Source of Dietary Fat in Pig Diet Affects Adipose Expression of Genes Related to Cancer, Cardiovascular, and Neurodegenerative Diseases
Abstract
:1. Introduction
2. Materials and Methods
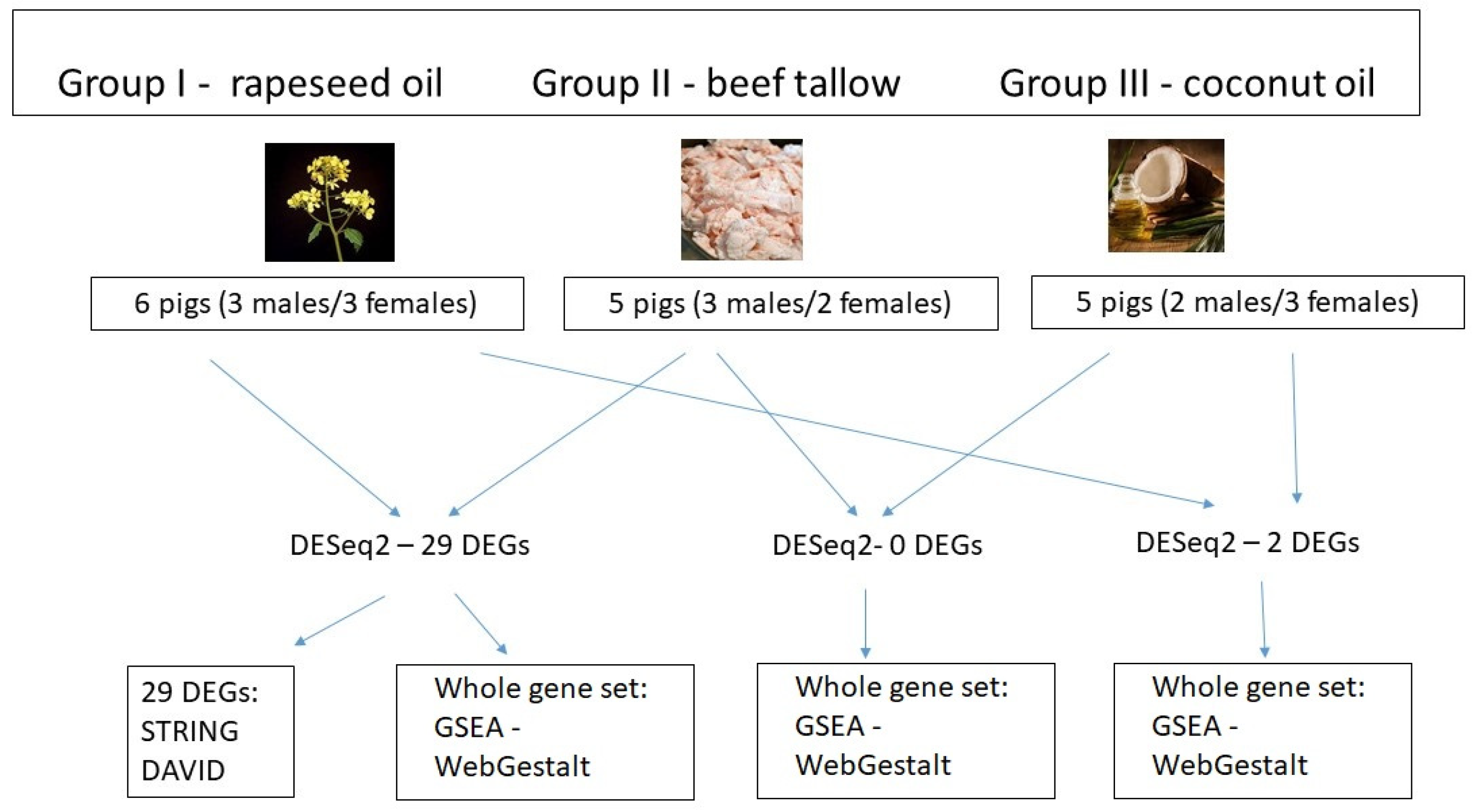
2.1. Animals and Diets
2.2. RNA Isolation, RNA-Seq, and qPCR Procedures
2.3. RNA-Seq Data Processing and Statistical Methods
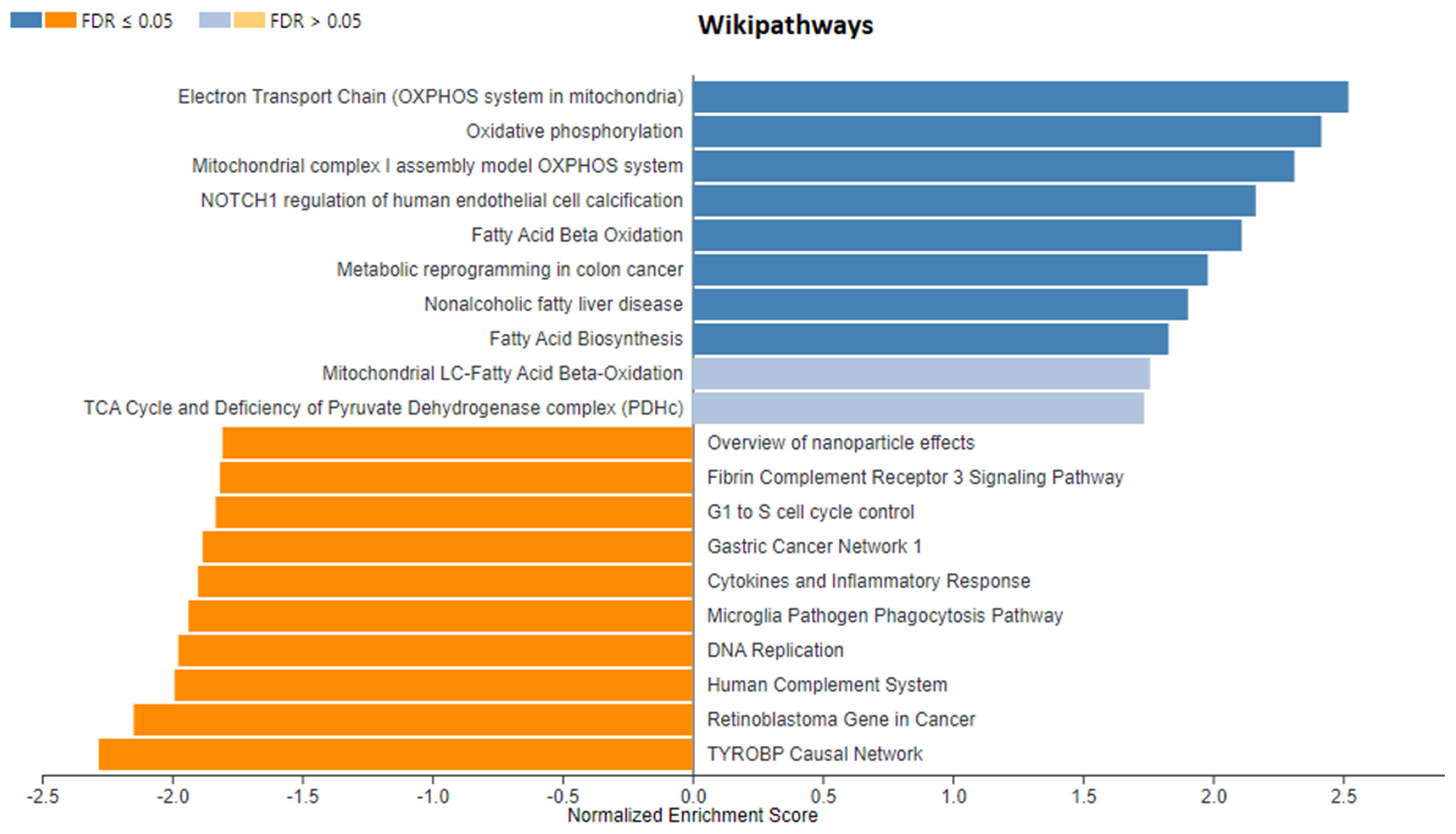
3. Results
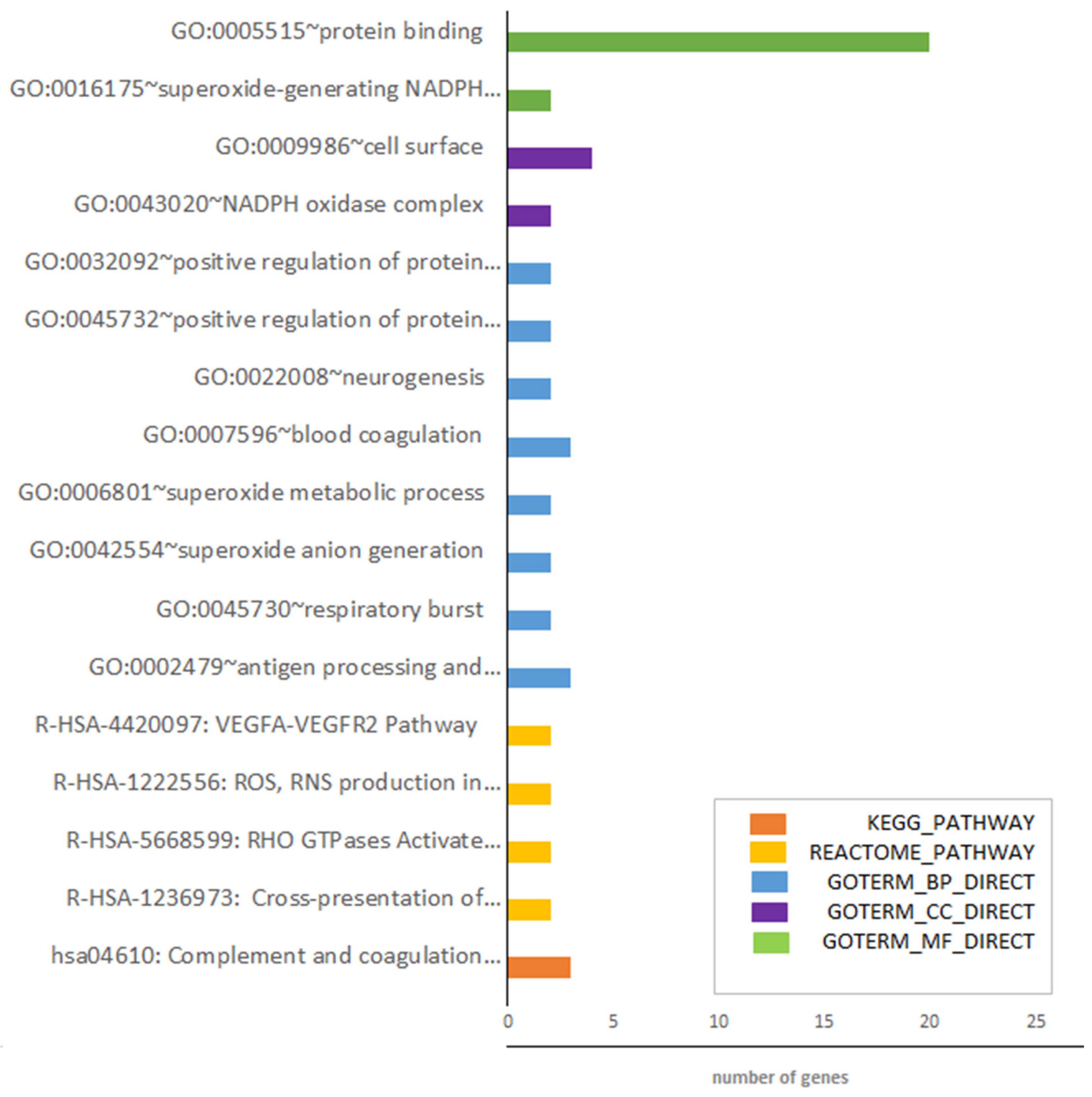
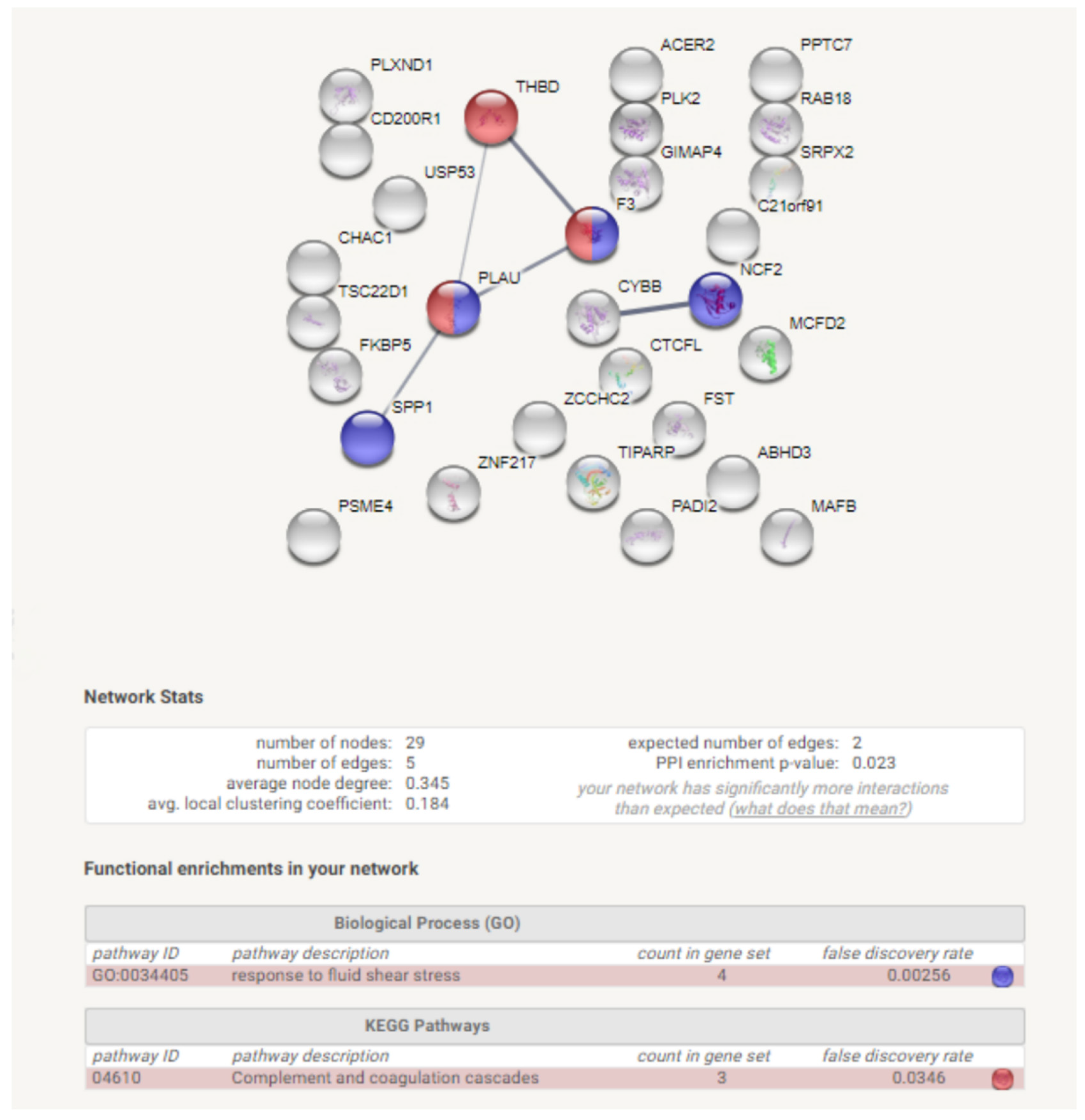
3.1. Identification of DEGs after RNA-Sequencing
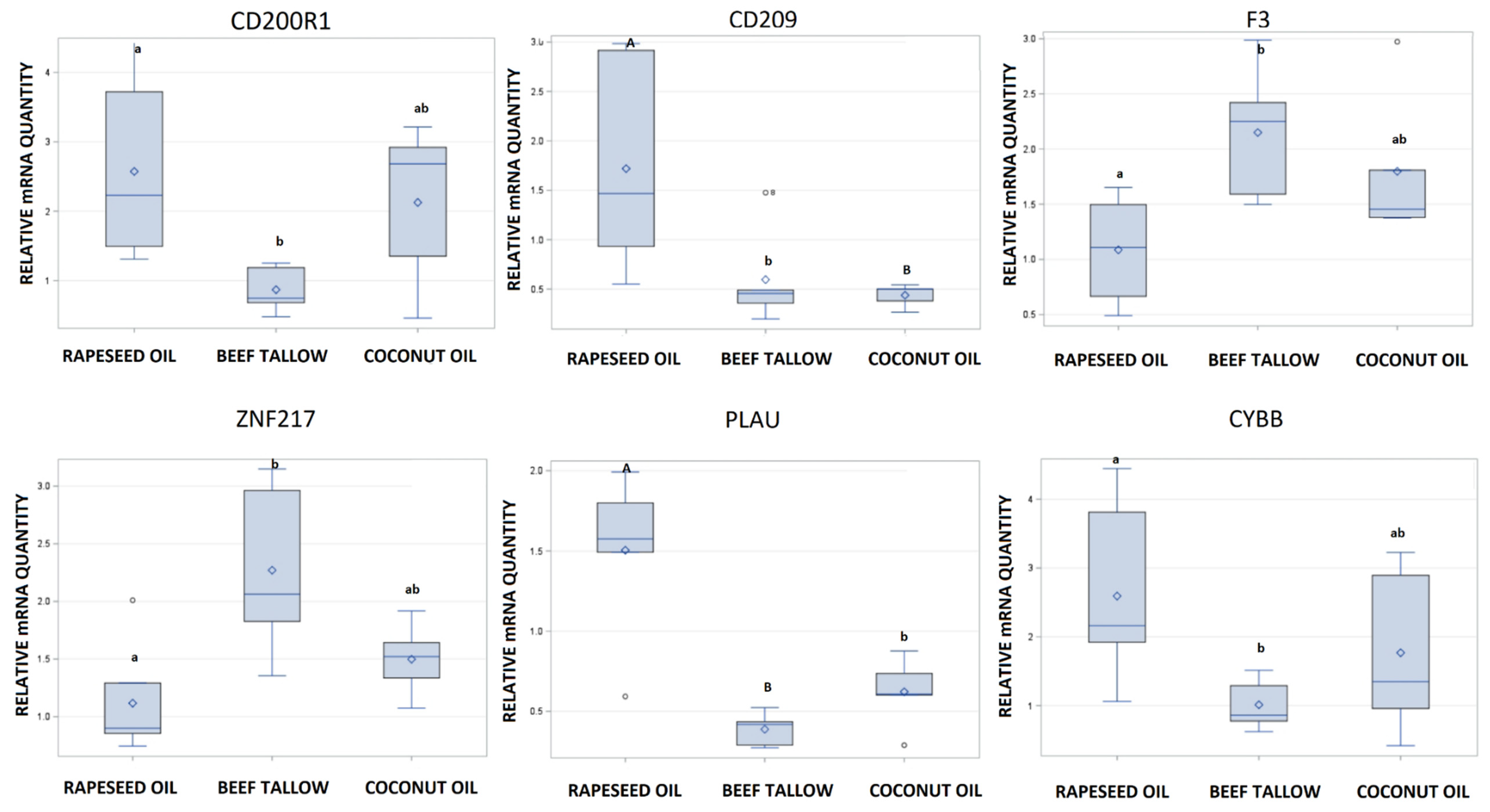
3.2. Validation of RNA-Sequencing by qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kinoshita, M.; Yokote, K.; Arai, H.; Iida, M.; Ishigaki, Y.; Ishibashi, S.; Umemoto, S.; Egusa, G.; Ohmura, H.; Okamura, T.; et al. Guidelines for Prevention of Atherosclerotic Cardiovascular Diseases 2017. Jpn. Atheroscler. Soc. 2018, 25, 846–984. [Google Scholar]
- Widmer, R.J.; Flammer, A.J.; Lerman, L.O.; Lerman, A. The Mediterranean diet, its components, and cardiovascular disease. Am. J. Med. 2015, 128, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Casas, R.; Sacanella, E.; Estruch, R. The Immune Protective Effect of the Mediterranean diet against chronic low-grade inflammatory diseases. Endocr. Metab. Immune Disord. Drug Targets 2016, 14, 245–254. [Google Scholar] [CrossRef]
- Hooper, L.; Summerbell, C.; Thompson, R.; Sills, D.; Roberts, F.; Moore, H.; Smith, G. Reduced or modified dietary fat for preventing cardiovascular disease. Cochrane Database Syst. Rev. 2011, 7, CD002137. [Google Scholar] [CrossRef]
