Evolution and Identification of the WRKY Gene Family in Quinoa (Chenopodium quinoa)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of WRKY Gene Family and Chromosomal Localizations
2.2. Phylogenetic Analysis, Gene Structure, Protein Conserved Motifs Identification and Gene Duplication
2.3. Interaction Network of CqWRKY Genes
2.4. Expression Profile Analysis in Various Tissues and under Different Abiotic Stresses of TaHDZ Genes
3. Results
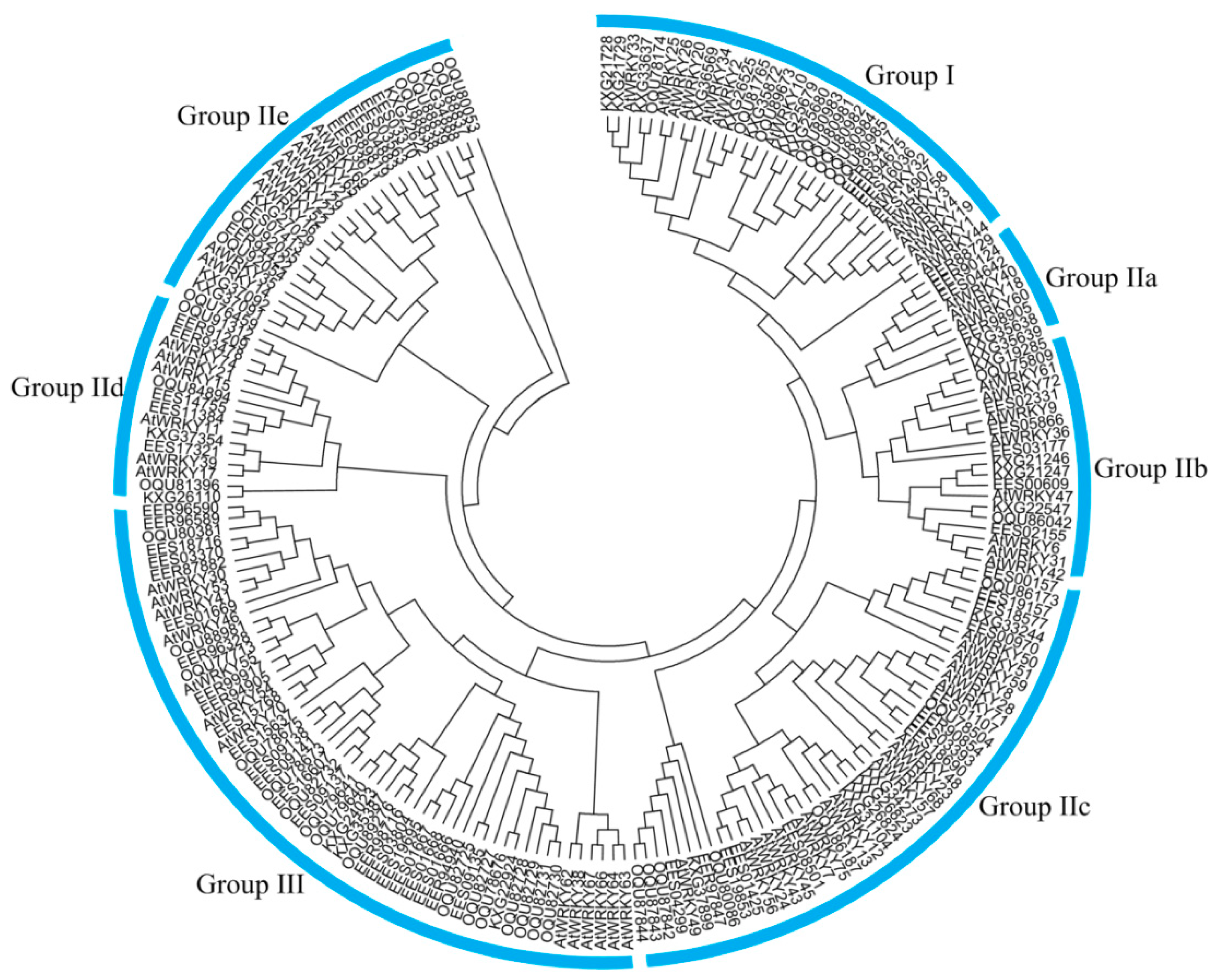
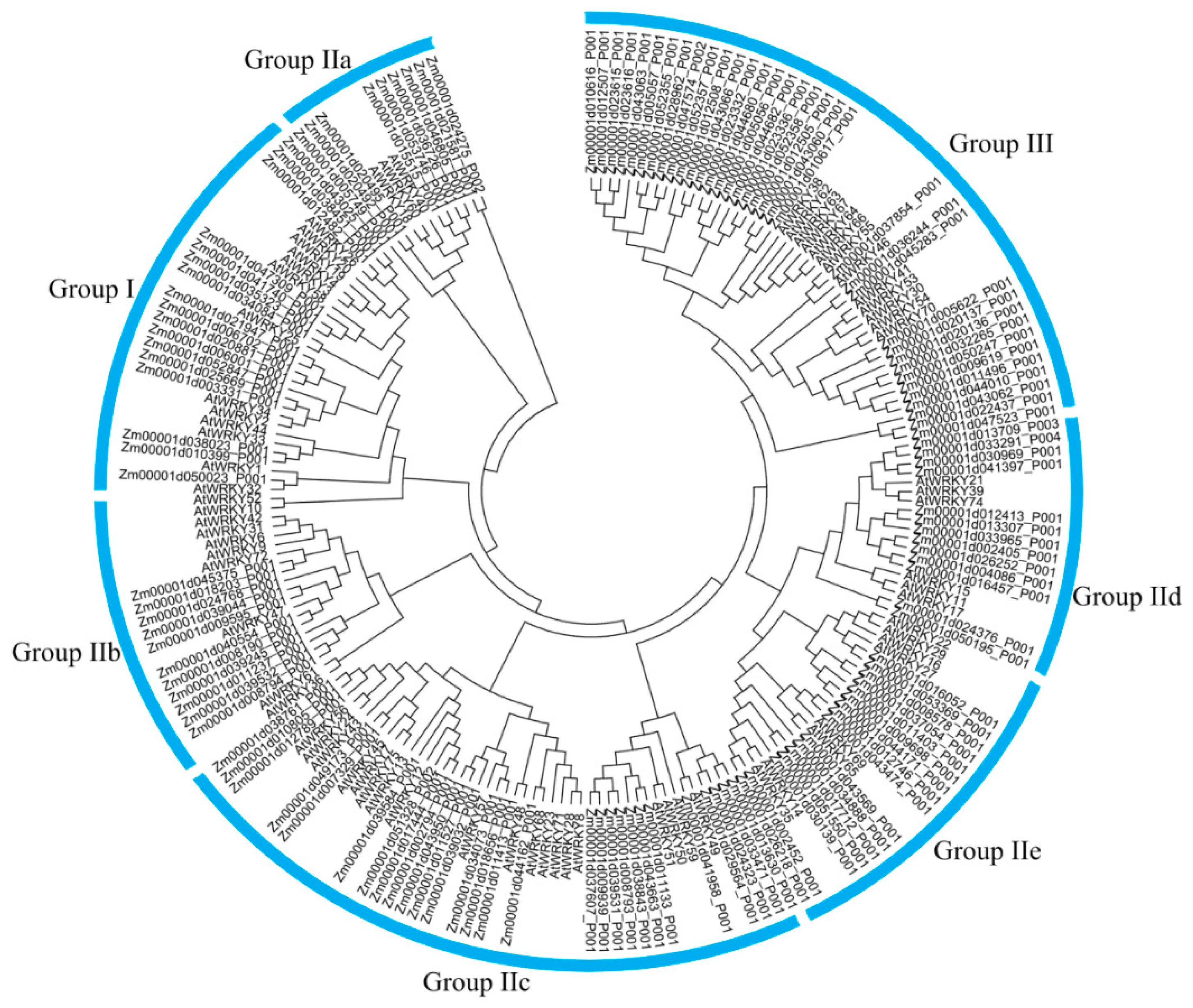
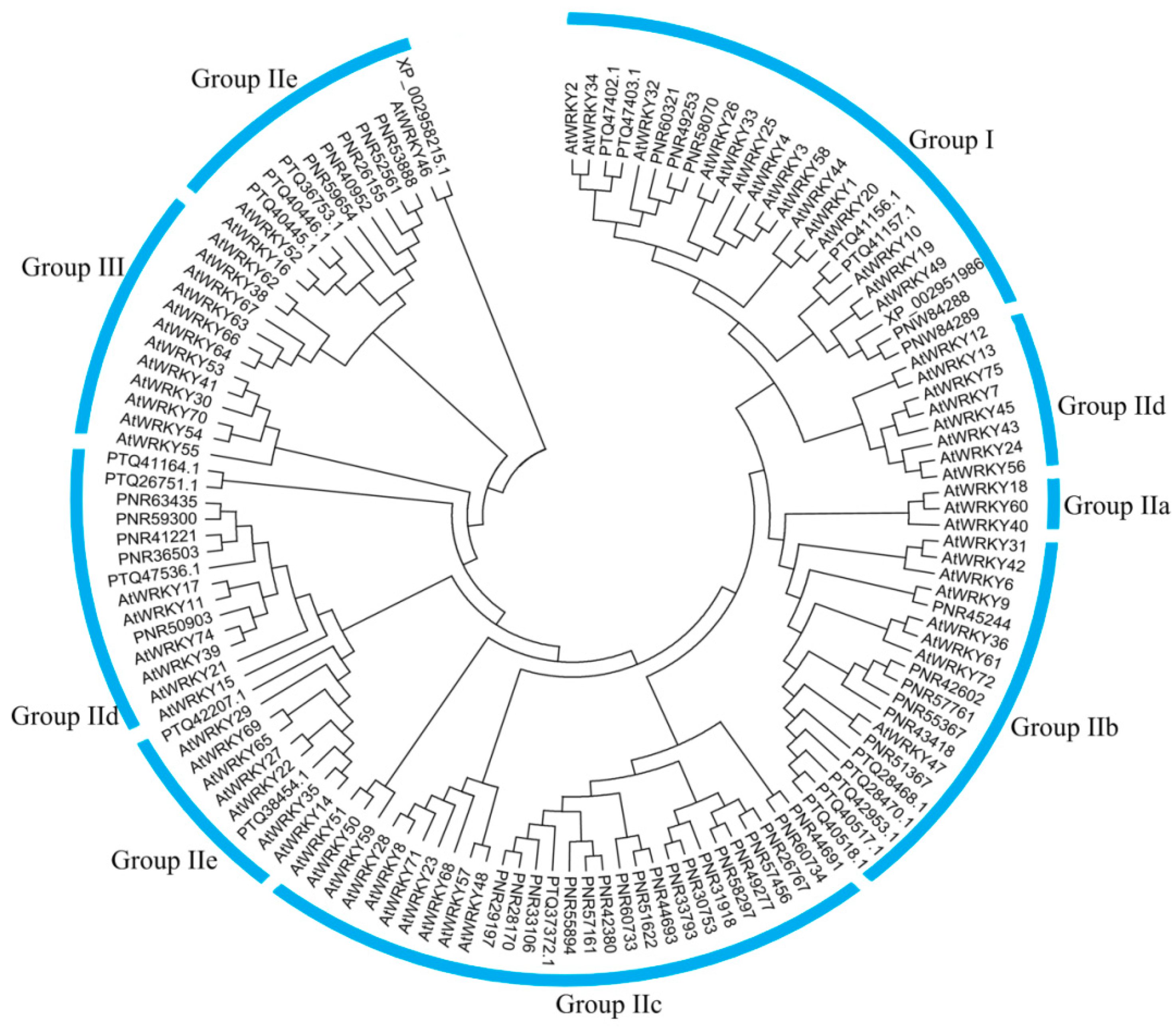
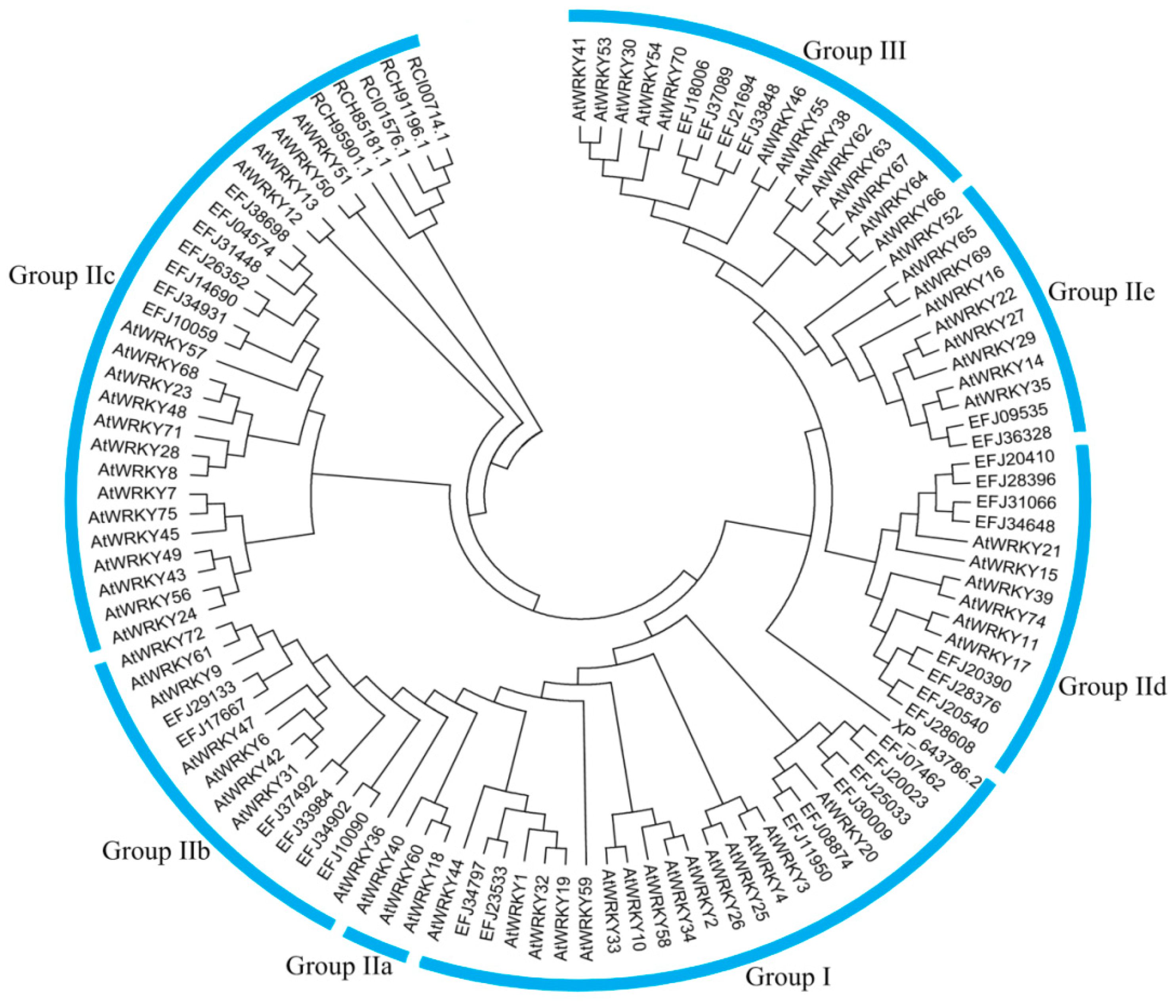
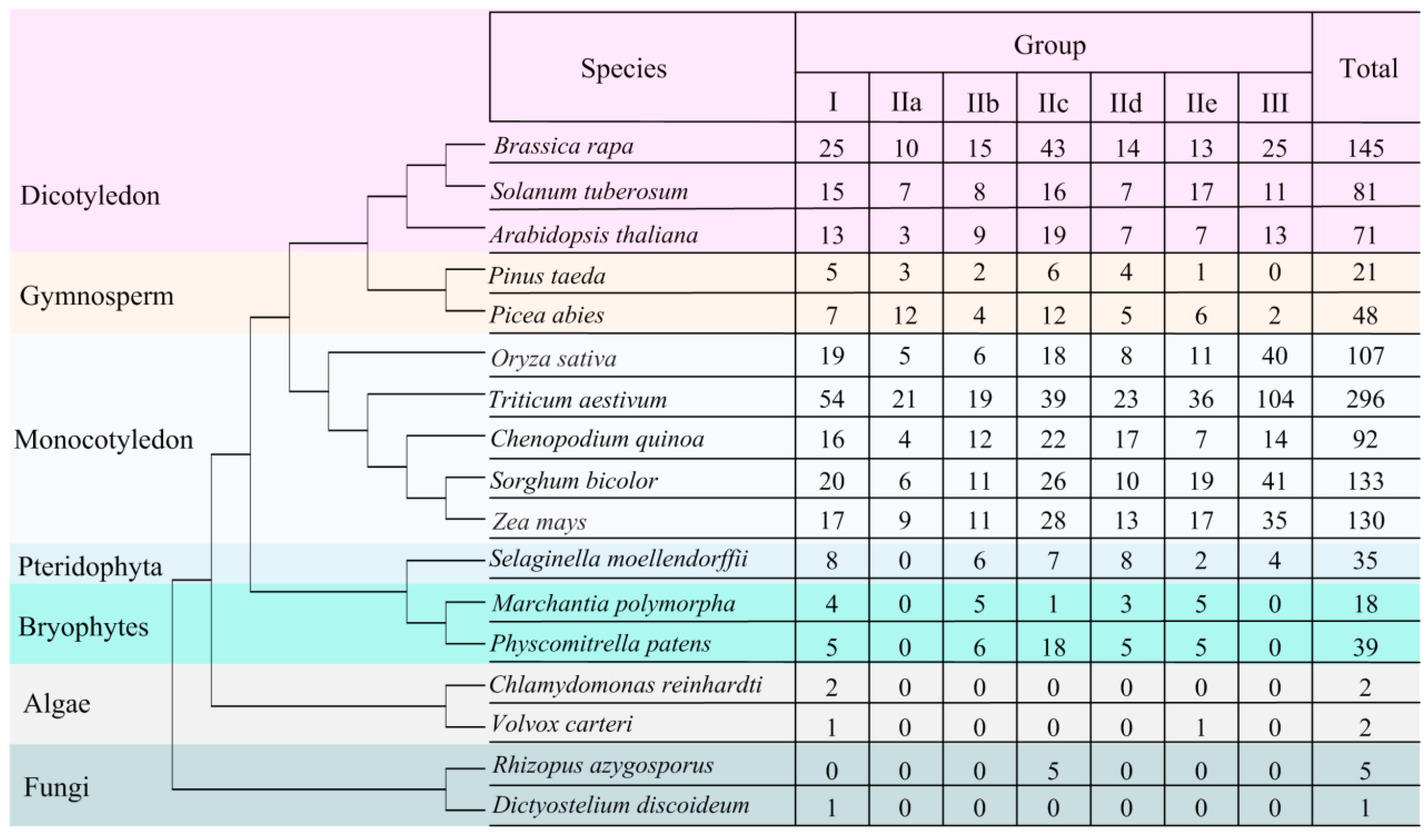
3.1. Global Identification and Evolution of WRKY Proteins from Eukaryotes to Plants
3.2. Identification of WRKY Genes in Quinoa
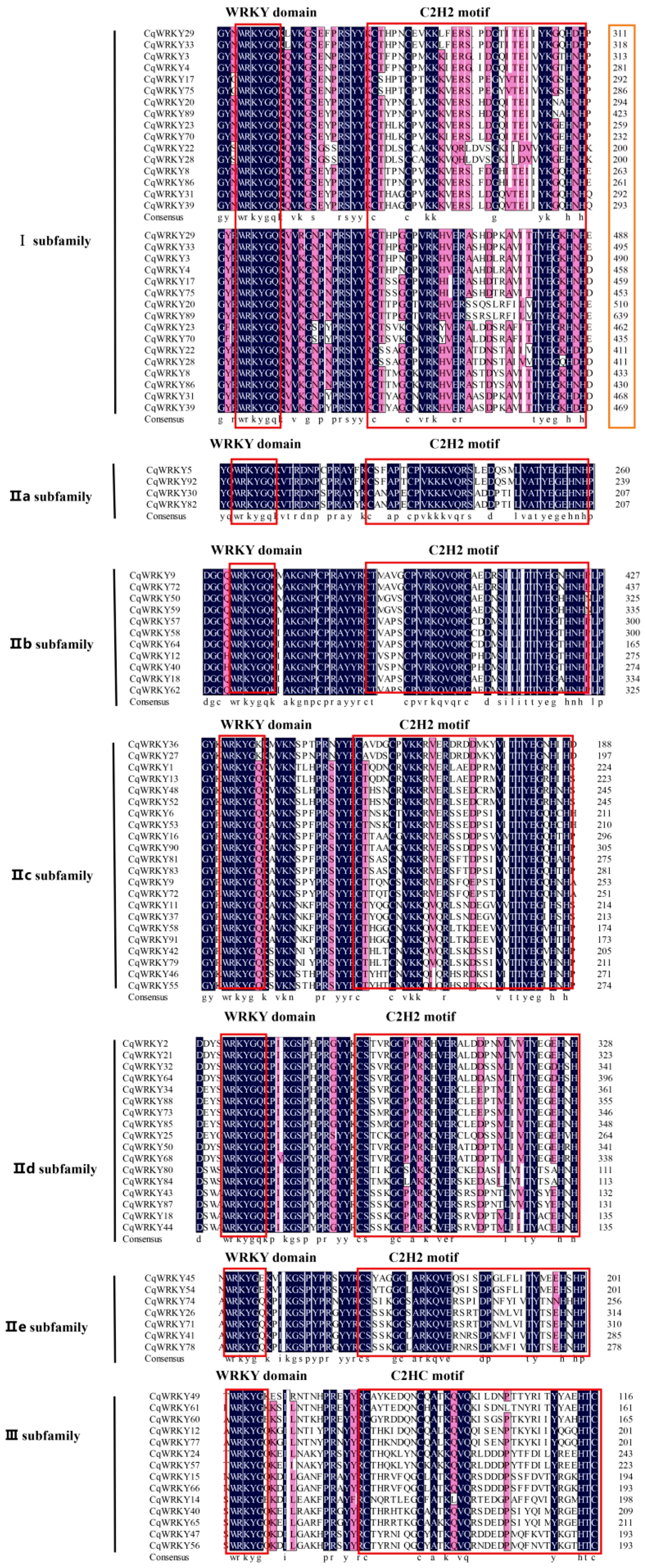
3.3. Multiple Sequence Alignment of CqWRKY
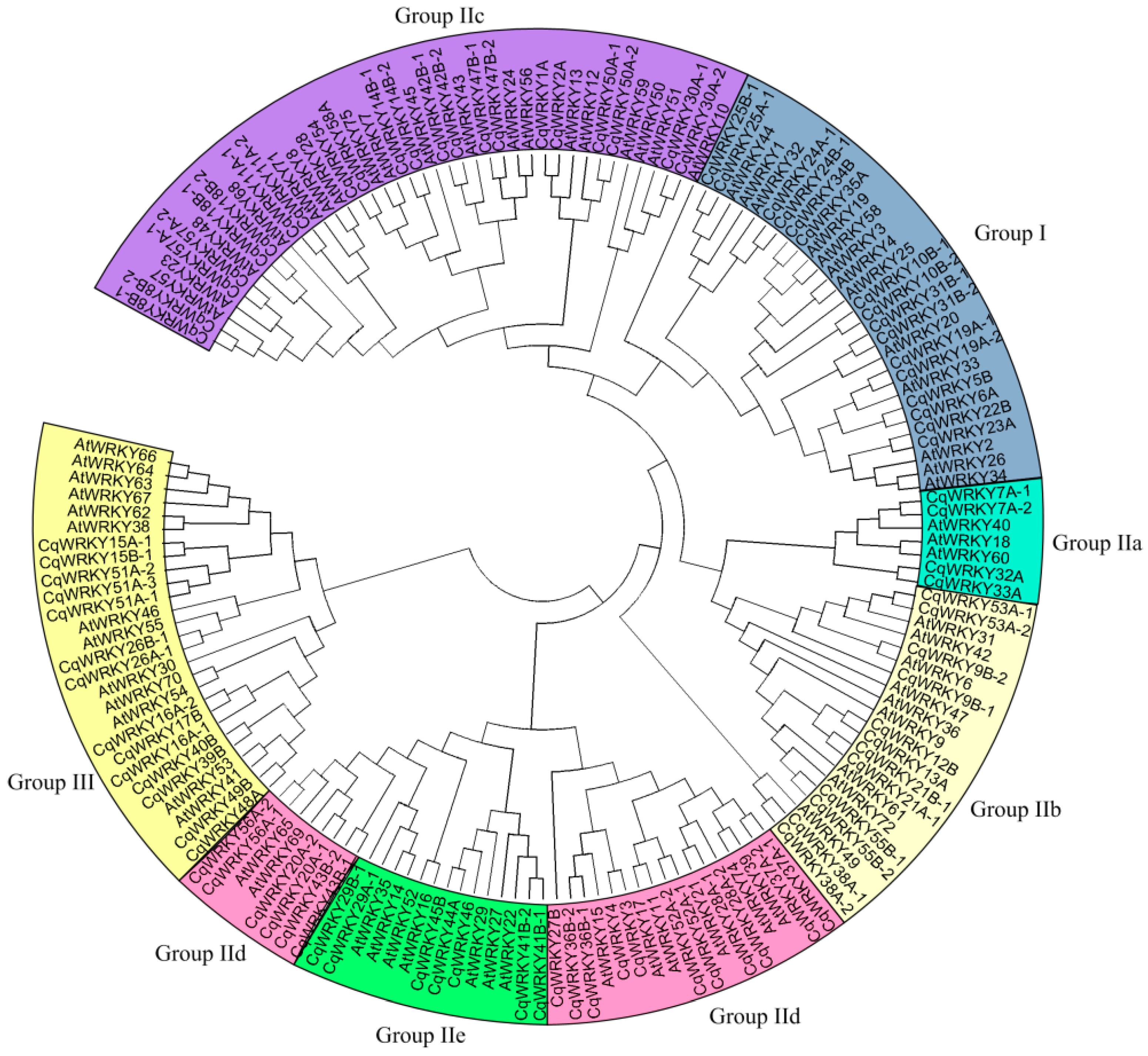
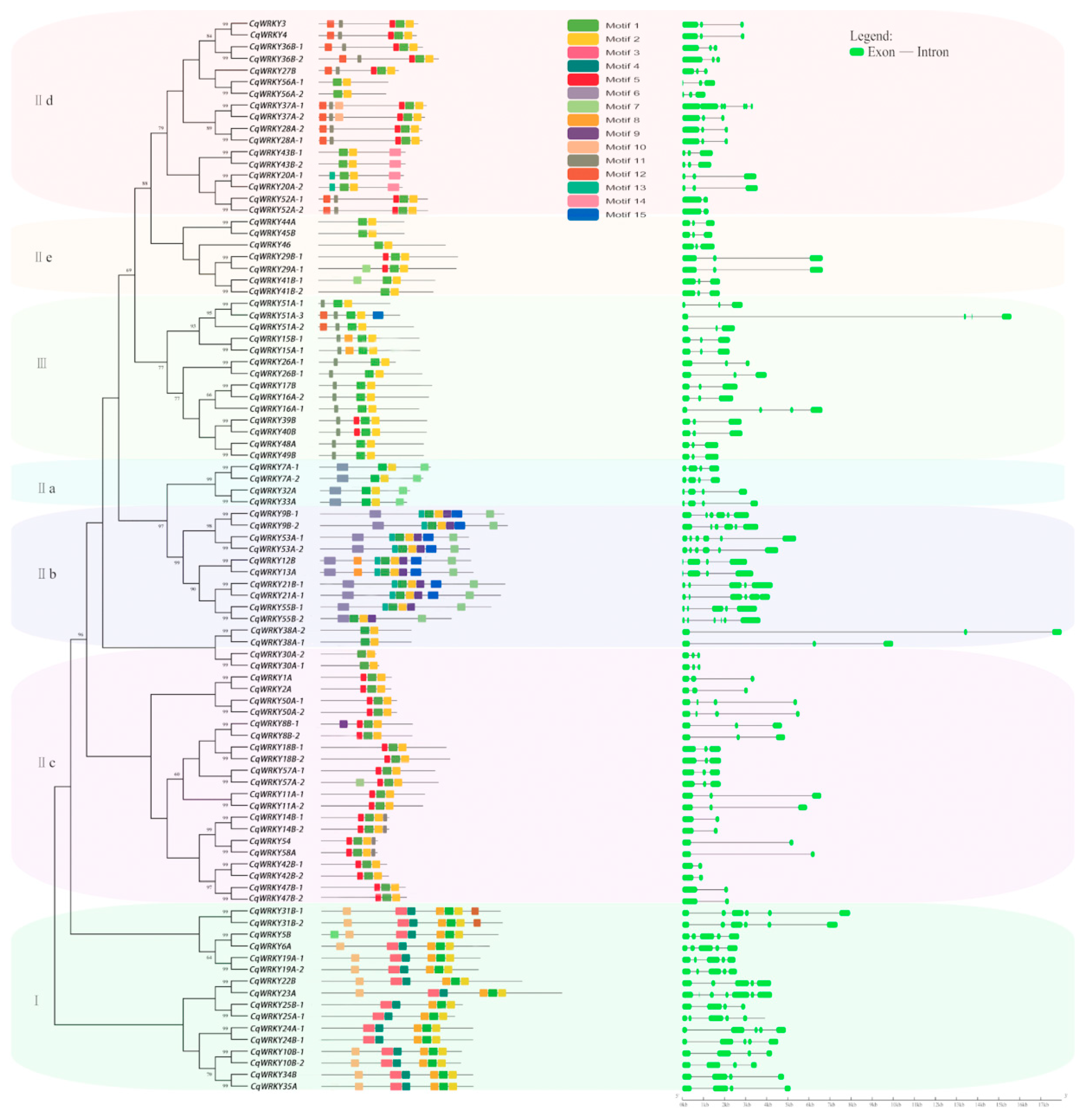
3.4. Phylogenetic Analysis of CqWRKY Genes
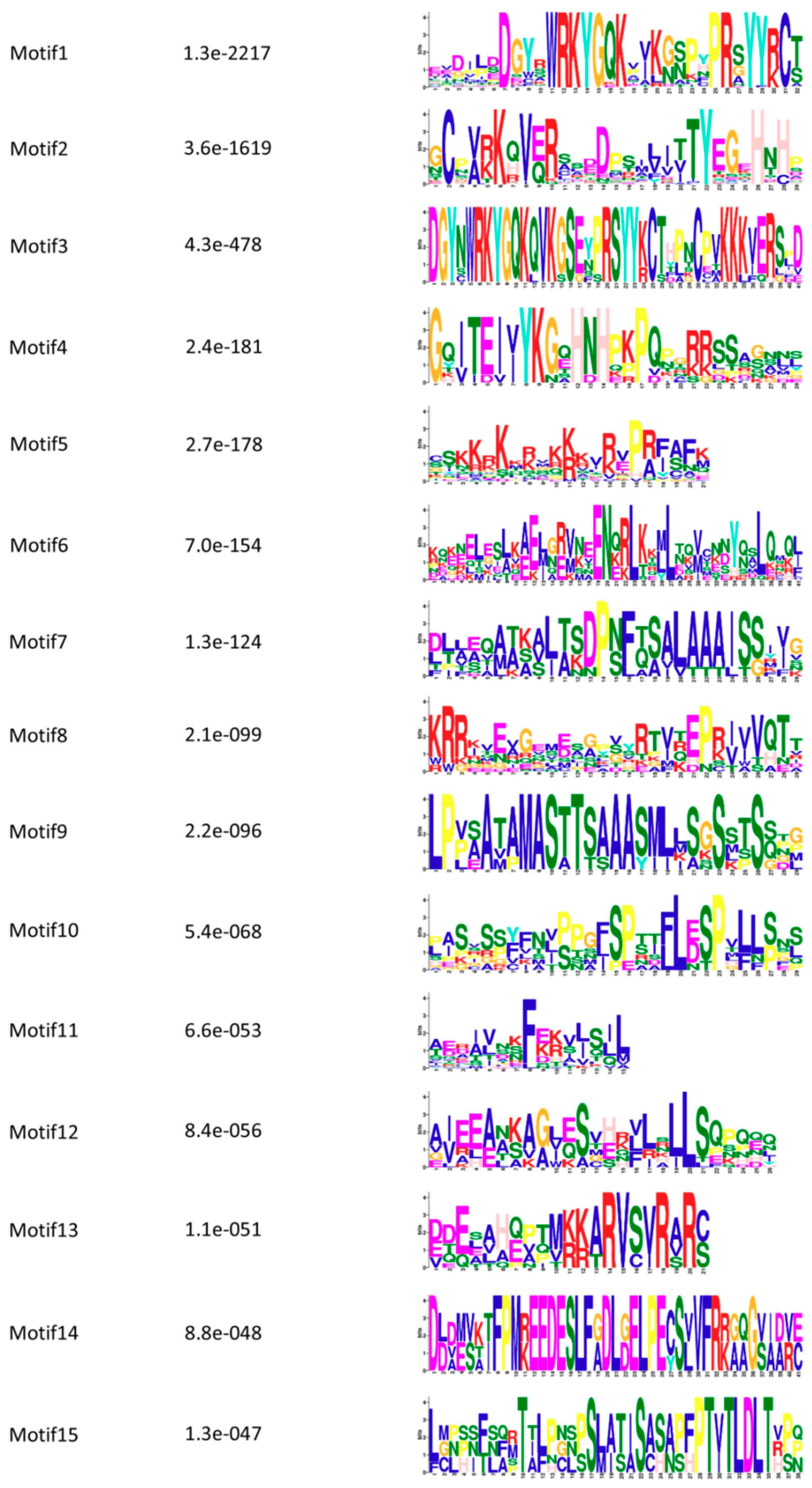
3.5. Conserved Motifs and Protein Structure Analysis
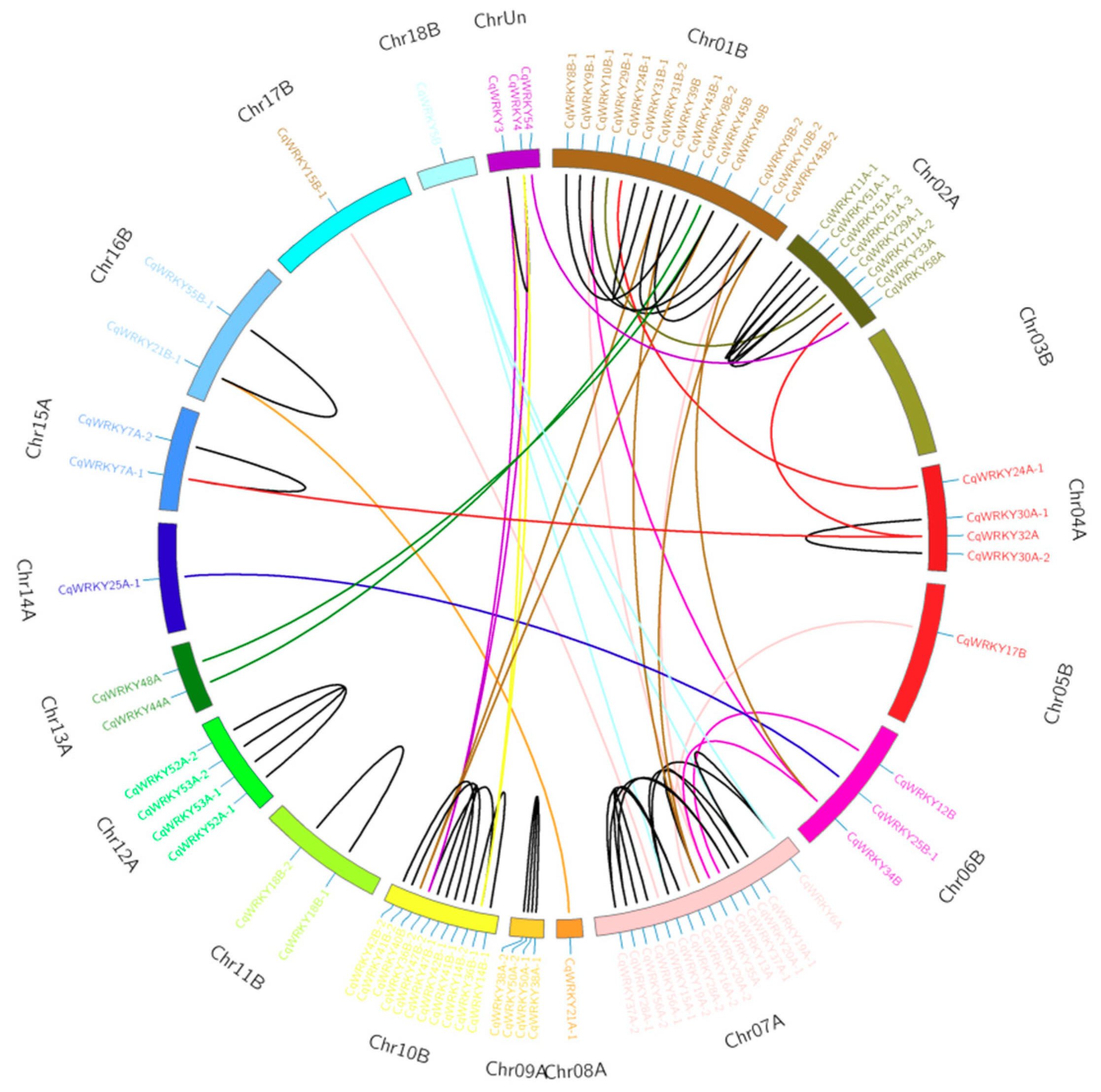
3.6. Gene Duplication Analysis of CqWRKY
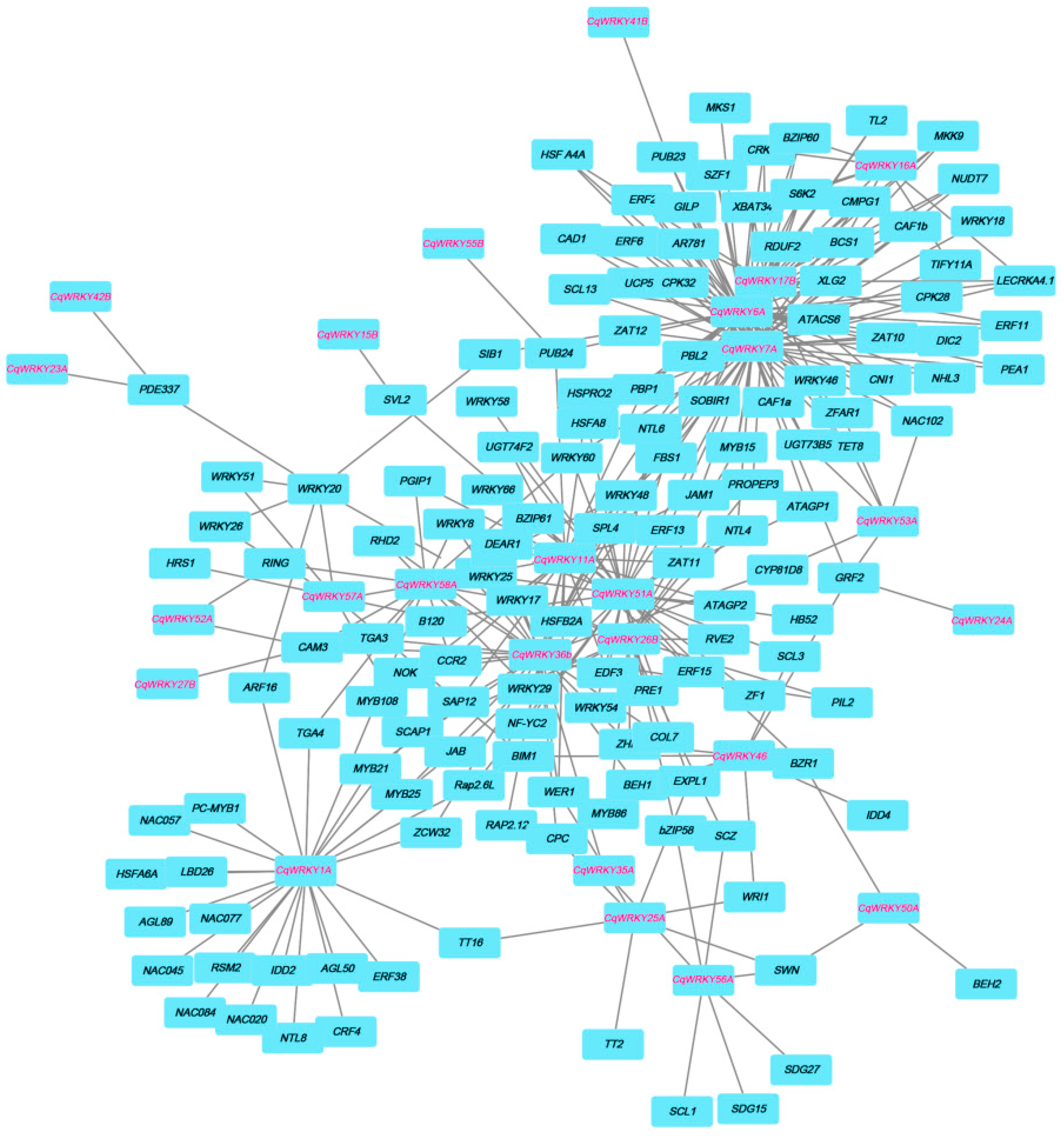
3.7. Interaction Network between CqWRKY Genes and Other Genes in Quinoa
3.8. Expression Profile Analysis of CqWRKY Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Group | Number | Name | Group | Number | Name |
|---|---|---|---|---|---|
| Animal | 1 | Echinococcus multilocularis | Plant | 1 | Marchantiapolymorpha |
| 2 | Caenorhabditis elegans | 2 | Physcomitrella patens | ||
| 3 | Drosophila melanogaster | 3 | Selaginellamoellendorffii | ||
| 4 | Lates calcarifer | 4 | Pinus taeda | ||
| 5 | Xenopus laevis | 5 | Piceaabies | ||
| 6 | Callithrix jacchus | 6 | Arabidopsis thaliana | ||
| 7 | Pan troglodytes | 7 | Solanum lycopersicum | ||
| Fungi | 1 | Rhizopus azygosporus | 8 | Brassica rapa | |
| 2 | Saccharomyces cerevisiae | 9 | Triticum aestivum | ||
| 3 | Dictyostelium discoideum | 10 | Zea mays | ||
| 4 | Gibberella | 11 | Oryza sativa | ||
| 5 | Penicillium chrysogenum | 12 | Sorghum bicolo | ||
| 6 | Ustilaginales | 13 | Chlamydomonas reinhardtii | ||
| 7 | Uredinales | 14 | Volvox carteri |
| Gene Name | Gene ID | Scaffold | Location | Protein Length | Subcellular localization | PI | MW(KDa) |
|---|---|---|---|---|---|---|---|
| CqWRKY1A | LOC110682067 | 2370 | Chr09A | 244 | Nuclear | 9.13 | 28.02363 |
| CqWRKY2A | LOC110695228 | 3086 | Chr07A | 242 | Nuclear | 9.2 | 27.80931 |
| CqWRKY3 | LOC110682343 | 3452 | ChrUn | 343 | Nuclear | 9.46 | 37.58446 |
| CqWRKY4 | LOC110701719 | 3452 | ChrUn | 338 | Nuclear | 9.53 | 37.01582 |
| CqWRKY5B | LOC110684199 | 2493 | Chr18B | 611 | Nuclear | 6.65 | 67.11943 |
| CqWRKY6A | LOC110684669 | 2528 | Chr07A | 580 | Nuclear | 7.18 | 63.94286 |
| CqWRKY7A-1 | LOC110684974 | 2314 | Chr15A | 391 | Nuclear | 8.15 | 43.08997 |
