DNA Replication Through Strand Displacement During Lagging Strand DNA Synthesis in Saccharomyces cerevisiae
Abstract
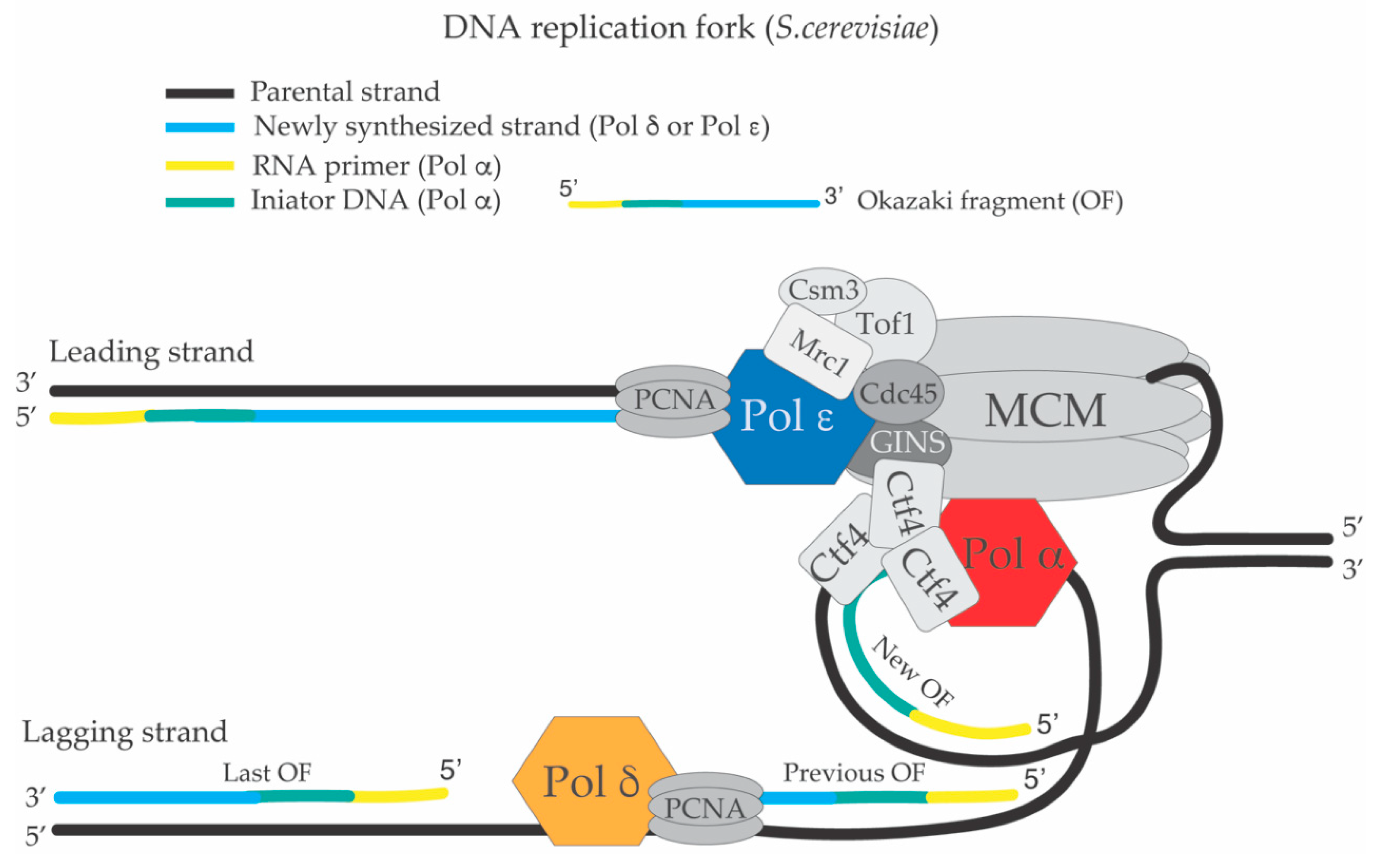
1. The Replication Fork and the DNA Synthesis Apparatus
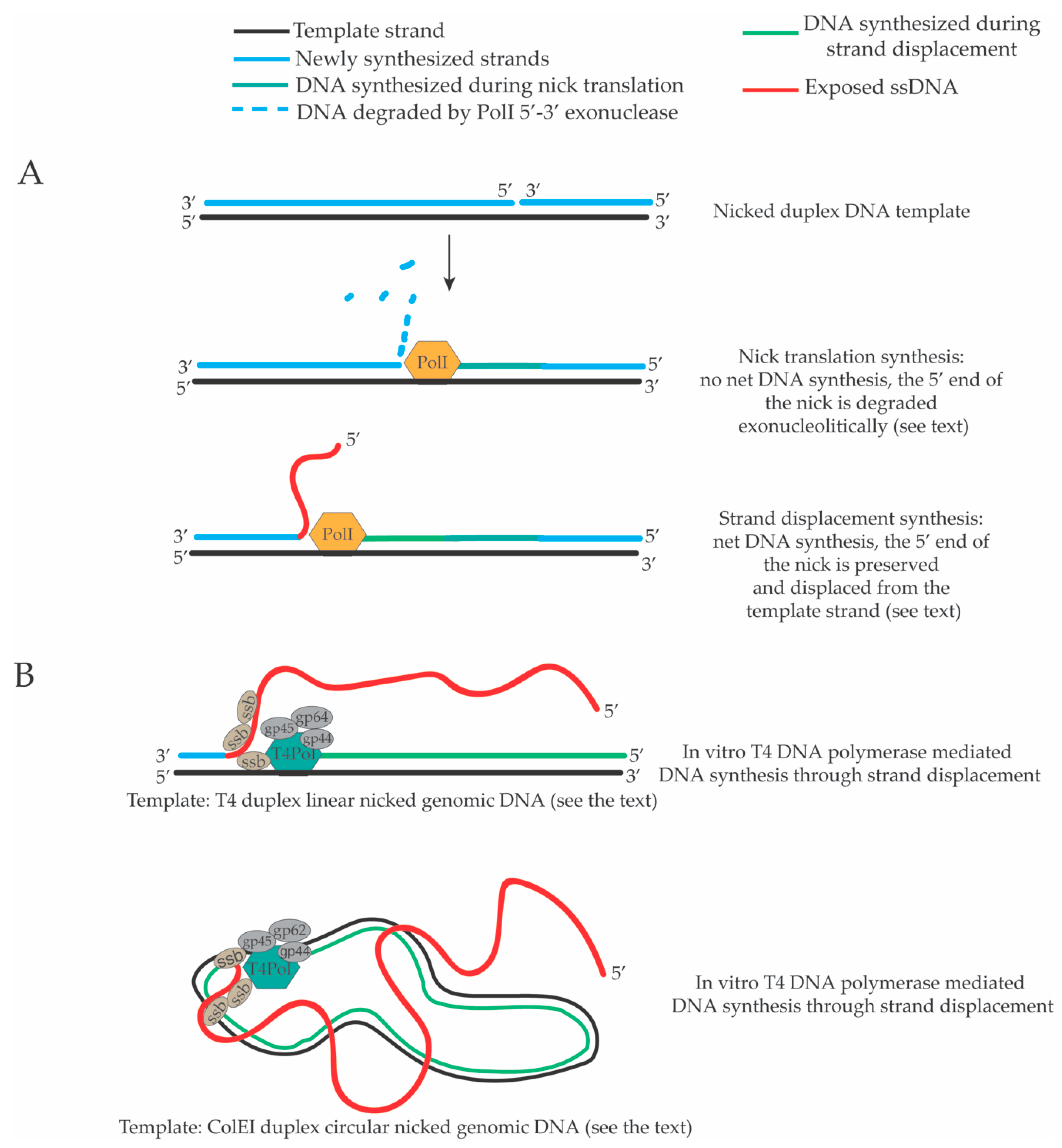
2. Strand Displacement Activity of Escherichia coli, Bacteriophage and Viral DNA Polymerases
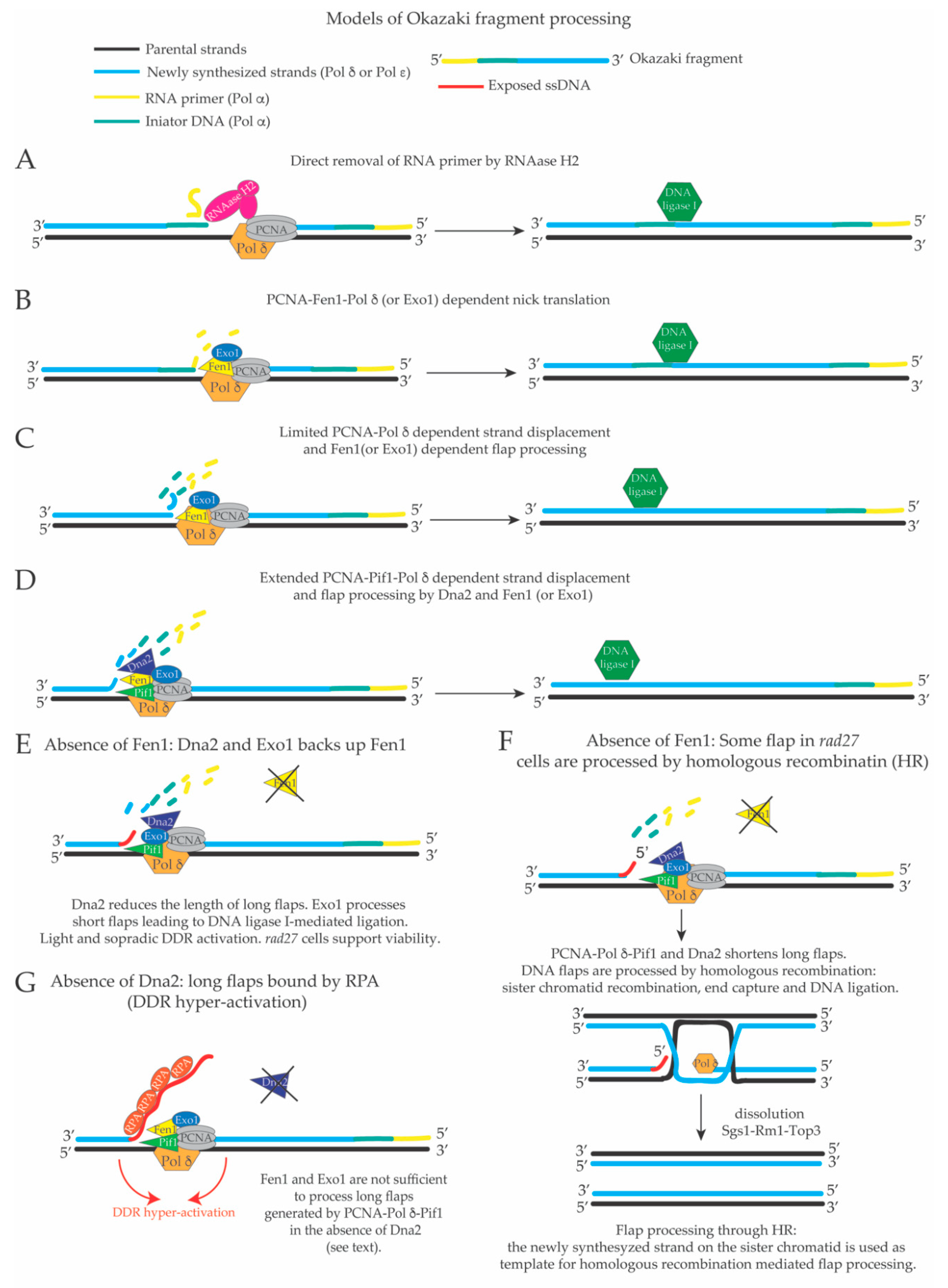
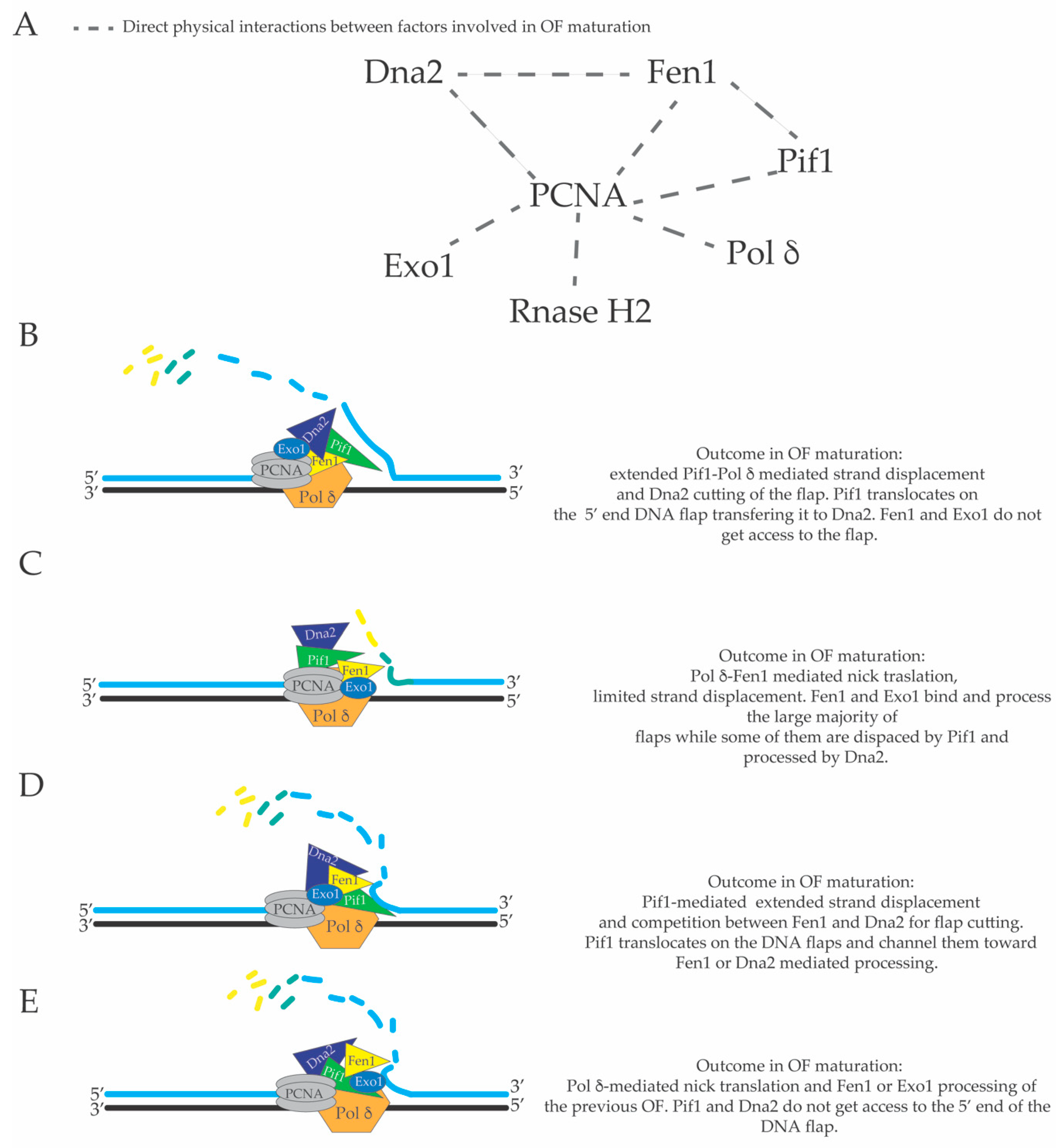
3. Limited Pol δ Strand Displacement Coupled with Short Flap Pathway of Okazaki Fragment Processing in S. cerevisiae
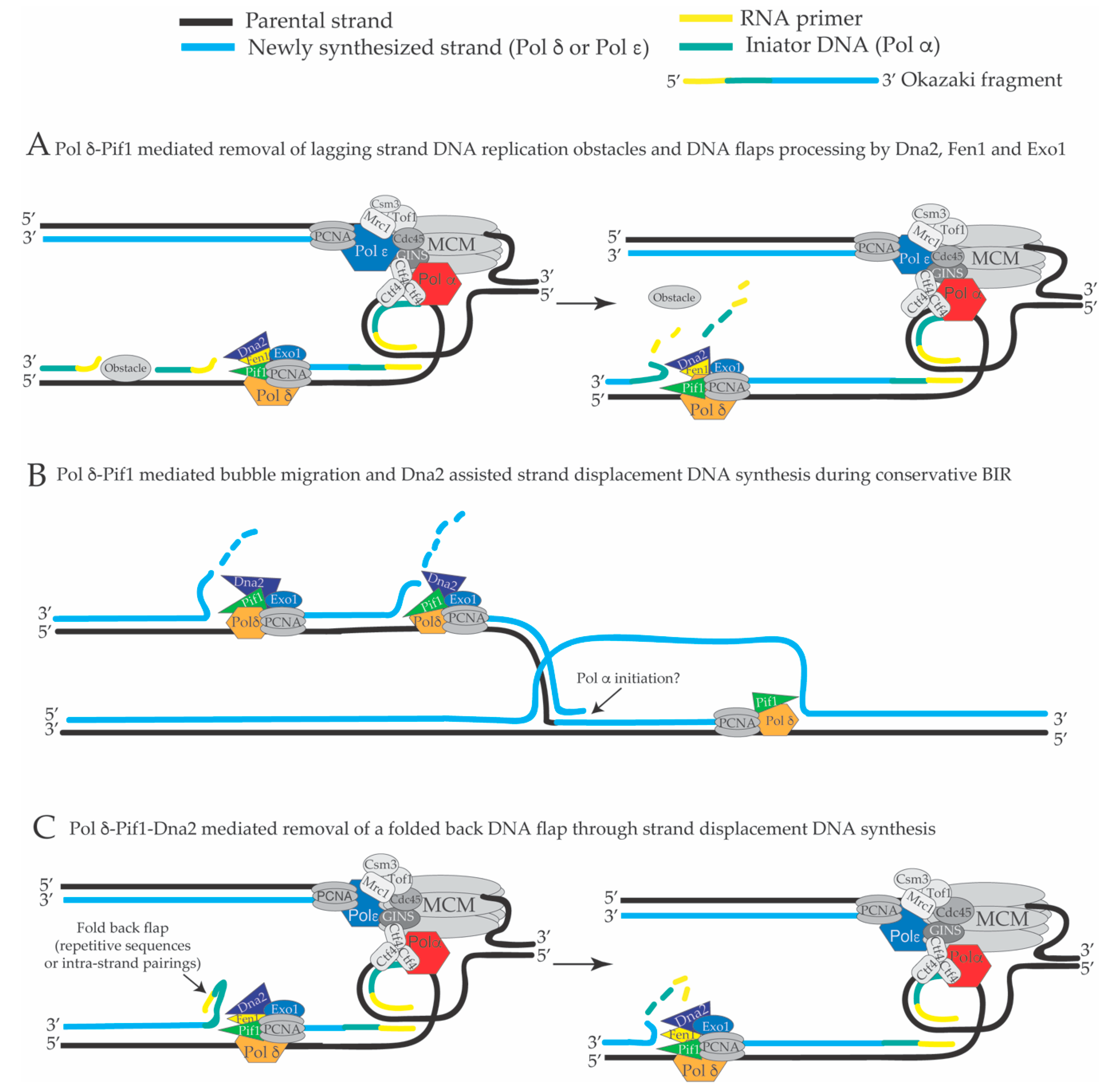
4. Extended Pol δ Strand Displacement and the Long Flap Pathway of Okazaki Fragment Processing in S. cerevisiae
Funding
Conflicts of Interest
References
- Reusswig, K.U.; Pfander, B. Control of Eukaryotic DNA Replication Initiation-Mechanisms to Ensure Smooth Transitions. Genes 2019, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; O’Donnell, M.E. The Eukaryotic CMG Helicase at the Replication Fork: Emerging Architecture Reveals an Unexpected Mechanism. Bioessays 2018, 40. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.P.; Labib, K. Chromosome Duplication in Saccharomyces cerevisiae. Genetics 2016, 203, 1027–1067. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Yuan, Z.; Sun, J.; Georgescu, R.; O’Donnell, M.E.; Li, H. Architecture of the Saccharomyces cerevisiae Replisome. Adv. Exp. Med. Biol. 2017, 1042, 207–228. [Google Scholar] [CrossRef] [PubMed]
- Deegan, T.D.; Diffley, J.F. MCM: One ring to rule them all. Curr. Opin. Struct. Biol. 2016, 37, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Waga, S.; Stillman, B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998, 67, 721–751. [Google Scholar] [CrossRef] [PubMed]
- Burgers, P.M.J.; Kunkel, T.A. Eukaryotic DNA Replication Fork. Annu. Rev. Biochem. 2017, 86, 417–438. [Google Scholar] [CrossRef] [PubMed]
- Stillman, B. Reconsidering DNA Polymerases at the Replication Fork in Eukaryotes. Mol. Cell 2015, 59, 139–141. [Google Scholar] [CrossRef]
- Zegerman, P.; Diffley, J.F. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 2007, 445, 281–285. [Google Scholar] [CrossRef]
- Lei, M.; Kawasaki, Y.; Young, M.R.; Kihara, M.; Sugino, A.; Tye, B.K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997, 11, 3365–3374. [Google Scholar] [CrossRef]
- Tanaka, S.; Umemori, T.; Hirai, K.; Muramatsu, S.; Kamimura, Y.; Araki, H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 2007, 445, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.; Deegan, T.D.; Janska, A.; Early, A.; Diffley, J.F. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 2015, 519, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Deegan, T.D.; Yeeles, J.T.; Diffley, J.F. Phosphopeptide binding by Sld3 links Dbf4-dependent kinase to MCM replicative helicase activation. EMBO J. 2016, 35, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Douglas, M.E.; Ali, F.A.; Costa, A.; Diffley, J.F.X. The mechanism of eukaryotic CMG helicase activation. Nature 2018, 555, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.C.; Zhou, J.C.; Perera, R.L.; van Deursen, F.; Evrin, C.; Ivanova, M.E.; Kilkenny, M.L.; Renault, L.; Kjaer, S.; Matak-Vinkovic, D.; et al. A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. Nature 2014, 510, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Ukomadu, C.; Jha, S.; Senga, T.; Dhar, S.K.; Wohlschlegel, J.A.; Nutt, L.K.; Kornbluth, S.; Dutta, A. Mcm10 and And-1/CTF4 recruit DNA polymerase α to chromatin for initiation of DNA replication. Genes Dev. 2007, 21, 2288–2299. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; De Falco, M.; Kamada, K.; Pisani, F.M.; Masai, H. The mini-chromosome maintenance (Mcm) complexes interact with DNA polymerase α-primase and stimulate its ability to synthesize RNA primers. PLoS ONE 2013, 8, e72408. [Google Scholar] [CrossRef]
- Georgescu, R.E.; Schauer, G.D.; Yao, N.Y.; Langston, L.D.; Yurieva, O.; Zhang, D.; Finkelstein, J.; O’Donnell, M.E. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. Elife 2015, 4, e04988. [Google Scholar] [CrossRef]
- Evrin, C.; Maman, J.D.; Diamante, A.; Pellegrini, L.; Labib, K. Histone H2A-H2B binding by Pol α in the eukaryotic replisome contributes to the maintenance of repressive chromatin. EMBO J. 2018, 37. [Google Scholar] [CrossRef]
- Gan, H.; Serra-Cardona, A.; Hua, X.; Zhou, H.; Labib, K.; Yu, C.; Zhang, Z. The Mcm2-Ctf4-Polalpha Axis Facilitates Parental Histone H3-H4 Transfer to Lagging Strands. Mol. Cell 2018, 72, 140–151. [Google Scholar] [CrossRef]