- Fuentes, E.; Fuentes, F.; Vilahur, G.; Badimon, L.; Palomo, I. Mechanisms of chronic state of inflammation as mediators that link obese adipose tissue and metabolic syndrome. Mediat. Inflamm. 2013, 2013, 136584. [Google Scholar] [CrossRef] [PubMed]
- Cevallos, W.H.; Holmes, W.L.; Myers, R.N.; Smink, R.D. Swine in atherosclerosis research: Development of an experimental animal model and study of the effect of dietary fats on cholesterol metabolism. Atherosclerosis 1979, 34, 303–317. [Google Scholar] [CrossRef]
- Oczkowicz, M.; Szmatoła, T.; Świątkiewicz, M.; Pawlina-Tyszko, K.; Gurgul, A.; Ząbek, T. Corn dried distillers grains with solubles (cDDGS) in the diet of pigs change the expression of adipose genes that are potential therapeutic targets in metabolic and cardiovascular diseases. BMC Genom. 2018, 19, 864. [Google Scholar] [CrossRef]
- Świątkiewicz, M.; Oczkowicz, M.; Ropka-Molik, K.; Hanczakowska, E. The effect of dietary fatty acid composition on adipose tissue quality and expression of genes related to lipid metabolism in porcine livers. Animal Feed Sci. Technol. 2016, 216, 204–215. [Google Scholar] [CrossRef]
- Kopec, A.K.; Abrahams, S.R.; Thornton, S.; Palumbo, J.S.; Mullins, E.S.; Divanovic, S.; Weiler, H.; Owens, A.P.; Mackman, N.; Goss, A.; et al. Thrombin promotes diet-induced obesity through fibrin-driven inflammation. J. Clin. Investig. 2017, 127, 3152–3166. [Google Scholar] [CrossRef]
- Lagrange, J.; Didelot, M.; Mohamadi, A.; Walton, L.A.; Bloemen, S.; de Laat, B.; Louis, H.; Thornton, S.N.; Derby, B.; Sherratt, M.J.; et al. Implication of free fatty acids in thrombin generation and fibrinolysis in vascular Inflammation in zucker rats and evolution with aging. Front. Physiol. 2017, 8, e949. [Google Scholar] [CrossRef]
