Comparative Transcriptome Analysis of Gonads for the Identification of Sex-Related Genes in Giant Freshwater Prawns (Macrobrachium Rosenbergii) Using RNA Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
2.3. RNA Extraction, Library Preparation, and Transcriptome Sequencing
2.4. Transcriptome De Novo Assembly and Functional Annotation
2.5. Identification of Differentially Expressed Genes (DEGs)
2.6. Detection of Simple Sequence Repeats (SSRs)
2.7. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
3. Results
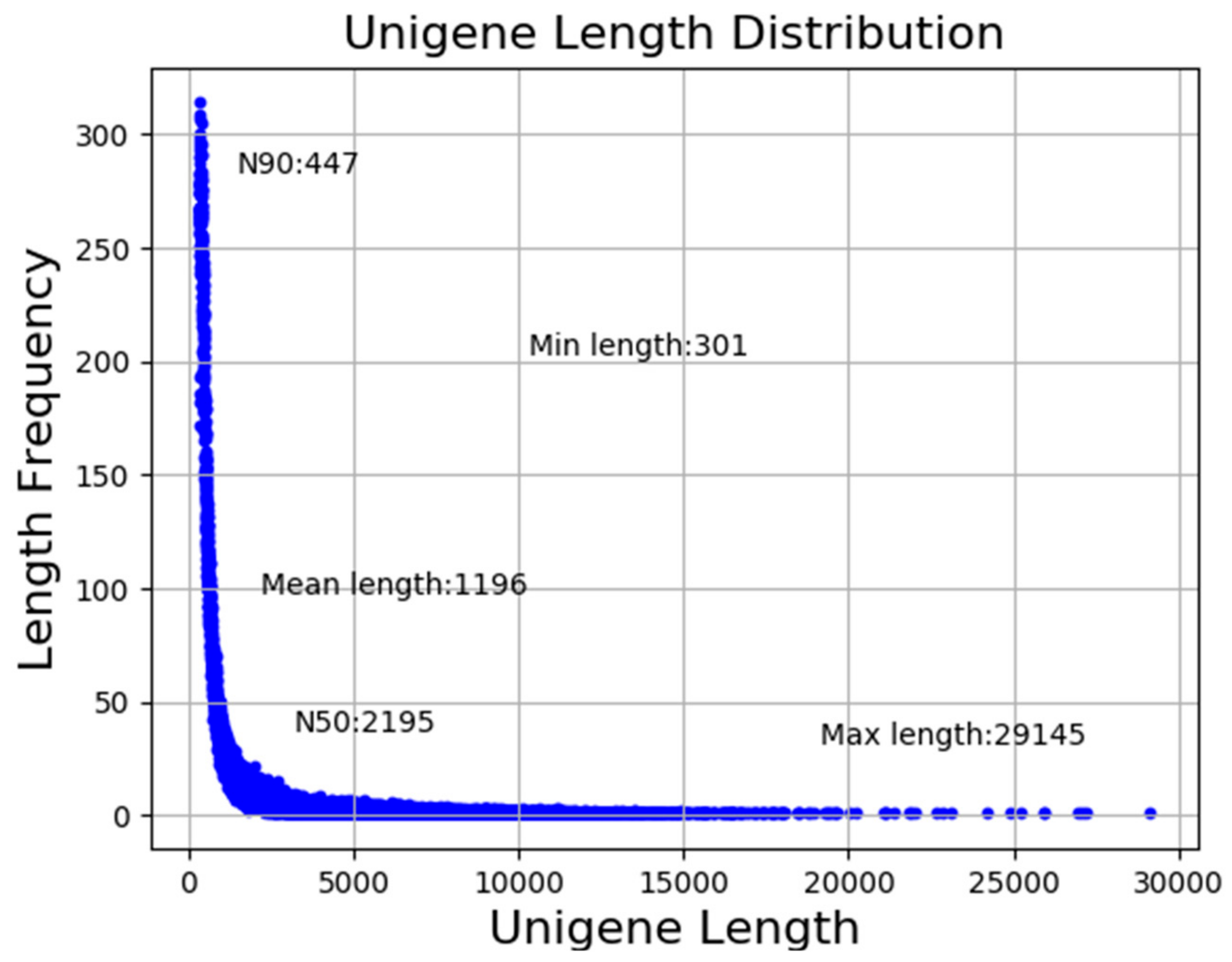
3.1. Sequencing Analysis and Transcriptome Assembly
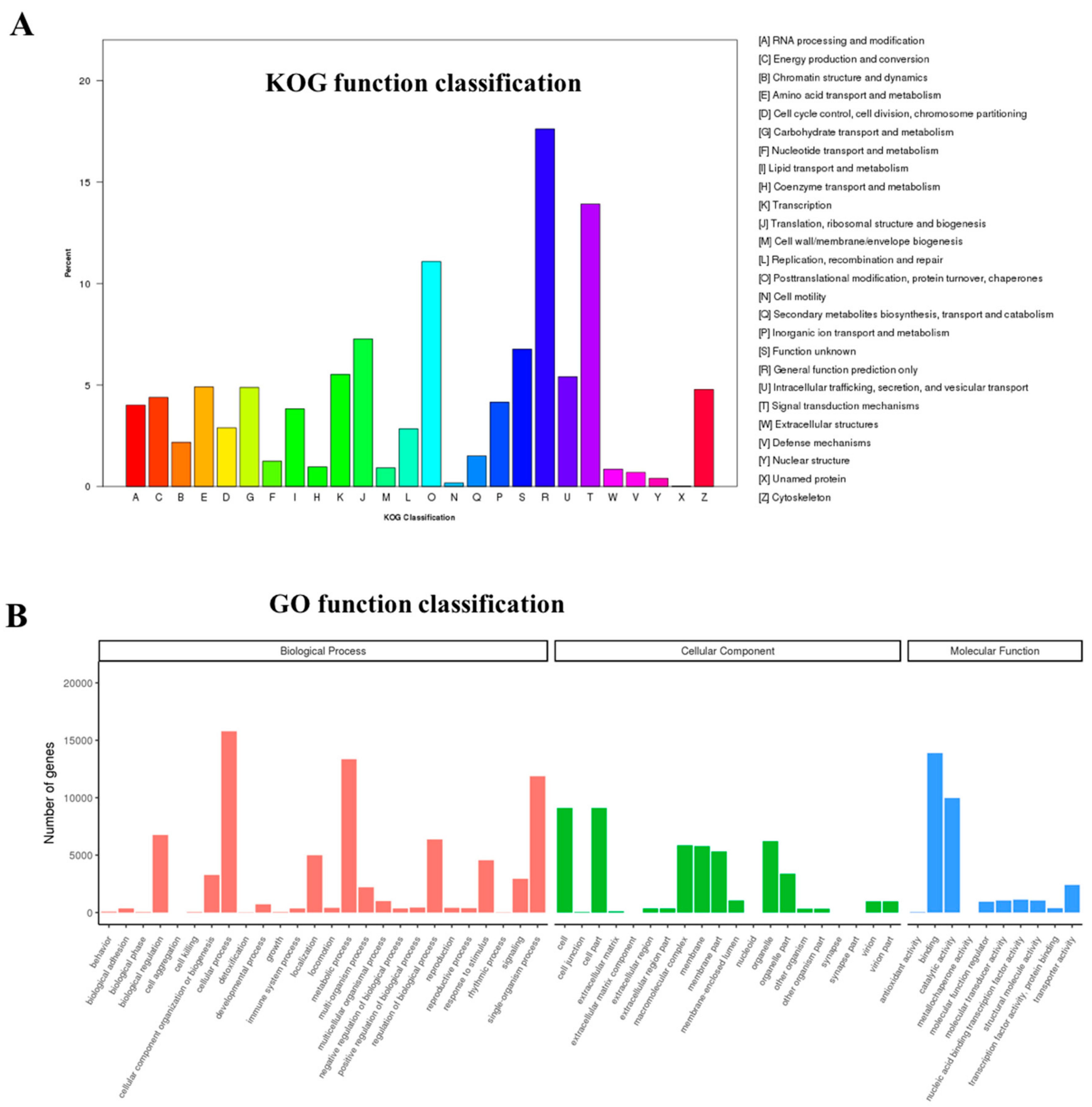
3.2. Sequence Annotation
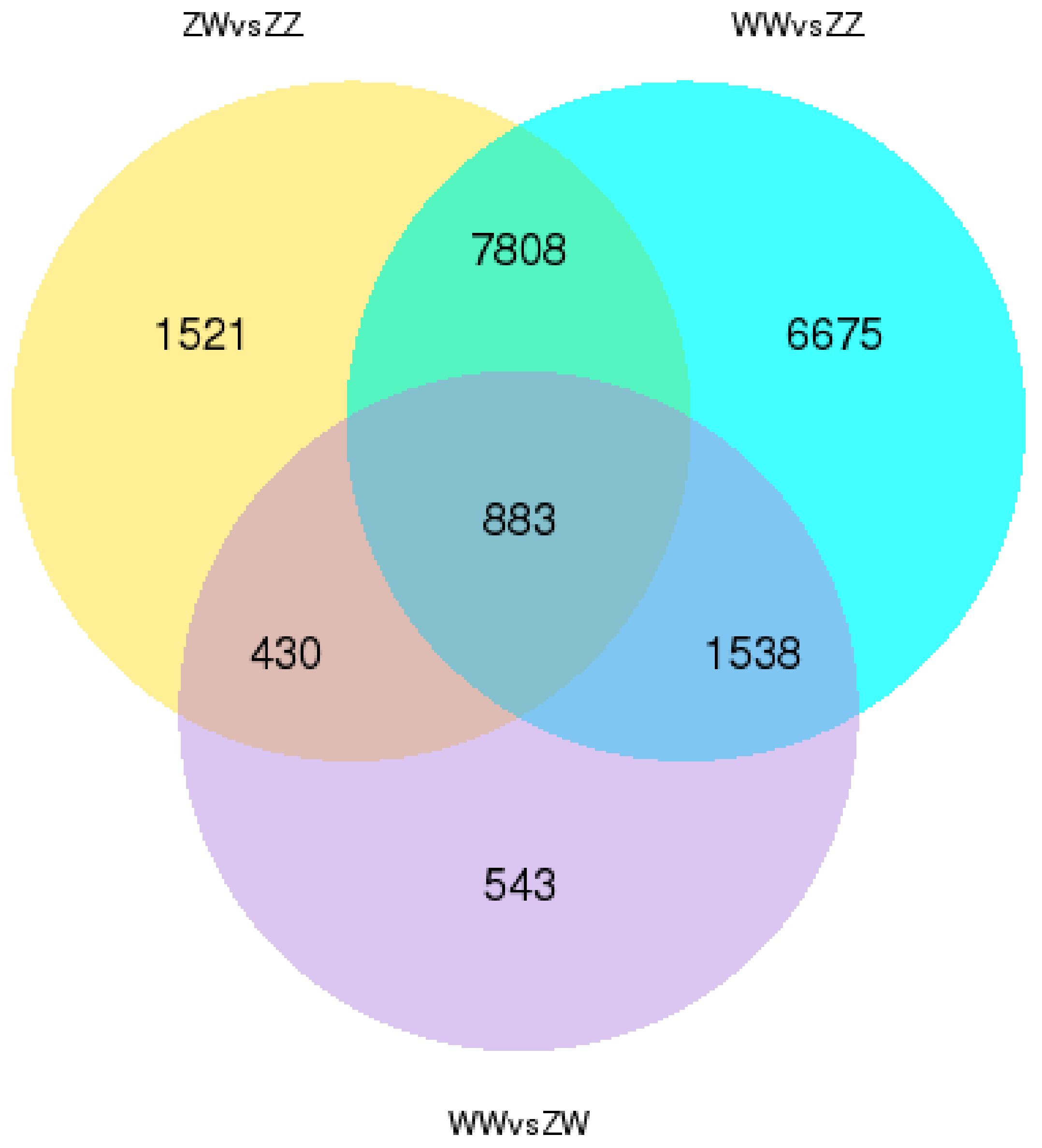
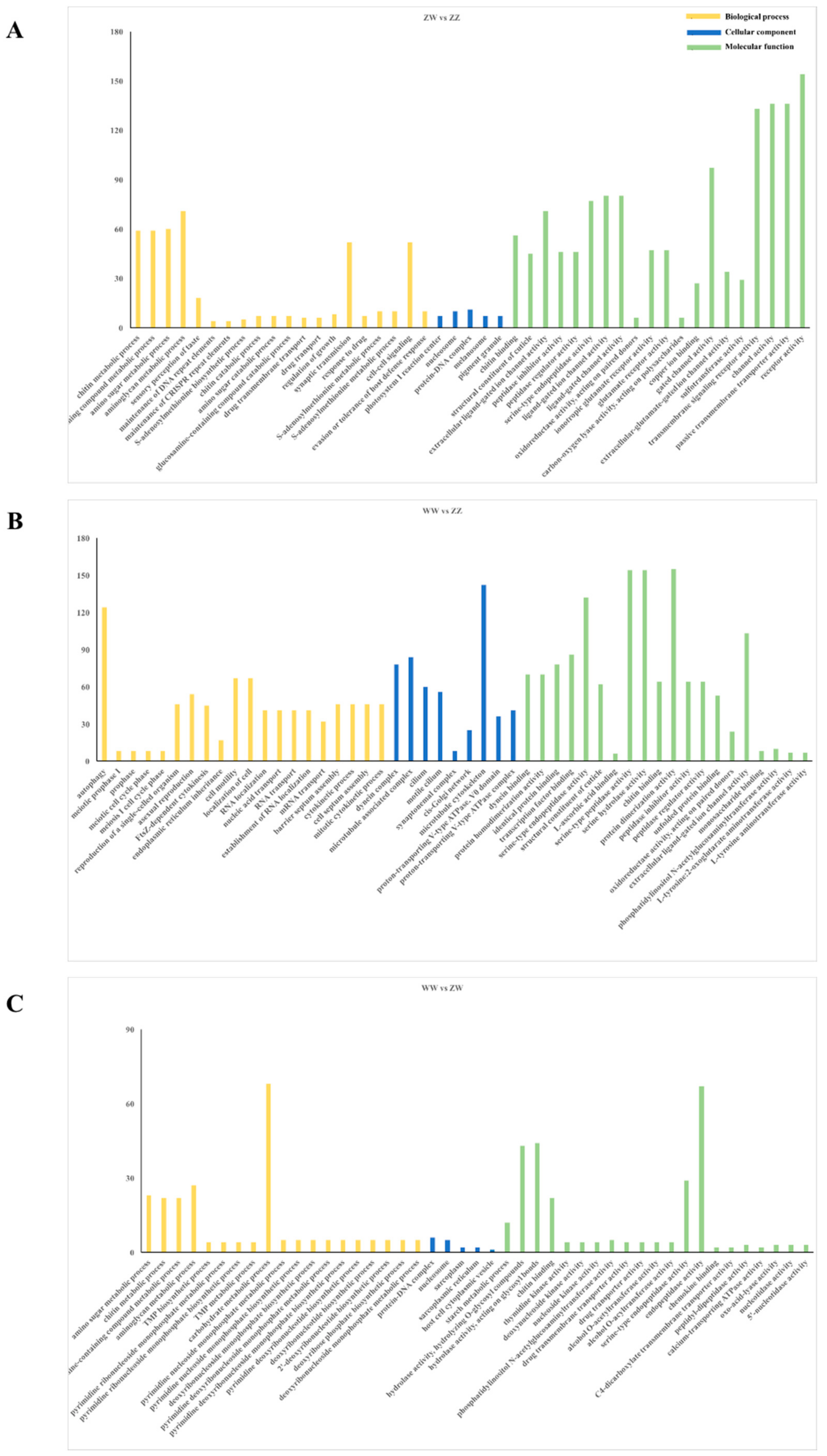
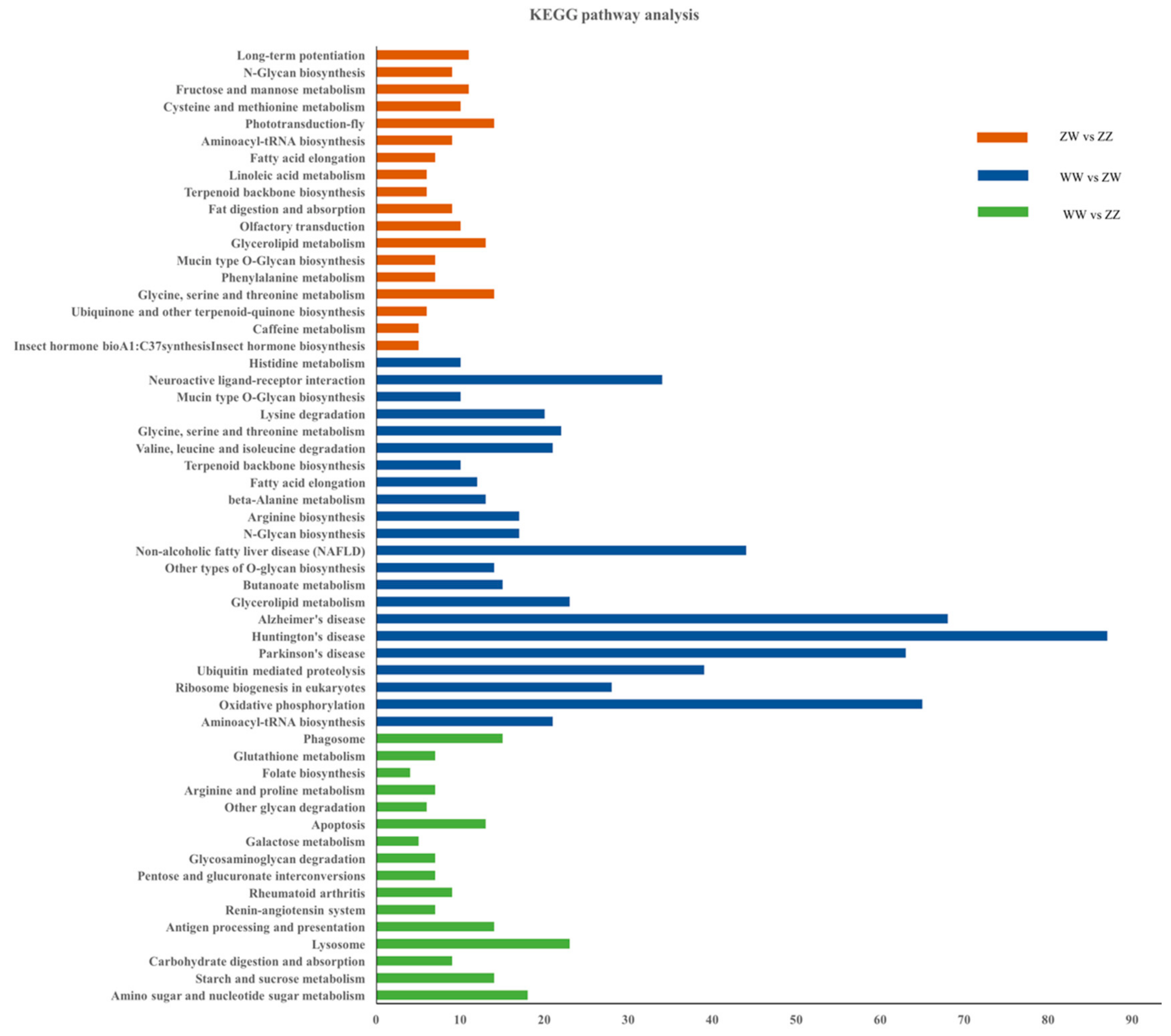
3.3. DEGs Identification and Enrichment Analysis
3.4. Genes of Interest Related to Sex
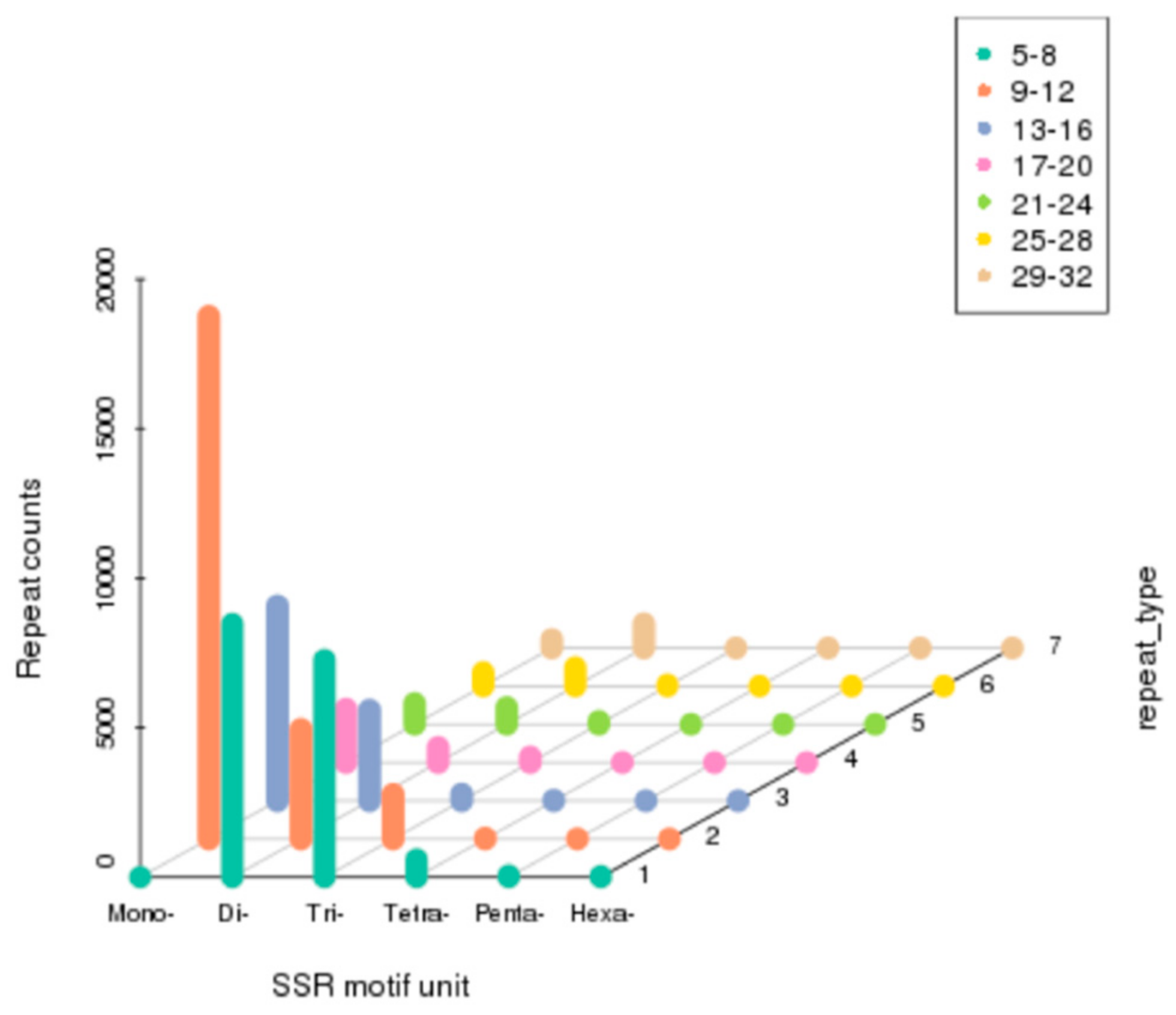
3.5. Discovery of Simple Sequence Repeats (SSRs)
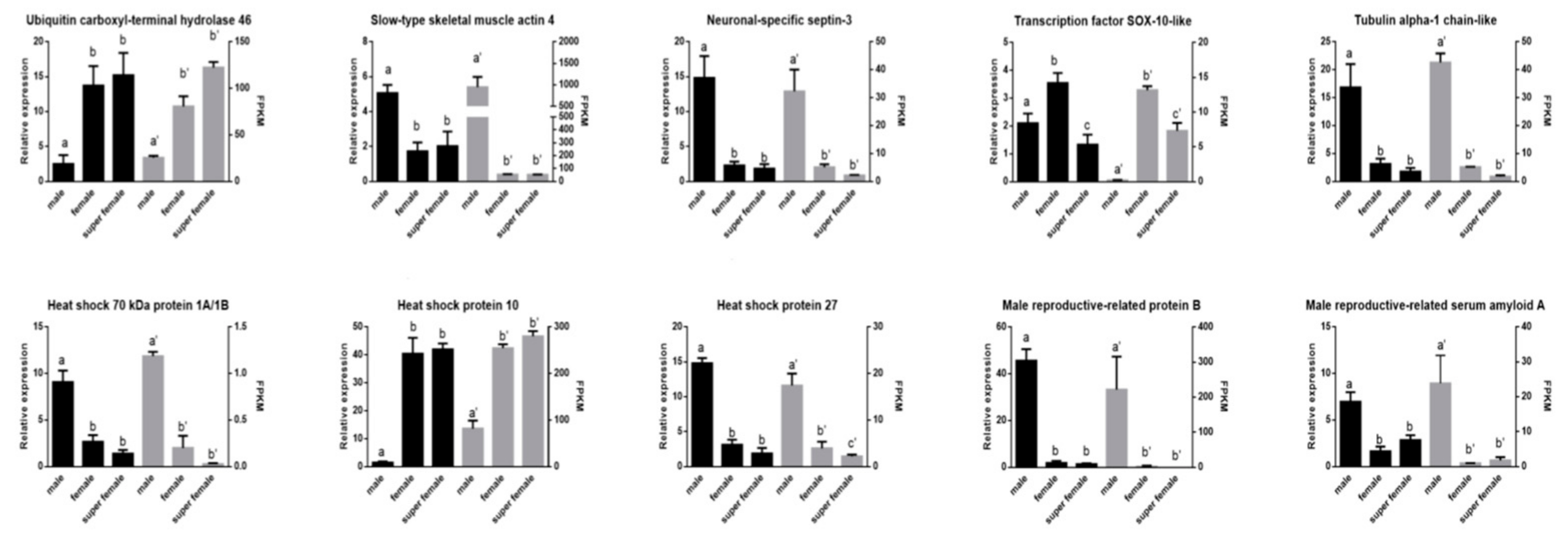
3.6. The qRT-PCR Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Aflalo, E.D.; Hoang, T.T.T.; Nguyen, V.; Lam, Q.; Nguyen, D.; Trinh, Q.; Raviv, S.; Sagi, A. A novel two-step procedure for mass production of all-male populations of the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 2006, 256, 468–478. [Google Scholar] [CrossRef]
- Malecha, S. The case for all-female freshwater prawn, M acrobrachium rosenbergii (D e M an), culture. Aquac. Res. 2012, 43, 1038–1048. [Google Scholar] [CrossRef]
- Waiho, K.; Shi, X.; Fazhan, H.; Li, S.K.; Zhang, Y.L.; Zheng, H.P.; Liu, W.H.; Fang, S.B.; Ikhwanuddin, M.; Ma, H.Y. High-Density Genetic Linkage Maps Provide Novel Insights Into ZW/ZZ Sex Determination System and Growth Performance in Mud Crab (Scylla paramamosain). Front. Genet. 2019, 10, 298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benzie, J.A.; Kenway, M.; Ballment, E. Growth of Penaeus monodon× Penaeus esculentus tiger prawn hybrids relative to the parental species. Aquaculture 2001, 193, 227–237. [Google Scholar] [CrossRef]
- Coman, F.E.; Sellars, M.J.; Norris, B.J.; Coman, G.J.; Preston, N.P. The effects of triploidy on Penaeus (Marsupenaeus) japonicus (Bate) survival, growth and gender when compared to diploid siblings. Aquaculture 2008, 276, 50–59. [Google Scholar] [CrossRef]
- Malecha, S.R.; Nevin, P.A.; Ha, P.; Barck, L.E.; Lamadrid-Rose, Y.; Masuno, S.; Hedgecock, D. Sex-ratios and sex-determination in progeny from crosses of surgically sex-reversed freshwater prawns, Macrobrachium rosenbergii. Aquaculture 1992, 105, 201–218. [Google Scholar] [CrossRef]
- Sagi, A. м-Pah Growth, maturation and progeny of sex-reversed. World Aquac. 1990, 21, 87. [Google Scholar]
- Ventura, T.; Aflalo, E.; Weil, S.; Kashkush, K.; Sagi, A. Isolation and characterization of a female-specific DNA marker in the giant freshwater prawn Macrobrachium rosenbergii. Heredity 2011, 107, 456. [Google Scholar] [CrossRef]