- Waga, S.; Stillman, B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature 1994, 369, 207–212. [Google Scholar] [CrossRef]
- Lujan, S.A.; Williams, J.S.; Kunkel, T.A. DNA Polymerases Divide the Labor of Genome Replication. Trends Cell Biol. 2016, 26, 640–654. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, T.A.; Burgers, P.M.J. Arranging eukaryotic nuclear DNA polymerases for replication: Specific interactions with accessory proteins arrange Pols α, δ, and ε in the replisome for leading-strand and lagging-strand DNA replication. Bioessays 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Aria, V.; Yeeles, J.T.P. Mechanism of Bidirectional Leading-Strand Synthesis Establishment at Eukaryotic DNA Replication Origins. Mol. Cell 2018. [Google Scholar] [CrossRef] [PubMed]
- Burgers, P.M.J.; Gordenin, D.; Kunkel, T.A. Who Is Leading the Replication Fork, Pol ε or Pol δ? Mol. Cell 2016, 61, 492–493. [Google Scholar] [CrossRef] [PubMed]
- Yeeles, J.T.P.; Janska, A.; Early, A.; Diffley, J.F.X. How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Mol. Cell 2017, 65, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Georgescu, R.E.; Langston, L.; Yao, N.Y.; Yurieva, O.; Zhang, D.; Finkelstein, J.; Agarwal, T.; O’Donnell, M.E. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat. Struct. Mol. Biol. 2014, 21, 664–670. [Google Scholar] [CrossRef]
- Devbhandari, S.; Jiang, J.; Kumar, C.; Whitehouse, I.; Remus, D. Chromatin Constrains the Initiation and Elongation of DNA Replication. Mol. Cell 2017, 65, 131–141. [Google Scholar] [CrossRef]
- Johnson, R.E.; Klassen, R.; Prakash, L.; Prakash, S. A Major Role of DNA Polymerase delta in Replication of Both the Leading and Lagging DNA Strands. Mol. Cell 2015, 59, 163–175. [Google Scholar] [CrossRef]
- Tsurimoto, T.; Stillman, B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 1991, 266, 1950–1960. [Google Scholar]
- Tsurimoto, T.; Stillman, B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase α and δ during initiation of leading and lagging strand synthesis. J. Biol. Chem. 1991, 266, 1961–1968. [Google Scholar] [PubMed]
- Katou, Y.; Kanoh, Y.; Bando, M.; Noguchi, H.; Tanaka, H.; Ashikari, T.; Sugimoto, K.; Shirahige, K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 2003, 424, 1078–1083. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Komata, M.; Katou, Y.; Guan, Z.; Reis, C.C.; Budd, M.; Shirahige, K.; Campbell, J.L. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol. Cell 2008, 32, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Giannattasio, M.; Branzei, D. S-phase checkpoint regulations that preserve replication and chromosome integrity upon dNTP depletion. Cell. Mol. Life Sci. 2017, 74, 2361–2380. [Google Scholar] [CrossRef]
- Schalbetter, S.A.; Mansoubi, S.; Chambers, A.L.; Downs, J.A.; Baxter, J. Fork rotation and DNA precatenation are restricted during DNA replication to prevent chromosomal instability. Proc. Natl. Acad. Sci. USA 2015, 112, E4565–E4570. [Google Scholar] [CrossRef] [PubMed]
- Bush, N.G.; Evans-Roberts, K.; Maxwell, A. DNA Topoisomerases. EcoSal Plus 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, R.; Okazaki, T.; Sakabe, K.; Sugimoto, K.; Sugino, A. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc. Natl. Acad. Sci. USA 1968, 59, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Kurosawa, Y.; Ogawa, T.; Hirose, S.; Okazaki, T.; Okazaki, R. Mechanism of DNA chain growth. XV. RNA-linked nascent DNA pieces in Escherichia coli strains assayed with spleen exonuclease. J. Mol. Biol. 1975, 96, 653–664. [Google Scholar] [CrossRef]
- Ayyagari, R.; Gomes, X.V.; Gordenin, D.A.; Burgers, P.M. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 AND DNA2. J. Biol. Chem. 2003, 278, 1618–1625. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.H.; Ayyagari, R.; Resnick, M.A.; Gordenin, D.A.; Burgers, P.M. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′–5′-exonuclease activities of Pol δ in the creation of a ligatable nick. J. Biol. Chem. 2003, 278, 1626–1633. [Google Scholar] [CrossRef] [PubMed]
- Kao, H.I.; Bambara, R.A. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit. Rev. Biochem. Mol. Biol. 2003, 38, 433–452. [Google Scholar] [CrossRef] [PubMed]
- Stodola, J.L.; Burgers, P.M. Mechanism of Lagging-Strand DNA Replication in Eukaryotes. Adv. Exp. Med. Biol. 2017, 1042, 117–133. [Google Scholar] [CrossRef] [PubMed]