- Sacks, F.M.; Lichtenstein, A.H.; Wu, J.H.Y.; Appel, L.J.; Creager, M.A.; Kris-Etherton, P.M.; Miller, M.; Rimm, E.B.; Rudel, L.L.; Robinson, J.G.; et al. Dietary fats and cardiovascular disease: A presidential advisory from the American Heart Association. Circulation 2017, 18, e136. [Google Scholar] [CrossRef] [PubMed]
- Khaw, K.T.; Sharp, S.J.; Finikarides, L.; Afzal, I.; Lentjes, M.; Luben, R.; Forouhi, N.G. Randomised trial of coconut oil, olive oil or butter on blood lipids and other cardiovascular risk factors in healthy men and women. BMJ Open 2018, 8, e020167. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Jiang, H.; Wang, M.; Zhang, D. Meta-analysis of the association between urokinase-plasminogen activator gene rs2227564 polymorphism and Alzheimer’s disease. Am. J. Alzheimer’s Dis. Other Dement. 2013, 28, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.-M.; van Groen, T.; Katre, A.; Cao, D.; Kadisha, I.; Ballinger, C.; Wang, L.; Carroll, S.L.; Li, L. Knockout of plasminogen activator inhibitor 1 gene reduces amyloid β peptide burden in a mouse model of Alzheimer’s disease. Neurobiol. Aging 2011, 32, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.S.; Xin, J.; Hu, Y.; Zhang, L.; Wang, J. Analyzing the genes related to Alzheimer’s disease via a network and pathway-based approach. Alzheimer’s Res. Ther. 2017, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.A.; Donini, C.F.; Nguyen, N.T.; Lincet, H.; Vendrell, J.A. The dark side of ZNF217, a key regulator of tumorigenesis with powerful biomarker value. Oncotarget 2015, 6, 41566–41581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.C.; Zheng, L.Q.; Pan, L.J.; Guo, J.X.; Yang, G.S. ZNF217 is overexpressed and enhances cell migration and invasion in colorectal carcinoma. Asian Pac. J. Cancer Prev. 2015, 16, 2459–2463. [Google Scholar] [CrossRef]