- Jiang, X.H.; Qiu, G.F. Female-only sex-linked amplified fragment length polymorphism markers support ZW/ZZ sex determination in the giant freshwater prawn Macrobrachium rosenbergii. Anim. Genet. 2013, 44, 782–785. [Google Scholar] [CrossRef]
- Ma, K.-Y.; Yu, S.-H.; Du, Y.-X.; Feng, S.-Q.; Qiu, L.-J.; Ke, D.-Y.; Luo, M.-Z.; Qiu, G.-F. Construction of a genomic bacterial artificial chromosome (BAC) library for the prawn Macrobrachium rosenbergii and initial analysis of ZW chromosome-derived BAC inserts. Mar. Biotechnol. 2019, 21, 206–216. [Google Scholar] [CrossRef]
- Cao, J.-X.; Yin, G.-L.; Yang, W.-J. Identification of a novel male reproduction-related gene and its regulated expression patterns in the prawn, Macrobrachium rosenbergii. Peptides 2006, 27, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.-X.; Dai, J.-Q.; Dai, Z.-M.; Yin, G.-L.; Yang, W.-J. A male reproduction-related kazal-type peptidase inhibitor gene in the prawn, Macrobrachium rosenbergii: Molecular characterization and expression patterns. Mar. Biotechnol. 2007, 9, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Raviv, S.; Glazer, L.; Sagi, A. Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 2008, 150, 1278–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phoungpetchara, I.; Tinikul, Y.; Poljaroen, J.; Changklungmoa, N.; Siangcham, T.; Sroyraya, M.; Chotwiwatthanakun, C.; Vanichviriyakit, R.; Hanna, P.J.; Sobhon, P. Expression of the male reproduction-related gene (Mar-Mrr) in the spermatic duct of the giant freshwater prawn, Macrobrachium rosenbergii. Cell Tissue Res. 2012, 348, 609–623. [Google Scholar] [CrossRef]
- Sharabi, O.; Manor, R.; Weil, S.; Aflalo, E.; Lezer, Y.; Levy, T.; Aizen, J.; Ventura, T.; Mather, P.; Khalaila, I. Identification and characterization of an insulin-like receptor involved in crustacean reproduction. Endocrinology 2015, 157, 928–941. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Luo, B.Y.; Feng, J.B.; Zhou, L.X.; Ma, K.Y.; Qiu, G.F. Identification and profiling of microRNAs during gonadal development in the giant freshwater prawn Macrobrachium rosenbergii. Sci. Rep. 2019, 9, 2406. [Google Scholar] [CrossRef]
- Jin, S.; Fu, H.; Sun, S.; Jiang, S.; Xiong, Y.; Gong, Y.; Qiao, H.; Zhang, W.; Wu, Y. Integrated analysis of microRNA and mRNA expression profiles during the sex-differentiation sensitive period in oriental river prawn, Macrobrachium nipponense. Sci. Rep. 2017, 7, 12011. [Google Scholar] [CrossRef] [Green Version]