- Gordenin, D.A.; Kunkel, T.A.; Resnick, M.A. Repeat expansion—All in a flap? Nat. Genet. 1997, 16, 116–118. [Google Scholar] [CrossRef] [PubMed]
- Johnston, L.H.; Nasmyth, K.A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature 1978, 274, 891–893. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Okazaki, T. Function of RNase H in DNA replication revealed by RNase H defective mutants of Escherichia coli. Mol. Gen. Genet. 1984, 193, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Qian, Y.; Frank, P.; Wintersberger, U.; Shen, B. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol. 1999, 19, 8361–8371. [Google Scholar] [CrossRef] [PubMed]
- Garg, P.; Stith, C.M.; Sabouri, N.; Johansson, E.; Burgers, P.M. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004, 18, 2764–2773. [Google Scholar] [CrossRef] [PubMed]
- Koc, K.N.; Stodola, J.L.; Burgers, P.M.; Galletto, R. Regulation of yeast DNA polymerase δ-mediated strand displacement synthesis by 5’-flaps. Nucleic Acids Res. 2015, 43, 4179–4190. [Google Scholar] [CrossRef] [PubMed]
- Stodola, J.L.; Burgers, P.M. Resolving individual steps of Okazaki-fragment maturation at a millisecond timescale. Nat. Struct. Mol. Biol. 2016, 23, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.L.; Pike, J.E.; Wang, W.; Burgers, P.M.; Campbell, J.L.; Bambara, R.A. Pif1 helicase directs eukaryotic Okazaki fragments toward the two-nuclease cleavage pathway for primer removal. J. Biol. Chem. 2008, 283, 27483–27493. [Google Scholar] [CrossRef] [PubMed]
- Pike, J.E.; Burgers, P.M.; Campbell, J.L.; Bambara, R.A. Pif1 helicase lengthens some Okazaki fragment flaps necessitating Dna2 nuclease/helicase action in the two-nuclease processing pathway. J. Biol. Chem. 2009, 284, 25170–25180. [Google Scholar] [CrossRef] [PubMed]
- Pike, J.E.; Henry, R.A.; Burgers, P.M.; Campbell, J.L.; Bambara, R.A. An alternative pathway for Okazaki fragment processing: Resolution of fold-back flaps by Pif1 helicase. J. Biol. Chem. 2010, 285, 41712–41723. [Google Scholar] [CrossRef] [PubMed]
- Budd, M.E.; Reis, C.C.; Smith, S.; Myung, K.; Campbell, J.L. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol. Cell. Biol. 2006, 26, 2490–2500. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 1998, 26, 5589–5595. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef] [PubMed]
- Budd, M.E.; Antoshechkin, I.A.; Reis, C.; Wold, B.J.; Campbell, J.L. Inviability of a DNA2 deletion mutant is due to the DNA damage checkpoint. Cell Cycle 2011, 10, 1690–1698. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.E.; Foiani, M.; Giannattasio, M. Dna2 processes behind the fork long ssDNA flaps generated by Pif1 and replication-dependent strand displacement. Nat. Commun. 2018, 9, 4830. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Ryu, G.H.; Seo, Y.S.; Tanaka, K.; Okayama, H.; MacNeill, S.A.; Yuasa, Y. The fission yeast pfh1+ gene encodes an essential 5′ to 3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res. 2002, 30, 4728–4739. [Google Scholar] [CrossRef]
- Reagan, M.S.; Pittenger, C.; Siede, W.; Friedberg, E.C. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol. 1995, 177, 364–371. [Google Scholar] [CrossRef]
- Siegal, G.; Turchi, J.J.; Myers, T.W.; Bambara, R.A. A 5′ to 3′ exonuclease functionally interacts with calf DNA polymerase epsilon. Proc. Natl. Acad. Sci. USA 1992, 89, 9377–9381. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Thrower, D.; Qiu, J.; Wu, P.; Zheng, L.; Zhou, M.; Bachant, J.; Wilson, D.M., III; Shen, B. Complementary functions of the Saccharomyces cerevisiae Rad2 family nucleases in Okazaki fragment maturation, mutation avoidance, and chromosome stability. DNA Repair 2003, 2, 925–940. [Google Scholar] [CrossRef]
- Balakrishnan, L.; Bambara, R.A. Flap endonuclease 1. Annu. Rev. Biochem. 2013, 82, 119–138. [Google Scholar] [CrossRef] [PubMed]
- Gary, R.; Ludwig, D.L.; Cornelius, H.L.; MacInnes, M.A.; Park, M.S. The DNA repair endonuclease XPG binds to proliferating cell nuclear antigen (PCNA) and shares sequence elements with the PCNA-binding regions of FEN-1 and cyclin-dependent kinase inhibitor p21. J. Biol. Chem. 1997, 272, 24522–24529. [Google Scholar] [CrossRef] [PubMed]
- Warbrick, E.; Lane, D.P.; Glover, D.M.; Cox, L.S. Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: A potential mechanism to co-ordinate DNA replication and repair. Oncogene 1997, 14, 2313–2321. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.; Harrington, J.; Lieber, M.R.; Burgers, P.M. Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem. 1995, 270, 22109–22112. [Google Scholar] [CrossRef]
