Abstract
The giant freshwater prawn (Macrobrachium rosenbergii) exhibits sex dimorphism between the male and female individuals. To date, the molecular mechanism governing gonadal development was unclear, and limited data were available on the gonad transcriptome of M. rosenbergii. Here, we conducted comprehensive gonadal transcriptomic analysis of female (ZW), super female (WW), and male (ZZ) M. rosenbergii for gene discovery. A total of 70.33 gigabases (Gb) of sequences were generated. There were 115,338 unigenes assembled with a mean size of 1196 base pair (bp) and N50 of 2195 bp. Alignment against the National Center for Biotechnology Information (NCBI) non-redundant nucleotide/protein sequence database (NR and NT), the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, SwissProt database, Protein family (Pfam), Gene ontology (GO), and the eukaryotic orthologous group (KOG) database, 36,282 unigenes were annotated at least in one database. Comparative transcriptome analysis observed that 10,641, 16,903, and 3393 genes were significantly differentially expressed in ZW vs. ZZ, WW vs. ZZ, and WW vs. ZW samples, respectively. Enrichment analysis of differentially expressed genes (DEGs) resulted in 268, 153, and 42 significantly enriched GO terms, respectively, and a total of 56 significantly enriched KEGG pathways. Additionally, 23 putative sex-related genes, including Gtsf1, IR, HSP21, MRPINK, Mrr, and other potentially promising candidate genes were identified. Moreover, 56,241 simple sequence repeats (SSRs) were identified. Our findings provide a valuable archive for further functional analyses of sex-related genes and future discoveries of underlying molecular mechanisms of gonadal development and sex determination.
1. Introduction
The giant freshwater prawn, Macrobrachium rosenbergii (M. rosenbergii), as an important commercial freshwater prawn species, is widely distributed in China and Southeast Asian countries due to its delicious flesh and high value nutrition. Like other crustaceans, M. rosenbergii displays sexual dimorphic growth pattern: The male individuals grow much larger and faster than the females [1,2]. Thus, based on the economic impact, developing the monosex population culture as the efficient approach to boost production has received great attention from researchers in M. rosenbergii. Nevertheless, the premise of monosex culture is to understand the molecular mechanism of sex determination, as well as to unravel sex-related genes.
Studies on sex determination have proved that many macruran species exhibit the ZW/ZZ sex determination system [3,4,5]. In M. rosenbergii, sex reversal experiments proposed that the female prawns bear the ZW sex chromosomes, and the male individuals bear the ZZ chromosomes [1,6,7]. Subsequently, isolation of sex-specific markers further deepened the insight of the ZW/ZZ sex determination mode [8,9]. Recently, Ma et al. have confirmed that M. rosenbergii has the ZW/ZZ sex determination system based on the bacterial artificial chromosome (BAC) library, and nine genes were unraveled to be the key sex-linked genes by PacBio sequencing. Amongst, three genes, including zinc knuckle domain (ZKD), reverse transcriptase, and ANCDUO, protein homologs were known genes without characterization of roles in sex determination. Although the Z and W chromosomes have not been identified, the results still provided a useful resource and comprehensive knowledge of genomic structure of M. rosenbergii [10]. However, sex-related genes and a list of biological pathways involved in gonads development have been revealed [11,12,13,14,15,16], the potential molecular mechanisms of sex determination in M. rosenbergii are still not well understood.
Currently, along with the emergence of next generation sequencing (NGS), RNA-sequencing (RNA-seq) was widely applied to detect the expression profiles of genes and identify the sex-related genes in aquaculture species, especially those lacking genomic sequences, due to its high throughput and low cost. To date, some progresses have been made in the characterization of sex-related genes and pathways in M. rosenbergii [16], Macrobrachium nipponense [17], Silver Sillago [18], Olive Flounder [19], Pacific abalone [20], Pacific white shrimp [21], catfish [22], and Acipenser dabryanus [23]. Hence, the purpose of this study was to perform gonadal transcriptomic sequencing of females, super females, and males to investigate the gene expression profiles, and to identify the sex-related candidate genes. These data would lay the foundation for better illustrating the molecular mechanisms of sex determination.
2. Materials and Methods
2.1. Ethics Statement
All procedures were in compliance with the institutional guidelines and under a protocol approved by the Animal Experimental Ethical Inspection Form of Guangxi University (GXU2019-072).