- Shiraishi, R.; Fujise, T.; Kuroki, T.; Kakimoto, T.; Miao, L.; Sakata, Y.; Tsunada, S.; Noda, T.; Iwakiri, R.; Fujimoto, K. Long-term ingestion of reduced glutathione suppressed an accelerating effect of beef tallow diet on colon carcinogenesis in rats. J. Gastroenterol. 2009, 44, 1026–1035. [Google Scholar] [CrossRef]
- Saliba, W.; Rennert, H.S.; Gronich, N.; Gruber, S.B.; Rennert, G. Red meat and processed meat intake and risk of colorectal cancer: A population-based case-control study. Eur. J. Cancer Prev. 2019, 28, 287–293. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, T.; Wang, T.; Wang, B. Suppression of lncRNA-ATB prevents amyloid-β-inducedneurotoxicity in PC12 cells via regulating miR-200/ZNF217 axis. Biomed. Pharmacother. 2018, 108, 707–715. [Google Scholar] [CrossRef]
- Gaucher, C.; Boudier, A.; Bonetti, J.; Clarot, I.; Leroy, P.; Parent, M. Glutathione: Antioxidant Properties Dedicated to Nanotechnologies. Antioxidants 2018, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Saharan, S.; Mandal, P.K. The emerging role of glutathione in Alzheimer’s disease. J. Alzheimer’s Dis. 2014, 40, 519–529. [Google Scholar] [CrossRef]
- Teskey, G.; Abrahem, R.; Cao, R.; Gyurjian, K.; Islamoglu, H.; Lucero, M.; Martinez, A.; Paredes, E.; Salaiz, O.; Robinson, B.; et al. Glutathione as a marker for human disease. Adv. Clin. Chem. 2018, 87, 141–159. [Google Scholar]