- Gomes, X.V.; Burgers, P.M. Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J. 2000, 19, 3811–3821. [Google Scholar] [CrossRef] [PubMed]
- Tom, S.; Henricksen, L.A.; Bambara, R.A. Mechanism whereby proliferating cell nuclear antigen stimulates flap endonuclease 1. J. Biol. Chem. 2000, 275, 10498–10505. [Google Scholar] [CrossRef] [PubMed]
- Ishimi, Y.; Claude, A.; Bullock, P.; Hurwitz, J. Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J. Biol. Chem. 1988, 263, 19723–19733. [Google Scholar] [PubMed]
- Goulian, M.; Richards, S.H.; Heard, C.J.; Bigsby, B.M. Discontinuous DNA synthesis by purified mammalian proteins. J. Biol. Chem. 1990, 265, 18461–18471. [Google Scholar] [PubMed]
- Waga, S.; Bauer, G.; Stillman, B. Reconstitution of complete SV40 DNA replication with purified replication factors. J. Biol. Chem. 1994, 269, 10923–10934. [Google Scholar] [PubMed]
- Budd, M.E.; Campbell, J.L. A yeast gene required for DNA replication encodes a protein with homology to DNA helicases. Proc. Natl. Acad. Sci. USA 1995, 92, 7642–7646. [Google Scholar] [CrossRef]
- Budd, M.E.; Choe, W.; Campbell, J.L. The nuclease activity of the yeast DNA2 protein, which is related to the RecB-like nucleases, is essential in vivo. J. Biol. Chem. 2000, 275, 16518–16529. [Google Scholar] [CrossRef] [PubMed]
- Levikova, M.; Pinto, C.; Cejka, P. The motor activity of DNA2 functions as an ssDNA translocase to promote DNA end resection. Genes Dev. 2017, 31, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Fortini, B.K.; Pokharel, S.; Polaczek, P.; Balakrishnan, L.; Bambara, R.A.; Campbell, J.L. Characterization of the endonuclease and ATP-dependent flap endo/exonuclease of Dna2. J. Biol. Chem. 2011, 286, 23763–23770. [Google Scholar] [CrossRef] [PubMed]
- Budd, M.E.; Campbell, J.L. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 1997, 17, 2136–2142. [Google Scholar] [CrossRef]
- Kang, H.Y.; Choi, E.; Bae, S.H.; Lee, K.H.; Gim, B.S.; Kim, H.D.; Park, C.; MacNeill, S.A.; Seo, Y.S. Genetic analyses of Schizosaccharomyces pombe dna2+ reveal that dna2 plays an essential role in Okazaki fragment metabolism. Genetics 2000, 155, 1055–1067. [Google Scholar]
- Levikova, M.; Cejka, P. The Saccharomyces cerevisiae Dna2 can function as a sole nuclease in the processing of Okazaki fragments in DNA replication. Nucleic Acids Res. 2015, 43, 7888–7897. [Google Scholar] [CrossRef]
- Tran, P.T.; Erdeniz, N.; Symington, L.S.; Liskay, R.M. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair 2004, 3, 1549–1559. [Google Scholar] [CrossRef]
- Budd, M.E.; Choe, W.C.; Campbell, J.L. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 1995, 270, 26766–26769. [Google Scholar] [CrossRef]
- Jeong, H.S.; Backlund, P.S.; Chen, H.C.; Karavanov, A.A.; Crouch, R.J. RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res. 2004, 32, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Frank, P.; Braunshofer-Reiter, C.; Wintersberger, U. Yeast RNase H(35) is the counterpart of the mammalian RNase HI, and is evolutionarily related to prokaryotic RNase HII. FEBS Lett. 1998, 421, 23–26. [Google Scholar] [CrossRef]
- Itaya, M. Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc. Natl. Acad. Sci. USA 1990, 87, 8587–8591. [Google Scholar] [CrossRef] [PubMed]
- Frank, P.; Braunshofer-Reiter, C.; Wintersberger, U.; Grimm, R.; Busen, W. Cloning of the cDNA encoding the large subunit of human RNase HI, a homologue of the prokaryotic RNase HII. Proc. Natl. Acad. Sci. USA 1998, 95, 12872–12877. [Google Scholar] [CrossRef] [PubMed]
- Hausen, P.; Stein, H. Ribonuclease H: An enzyme degrading the RNA moiety of DNA-RNA hybrids. Eur. J. Biochem. 1970, 14, 278–283. [Google Scholar] [CrossRef]
- Chon, H.; Vassilev, A.; DePamphilis, M.L.; Zhao, Y.; Zhang, J.; Burgers, P.M.; Crouch, R.J.; Cerritelli, S.M. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 2009, 37, 96–110. [Google Scholar] [CrossRef]
- Kokoska, R.J.; Stefanovic, L.; Tran, H.T.; Resnick, M.A.; Gordenin, D.A.; Petes, T.D. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t). Mol. Cell. Biol. 1998, 18, 2779–2788. [Google Scholar] [CrossRef]
- Tishkoff, D.X.; Filosi, N.; Gaida, G.M.; Kolodner, R.D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell 1997, 88, 253–263. [Google Scholar] [CrossRef]
- Chen, X.; Paudyal, S.C.; Chin, R.I.; You, Z. PCNA promotes processive DNA end resection by Exo1. Nucleic Acids Res. 2013, 41, 9325–9338. [Google Scholar] [CrossRef]