| CqWRKY7A-2 | LOC110739966 | 2314 | Chr15A | 357 | Nuclear | 6.85 | 39.40957 |
| CqWRKY8B-1 | LOC110686140 | 1000 | Chr01B | 316 | Nuclear | 6.29 | 34.45764 |
| CqWRKY8B-2 | LOC110722117 | 1000 | Chr01B | 315 | Nuclear | 6.18 | 34.2994 |
| CqWRKY9B-1 | LOC110687395 | 1862 | Chr01B | 636 | Nuclear | 6.54 | 69.18332 |
| CqWRKY9B-2 | LOC110731310 | 1862 | Chr01B | 648 | Nuclear | 6.29 | 70.53042 |
| CqWRKY10B-1 | LOC110689002 | 2048 | Chr01B | 484 | Nuclear | 8.42 | 52.92818 |
| CqWRKY10B-2 | LOC110735049 | 2048 | Chr01B | 481 | Nuclear | 7.31 | 52.78703 |
| CqWRKY11A-1 | LOC110690337 | 1747 | Chr02A | 358 | Nuclear | 6.22 | 39.7074 |
| CqWRKY11A-2 | LOC110728618 | 1747 | Chr02A | 351 | Nuclear | 6.52 | 39.01257 |
| CqWRKY12B | LOC110690435 | 2837 | Chr06B | 521 | Nuclear | 7.64 | 56.85428 |
| CqWRKY13A | LOC110712087 | 1214 | Chr07A | 529 | Nuclear | 7.21 | 57.70618 |
| CqWRKY14B-1 | LOC110690523 | 1870 | Chr10B | 235 | Nuclear | 7.81 | 26.93543 |
| CqWRKY14B-2 | LOC110711270 | 1870 | Chr10B | 234 | Nuclear | 6.89 | 27.04852 |
| CqWRKY15A-1 | LOC110731563 | 1001 | Chr07A | 349 | Nuclear | 5.59 | 39.17953 |
| CqWRKY15B-1 | LOC110690718 | 2858 | Chr17B | 347 | Nuclear | 5.65 | 39.12759 |
| CqWRKY16A-1 | LOC110696174 | 1001 | Chr07A | 345 | Nuclear | 5.67 | 39.23355 |
| CqWRKY16A-2 | LOC110725677 | 1001 | Chr07A | 379 | Nuclear | 5.99 | 42.5395 |
| CqWRKY17B | LOC110696200 | 3107 | Chr05B | 390 | Nuclear | 6.09 | 43.81166 |
| CqWRKY18B-1 | LOC110697304 | 2177 | Chr11B | 432 | Nuclear | 6.78 | 47.09947 |
| CqWRKY18B-2 | LOC110738211 | 2177 | Chr11B | 445 | Nuclear | 6.3 | 48.47655 |
| CqWRKY19A-1 | LOC110698105 | 1001 | Chr07A | 548 | Nuclear | 6 | 60.64106 |
| CqWRKY19A-2 | LOC110730454 | 1001 | Chr07A | 542 | Nuclear | 6 | 59.99832 |
| CqWRKY20A-1 | LOC110699703 | 3389 | Chr07A | 293 | Nuclear | 5.99 | 32.19663 |
| CqWRKY20A-2 | LOC110719121 | 3389 | Chr07A | 289 | Nuclear | 5.89 | 31.60399 |
| CqWRKY21A-1 | LOC110726496 | 1675 | Chr08A | 622 | Nuclear | 6.61 | 67.91068 |
| CqWRKY21B-1 | LOC110700550 | 3422 | Chr16B | 639 | Nuclear | 6.33 | 69.6783 |
| CqWRKY22B | LOC110701089 | 3429 | Chr05B | 693 | Nuclear | 5.64 | 75.49235 |
| CqWRKY23A | LOC110737209 | 2088 | Chr07A | 831 | Nuclear | 6.02 | 91.02864 |
| CqWRKY24A-1 | LOC110702356 | 1189 | Chr04A | 523 | Nuclear | 8.11 | 57.79983 |
| CqWRKY24B-1 | LOC110705651 | 3674 | Chr01B | 523 | Nuclear | 7.86 | 57.66874 |
| CqWRKY25A-1 | LOC110727210 | 1699 | Chr14A | 460 | Nuclear | 9.48 | 50.9117 |
| CqWRKY25B-1 | LOC110702616 | 3489 | Chr06B | 487 | Nuclear | 8.98 | 53.90871 |
| CqWRKY26A-1 | LOC110704369 | 3631 | Chr04A | 264 | Nuclear | 6.39 | 29.58899 |
| CqWRKY26B-1 | LOC110722941 | 1516 | Chr01B | 356 | Nuclear | 6.16 | 40.08192 |
| CqWRKY27B | LOC110704656 | 3651 | Chr16B | 276 | Nuclear | 9.49 | 30.8523 |
| CqWRKY28A-1 | LOC110734403 | 2008 | Chr07A | 358 | Nuclear | 9.62 | 39.61248 |