- Homma, T.; Fujii, J. Application of glutathione as anti-oxidative and anti-aging drugs. Curr. Drug Metab. 2015, 16, 560–571. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.; Shen, W.L.; Shih, C.M.; Ho, K.H.; Cheng, C.H.; Lin, C.W.; Lee, C.C.; Liu, A.J.; Chen, K.C. The CHAC1-inhibited Notch3 pathway is involved in temozolomide-induced glioma cytotoxicity. Neuropharmacology 2017, 116, 300–314. [Google Scholar] [CrossRef] [PubMed]
- Dahmene, M.; Bérard, M.; Oueslati, A. Dissecting the Molecular Pathway Involved in PLK2 Kinase-mediated α-Synuclein-selective Autophagic Degradation. J. Biol. Chem. 2017, 292, 3919–3928. [Google Scholar] [CrossRef]
- Kofoed, R.H.; Zheng, J.; Ferreira, N.; Lykke-Andersen, S.; Salvi, M.; Betzer, C.; Reimer, L.; Jensen, T.H.; Fog, K.; Jensen, P.H. Polo-like kinase 2 modulates α-synuclein protein levels by regulating its mRNA production. Neurobiol. Dis. 2017, 106, 49–62. [Google Scholar] [CrossRef]
- Mahmoud, A.M.; Ali, M.M.; Miranda, E.R.; Mey, J.T.; Blackburn, B.K.; Haus, J.M.; Phillips, S.A. Nox2 contributes to hyperinsulinemia-induced redox imbalance and impaired vascular function. Redox Biol. 2017, 13, 288–300. [Google Scholar] [CrossRef]
- Violi, F.; Carnevale, R.; Loffredo, L.; Pignatelli, P.; Gallin, J.I. NADPH oxidase-2 and atherothrombosis: Insight from chronic granulomatous disease. Arterioscler. Thrombosis Vasc. Biol. 2017, 37, 218–225. [Google Scholar] [CrossRef]
- Kojima, Y.; Volkmer, J.P.; McKenna, K.; Civelek, M.; Lusis, A.J.; Miller, C.L.; Direnzo, D.; Nanda, V.; Ye, J.; Connolly, A.J.; et al. CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis. Nature 2016, 536, 86–90. [Google Scholar] [CrossRef]
- Dupré-Crochet, S.; Erard, M.; Nüβe, O. ROS production in phagocytes: Why, when, and where? J. Leukoc. Biol. 2013, 94, 657–670. [Google Scholar] [CrossRef]