- Tian, C.; Li, Z.; Dong, Z.; Huang, Y.; Du, T.; Chen, H.; Jiang, D.; Deng, S.; Zhang, Y.; Wanida, S.; et al. Transcriptome Analysis of Male and Female Mature Gonads of Silver Sillago (Sillago sihama). Genes 2019, 10, 129. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.; You, F.; Wang, L.; Weng, S.; Wu, Z.; Hu, J.; Zou, Y.; Tan, X.; Zhang, P. Gonadal transcriptome analysis of male and female olive flounder (Paralichthys olivaceus). BioMed Res. Int. 2014, 2014, 291067. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Xu, D.; Ye, H.; Yue, H.; Ooka, S.; Kondo, H.; Yazawa, R.; Takeuchi, Y. Gonadal Transcriptome Analysis of Pacific Abalone Haliotis discus discus: Identification of Genes Involved in Germ Cell Development. Mar. Biotechnol. 2018, 20, 467–480. [Google Scholar] [CrossRef]
- Peng, J.; Wei, P.; Zhang, B.; Zhao, Y.; Zeng, D.; Chen, X.; Li, M.; Chen, X. Gonadal transcriptomic analysis and differentially expressed genes in the testis and ovary of the Pacific white shrimp (Litopenaeus vannamei). BMC Genom. 2015, 16, 1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.-J.; Zhou, Y.-L.; Wang, Z.-W.; Li, G.-H.; Jin, F.-P.; Cui, L.-L.; Gao, H.-T.; Li, X.-P.; Zhou, L.; Gui, J.-F. Comparative Transcriptome Analysis Reveals Differentially Expressed Genes and Signaling Pathways Between Male and Female Red-Tail Catfish (Mystus wyckioides). Mar. Biotechnol. 2019, 21, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Y.; Gong, Q.; Lai, J.; Song, M.; Du, J.; Deng, X. Gonadal transcriptome sequencing of the critically endangered Acipenser dabryanus to discover candidate sex-related genes. PeerJ 2018, 6, e5389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, T.; Rosen, O.; Eilam, B.; Azulay, D.; Aflalo, E.D.; Manor, R.; Shechter, A.; Sagi, A. A Single Injection of Hypertrophied Androgenic Gland Cells Produces All-Female Aquaculture. Mar. Biotechnol. 2016, 18, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Ai, C.; Kong, L. CGPS: A machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. J. Genet. Genom. 2018, 45, 489–504. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.-M.; Tao, Z.; Shen, C.; Qian, D.; Wang, C.-L.; Zhou, Q.-C.; Jin, S. β-actin gene expression is variable among individuals and not suitable for normalizing mRNA levels in Portunus trituberculatus. Fish Shellfish. Immunol. 2018, 81, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V.; Tellabati, M.; Nelli, R.K.; White, G.A.; Perez, B.B.; Sebastian, S.; Slomka, M.J.; Brookes, S.M.; Brown, I.H.; Dunham, S.P. 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol. J. 2012, 9, 230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sagi, A.; Ra’anan, Z.; Cohen, D.; Wax, Y. Production of Macrobrachium rosenbergii in monosex populations: Yield characteristics under intensive monoculture conditions in cages. Aquaculture 1986, 51, 265–275. [Google Scholar] [CrossRef]
- Cao, J.; Wu, L.; Jin, M.; Li, T.; Hui, K.; Ren, Q. Transcriptome profiling of the Macrobrachium rosenbergii lymphoid organ under the white spot syndrome virus challenge. Fish Shellfish. Immunol. 2017, 67, 27–39. [Google Scholar] [CrossRef]
- Nguyen Thanh, H.; Zhao, L.; Liu, Q. De novo transcriptome sequencing analysis and comparison of differentially expressed genes (DEGs) in Macrobrachium rosenbergii in China. PLoS ONE 2014, 9, e109656. [Google Scholar] [CrossRef]