| CqWRKY28A-2 | LOC110728935 | 2008 | Chr07A | 356 | Nuclear | 9.66 | 39.37821 |
| CqWRKY29A-1 | LOC110728527 | 1747 | Chr02A | 476 | Nuclear | 6.72 | 51.73296 |
| CqWRKY29B-1 | LOC110704717 | 1000 | Chr01B | 481 | Nuclear | 6.13 | 52.06432 |
| CqWRKY30A-1 | LOC110705289 | 3966 | Chr04A | 200 | Nuclear | 5.83 | 22.90195 |
| CqWRKY30A-2 | LOC110710397 | 3966 | Chr04A | 191 | Extracellular | 5.61 | 21.83396 |
| CqWRKY31B-1 | LOC110705952 | 3686 | Chr01B | 618 | Nuclear | 6.26 | 66.68202 |
| CqWRKY31B-2 | LOC110706951 | 3686 | Chr01B | 625 | Nuclear | 6.17 | 67.76825 |
| CqWRKY32A | LOC110705979 | 1206 | Chr04A | 310 | Nuclear | 8.06 | 35.02528 |
| CqWRKY33A | LOC110733669 | 1995 | Chr02A | 299 | Nuclear | 6.33 | 33.78876 |
| CqWRKY34B | LOC110706383 | 3751 | Chr06B | 523 | Nuclear | 6.68 | 56.45296 |
| CqWRKY35A | LOC110713207 | 1214 | Chr07A | 524 | Nuclear | 6.93 | 56.65928 |
| CqWRKY36B-1 | LOC110706462 | 1559 | Chr10B | 359 | Nuclear | 9.6 | 39.65634 |
| CqWRKY36B-2 | LOC110724341 | 1559 | Chr10B | 414 | Nuclear | 9.28 | 46.11367 |
| CqWRKY37A-1 | LOC110708385 | 2088 | Chr07A | 372 | Nuclear | 9.8 | 41.4642 |
| CqWRKY37A-2 | LOC110737126 | 2088 | Chr07A | 366 | Nuclear | 9.74 | 40.64239 |
| CqWRKY38A-1 | LOC110710092 | 2646 | Chr09A | 313 | Nuclear | 6.55 | 34.80783 |
| CqWRKY38A-2 | LOC110724156 | 2646 | Chr09A | 313 | Nuclear | 7.66 | 34.67071 |
| CqWRKY39B | LOC110714054 | 4250 | Chr01B | 373 | Nuclear | 5.84 | 41.81242 |
| CqWRKY40B | LOC110725171 | 1606 | Chr10B | 371 | Nuclear | 5.6 | 41.73932 |
| CqWRKY41B-1 | LOC110714254 | 1870 | Chr10B | 403 | Nuclear | 6.21 | 43.4019 |
| CqWRKY41B-2 | LOC110731586 | 1870 | Chr10B | 396 | Nuclear | 6.08 | 42.46192 |
| CqWRKY42B-1 | LOC110714280 | 1870 | Chr10B | 227 | Nuclear | 6.99 | 25.88965 |
| CqWRKY42B-2 | LOC110731590 | 1870 | Chr10B | 233 | Nuclear | 6.95 | 26.65745 |
| CqWRKY43B-1 | LOC110716147 | 4338 | Chr01B | 299 | Nuclear | 5.94 | 33.68204 |
| CqWRKY43B-2 | LOC110736290 | 4338 | Chr01B | 299 | Nuclear | 6 | 33.64207 |
| CqWRKY44A | LOC110719591 | 1280 | Chr13A | 297 | Nuclear | 5.46 | 33.49528 |
| CqWRKY45B | LOC110722558 | 1480 | Chr01B | 296 | Nuclear | 6.26 | 33.15403 |
| CqWRKY46 | LOC110729570 | 1783 | ChrUn | 438 | Nuclear | 5.92 | 48.43488 |
| CqWRKY47B-1 | LOC110719592 | 1870 | Chr10B | 292 | Peroxisome | 8.51 | 32.94533 |
| CqWRKY47B-2 | LOC110722559 | 1870 | Chr10B | 295 | Peroxisome | 8.11 | 33.28853 |
| CqWRKY48A | LOC110719657 | 1280 | Chr13A | 361 | Nuclear | 5.83 | 40.4269 |
| CqWRKY49B | LOC110722668 | 1480 | Chr01B | 361 | Nuclear | 5.79 | 40.31987 |
| CqWRKY50A-1 | LOC110720500 | 2370 | Chr09A | 262 | Nuclear | 8.71 | 29.83144 |
| CqWRKY50A-2 | LOC110721539 | 2370 | Chr09A | 262 | Nuclear | 8.26 | 29.9865 |
| CqWRKY51A-1 | LOC110720675 | 1529 | Chr02A | 247 | Extracellular | 5.37 | 28.26295 |
| CqWRKY51A-2 | LOC110723602 | 1529 | Chr02A | 329 | Nuclear | 5.84 | 37.03055 |