- Tran, P.T.; Erdeniz, N.; Dudley, S.; Liskay, R.M. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair 2002, 1, 895–912. [Google Scholar] [CrossRef]
- Balk, B.; Maicher, A.; Dees, M.; Klermund, J.; Luke-Glaser, S.; Bender, K.; Luke, B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 2013, 20, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Allen-Soltero, S.; Martinez, S.L.; Putnam, C.D.; Kolodner, R.D. A Saccharomyces cerevisiae RNase H2 interaction network functions to suppress genome instability. Mol. Cell. Biol. 2014, 34, 1521–1534. [Google Scholar] [CrossRef] [PubMed]
- Budd, M.E.; Tong, A.H.; Polaczek, P.; Peng, X.; Boone, C.; Campbell, J.L. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 2005, 1, e61. [Google Scholar] [CrossRef] [PubMed]
- Koc, K.N.; Singh, S.P.; Stodola, J.L.; Burgers, P.M.; Galletto, R. Pif1 removes a Rap1-dependent barrier to the strand displacement activity of DNA polymerase δ. Nucleic Acids Res. 2016, 44, 3811–3819. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Bochman, M.L.; Garcia, P.D.; Cejka, P.; Friedman, K.L.; Kowalczykowski, S.C.; Zakian, V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 2013, 497, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Ribeyre, C.; Lopes, J.; Boule, J.B.; Piazza, A.; Guedin, A.; Zakian, V.A.; Mergny, J.L.; Nicolas, A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009, 5, e1000475. [Google Scholar] [CrossRef]
- Tran, P.L.T.; Pohl, T.J.; Chen, C.F.; Chan, A.; Pott, S.; Zakian, V.A. PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat. Commun. 2017, 8, 15025. [Google Scholar] [CrossRef]
- Buzovetsky, O.; Kwon, Y.; Pham, N.T.; Kim, C.; Ira, G.; Sung, P.; Xiong, Y. Role of the Pif1-PCNA Complex in Pol δ-Dependent Strand Displacement DNA Synthesis and Break-Induced Replication. Cell Rep. 2017, 21, 1707–1714. [Google Scholar] [CrossRef]
- Galletto, R.; Tomko, E.J. Translocation of Saccharomyces cerevisiae Pif1 helicase monomers on single-stranded DNA. Nucleic Acids Res. 2013, 41, 4613–4627. [Google Scholar] [CrossRef]
- Henry, R.A.; Balakrishnan, L.; Ying-Lin, S.T.; Campbell, J.L.; Bambara, R.A. Components of the secondary pathway stimulate the primary pathway of eukaryotic Okazaki fragment processing. J. Biol. Chem. 2010, 285, 28496–28505. [Google Scholar] [CrossRef]
- Tanaka, H.; Ryu, G.H.; Seo, Y.S.; MacNeill, S.A. Genetics of lagging strand DNA synthesis and maturation in fission yeast: Suppression analysis links the Dna2-Cdc24 complex to DNA polymerase δ. Nucleic Acids Res. 2004, 32, 6367–6377. [Google Scholar] [CrossRef]
- Zhang, H.; Hua, Y.; Li, R.; Kong, D. Cdc24 Is Essential for Long-range End Resection in the Repair of Double-stranded DNA Breaks. J. Biol. Chem. 2016, 291, 24961–24973. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Wang, J.; Li, S.; Kamiya, K.; Chen, X.; Zhou, P. Determination of the biochemical properties of full-length human PIF1 ATPase. Prion 2013, 7, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rodriguez, N.; Wong, R.P.; Ulrich, H.D. The helicase Pif1 functions in the template switching pathway of DNA damage bypass. Nucleic Acids Res. 2018, 46, 8347–8356. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Hu, J.; Wang, J.; Kong, D. Direct Visualization of RNA-DNA Primer Removal from Okazaki Fragments Provides Support for Flap Cleavage and Exonucleolytic Pathways in Eukaryotic Cells. J. Biol. Chem. 2017, 292, 4777–4788. [Google Scholar] [CrossRef] [PubMed]
- Kahli, M.; Osmundson, J.S.; Yeung, R.; Smith, D.J. Processing of eukaryotic Okazaki fragments by redundant nucleases can be uncoupled from ongoing DNA replication in vivo. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Maga, G.; Villani, G.; Tillement, V.; Stucki, M.; Locatelli, G.A.; Frouin, I.; Spadari, S.; Hubscher, U. Okazaki fragment processing: Modulation of the strand displacement activity of DNA polymerase δ by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc. Natl. Acad. Sci. USA 2001, 98, 14298–14303. [Google Scholar] [CrossRef]
- Jin, Y.H.; Obert, R.; Burgers, P.M.; Kunkel, T.A.; Resnick, M.A.; Gordenin, D.A. The 3′→5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. USA 2001, 98, 5122–5127. [Google Scholar] [CrossRef]
- Einolf, H.J.; Guengerich, F.P. Kinetic analysis of nucleotide incorporation by mammalian DNA polymerase delta. J. Biol. Chem. 2000, 275, 16316–16322. [Google Scholar] [CrossRef]