- Jung, H.; Yoon, B.-H.; Kim, W.-J.; Kim, D.-W.; Hurwood, D.; Lyons, R.; Salin, K.; Kim, H.-S.; Baek, I.; Chand, V. Optimizing hybrid de novo transcriptome assembly and extending genomic resources for giant freshwater prawns (Macrobrachium rosenbergii): The identification of genes and markers associated with reproduction. Int. J. Mol. Sci. 2016, 17, 690. [Google Scholar] [CrossRef]
- Bose, U.; Kruangkum, T.; Wang, T.; Zhao, M.; Ventura, T.; Mitu, S.A.; Hodson, M.P.; Shaw, P.N.; Sobhon, P.; Cummins, S.F. Biomolecular changes that occur in the antennal gland of the giant freshwater prawn (Machrobrachium rosenbergii). PLoS ONE 2017, 12, e0177064. [Google Scholar] [CrossRef] [Green Version]
- Huntriss, J.; Lu, J.; Hemmings, K.; Bayne, R.; Anderson, R.; Rutherford, A.; Balen, A.; Elder, K.; Picton, H.M. Isolation and expression of the human gametocyte-specific factor 1 gene (GTSF1) in fetal ovary, oocytes, and preimplantation embryos. J. Assist. Reprod. Genet. 2017, 34, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Takemoto, N.; Yoshimura, T.; Miyazaki, S.; Tashiro, F.; Miyazaki, J.-I. Gtsf1l and Gtsf2 are specifically expressed in gonocytes and spermatids but are not essential for spermatogenesis. PLoS ONE 2016, 11, e0150390. [Google Scholar] [CrossRef]
- Jiang, D.N.; Yang, H.H.; Li, M.H.; Shi, H.J.; Zhang, X.B.; Wang, D.S. gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 2016, 83, 497–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, W.; Chen, J.; Tan, D.; Yang, J.; Sun, L.; Wei, J.; Conte, M.A.; Kocher, T.D.; Wang, D. Transcriptome display during tilapia sex determination and differentiation as revealed by RNA-Seq analysis. BMC Genom. 2018, 19, 363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neill, J.D.; Duck, L.W.; Sellers, J.C.; Musgrove, L.C. A gonadotropin-releasing hormone (GnRH) receptor specific for GnRH II in primates. Biochem. Biophys. Res. Commun. 2001, 282, 1012–1018. [Google Scholar] [CrossRef] [PubMed]
- Millar, R.; Lowe, S.; Conklin, D.; Pawson, A.; Maudsley, S.; Troskie, B.; Ott, T.; Millar, M.; Lincoln, G.; Sellar, R.; et al. A novel mammalian receptor for the evolutionarily conserved type II GnRH. Proc. Natl. Acad. Sci. USA 2001, 98, 9636–9641. [Google Scholar] [CrossRef] [Green Version]
- Desaulniers, A.T.; Cederberg, R.A.; Lents, C.A.; White, B.R. Expression and Role of Gonadotropin-Releasing Hormone 2 and Its Receptor in Mammals. Front. Endocrinol. 2017, 8, 269. [Google Scholar] [CrossRef] [Green Version]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Rosen, O.; Sagi, A. Timing sexual differentiation: Full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol. Reprod. 2012, 86, 91–96. [Google Scholar] [CrossRef]
- Fu, C.; Zeng, Q.; Li, F.; Wang, H.; Sun, J.; Wang, H. Comparative Transcriptome Analysis Reveals Related Regulatory Mechanisms of Androgenic Gland in Eriocheir sinensis. BioMed. Res. Int. 2017, 2017, 4956216. [Google Scholar] [CrossRef] [Green Version]