| CqWRKY51A-3 | LOC110723659 | 1529 | Chr02A | 281 | Nuclear | 5.27 | 32.04714 |
| CqWRKY52A-1 | LOC110720830 | 1373 | Chr12A | 376 | Nuclear | 9.58 | 41.44137 |
| CqWRKY52A-2 | LOC110726911 | 1373 | Chr12A | 377 | Nuclear | 9.54 | 41.6516 |
| CqWRKY53A-1 | LOC110720931 | 1373 | Chr12A | 513 | Nuclear | 5.55 | 56.43854 |
| CqWRKY53A-2 | LOC110723870 | 1373 | Chr12A | 517 | Nuclear | 5.46 | 57.13047 |
| CqWRKY54 | LOC110723391 | 3819 | ChrUn | 195 | chloroplast | 9.43 | 22.49956 |
| CqWRKY55B-1 | LOC110723405 | 1526 | Chr16B | 589 | Nuclear | 6.13 | 64.4822 |
| CqWRKY55B-2 | LOC110727007 | 1526 | Chr16B | 452 | Nuclear | 7.98 | 49.25958 |
| CqWRKY56A-1 | LOC110732902 | 2008 | Chr07A | 239 | Nuclear | 9.05 | 27.35754 |
| CqWRKY56A-2 | LOC110734287 | 2008 | Chr07A | 232 | Nuclear | 8.94 | 26.56771 |
| CqWRKY57A-1 | LOC110733008 | 1992 | Chr09A | 393 | Nuclear | 6.42 | 42.77934 |
| CqWRKY57A-2 | LOC110734264 | 1992 | Chr09A | 405 | Nuclear | 6.26 | 43.97958 |
| CqWRKY58A | LOC110739691 | 2306 | Chr02A | 194 | Cytoplasm | 9.5 | 22.37437 |

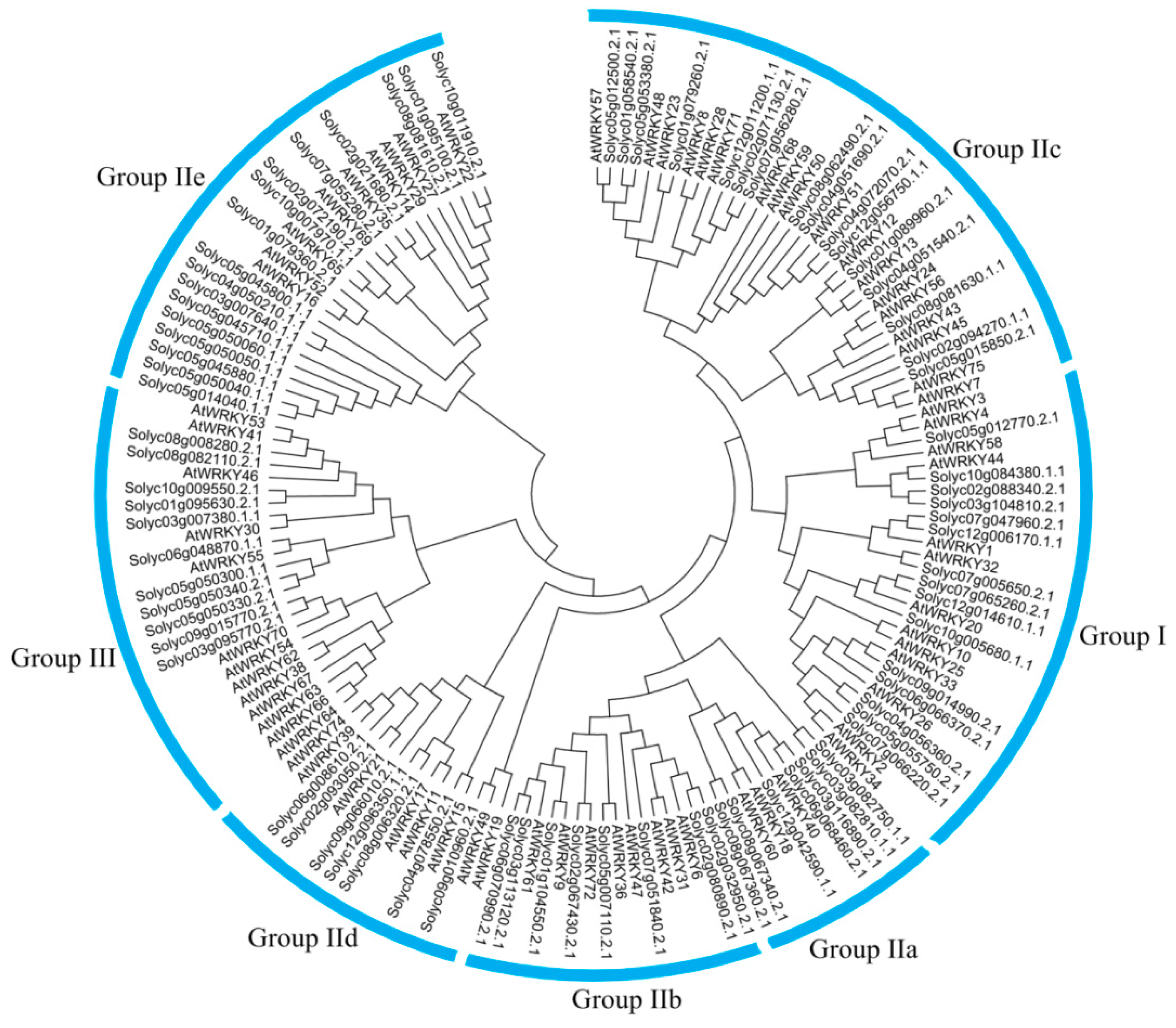
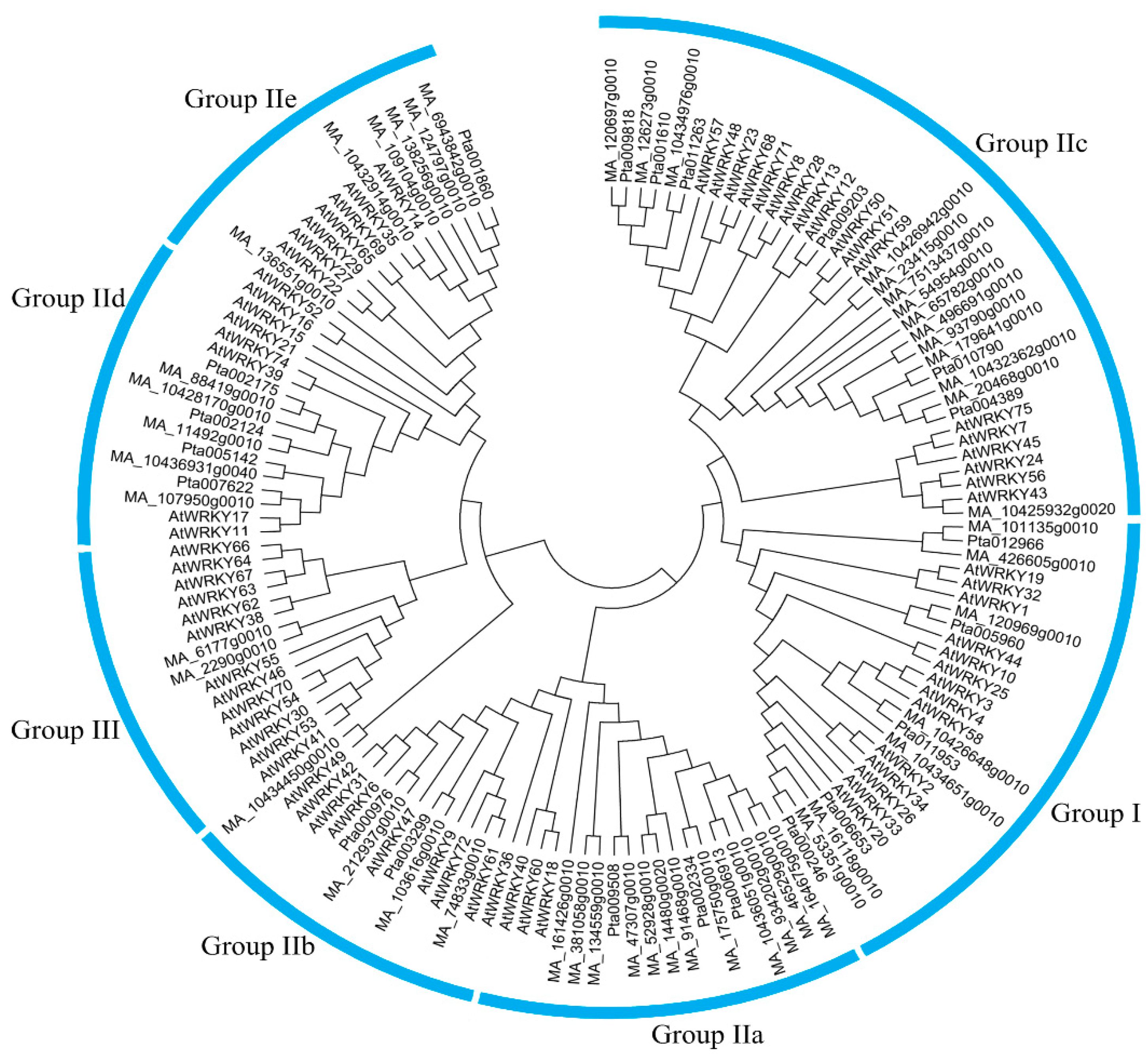
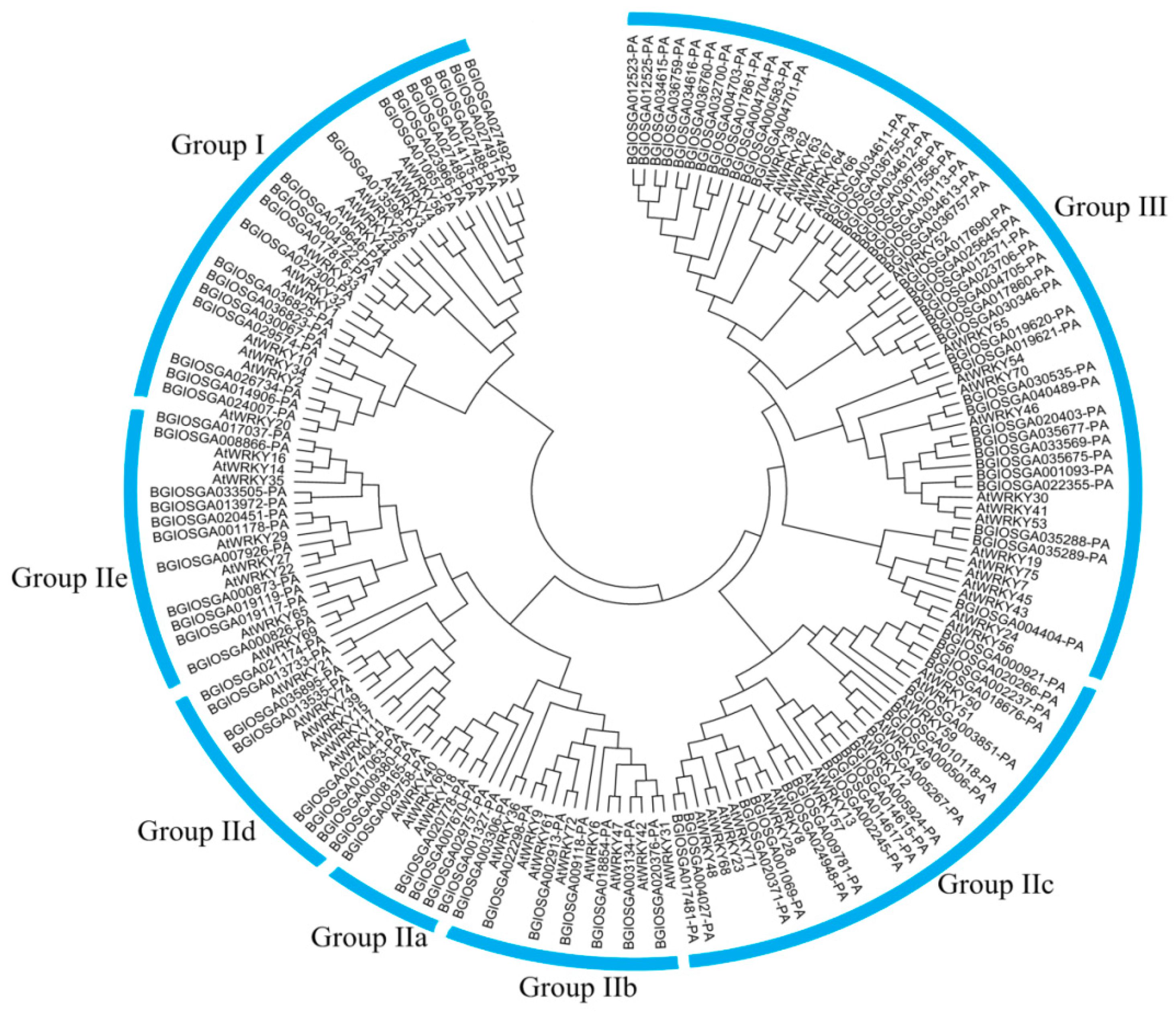
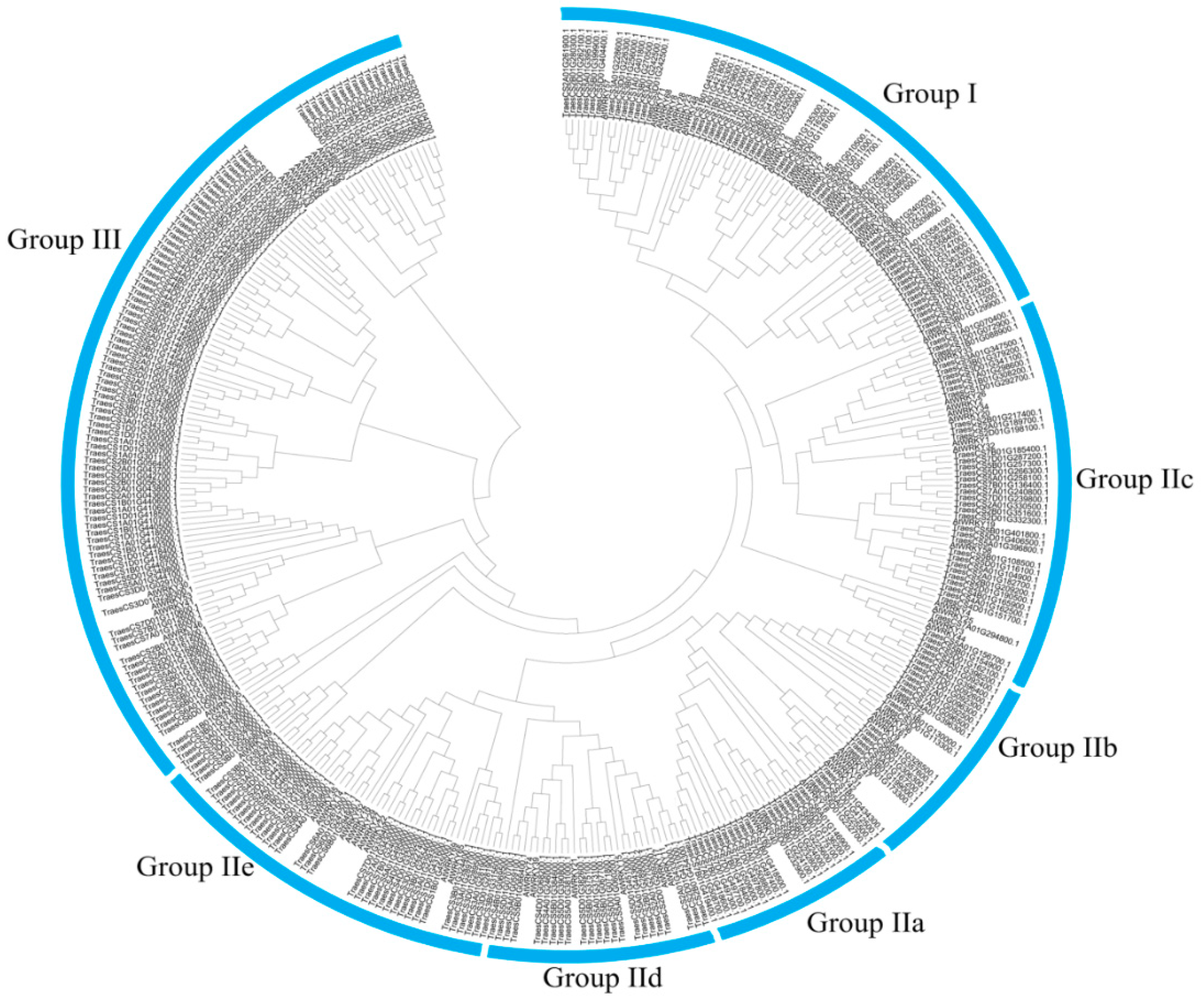
References
- Yue, H.; Wang, M.; Liu, S.; Du, X.; Song, W.; Nie, X. Transcriptome-wide identification and expression profiles of the WRKY transcription factor family in Broomcorn millet (Panicum miliaceum L.). BMC Genom. 2016, 17, 343. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Cui, M.-Y.; Hu, Y.; Gao, K.; Xie, Y.-G.; Jiang, Y.; Feng, J.-Y. Ectopic expression of FvWRKY42, a WRKY transcription factor from the diploid woodland strawberry (Fragaria vesca), enhances resistance to powdery mildew, improves osmotic stress resistance, and increases abscisic acid sensitivity in Arabidopsis. Plant Sci. 2018, 275, 60–74. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Hou, L.; Xiao, P.; Guo, Y.; Deyholos, M.K.; Liu, X. VvWRKY30, a grape WRKY transcription factor, plays a positive regulatory role under salinity stress. Plant Sci. 2019, 280, 132–142. [Google Scholar] [CrossRef]
- Wang, C.-T.; Ru, J.-N.; Liu, Y.-W.; Yang, J.-F.; Li, M.; Xu, Z.-S.; Fu, J.-D. The Maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2580. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-T.; Ru, J.-N.; Liu, Y.-W.; Li, M.; Zhao, D.; Yang, J.-F.; Fu, J.-D.; Xu, Z.-S. Maize WRKY Transcription Factor ZmWRKY106 Confers Drought and Heat Tolerance in Transgenic Plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef] [PubMed]
- Hui, S.; Wang, P.; Lin, J.Y.; Zhao, C.; Bi, Y.; Wang, X. Genome-Wide Identification and Characterization of WRKY Gene Family in Peanut. Front. Plant Sci. 2016, 7, 9. [Google Scholar]
- Gao, H.; Wang, Y.; Xu, P.; Zhang, Z. Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 2018, 9, 997. [Google Scholar] [CrossRef]