- Masoud, R.; Bizouarn, T.; Houée-Levin, C. Cholesterol: A modulator of the phagocyte NADPH oxidase activity—A cell-free study. Redox Biol. 2014, 3, 16–24. [Google Scholar] [CrossRef]
- Denke, M.A. Role of beef and beef tallow, an enriched source of stearic acid, in a cholesterol-lowering diet. Am. J. Clin. Nutr. 1994, 60, 1044–1049. [Google Scholar] [CrossRef] [Green Version]
- Varnum, M.M.; Kiyota, T.; Ingraham, K.L.; Ikezu, S.; Ikezu, T. The anti-inflammatory glycoprotein, cd200, restores neurogenesis and enhances amyloid phagocytosis in a mouse model of alzheimer’s disease. Neurobiol. Aging 2015, 36, 2995–3007. [Google Scholar] [CrossRef] [Green Version]
- Atri, C.; Guerfali, F.Z.; Laouini, D. Role of human macrophage polarization in inflammation during infectious diseases. Int. J. Mol. Sci. 2018, 19, 1801. [Google Scholar] [CrossRef] [Green Version]
- Ingersoll, M.A. Sex differences shape the response to infectious diseases. PLoS Pathog. 2017, 13, 1006688. [Google Scholar] [CrossRef]
- Martin-Kleiner, I. BORIS in human cancers—A review. Eur. J. Cancer 2012, 48, 929–935. [Google Scholar] [CrossRef]
- Loukinov, D. Targeting CTCFL/BORIS for the immunotherapy of cancer. Cancer Immunol. Immunother. 2018, 67, 1955–1965. [Google Scholar] [CrossRef]
- Bolton, J.; Montastier, E.; Carayol, J.; Bonnel, S.; Mir, L.; Marques, M.A.; Astrup, A.; Saris, W.; Iacovoni, J.; Villa-Vialaneix, N.; et al. Molecular biomarkers for weight control in obese individuals subjected to a multiphase dietary intervention. J. Clin. Endocrinol. Metab. 2017, 102, 2751–2761. [Google Scholar] [CrossRef] [Green Version]
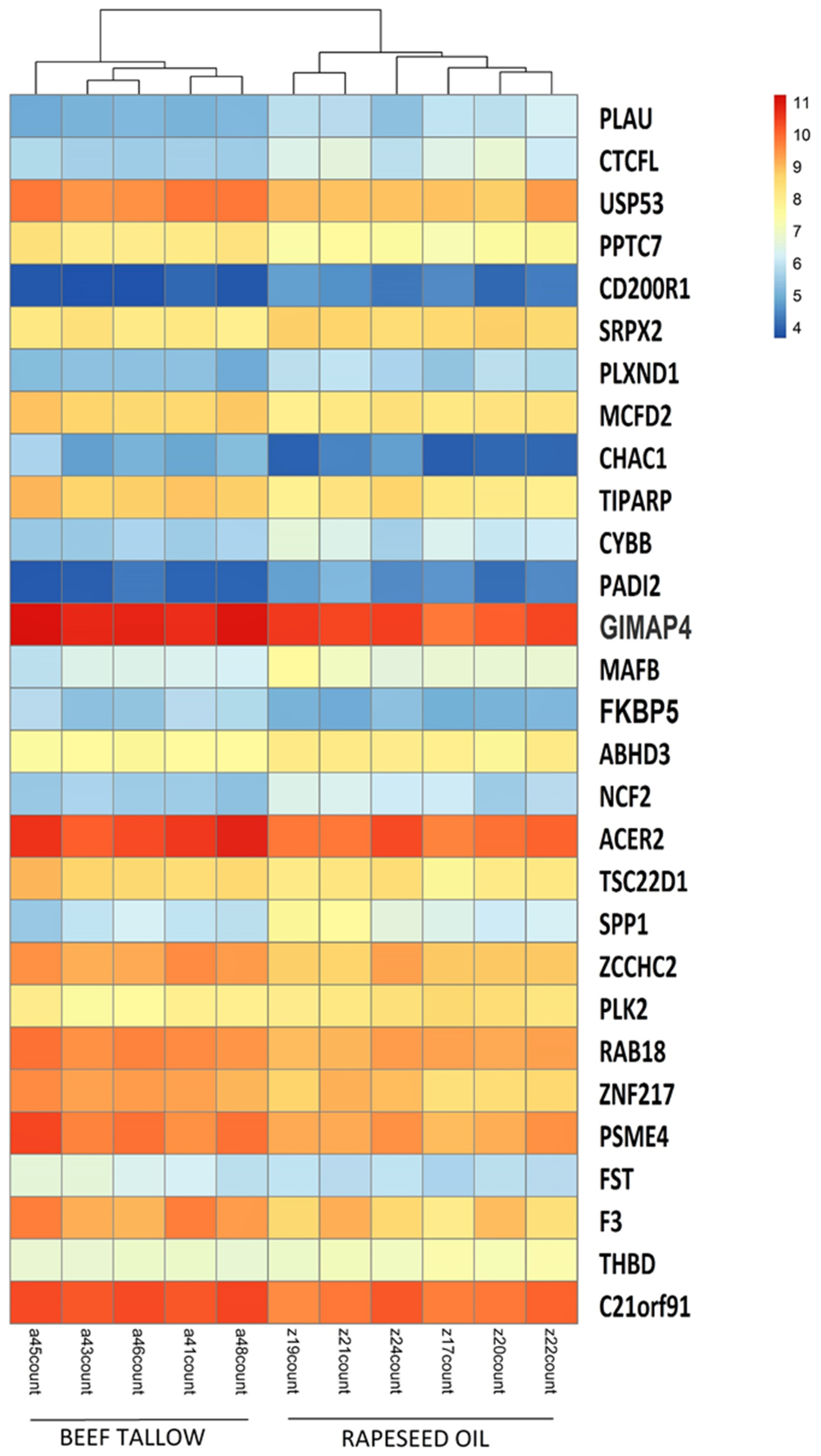

| Rapeseed Oil Group vs. Beef Tallow Group | ||||
|---|---|---|---|---|