2.2. Sample Collection
The super female (WW genotype) individuals were purchased from the Enzootic Company from Israel (https://enzootic.com), and reared in a national M. rosenbergii seed multiplication farm in Nanning, Guangxi, China. The male (ZZ genotype) and female (ZW genotype) prawns from the same family were collected from this farm. For the WW individuals, they were achieved through a novel biotechnology with two steps: Females were sex-reversed into neo-males by injecting the suspended hypertrophied androgenic gland cells, crossing neo-males with normal females to validated WW females. Notably, the phenotypes of super females were not different from normal females and the fecundity of WW individuals has shown no significant deviation from the ZW individuals [24]. All the healthy experimental individuals used in this study were five months of age, and were maintained in aerated freshwater at 26 ± 2 °C in the previously mentioned farm. Ovaries from females (ZW) and super females (WW), and testes from males (ZZ), were collected and immediately frozen in liquid nitrogen, then stored at 80 °C until RNA extraction. For each of three groups, three biological replicates were used for RNA-seq, with each replicate being pooled by three prawns. The growth performance of the sample individuals is listed in Table 1 and Table S1.

Table 1.
Growth performance of the 27 samples among the three groups.
2.3. RNA Extraction, Library Preparation, and Transcriptome Sequencing
Three individuals of each sample were pooled to extract the total RNA using Trizol Reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions, and then treated with RNase-free DNase I (TianGen, Beijing, China) to eliminate genomic DNA contamination. The integrity and concentration of RNA was determined using the Agilent Bioanalyzer 2100 system (Agilent Technologies, California, USA) and the Qubit® 2.0 Flurometer (Life Technologies, Carlsbad, USA), respectively. High-quality RNA samples (OD260/280 = 1.8–2.2, OD260/230 > 2.0, RNA integrity number (RIN) > 7.5, 28S: 18S > 1) was stored at −80 °C and used for library construction. A total of nine sequencing libraries were constructed with NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, Beijing, China) following the manufacturer’s recommendations. Finally, the libraries were sequenced with the Illumina Hiseq platform using paired-end strategy.
2.4. Transcriptome De Novo Assembly and Functional Annotation
The raw sequencing reads were processed through a Perl program for quality control. Clean reads were obtained by removing reads with adaptors and poly-N, removing ambiguous reads containing more than 10% unknown bases and the low-quality reads (the rate of reads in which the quality value ≤20 was more than 50%). Transcriptome assembly was accomplished by Trinity [25] with min_kmer_cov set to 2 by default and all other parameters set to default values. Then, a single set of nonredundant unigenes were acquired with the GICL clustering software [26].
Functional annotation of all assembled unigenes was performed with the following seven databases and four softwares: National Center for Biotechnology Information (NCBI) nonredundant nucleotide sequence database (Nt) based on the NCBI Blast (v2.2.28+) (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (E-value < 1 × 10−5) NCBI nonredundant protein sequence database (Nr), SwissProt database, Kyoto Encyclopedia of Genes and Genomes (KEGG) (E-value < 1 × 10−10) using KEGG Automatic Annotation Server (KAAS) and eukaryotic orthologous group (KOG) database based on the diamond (version 0.8.22) (E-value < 1 × 10−3), Protein family (Pfam) by Hmmscan with the E-value less than 0.01, and Gene Ontology (GO) through Blast2GO (version 3.0) (https://www.blast2go.com/) (E-value < 1 × 10−6), respectively. The unigenes annotated by at least one database were considered as annotated successfully based on the corresponding threshold. When unigenes were different in the cases where searches against different databases disagreed with each other, the desired annotation information was according to our experiment objective and the reliability of different databases (NR > GO > KEGG > SwissProt > Pfam > KOG > NT). Thus, in our study, we preferred the annotated results of NR database.
2.5. Identification of Differentially Expressed Genes (DEGs)
RSEM software (version v1.2.15) was used for reads mapping to the assembled reference transcriptome with default parameters [27]. Gene expression levels were calculated based on the expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced (FPKM). Differential gene expression analysis among the three groups was performed with the DESeq2 [28] within q-value < 0.01, |log2 (fold change)| > 2. Additionally, GO enrichment and KEGG pathway analysis of DEGs was implemented with the GOseq tool and online software KOBAS (Version 3.0) (http://kobas.cbi.pku.edu.cn/index.php) [29,30], respectively. Among them, those with p-value < 0.05 were considered to be significantly enriched.