- Raineri, J.; Wang, S.; Peleg, Z.; Blumwald, E.; Chan, R.L. The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Mol. Biol. 2015, 88, 401–413. [Google Scholar] [CrossRef]
- Dai, X.; Wang, Y.; Zhang, W.-H. OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 2015, 67, 947–960. [Google Scholar] [CrossRef]
- Wu, J.; Chen, J.; Wang, L.; Wang, S. Genome-Wide Investigation of WRKY Transcription Factors Involved in Terminal Drought Stress Response in Common Bean. Front. Plant Sci. 2017, 8, 380. [Google Scholar] [CrossRef]
- Knoth, C.; Ringler, J.; Dangl, J.L.; Eulgem, T. Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant Microbe Interact. 2007, 20, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, D.; Yang, C.; Kong, N.; Shi, Z.; Zhao, P.; Nan, Y.; Nie, T.; Wang, R.; Ma, H. Genome-wide identification of the potato WRKY transcription factor family. PLoS ONE 2017, 12, e0181573. [Google Scholar] [CrossRef] [PubMed]
- Hulse, S. In silico genome-wide identification and phylogenetic analysis of the WRKY transcription factor family in sweet orange (Citrus sinensis). Aust. J. Crop Sci. 2017, 11, 716–726. [Google Scholar]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 Transcription Factor Is a Modulator of Phosphate Acquisition and Root Development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Song, Y.; Xing, F.; Wang, N.; Wen, F.; Zhu, C. GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma 2015, 253, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Abid, G.; Muhovski, Y.; Mingeot, D.; Saidi, M.; Aouida, M.; Aroua, I.; M’hamdi, M.; Barhoumi, F.; Rezgui, S.; Jebara, M. Identification and characterization of two faba bean (Vicia faba L.) WRKY transcription factors and their expression analysis during salt and drought stress. J. Agric. Sci. 2017, 155, 791–803. [Google Scholar] [CrossRef]
- Wang, Y.; Shu, Z.; Wang, W.; Jiang, X.; Li, D.; Pan, J.; Li, X. CsWRKY2, a novel WRKY gene from Camellia sinensis, is involved in cold and drought stress responses. Biol. Plant. 2016, 60, 1–9. [Google Scholar] [CrossRef]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]
- Gu, L.; Li, L.; Wei, H.; Wang, H.; Su, J.; Guo, Y.; Yu, S. Identification of the group IIa WRKY subfamily and the functional analysis ofGhWRKY17in upland cotton (Gossypium hirsutum L.). PLoS ONE 2018, 13, e0191681. [Google Scholar] [CrossRef]
- Huang, Y.; Li, M.-Y.; Wu, P.; Xu, Z.-S.; Que, F.; Wang, F.; Xiong, A.-S. Members of WRKY Group III transcription factors are important in TYLCV defense signaling pathway in tomato (Solanum lycopersicum). BMC Genom. 2016, 17, 788. [Google Scholar] [CrossRef]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N. The genome of Chenopodium quinoa. Nature 2017, 542, 307. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.R.; Parker, M.L. Quinoa. In Pseudocereals and Less Common Cereals; Springer: Berlin/Heidelberg, Germany, 2002; pp. 93–122. [Google Scholar]