| Ensemble ID | Base Mean | Fold Change | p Adjusted | Annotation |
| ENSSSCG00000010312 | 50.41389333 | −2.3625150 | 1.13E−06 | PLAU |
| ENSSSCG00000011925 | 19.57714649 | −2.2069404 | 0.000615734 | CD200R1 |
| ENSSSCG00000007505 | 72.29712452 | −2.2061260 | 5.68E−06 | CTCFL |
| ENSSSCG00000026478 | 23.05080559 | −1.9462864 | 0.024563058 | PADI2 |
| ENSSSCG00000009216 | 108.6558323 | −1.8750910 | 0.039998048 | SPP1 |
| ENSSSCG00000012482 | 48.83657882 | −1.8703811 | 0.003229498 | SRPX2 |
| ENSSSCG00000012229 | 65.93558552 | −1.8667126 | 0.007760072 | CYBB |
| ENSSSCG00000027826 | 111.3101373 | −1.7931908 | 0.026147634 | GIMAP4 |
| ENSSSCG00000015559 | 61.23829631 | −1.7816443 | 0.029057113 | NCF2 |
| ENSSSCG00000011592 | 355.9228073 | −1.5539315 | 0.003229498 | PLXND1 |
| ENSSSCG00000016925 | 283.1958901 | −1.5317017 | 0.04197925 | PLK2 |
| ENSSSCG00000007115 | 131.5794279 | −1.4363220 | 0.0489776 | THBD |
| ENSSSCG00000030898 | 234.0696594 | −1.4207480 | 0.026147634 | MAFB |
| ENSSSCG00000028655 | 698.3266126 | 1.4352279 | 0.04197925 | RAB18 |
| ENSSSCG00000021429 | 1122.072235 | 1.4361345 | 0.049411475 | C21orf91 |
| ENSSSCG00000004897 | 580.8230106 | 1.5350072 | 0.041653549 | ZCCHC2 |
| ENSSSCG00000021325 | 359.9374829 | 1.5520128 | 0.003437732 | MCFD2 |
| ENSSSCG00000009422 | 352.6278443 | 1.5912329 | 0.038291764 | TSC22D1 |
| ENSSSCG00000007484 | 548.372453 | 1.6200279 | 0.047085346 | ZNF217 |
| ENSSSCG00000026955 | 1269.38782 | 1.6265332 | 0.036381897 | ACER2 |
| ENSSSCG00000001549 | 1661.832725 | 1.6413418 | 0.026147634 | FKBP5 |
| ENSSSCG00000008412 | 812.4588633 | 1.6462623 | 0.047684732 | PSME4 |
| ENSSSCG00000016892 | 71.97654154 | 1.6559401 | 0.048396997 | FST |