2.6. Detection of Simple Sequence Repeats (SSRs)
MIcroSAtellite (MISA) software (http://pgrc.ipk-gatersleben.de/misa/misa.html) was used to identify the simple sequence repeats (SSRs) of the transcriptome. Six types of SSR were investigated, including mono-, di-, tri-, tetra-, penta-, and hexa-nucleotide motifs. The Primer 3 software (http://primer3.sourceforge.net/releases.php) was then used for primer design based on the flanking regions of SSRs for further validation.
2.7. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
To validate the accuracy of gene expression data obtained by RNA-seq, 10 DEGs were selected to be verified by qRT-PCR using the same samples for RNA-seq. The 10 selected DEGs consisted of 6 sex-related candidate genes (ubiquitin carboxyl-terminal hydrolase 46, transcription factor SOX-10-like, heat shock protein 70, heat shock 70 kDa protein 1A/1B, male reproductive-related protein B, and male reproductive-related serum amyloid A), one DEG (heat shock protein 10) from the heat shock protein family, one DEG (slow-type skeletal muscle actin 4) with the highest expression in ZZ gonads, and the two DEGs (neuronal-specific septin-3 and tubulin α-1 chain-like) with two-fold different expression between WW gonads and ZW gonads. PCR Primers were designed with Primer 3 and Oligo 7.0 (Table S2). Then 18S was used as a housekeeping gene to normalize the mRNA levels of DEGs [31,32]. The qRT-PCR was performed in triplicate by using the Lightcycler 480 II (Roche Applied Science, Penzberg, Germany) with the following reaction conditions: Pre-denaturation at 95 °C for 10 s; amplification 45 cycles of 95 °C for 10 s, 59 °C for 10 s, and 72 °C for 10 s, and using the following program: 95 °C for 10 min; 45 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 10 s; 72 °C for 6 min. The relative expression levels were calculated with the method of 2−ΔΔCt as described previously [33].
3. Results
3.1. Sequencing Analysis and Transcriptome Assembly
In the present study, a total of 70.33 gigabases (Gb) of clean bases were generated. The clean reads ranging from 47,066,162 to 60,480,354 for nine libraries with the GC% of approximately 42% per library were used for further analysis. In addition, Q20 and Q30 of each library were higher than 97% and 93%, respectively (Table 2). As illustrated in Figure 1, a total of 115,338 unigenes were assembled with a mean size of 1196 bp and N50 of 2195 bp.

Table 2.
Summary statistics of gonadal transcriptome sequencing data for Macrobrachium rosenbergii (M. rosenbergii).
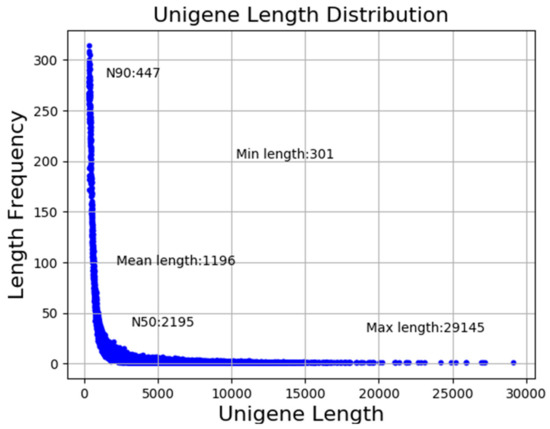
Figure 1.
Length distribution of unigenes.
3.2. Sequence Annotation
All unigenes were searched against the Nr, Nt, KEGG, SwissProt, Pfam, GO, and KOG database, among them, most of the unigenes were annotated in Pfam and GO databases, while 2543 unigenes were annotated in all seven databases. Meanwhile, 36,282 unigenes founded as putative homologues were annotated at least in one database. The annotation success rate of unigenes was 23,483 in Nr (20.36%), 6556 in Nt (5.68%), 9390 in KEGG (8.14%), 15,315 in SwissProt (13.27%), 27,160 in Pfam (23.54%), 27,160 in GO (23.54%), and 8158 in KOG (7.07%) (Table 3).

Table 3.
Statistics of annotation results.
Through the KOG function classification, 8158 unigenes were classified into 26 functional categories, the top 3 of which were as follows: General function prediction only (1437 unigenes), signal transduction mechanisms (1135 unigenes), and posttranslational modification, protein turnover, chaperones (904 unigenes) (Figure 2A).