- Bhathal, S.K.; Kaur, N.; Gill, J. Effect of processing on the nutritional composition of quinoa (Chenopodium quinoa Willd). Agric. Res. J. 2017, 54, 90–93. [Google Scholar] [CrossRef]
- Nowak, V.; Du, J.; Charrondière, U.R. Assessment of the nutritional composition of quinoa (Chenopodium quinoa Willd.). Food Chem. 2016, 193, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Chen, C.; Li, C.; Liu, J.; Liu, C.; He, Y. Genome-wide investigation of WRKY gene family in pineapple: Evolution and expression profiles during development and stress. BMC Genom. 2018, 19, 490. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yue, C.; Chen, D.; Zheng, Y.; Zhang, Q.; Yang, J.; Ye, N. Genome-wide identification of WRKY family genes and their response to abiotic stresses in tea plant (Camellia sinensis). Genes Genom. 2018, 41, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.L.; Guo, Z.J.; Wang, H.H.; Li, J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005, 12, 9–26. [Google Scholar] [CrossRef]
- Kersey, P.J.; Allen, J.E.; Armean, I.; Boddu, S.; Bolt, B.J.; Carvalho-Silva, D.; Christensen, M.; Davis, P.; Falin, L.J.; Grabmueller, C. Ensembl Genomes 2016: More genomes, more complexity. Nucleic Acids Res. 2015, 44, D574–D580. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Guo, A.; Zhu, Q.; Chen, X.; Luo, J. GSDS: A gene structure display server. Yi Chuan = Hered. 2007, 29, 1023–1026. [Google Scholar] [CrossRef]
- Wang, M.; Yue, H.; Feng, K.; Deng, P.; Song, W.; Nie, X. Genome-wide identification, phylogeny and expressional profiles of mitogen activated protein kinase kinase kinase (MAPKKK) gene family in bread wheat (Triticum aestivum L.). BMC Genom. 2016, 17, 668. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Yang, S.; Kim, E.; Ko, Y.; Hwang, S.; Shin, J.; Shim, J.E.; Shim, H.; Kim, H.; Kim, C. AraNet v2: An improved database of co-functional gene networks for the study of Arabidopsis thaliana and 27 other nonmodel plant species. Nucleic Acids Res. 2015, 43, D996–D1002. [Google Scholar] [CrossRef] [PubMed]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape Plugin to Assess Overrepresentation of Gene Ontology Categories in Biological Networks; Oxford University Press: Oxford, UK, 2005; pp. 3448–3449. [Google Scholar]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L. The WRKY transcription factor superfamily: Its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005, 5, 1. [Google Scholar]
- Li, D.; Liu, P.; Yu, J.; Wang, L.; Dossa, K.; Zhang, Y.; Zhou, R.; Wei, X.; Zhang, X. Genome-wide analysis of WRKY gene family in the sesame genome and identification of the WRKY genes involved in responses to abiotic stresses. BMC Plant Biol. 2017, 17, 152. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, S.; Chen, M.; Qin, L.; Kong, L.; Xia, G. WRKY Transcription Factors in Wheat and Their Induction by Biotic and Abiotic Stress. Plant Mol. Biol. Rep. 2013, 31, 1053–1067. [Google Scholar] [CrossRef]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef]
