31500D | 0.80 | Pectinacetylesterase | - | - | - | - | ES | - | |
| BnaA07g32180D | 0.80 | VPS35 homolog B | RI | - | RI | - | - | - | |
| BnaA01g30320D | 0.80 | Phosphoglycerate kinase | A3SS | - | - | - | - | - | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, J.; Li, W.; Wang, S.; Zhang, X.; Coules, A.; Ding, G.; Xu, F.; Ren, J.; Lu, C.; Shi, L. Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus. Genes 2019, 10, 224. https://doi.org/10.3390/genes10030224
Gu J, Li W, Wang S, Zhang X, Coules A, Ding G, Xu F, Ren J, Lu C, Shi L. Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus. Genes. 2019; 10(3):224. https://doi.org/10.3390/genes10030224
Chicago/Turabian StyleGu, Jin, Wei Li, Sheliang Wang, Xiaoyan Zhang, Anne Coules, Guangda Ding, Fangsen Xu, Jian Ren, Chungui Lu, and Lei Shi. 2019. "Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus" Genes 10, no. 3: 224. https://doi.org/10.3390/genes10030224
APA StyleGu, J., Li, W., Wang, S., Zhang, X., Coules, A., Ding, G., Xu, F., Ren, J., Lu, C., & Shi, L. (2019). Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus. Genes, 10(3), 224. https://doi.org/10.3390/genes10030224