- Von Philipsborn, A.C.; Jörchel, S.; Tirian, L.; Demir, E.; Morita, T.; Stern, D.L.; Dickson, B.J. Cellular and behavioral functions of fruitless isoforms in Drosophila courtship. Curr. Biol. 2014, 24, 242–251. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Hui, M.; Cui, Z.; Luo, D.; Song, C.; Li, Y.; Liu, L. Comparative transcriptome analysis reveals sex-biased gene expression in juvenile Chinese mitten crab Eriocheir sinensis. PLoS ONE 2015, 10, e0133068. [Google Scholar] [CrossRef] [Green Version]
- Forstbauer, L.M.; Brockschmidt, F.F.; Moskvina, V.; Herold, C.; Redler, S.; Herzog, A.; Hillmer, A.M.; Meesters, C.; Heilmann, S.; Albert, F. Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for alopecia areata. Eur. J. Hum. Genet. 2012, 20, 326. [Google Scholar] [CrossRef]
- Ge, L.-Q.; Xia, T.; Huang, B.; Song, Q.-S.; Zhang, H.-W.; Stanley, D.; Yang, G.-Q.; Wu, J.-C. Suppressing male spermatogenesis-associated protein 5-like gene expression reduces vitellogenin gene expression and fecundity in Nilaparvata lugens Stål. Sci. Rep. 2016, 6, 28111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, J.; Wang, Y.-L.; Setsuie, R.; Sekiguchi, S.; Sakurai, M.; Sato, Y.; Lee, W.-W.; Ishii, Y.; Kyuwa, S.; Noda, M. Developmental regulation of ubiquitin C-terminal hydrolase isozyme expression during spermatogenesis in mice. Biol. Reprod. 2004, 71, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J. The new function of two ubiquitin C-terminal hydrolase isozymes as reciprocal modulators of germ cell apoptosis. Exp. Anim. 2007, 56, 71–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warren, J.T.; Petryk, A.; Marqués, G.; Jarcho, M.; Parvy, J.-P.; Dauphin-Villemant, C.; O’Connor, M.B.; Gilbert, L.I. Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2002, 99, 11043–11048. [Google Scholar] [CrossRef] [Green Version]
- Ranson, H.; Claudianos, C.; Ortelli, F.; Abgrall, C.; Hemingway, J.; Sharakhova, M.V.; Unger, M.F.; Collins, F.H.; Feyereisen, R. Evolution of supergene families associated with insecticide resistance. Science 2002, 298, 179–181. [Google Scholar] [CrossRef]
- Chávez, V.M.; Marqués, G.; Delbecque, J.P.; Kobayashi, K.; Hollingsworth, M.; Burr, J.; Natzle, J.E.; O’Connor, M.B. The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development 2000, 127, 4115–4126. [Google Scholar]
- Zhu, J.; Chen, Y.; Zhao, Q.; Tang, S.; Huang, J.; Shen, X. Expression profile of several genes on ecdysteroidogenic pathway related to diapause in pupal stage of Bombyx mori bivoltine strain. Gene 2019, 707, 109–116. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schütz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835. [Google Scholar] [CrossRef] [Green Version]