- Okay, S.; Derelli, E.; Unver, T. Transcriptome-wide identification of bread wheat WRKY transcription factors in response to drought stress. Mol. Genet. Genom. 2014, 289, 765–781. [Google Scholar] [CrossRef]
- Xu, H.; Watanabe, K.A.; Zhang, L.; Shen, Q.J. WRKY transcription factor genes in wild rice Oryza nivara. DNA Res. 2016, 23, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.; Shu, D.; Wang, M.; Xing, G.; Zhan, H.; Du, X.; Song, W.; Nie, X. Genome-Wide Identification and Expression Analysis of the HD-Zip Gene Family in Wheat (Triticum aestivum L.). Genes 2018, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Shao, F.; Macmillan, C.; Wilson, I.W.; Merwe, K.V.D.; Hussey, S.G.; Myburg, A.A.; Dong, X.; Qiu, D. Genomewide analysis of the lateral organ boundaries domain gene family in Eucalyptus grandisreveals members that differentially impact secondary growth. Plant Biotechnol. J. 2018, 16, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef]
- Thirugnanasambantham, K.; Durairaj, S.; Saravanan, S.; Karikalan, K.; Muralidaran, S.; Islam, V.I.H. Role of ethylene response transcription factor (ERF) and its regulation in response to stress encountered by plants. Plant Mol. Biol. Rep. 2015, 33, 347–357. [Google Scholar] [CrossRef]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ng, C.K.-Y.; Fan, L.-M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Wang, M.; Vannozzi, A.; Wang, G.; Liang, Y.H.; Tornielli, G.B.; Zenoni, S.; Cavallini, E.; Pezzotti, M.; Cheng, Z.M. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 2014, 1, 14016. [Google Scholar] [CrossRef]
- Li, H.L.; Guo, D.; Yang, Z.P.; Tang, X.; Peng, S.Q. Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics 2014, 104, 14–23. [Google Scholar] [CrossRef]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef]
- Ling, J.; Jiang, W.; Zhang, Y.; Yu, H.; Mao, Z.; Gu, X.; Huang, S.; Xie, B. Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genom. 2011, 12, 471. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wang, F.; Hou, X.L.; Wang, Z.; Huang, Z.N. Genome-Wide Fractionation and Identification of WRKY Transcription Factors in Chinese Cabbage (Brassica rapa ssp. pekinensis ) Reveals Collinearity and Their Expression Patterns Under Abiotic and Biotic Stresses. Plant Mol. Biol. Rep. 2014, 32, 781–795. [Google Scholar] [CrossRef]
- Satapathy, L.; Singh, D.; Ranjan, P.; Kumar, D.; Kumar, M.; Prabhu, K.V.; Mukhopadhyay, K. Transcriptome-wide analysis of WRKY transcription factors in wheat and their leaf rust responsive expression profiling. Mol. Genet. Genom. 2014, 289, 1289–1306. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lin, J.; Li, X.G.; Chang, Y. Genome-wide identification of pear HD-Zip gene family and expression patterns under stress induced by drought, salinity, and pathogen. Acta Physiol. Plant. 