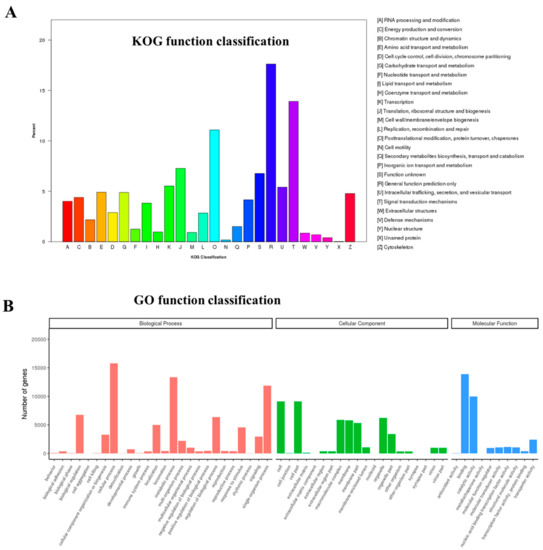
Figure 2.
Functional classifications of unigenes in M. rosenbergii gonadal transcriptome. (A) eukaryotic orthologous group (KOG) classification. (B) Gene Ontonogy (GO) classification.
Aligning to the GO database, a total of 27,160 unigenes were sorted into three major GO categories: Biological processes, molecular functions, and cellular components. Among them, “cellular process” (15,773), “cell part” (9096), and “cell” (9096), as well as “binding” (13,890), were the main terms, respectively (Figure 2B).
As for KEGG assignment, 9390 unigenes were mapped to 230 KEGG pathways, 4.12% (387 unigenes) of which were mapped to “Ribosome”, followed by “Lysosome”, and “Phagosome”. In addition, we identified several sex-related KEGG pathways, such as MAPK signaling pathway (ko04010, 134 unigenes), GnRH signaling pathway (ko04912, 75 unigenes), progesterone-mediated oocyte maturation (ko04914, 77 unigenes), focal adhesion (ko04510, 176 unigenes), calcium signaling pathway (ko04020, 130 unigenes), ubiquitin mediated proteolysis (ko04120, 105 unigenes), and wnt signaling pathway (ko04310, 81 unigenes) (Table S3).
3.3. DEGs Identification and Enrichment Analysis
As a result, 10,641 genes were significantly differentially expressed in ZW vs. ZZ and 16,903 genes were found differentially expressed in WW vs. ZZ. Between WW vs. ZW, only 3393 genes were revealed as DEGs (Figure 3 and Table S4).
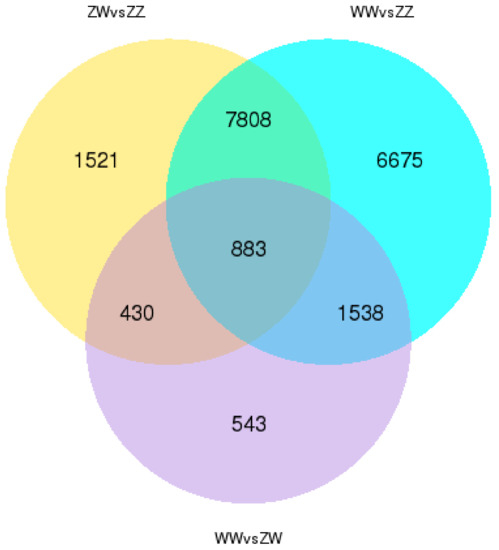
Figure 3.
Venn diagram showing the numbers of expressed genes and differentially expressed genes (DEGs) in ZW, WW, and ZZ.
GO annotation was performed to classify the DEGs among the three groups. As a result, 268, 153, and 42 GO terms were significantly enriched, respectively (Figure 4). Amongst them, the most enriched GO terms at the level of biological processes were “chitin metabolic process” and “glucosamine-containing compound metabolic process”. At the molecular function level, “dynein binding” was the dominant term. At the cellular component, “dynein complex” was the most significantly enriched term.
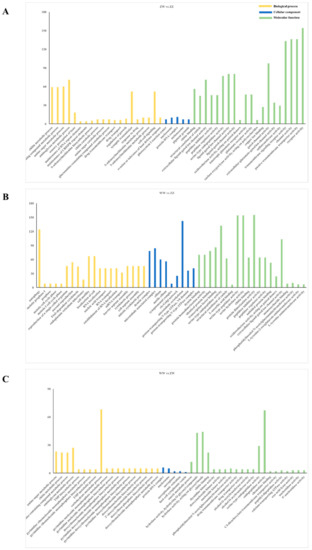
Figure 4.