- Mozzherin, D.J.; Tan, C.K.; Downey, K.M.; Fisher, P.A. Architecture of the active DNA polymerase δ·proliferating cell nuclear antigen·template-primer complex. J. Biol. Chem. 1999, 274, 19862–19867. [Google Scholar] [CrossRef]
- Kelly, R.B.; Cozzarelli, N.R.; Deutscher, M.P.; Lehman, I.R.; Kornberg, A. Enzymatic synthesis of deoxyribonucleic acid. XXXII. Replication of duplex deoxyribonucleic acid by polymerase at a single strand break. J. Biol. Chem. 1970, 245, 39–45. [Google Scholar] [PubMed]
- Masamune, Y.; Richardson, C.C. Strand displacement during deoxyribonucleic acid synthesis at single strand breaks. J. Biol. Chem. 1971, 246, 2692–2701. [Google Scholar] [PubMed]
- Nossal, N.G. DNA synthesis on a double-stranded DNA template by the T4 bacteriophage DNA polymerase and the T4 gene 32 DNA unwinding protein. J. Biol. Chem. 1974, 249, 5668–5676. [Google Scholar] [PubMed]
- Nossal, N.G.; Peterlin, B.M. DNA replication by bacteriophage T4 proteins. The T4 43, 32, 44--62, and 45 proteins are required for strand displacement synthesis at nicks in duplex DNA. J. Biol. Chem. 1979, 254, 6032–6037. [Google Scholar] [PubMed]
- Lechner, R.L.; Engler, M.J.; Richardson, C.C. Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J. Biol. Chem. 1983, 258, 11174–11184. [Google Scholar] [PubMed]
- Dong, F.; Weitzel, S.E.; von Hippel, P.H. A coupled complex of T4 DNA replication helicase (gp41) and polymerase (gp43) can perform rapid and processive DNA strand-displacement synthesis. Proc. Natl. Acad. Sci. USA 1996, 93, 14456–14461. [Google Scholar] [CrossRef] [PubMed]
- Salas, M. Protein-priming of DNA replication. Annu. Rev. Biochem. 1991, 60, 39–71. [Google Scholar] [CrossRef] [PubMed]
- Blanco, L.; Salas, M. Characterization and purification of a phage phi 29-encoded DNA polymerase required for the initiation of replication. Proc. Natl. Acad. Sci. USA 1984, 81, 5325–5329. [Google Scholar] [CrossRef]
- Soengas, M.S.; Gutierrez, C.; Salas, M. Helix-destabilizing activity of phi 29 single-stranded DNA binding protein: Effect on the elongation rate during strand displacement DNA replication. J. Mol. Biol. 1995, 253, 517–529. [Google Scholar] [CrossRef]
- Inciarte, M.R.; Salas, M.; Sogo, J.M. Structure of replicating DNA molecules of Bacillus subtilis bacteriophage phi 29. J. Virol. 1980, 34, 187–199. [Google Scholar]
- Sogo, J.M.; Garcia, J.A.; Penalva, M.A.; Salas, M. Structure of protein-containing replicative intermediates of Bacillus subtilis phage phi 29 DNA. Virology 1982, 116, 1–18. [Google Scholar] [CrossRef]
- Tsurumi, T.; Yamada, H.; Daikoku, T.; Yamashita, Y.; Nishiyama, Y. Strand displacement associated DNA synthesis catalyzed by the Epstein-Barr virus DNA polymerase. Biochem. Biophys. Res. Commun. 1997, 238, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Nowak, R.; Kulik, J.; Siedlecki, J.A. The ability of DNA polymerase beta to synthesize DNA beyond the gap with displacement of the non-replicated strand. Acta Biochim. Pol. 1987, 34, 205–215. [Google Scholar] [PubMed]
- Prasad, R.; Lavrik, O.I.; Kim, S.J.; Kedar, P.; Yang, X.P.; Vande Berg, B.J.; Wilson, S.H. DNA polymerase β-mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 2001, 276, 32411–32414. [Google Scholar] [CrossRef] [PubMed]
- Langelier, M.F.; Eisemann, T.; Riccio, A.A.; Pascal, J.M. PARP family enzymes: Regulation and catalysis of the poly(ADP-ribose) posttranslational modification. Curr. Opin. Struct. Biol. 2018, 53, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Maya-Mendoza, A.; Moudry, P.; Merchut-Maya, J.M.; Lee, M.; Strauss, R.; Bartek, J. High speed of fork progression induces DNA replication stress and genomic instability. Nature 2018, 559, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Canceill, D.; Viguera, E.; Ehrlich, S.D. Replication slippage of different DNA polymerases is inversely related to their strand displacement efficiency. J. Biol. Chem. 1999, 274, 27481–27490. [Google Scholar] [CrossRef] [PubMed]
- Weston-Hafer, K.; Berg, D.E. Deletions in plasmid pBR322: Replication slippage involving leading and lagging strands. Genetics 1991, 127, 649–655. [Google Scholar] [PubMed]
- Carvalho, C.M.; Pehlivan, D.; Ramocki, M.B.; Fang, P.; Alleva, B.; Franco, L.M.; Belmont, J.W.; Hastings, P.J.; Lupski, J.R. Replicative mechanisms for CNV formation are error prone. Nat. Genet. 2013, 45, 1319–1326. [Google Scholar] [CrossRef] [PubMed]
- Stith, C.M.; Sterling, J.; Resnick, M.A.; Gordenin, D.A.; Burgers, P.M. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand displacement synthesis. J. Biol. Chem. 2008, 283, 34129–34140. [Google Scholar] [CrossRef] [PubMed]