| ENSSSCG00000022447 | 568.7564471 | 1.7298209 | 0.048396997 | F3 |
| ENSSSCG00000027646 | 386.8914671 | 1.7299692 | 0.007003091 | TIPARP |
| ENSSSCG00000028010 | 42.58112492 | 1.7664172 | 0.026147634 | ABHD3 |
| ENSSSCG00000009825 | 239.1954653 | 1.7791477 | 2.93E−05 | PPTC7 |
| ENSSSCG00000023174 | 679.5951006 | 1.8861755 | 5.68E−06 | USP53 |
| ENSSSCG00000004754 | 30.2851475 | 2.0652702 | 0.005616471 | CHAC1 |
| Rapeseed Oil Group vs. Coconut Oil Group | ||||
| ENSSSCG00000013579 | 105.0542477 | −2.1693396 | 0.000180283 | CD209 |
| ENSSSCG00000020756 | 25.934304 | 1.9393441 | 0.00330069 | FAM101A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oczkowicz, M.; Szmatoła, T.; Świątkiewicz, M. Source of Dietary Fat in Pig Diet Affects Adipose Expression of Genes Related to Cancer, Cardiovascular, and Neurodegenerative Diseases. Genes 2019, 10, 948. https://doi.org/10.3390/genes10120948
Oczkowicz M, Szmatoła T, Świątkiewicz M. Source of Dietary Fat in Pig Diet Affects Adipose Expression of Genes Related to Cancer, Cardiovascular, and Neurodegenerative Diseases. Genes. 2019; 10(12):948. https://doi.org/10.3390/genes10120948
Chicago/Turabian StyleOczkowicz, Maria, Tomasz Szmatoła, and Małgorzata Świątkiewicz. 2019. "Source of Dietary Fat in Pig Diet Affects Adipose Expression of Genes Related to Cancer, Cardiovascular, and Neurodegenerative Diseases" Genes 10, no. 12: 948. https://doi.org/10.3390/genes10120948
APA StyleOczkowicz, M., Szmatoła, T., & Świątkiewicz, M. (2019). Source of Dietary Fat in Pig Diet Affects Adipose Expression of Genes Related to Cancer, Cardiovascular, and Neurodegenerative Diseases. Genes, 10(12), 948. https://doi.org/10.3390/genes10120948