- Uhlenhaut, N.H.; Jakob, S.; Anlag, K.; Eisenberger, T.; Sekido, R.; Kress, J.; Treier, A.C.; Klugmann, C.; Klasen, C.; Holter, N.I.; et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 2009, 139, 1130–1142. [Google Scholar] [CrossRef] [Green Version]
- Pisarska, M.D.; Barlow, G.; Kuo, F.T. Minireview: Roles of the forkhead transcription factor FOXL2 in granulosa cell biology and pathology. Endocrinology 2011, 152, 1199–1208. [Google Scholar] [CrossRef] [Green Version]
- Picard, D. Chaperoning steroid hormone action. Trends Endocrinol. Metab. TEM 2006, 17, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Kohno, S.; Katsu, Y.; Urushitani, H.; Ohta, Y.; Iguchi, T.; Guillette, L.J., Jr. Potential contributions of heat shock proteins to temperature-dependent sex determination in the American alligator. Sex. Dev. 2010, 4, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Kurata, S.; Fujimoto, H.; Hoshi, M. Haploid specific activations of protamine 1 and hsc70t genes in mouse spermatogenesis. Biochim. Biophys. Acta 1993, 1174, 274–278. [Google Scholar] [CrossRef]
- Dai, Z.-M.; Zhu, X.-J.; Yang, W.-J. Full-length normalization subtractive hybridization: A novel method for generating differentially expressed cDNAs. Mol. Biotechnol. 2009, 43, 257. [Google Scholar] [CrossRef] [PubMed]
- Sroyraya, M.; Hanna, P.J.; Changklungmoa, N.; Senarai, T.; Siangcham, T.; Tinikul, Y.; Sobhon, P. Expression of the male reproduction-related gene in spermatic ducts of the blue swimming crab, Portunus pelagicus, and transfer of modified protein to the sperm acrosome. Microsc. Res. Tech. 2013, 76, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.M.; Mathialagan, N.; Duffy, J.Y.; Smith, G.W. Regulation and regulatory role of proteinase inhibitors. Crit. Rev. Eukaryot. Gene Expr. 1995, 5, 385–436. [Google Scholar] [CrossRef] [PubMed]
- Coleman, S.; DrÄhn, B.; Petersen, G.; Stolorov, J.; Kraus, K. A Drosophila male accessory gland protein that is a member of the serpin superfamily of proteinase inhibitors is transferred to females during mating. Insect Biochem. Mol. Biol. 1995, 25, 203–207. [Google Scholar] [CrossRef]
- Li, Y.; Qian, Y.-Q.; Ma, W.-M.; Yang, W.-J. Inhibition mechanism and the effects of structure on activity of male reproduction-related peptidase inhibitor Kazal-type (MRPINK) of Macrobrachium rosenbergii. Mar. Biotechnol. 2009, 11, 252–259. [Google Scholar] [CrossRef]
- Qian, Y.Q.; Li, Y.; Yang, F.; Yu, Y.Q.; Yang, J.S.; Yang, W.J. Two Kazal-type protease inhibitors from Macrobrachium nipponense and Eriocheir sinensis: Comparative analysis of structure and activities. Fish Shellfish. Immunol. 2012, 32, 446–458. [Google Scholar] [CrossRef]