- Kao, H.I.; Veeraraghavan, J.; Polaczek, P.; Campbell, J.L.; Bambara, R.A. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 2004, 279, 15014–15024. [Google Scholar] [CrossRef] [PubMed]
- Reid, A. Nick translation. Methods Mol. Biol. 2002, 179, 23–25. [Google Scholar] [PubMed]
- Swan, M.K.; Johnson, R.E.; Prakash, L.; Prakash, S.; Aggarwal, A.K. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat. Struct. Mol. Biol. 2009, 16, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Gerik, K.J.; Li, X.; Pautz, A.; Burgers, P.M. Characterization of the two small subunits of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem. 1998, 273, 19747–19755. [Google Scholar] [CrossRef] [PubMed]
- Acharya, N.; Klassen, R.; Johnson, R.E.; Prakash, L.; Prakash, S. PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc. Natl. Acad. Sci. USA 2011, 108, 17927–17932. [Google Scholar] [CrossRef]
- Dahan, D.; Tsirkas, I.; Dovrat, D.; Sparks, M.A.; Singh, S.P.; Galletto, R.; Aharoni, A. Pif1 is essential for efficient replisome progression through lagging strand G-quadruplex DNA secondary structures. Nucleic Acids Res. 2018, 46, 11847–11857. [Google Scholar] [CrossRef]
- Stewart, J.A.; Campbell, J.L.; Bambara, R.A. Flap endonuclease disengages Dna2 helicase/nuclease from Okazaki fragment flaps. J. Biol. Chem. 2006, 281, 38565–38572. [Google Scholar] [CrossRef]
- Stewart, J.A.; Miller, A.S.; Campbell, J.L.; Bambara, R.A. Dynamic removal of replication protein A by Dna2 facilitates primer cleavage during Okazaki fragment processing in Saccharomyces cerevisiae. J. Biol. Chem. 2008, 283, 31356–31365. [Google Scholar] [CrossRef]
- Kunkel, T.A.; Hamatake, R.K.; Motto-Fox, J.; Fitzgerald, M.P.; Sugino, A. Fidelity of DNA polymerase I and the DNA polymerase I-DNA primase complex from Saccharomyces cerevisiae. Mol. Cell. Biol. 1989, 9, 4447–4458. [Google Scholar] [CrossRef]
- Fu, Y.V.; Yardimci, H.; Long, D.T.; Ho, T.V.; Guainazzi, A.; Bermudez, V.P.; Hurwitz, J.; van Oijen, A.; Scharer, O.D.; Walter, J.C. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 2011, 146, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Belotserkovskii, B.P.; Tornaletti, S.; D’Souza, A.D.; Hanawalt, P.C. R-loop generation during transcription: Formation, processing and cellular outcomes. DNA Repair 2018, 71, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Bermejo, R.; Lai, M.S.; Foiani, M. Preventing replication stress to maintain genome stability: Resolving conflicts between replication and transcription. Mol. Cell 2012, 45, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Achar, Y.J.; Foiani, M. Coordinating Replication with Transcription. Adv. Exp. Med. Biol. 2017, 1042, 455–487. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, A.M.; Newlon, C.S. DNA replication fork pause sites dependent on transcription. Science 1996, 272, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Azvolinsky, A.; Giresi, P.G.; Lieb, J.D.; Zakian, V.A. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol. Cell 2009, 34, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Brambati, A.; Zardoni, L.; Achar, Y.J.; Piccini, D.; Galanti, L.; Colosio, A.; Foiani, M.; Liberi, G. Dormant origins and fork protection mechanisms rescue sister forks arrested by transcription. Nucleic Acids Res. 2018, 46, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Alzu, A.; Bermejo, R.; Begnis, M.; Lucca, C.; Piccini, D.; Carotenuto, W.; Saponaro, M.; Brambati, A.; Cocito, A.; Foiani, M.; et al. Senataxin associates with replication forks to protect fork integrity across RNA-polymerase-II-transcribed genes. Cell 2012, 151, 835–846. [Google Scholar] [CrossRef]
- Wilson, M.A.; Kwon, Y.; Xu, Y.; Chung, W.H.; Chi, P.; Niu, H.; Mayle, R.; Chen, X.; Malkova, A.; Sung, P.; et al. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature 2013, 502, 393–396. [Google Scholar] [CrossRef]
- Donnianni, R.A.; Symington, L.S. Break-induced replication occurs by conservative DNA synthesis. Proc. Natl. Acad. Sci. USA 2013, 110, 13475–13480. [Google Scholar] [CrossRef]
- Kramara, J.; Osia, B.; Malkova, A. Break-Induced Replication: The Where, the Why, and the How. Trends Genet. 2018. [Google Scholar] [CrossRef] [PubMed]
- Llorente, B.; Smith, C.E.; Symington, L.S. Break-induced replication: What is it and what is it for? Cell Cycle 2008, 7, 859–864. [Google Scholar] [CrossRef]