GO enrichment analysis of DEGs. (A) GO enrichment analysis of DEGs in ZW vs. ZZ. (B) GO enrichment analysis of DEGs in WW vs. ZZ. (C) GO enrichment analysis of DEGs in WW vs. ZW.
Meanwhile, enrichment analysis of KEGG pathway was conducted. Totally, 56 KEGG pathways were significantly enriched. The top two signaling pathways were “amino sugar and nucleotide sugar metabolism” and “starch and sucrose metabolism” (Figure 5).
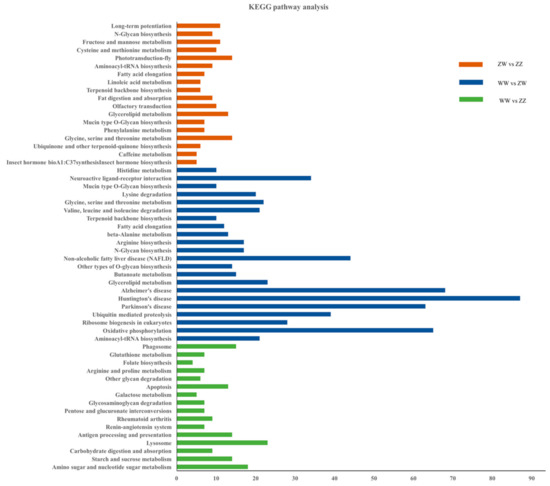
Figure 5.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of DEGs.
3.4. Genes of Interest Related to Sex
Based on the functional annotation of unigenes, along with the previous publications, 23 sex-related candidate genes implicated in gonadal development and sex determination were identified (Table 4). Notably, among them, two genes (gametocyte-specific factor 1 and insulin-like receptor) showed specific expression in ovaries between female and super female. And seven genes, including heat shock protein 21, heat shock protein isoform 12Ai1, male reproductive tract-specific Kazal-type proteinase inhibitor, male reproductive-related protein, male reproductive-related protein B, male reproductive-related protein A, and male reproductive-related protein Mar-Mrr, showed male-specific expression patterns. These genes could be considered as the most promising sex-related candidates.

Table 4.
List of sex-related candidate genes in M. rosenbergii.
3.5. Discovery of Simple Sequence Repeats (SSRs)
In the present study, MicroSAtellite (MISA) software was used to detect the SSRs of M. rosenbergii. As a result, a total of 56,241 SSRs were identified in 33,189 SSR-containing sequences (Figure 6). Among them, mononucleotide (27,574, 49.03%) was the dominant, followed by dinucleotide (18,598, 33.07%), trinucleotide (9311, 16.56%), tetranucleotide (655, 1.16%), pentanucleotide (63, 0.11%), and hexanucleotide (40, 0.07%).
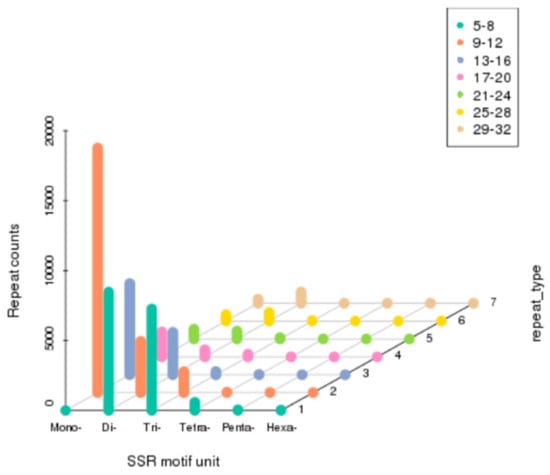
Figure 6.
Distribution of identified simple sequence repeats (SSRs) according to motif types in the sequences of M. rosenbergii.
3.6. The qRT-PCR Validation
Furthermore, 10 DEGs were selected for qRT-PCR validation. As a result, all the detected genes showed similar expression patterns (Figure 7), which indicated the reliability and accuracy of our transcriptome analysis.
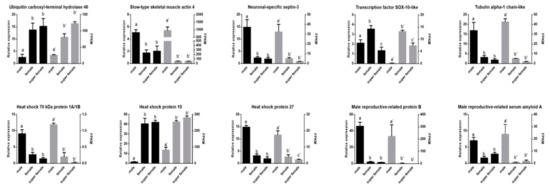
Figure 7.
Verification of the expression patterns both in real-time quantitative reverse transcription PCR and RNA-seq. Groups with different letters were significantly different (p < 0.05).