- Li, M.; Yang, H.; Zhao, J.; Fang, L.; Shi, H.; Li, M.; Sun, Y.; Zhang, X.; Jiang, D.; Zhou, L.; et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 2014, 197, 591–599. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Sun, Y.; Zhao, J.; Shi, H.; Zeng, S.; Ye, K.; Jiang, D.; Zhou, L.; Sun, L.; Tao, W. A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile tilapia, Oreochromis niloticus. PLoS Genet. 2015, 11, e1005678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- See, L.M.; Hassan, R.; Tan, S.G.; Bhassu, S. Novel polymorphic microsatellite DNA markers from Malaysian giant freshwater prawn, Macrobrachium rosenbergii. Genetika 2011, 47, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Jin, M.; Ren, Q. Transcriptome analysis of Macrobrachium rosenbergii intestines under the white spot syndrome virus and poly (I:C) challenges. PLoS ONE 2018, 13, e0204626. [Google Scholar] [CrossRef] [PubMed]
| Sample | Body Weight (g) | Body Length (cm) |
|---|---|---|
| ZW | 9.61 ± 0.44 | 6.31 ± 0.16 |
| WW | 9.65 ± 0.28 | 7.62 ± 0.12 |
| ZZ | 13.09 ± 0.32 | 7.02 ± 0.11 |
| Sample | Raw Reads | Clean Reads | Clean Bases | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|
| ZW_1 | 52,214,594 | 47,435,802 | 7.12G | 97.84 | 93.90 | 42.00 |
| ZW_2 | 57,714,006 | 52,410,538 | 7.86G | 97.81 | 93.82 | 41.88 |
| ZW_3 | 52,276,492 | 47,066,162 | 7.06G | 97.77 | 93.74 | 41.75 |
| WW_1 | 60,924,940 | 54,729,552 | 8.21G | 98.04 | 94.31 | 41.81 |
| WW_2 | 62,970,576 | 57,154,534 | 8.57G | 97.99 | 94.18 | 41.85 |
| WW_3 | 66,416,082 | 60,480,354 | 9.07G | 97.94 | 94.10 | 41.69 |
| ZZ_1 | 54,655,372 | 49,757,368 | 7.46G | 97.68 | 93.52 | 41.85 |
| ZZ_2 | 57,466,470 | 52,158,488 | 7.82G | 97.71 | 93.62 | 41.93 |
| ZZ_3 | 52,503,322 | 47,730,328 | 7.16G | 97.91 | 94.07 | 43.53 |
| Database | Number of Unigenes | Percentage (%) |
|---|---|---|
| Nr | 23,483 | 20.36 |
| Nt | 6,556 | 5.68 |
| KEGG | 9,390 | 8.14 |
| SwissProt | 15,315 | 13.27 |
| Pfam | 27,160 | 23.54 |
| GO | 27,160 | 23.54 |
| KOG | 8,158 | 7.07 |
| all database | 2,543 | 2.20 |
| at least one database | 36,282 | 31.45 |
| Gene Description | Hit Species | Mean FPKM (ZW) | Mean FPKM (WW) | Mean FPKM (ZZ) | NR ID | NR E-Value |
|---|---|---|---|---|---|---|
| gametocyte-specific factor 1 | Macrobrachium nipponense | 3.58 | 7.12 | 0.02 | AMY62701.1 | 2.20 × 10−6 |
| gonadotropin-releasing hormone II receptor | Orussus abietinus | 0.87 | 0.71 | 0.16 | XP_012272108.1 | 8.8 × 10−95 |
| insulin-like receptor | Trachymyrmex zeteki | 2.35 | 16.93 | 0.00 | KYQ55071.1 | 4.60 × 10−183 |
| PREDICTED: sex determination protein fruitless-like | Fopius arisanus | 11.21 | 6.76 | 0.99 | XP_011299024.1 | 3.70 × 10−31 |
| spermatogenesis-associated protein 5-like | Planoprotostelium fungivorum | 7.12 | 16.61 | 2.50 | PRP83878.1 | 3.30 × 10−56 |
| transcription factor SOX-10-like | Zootermopsis nevadensis | 13.15 | 7.33 | 0.21 | XP_021918795.1 | 1.60 × 10−49 |
| ubiquitin carboxyl-terminal hydrolase 46 | Aedes aegypti | 80.51 | 122.33 | 25.60 | XP_021706106.1 | 3.70 × 10−147 |
| Ubiquitin carboxyl-terminal hydrolase isozyme L5 | Scylla paramamosain | 24.24 | 77.3 | 26.76 | ACM43511.1 | 6.60 × 10−150 |
| cytochrome P450 CYP302a1 | Portunus trituberculatus | 249.96 | 271.16 | 0.86 | AJA06113.1 | 3.80 × 10−152 |