- Malkova, A.; Ivanov, E.L.; Haber, J.E. Double-strand break repair in the absence of RAD51 in yeast: A possible role for break-induced DNA replication. Proc. Natl. Acad. Sci. USA 1996, 93, 7131–7136. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.S.; McKenna, A.E.; Motycka, T.A.; Matsumoto, Y.; Tomkinson, A.E. Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr. Biol. 2000, 10, 919–922. [Google Scholar] [CrossRef]
- Lu, X.; Tan, C.K.; Zhou, J.Q.; You, M.; Carastro, L.M.; Downey, K.M.; So, A.G. Direct interaction of proliferating cell nuclear antigen with the small subunit of DNA polymerase delta. J. Biol. Chem. 2002, 277, 24340–24345. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Whitehouse, I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. Nature 2012, 483, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Ivessa, A.S.; Lenzmeier, B.A.; Bessler, J.B.; Goudsouzian, L.K.; Schnakenberg, S.L.; Zakian, V.A. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 2003, 12, 1525–1536. [Google Scholar] [CrossRef]
- Osmundson, J.S.; Kumar, J.; Yeung, R.; Smith, D.J. Pif1-family helicases cooperatively suppress widespread replication-fork arrest at tRNA genes. Nat. Struct. Mol. Biol. 2017, 24, 162–170. [Google Scholar] [CrossRef]
- Ivessa, A.S.; Zhou, J.Q.; Zakian, V.A. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 2000, 100, 479–489. [Google Scholar] [CrossRef]
- Sakurai, S.; Kitano, K.; Yamaguchi, H.; Hamada, K.; Okada, K.; Fukuda, K.; Uchida, M.; Ohtsuka, E.; Morioka, H.; Hakoshima, T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005, 24, 683–693. [Google Scholar] [CrossRef]
- Zhou, X.; Ren, W.; Bharath, S.R.; Tang, X.; He, Y.; Chen, C.; Liu, Z.; Li, D.; Song, H. Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1. Cell Rep. 2016, 14, 2030–2039. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.Y.; Chen, W.F.; Rety, S.; Liu, N.N.; Wu, W.Q.; Dai, Y.X.; Li, D.; Ma, H.Y.; Dou, S.X.; Xi, X.G. Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase. Nucleic Acids Res. 2018, 46, 1486–1500. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Pourmal, S.; Pavletich, N.P. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. Elife 2015, 4. [Google Scholar] [CrossRef]
- Levikova, M.; Klaue, D.; Seidel, R.; Cejka, P. Nuclease activity of Saccharomyces cerevisiae Dna2 inhibits its potent DNA helicase activity. Proc. Natl. Acad. Sci. USA 2013, 110, E1992–E2001. [Google Scholar] [CrossRef]
- Giannattasio, M.; Zwicky, K.; Follonier, C.; Foiani, M.; Lopes, M.; Branzei, D. Visualization of recombination-mediated damage bypass by template switching. Nat. Struct. Mol. Biol. 2014, 21, 884–892. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Szakal, B. Building up and breaking down: Mechanisms controlling recombination during replication. Crit. Rev. Biochem. Mol. Biol. 2017, 52, 381–394. [Google Scholar] [CrossRef] [PubMed]
| Saccharomyce Cerevisiae | Schizosaccharomyces Pombe | Human | |||
|---|---|---|---|---|---|
| Protein(s) | Gene(s) | Protein(s) | Gene(s) | Protein(s) | Gene(s) |
| PCNA | POL 30 | PCNA | pcn1+ | PCNA | PCNA |
| DNA Polymerase δ (Pol 3-Pol 31-Pol 32) | POL 3, POL 31 and POL 32 | DNA Polymerase δ (cdc6-cdc1-cdc27) | cdc6+, cdc1+ and cdc27+ | DNA Polymerase δ (p125-p50-p68-p12) | POLD1, POLD2, POLD3 and POLD4 |
| Pif1 | PIF1 | pfh1 | pfh1+ | Pif1 | PIF1 |
| Dna2 | DNA2 | dna2 | dna2+ | Dna2 | DNA2 |
| Exo1 | EXO1 | exo1 | exo1+ | Exo1 | EXO1 |
| Rad27 | RAD27 | rad2 | rad2+ | FEN1 | FEN1 |
| RNase H2 (Rnh201-Rnh202-Rnh203) | RNH201, RNH202 and RNH203 | RNase H2 (Rnh201-Rnh202-Rnh203) | rnh201+, rnh202+ and rnh203+ | RNase H1 | RNASEH1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannattasio, M.; Branzei, D. DNA Replication Through Strand Displacement During Lagging Strand DNA Synthesis in Saccharomyces cerevisiae. Genes 2019, 10, 167. https://doi.org/10.3390/genes10020167
Giannattasio M, Branzei D. DNA Replication Through Strand Displacement During Lagging Strand DNA Synthesis in Saccharomyces cerevisiae. Genes. 2019; 10(2):167. https://doi.org/10.3390/genes10020167
Chicago/Turabian StyleGiannattasio, Michele, and Dana Branzei. 2019. "DNA Replication Through Strand Displacement During Lagging Strand DNA Synthesis in Saccharomyces cerevisiae" Genes 10, no. 2: 167. https://doi.org/10.3390/genes10020167
APA StyleGiannattasio, M., & Branzei, D. (2019). DNA Replication Through Strand Displacement During Lagging Strand DNA Synthesis in Saccharomyces cerevisiae. Genes, 10(2), 167. https://doi.org/10.3390/genes10020167