2015, 37, 189. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, R.; Luo, X.; Jiang, Z.; Shu, H. Genome-wide identification and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene 2013, 531, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Yu, L.; Li, Z.; Liu, H.; Han, R. Molecular evolution and gene expression differences within the HD-Zip transcription factor family of Zea mays L. Genetica 2016, 144, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, Z.; Zhao, H.; Zhao, Y.; Cheng, B.; Xiang, Y. Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 2014, 9, e87156. [Google Scholar] [CrossRef]
- Tashi, G.; Zhan, H.; Xing, G.; Chang, X.; Zhang, H.; Nie, X.; Ji, W. Genome-wide identification and expression analysis of heat shock transcription factor family in Chenopodium quinoa Willd. Agronomy 2018, 8, 103. [Google Scholar] [CrossRef]
- Schmöckel, S.M.; Lightfoot, D.J.; Razali, R.; Tester, M.; Jarvis, D.E. Identification of putative transmembrane proteins involved in salinity tolerance in Chenopodium quinoa by integrating physiological data, RNAseq, and SNP analyses. Front. Plant Sci. 2017, 8, 1023. [Google Scholar] [CrossRef]
- Bi, C.; Xu, Y.; Ye, Q.; Yin, T.; Ning, Y. Genome-wide identification and characterization of WRKY gene family in Salix suchowensis. PeerJ 2016, 4, e2437. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, N.-N.; Kong, L.; Gong, S.-Y.; Li, Y.; Li, X.-B. Molecular characterization of 26 cotton WRKY genes that are expressed differentially in tissues and are induced in seedlings under high salinity and osmotic stress. Plant Cell Tissue Organ Cult. 2014, 119, 141–156. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.; Yao, W.; Zhou, B.; Li, R.; Jiang, T. Expression patterns of WRKY genes in di-haploid Populus simonii × P. nigra in response to salinity stress revealed by quantitative real-time PCR and RNA sequencing. Plant Cell Rep. 2014, 33, 1687–1696. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, H.; Chang, X.; Zhi, Y.; Wang, L.; Xing, G.; Song, W.; Nie, X. Evolution and Identification of the WRKY Gene Family in Quinoa (Chenopodium quinoa). Genes 2019, 10, 131. https://doi.org/10.3390/genes10020131
Yue H, Chang X, Zhi Y, Wang L, Xing G, Song W, Nie X. Evolution and Identification of the WRKY Gene Family in Quinoa (Chenopodium quinoa). Genes. 2019; 10(2):131. https://doi.org/10.3390/genes10020131
Chicago/Turabian StyleYue, Hong, Xi Chang, Yongqiang Zhi, Lan Wang, Guangwei Xing, Weining Song, and Xiaojun Nie. 2019. "Evolution and Identification of the WRKY Gene Family in Quinoa (Chenopodium quinoa)" Genes 10, no. 2: 131. https://doi.org/10.3390/genes10020131
APA StyleYue, H., Chang, X., Zhi, Y., Wang, L., Xing, G., Song, W., & Nie, X. (2019). Evolution and Identification of the WRKY Gene Family in Quinoa (Chenopodium quinoa). Genes, 10(2), 131. https://doi.org/10.3390/genes10020131