4. Discussion
As an economically important aquaculture species, a better understanding of the genetic and biological mechanisms underlying the complex ZW/ZZ sex determination system of M. rosenbergii is important, yet it is poorly elucidated. Sagi et al. firstly reported the sex dimorphism in M. rosenbergii [34]. Subsequently, two sex reversal experiments uncovered that M. rosenbergii bears the ZW/ZZ sex determination system. Mating of sex-reversed females (neo-males) with normal females (ZW) could increase a higher ratio of females, while mating of sex-reversed males (neo-females) with normal males could produce all-male (ZZ × ZZ) progeny [1,6,7]. Recently, along with the emergence of high-throughput sequencing, RNA-seq as a most powerful tool is available for illustrating the mechanisms of immune response [35], determining the expression pattern of reproduction, growth, and pheromone communication in hepatopancreas, gill, muscle, and antennal gland [36,37,38]. In the present study, we profiled to detect the transcriptomes of gonads in females, super females, and the males using RNA-seq, with the aim of interpreting the molecular mechanism involved in the sex determination, and identifying sex-related candidate genes.
The general consensus in the prawn (M. rosenbergii) culture industry is that the major approach to potentially boost the prawn production is to produce all females, due to their normal size distribution, higher production, and product values compared to all males [2]. Hence, in our study, to better illustrate the molecular mechanism of sex-determination, we used the normal females, super females, and males to detect the gonadal expression profile. Therein the male individuals and normal female individuals were from one full-sib family, and, along with the super females, they were reared in the same conditions and environments in the national M. rosenbergii seed multiplication farm. Moreover, all the healthy samples were five months; meanwhile, the body weight and body length of samples, as well as the development of ovaries for females and super females, were similar, thus reducing the background effects as far as possible.
We identified 23 sex-related candidate genes. Among them, two and seven genes showed specific expression patterns in ovaries and testes. Gametocyte-specific factor 1 (Gtsf1) as a member of unknown protein family 0224 (UPF0224) was involved in Piwi-interacting RNA (piRNA) pathway [39]. Some evidences indicated that Gtsf1 participated in spermatogenesis, and had significant impact on male germ cells [39,40]. Moreover, in teleost, Gtsf1 as a key candidate gene was involved in sex differentiation [41,42]. Intriguingly, in this study, Gtsf1 displayed sexual dimorphic expression and no expression was detected in testes. Furthermore, the expression in super females was more than two-fold higher than that in females. The current data insinuated that Gtsf1 could be considered as a key candidate gene related to gonadal development and sex determination.
Gonadotropin-releasing hormone II receptor (GnRHR2) was originally identified in humans [43]. Evidence has been accumulated that GnRH2 is required for sexual behavior [41]. Furthermore, initial studies have discovered that GnRHR2 together with its ligand GnRH2, were considered to be novel regulators in driving the reproduction process in mammals [44,45]. Toward this end, our data detected that GnRHR2 was upregulated in ovaries. Taken together, we conjectured that GnRHR2 has essential functions in female reproduction.
Insulin-like receptor (IR) functions as the pivotal member of insulin family signaling pathway and directs the male sexual differentiation in mammals [46]. Earlier evidence has shown that IR interacts with the insulin-like androgenic gland hormone (IAG) to regulate sex differentiation and spermatogenesis in crustaceans [13,15,46,47]. In our study, IR has shown higher expression in ovaries than in testes. Notably, the expression of IR in super females was more than two-fold than that in females. The results presented here suggest that IR could be a promising candidate gene for sex differentiation between females and super females.
Sex determination protein fruitless-like (Fru) was specially displayed in regulating the sexual orientation and sex behavior in Drosophila melanogaster [48,49]. In our transcriptome database, Fru showed sexual dimorphism in ovaries and testes, which displayed upregulated expression in ovaries. As observed in the Chinese mitten crab [50], Fru was implicated in sex determination pathway-like in Drosophila melanogaster.
Spermatogenesis-associated protein 5-like (SPATA5L) belonging to the ATPase family protein 2 homolog was first identified in mice and has essential functions in spermatogenesis, alopecia areata, intellectual disability, and other serious disorders [50]. Ge et al. have shown that down regulation of the expression of SPATA5L in males could decrease the fecundity of females during mating [51]. In our research, SPATA5L was expressed at higher levels in ovaries. Taken together, the current data provides a hint that SPATA5L might be a promising candidate for sex differentiation.