| cytochrome P450 CYP315a1 | Portunus trituberculatus | 2.61 | 1.25 | 83.75 | AJF94636.1 | 5.00 × 10−147 |
| forkhead box L2 | Procambarus clarkii | 6.64 | 1.83 | 29.28 | ALD48735.1 | 3.70 × 10−99 |
| heat shock protein 27 | Procambarus clarkii | 3.93 | 2.21 | 17.4 | AGZ84434.1 | 7.60 × 10−53 |
| heat shock protein 70 | Portunus trituberculatus | 2.02 | 0.23 | 10.24 | ACZ02405.1 | 5.00 × 10−308 |
| heat shock protein 70 cognate 3 | Eriocheir sinensis | 0.67 | 0.04 | 2.51 | AHA61465.1 | 8.30 × 10−300 |
| heat shock 70 kDa protein 1A/1B | Toxocara canis | 0.20 | 0.02 | 1.18 | KHN76385.1 | 8.60 × 10−153 |
| heat shock protein 21 | M. rosenbergi | 0.11 | 0.10 | 2.05 | AET34915.1 | 8.80 × 10−31 |
| heat shock protein isoform 12Ai1 | Cherax destructor | 0.10 | 0.05 | 0.95 | AKB96220.1 | 2.40 × 10−97 |
| male reproductive-related protein B | M. rosenbergi | 1.91 | 0.01 | 221.43 | ABQ41229.1 | 1.00 × 10−32 |
| male reproductive-related protein | M. rosenbergi | 0.00 | 0.00 | 8.97 | ABQ41231.1 | 3.60 × 10−40 |
| male reproductive-related serum amyloid A | M. rosenbergi | 0.96 | 1.76 | 23.86 | ABQ41247.1 | 2.40 × 10−39 |
| male reproductive-related protein A | M. rosenbergi | 0.00 | 0.00 | 3.04 | ABQ41213.1 | 6.80 × 10−22 |
| male reproductive tract-specific Kazal-type proteinase inhibitor | M. rosenbergi | 0.20 | 0.00 | 195.26 | AAX83134.1 | 8.40 × 10−76 |
| male reproductive-related protein Mar-Mrr | M. rosenbergi | 0.33 | 0.00 | 120.44 | ABQ41239.1 | 8.20 × 10−32 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Yuan, X.; Qiu, Q.; Huang, G.; Jiang, Q.; Fu, P.; Zhang, Y.; Jia, Y.; Yang, X.; Jiang, H. Comparative Transcriptome Analysis of Gonads for the Identification of Sex-Related Genes in Giant Freshwater Prawns (Macrobrachium Rosenbergii) Using RNA Sequencing. Genes 2019, 10, 1035. https://doi.org/10.3390/genes10121035
Jiang J, Yuan X, Qiu Q, Huang G, Jiang Q, Fu P, Zhang Y, Jia Y, Yang X, Jiang H. Comparative Transcriptome Analysis of Gonads for the Identification of Sex-Related Genes in Giant Freshwater Prawns (Macrobrachium Rosenbergii) Using RNA Sequencing. Genes. 2019; 10(12):1035. https://doi.org/10.3390/genes10121035
Chicago/Turabian StyleJiang, Jianping, Xiang Yuan, Qingqing Qiu, Guanghua Huang, Qinyang Jiang, Penghui Fu, Yu Zhang, Yinhai Jia, Xiurong Yang, and Hesheng Jiang. 2019. "Comparative Transcriptome Analysis of Gonads for the Identification of Sex-Related Genes in Giant Freshwater Prawns (Macrobrachium Rosenbergii) Using RNA Sequencing" Genes 10, no. 12: 1035. https://doi.org/10.3390/genes10121035
APA StyleJiang, J., Yuan, X., Qiu, Q., Huang, G., Jiang, Q., Fu, P., Zhang, Y., Jia, Y., Yang, X., & Jiang, H. (2019). Comparative Transcriptome Analysis of Gonads for the Identification of Sex-Related Genes in Giant Freshwater Prawns (Macrobrachium Rosenbergii) Using RNA Sequencing. Genes, 10(12), 1035. https://doi.org/10.3390/genes10121035