Transcription factor SOX-10-like as a member of SRY-related transcription factors from the Sox (Sry-type HMG box) E subfamily was reported to play a critical role in testis development [18,52]. In our research, it was expressed higher in ovaries than that in testes.
The ubiquitin-related homologous genes have been investigated and shown higher expression in testis and ovary, and further involved in reproductive process [53]. In our gonadal transcriptome database, two ubiquitin-related homologous genes (ubiquitin carboxyl-terminal hydrolase 46 and ubiquitin carboxyl-terminal hydrolase isozyme L5) were identified. It is important to further characterize these genes in M. rosenbergii to illustrate their potential role in sex differentiation and sex determination.
Cytochrome P450 302A1 (CYP302a1) was a new member of the cytochrome P450 (CYP) super-family. It encodes 22-hydroxylase and participates in ecdysteroid biosynthesis [54,55]. In drosophila and mosquito, CYP302a1 was highly expressed in females, which further suggested that CYP302a1 has an important role in the ovaries, as described by Chavez et al. [56] and Warren et al. [54]. Thus, our data have shown that CYP302a1 was also expressed predominantly in the ovaries, which was in accordance with previous studies and further supporting our finding of the function of CYP302a1 in the ovary.
Cytochrome P450 CYP315a1 (CYP315a1), another a member of the CYP super-family gene, was reported to be implicated in the ecdysteroidogenic pathway in Bombyx mori [57] and Drosophila melanogaster [54]. As we all know, steroid hormones were responsible for controlling reproduction and development in higher organisms and arthropods [58]. Thus, we deduced that CYP315a1 may be involved in testicular development.
Forkhead box l2 (Foxl2), a member of Fox gene family, encodes a conserved transcription factor and is a special marker of ovarian differentiation [59]. Moreover, Foxl2 plays a critical role in ovarian differentiation and maintenance [60]. In our study, interestingly, Foxl2 displayed upregulated expression in testes, and has shown similar expression pattern in Litopenaeus vannamei [21]. However, the corresponding role in M. rosenbergii remains to be determined.
Heat shock proteins (HSPs) contribute to the interaction with steroid hormone receptors, temperature, and estrogen signaling, etc. [61,62]. Furthermore, HSPs were considered as promising candidates with potential effects on temperature-dependent sex determination (TSD) [62]. In addition, Matsumoto et al. have verified that heat shock cognate 70 kDa proteins played a regulatory role in mouse spermatogenesis [63]. In the present study, all three HSPs (heat shock protein 27, heat shock protein 70, and heat shock protein 70 cognate3) exhibited upregulated expression pattern in the testes, while heat shock 70 kDa protein 1A/1B, heat shock protein 21, and heat shock protein isoform 12Ai1 displayed testis-specific expression patterns as well.
Some male reproductive-related genes were also identified. Among them, two male-biased genes (male reproductive-related protein B and male reproductive-related serum amyloid A) and two testis-specific genes (male reproductive-related protein and male reproductive-related protein A) were upregulated and showed a specific expression pattern in testes which was similar to the results of Dai et al. [64]. Meanwhile, the other two testis-specific genes, including male reproductive-related protein Mar-Mrr and male reproductive tract-specific Kazal-type proteinase inhibitor, showed a specific testicular expression pattern. Earlier studies had demonstrated that Mar-Mrr was specially expressed in the male reproductive tract, and was required for sperm function [11,14,65]. Thus, combining the current results indicates that Mar-Mrr is implicated in male-reproduction.
Peptidase inhibitors were not only functional on cell migration, signal transmission, wound healing, and tissue remodeling [66], but also on male-reproduction in Drosophila [67]. Previous studies have confirmed that the male reproduction-related peptidase inhibitor Kazal-type (MRPINK) gene has a potential effect on male reproductive processes in prawns and crab [12,68,69]. In our study, one peptidase inhibitor (male reproductive tract-specific Kazal-type proteinase inhibitor) was expressed at higher levels in male gonads, suggesting that the peptidase inhibitor plays a critical role in gonadal development.
Of note, these identified sex-related genes and RNA-seq data could promote our understanding on the molecular mechanism involved in the sex determination. In addition, RNA interference (RNAi) and clustered regularly interspaced short palindromic repeats/CRISPR associated (CRISPR/Cas9)-mediated genome editing has revolutionized the gene functional determination, and was applied to controlling of sex development in aquaculture species [15,70,71]. Definitely, the actual functional roles of these selected sex-related genes were urgently required to be investigated, and the sex-specific genes may provide strong support for further sex manipulation and monosex culture based on the genome editing strategy.
Notably, numerous SSRs were identified. Currently, SSRs, single nucleotide polymorphisms (SNPs), and insertions and deletions (indels) are widely used for genetic linkage mapping, quantitative trait locus (QTL) detection, genetic diversity assessment etc. In M. rosenbergii, See et al. have developed microsatellite markers to evaluate genetic diversity [72]. Using the high throughput sequencing, it is feasible to detect and discover the large numbers of SSRs [37,73]. Correspondingly, the SSRs identified in the present study could serve as genetic markers for further QTL mapping and marker-assisted selection (MAS) in M. rosenbergii.
5. Conclusions
This is the first comprehensive report on the gonadal transcriptome of the M. rosenbergii, and 115,338 unigenes were identified. By comparing ovary and testis transcriptomes, numerous DEGs were identified, and 23 sex-related genes were revealed. Moreover, 56,241 SSRs were discovered and could be used as genetic markers. These findings established a valuable database for future functional analyses of sex-related genes and understanding the molecular mechanisms of sex determination in M. rosenbergii.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/12/1035/s1: Table S1: Growth performance of the 27 samples among the three groups. Table S2: Primers of the 10 DEGs used for qRT-PCR. Table S3 KEGG pathway analysis of 9,390 unigenes. Table S4 DEGs in ZW vs. ZZ, WW vs. ZZ, and WW vs. ZW.
Author Contributions
Data curation, J.J., X.Y. (Xiang Yuan), Q.Q., P.F., Y.Z., and Y.J.; Formal analysis, J.J., X.Y. (Xiang Yuan), and Q.Q.; Funding acquisition, H.J.; Investigation, J.J. and X.Y. (Xiang Yuan), Methodology, J.J., X.Y. (Xiang Yuan), Q.Q., G.H., and Q.J.; Project administration, H.J.; Resources, G.H. and Q.J.; Supervision, H.J. and X.Y. (Xiurong Yang); Writing—original draft, J.J. and X.Y. (Xiang Yuan); Writing—review and editing, H.J. and X.Y. (Xiurong Yang)
Acknowledgments
We thank senior researcher Ilie Daniela Elena and Dr. Alexandru Eugeniu Mizeranschi (Research and Development Station for Bovine, Arad, Romania) for carefully proof-read the manuscript. This work was financially supported by the Science and Technology Major Project of Guangxi (Guike AA17204080-6), Guangxi Natural Science Foundation (2018GXNSFBA281209) and Guangxi Postdoctoral Fundamental Research Funds (Y201002037).
Conflicts of Interest
The authors declare no conflict of interest and the sponsors had no role in the design, execution, interpretation, or writing of the study.
References
- Aflalo, E.D.; Hoang, T.T.T.; Nguyen, V.; Lam, Q.; Nguyen, D.; Trinh, Q.; Raviv, S.; Sagi, A. A novel two-step procedure for mass production of all-male populations of the giant freshwater prawn Macrobrachium rosenbergii. Aquaculture 2006, 256, 468–478. [Google Scholar] [CrossRef]
- Malecha, S. The case for all-female freshwater prawn, M acrobrachium rosenbergii (D e M an), culture. Aquac. Res. 2012, 43, 1038–1048. [Google Scholar] [CrossRef]
- Waiho, K.; Shi, X.; Fazhan, H.; Li, S.K.; Zhang, Y.L.; Zheng, H.P.; Liu, W.H.; Fang, S.B.; Ikhwanuddin, M.; Ma, H.Y. High-Density Genetic Linkage Maps Provide Novel Insights Into ZW/ZZ Sex Determination System and Growth Performance in Mud Crab (Scylla paramamosain). Front. Genet. 2019, 10, 298. [Google Scholar] [CrossRef] [PubMed]
- Benzie, J.A.; Kenway, M.; Ballment, E. Growth of Penaeus monodon× Penaeus esculentus tiger prawn hybrids relative to the parental species. Aquaculture 2001, 193, 227–237. [Google Scholar] [CrossRef]
- Coman, F.E.; Sellars, M.J.; Norris, B.J.; Coman, G.J.; Preston, N.P. The effects of triploidy on Penaeus (Marsupenaeus) japonicus (Bate) survival, growth and gender when compared to diploid siblings. Aquaculture 2008, 276, 50–59. [Google Scholar] [CrossRef]
- Malecha, S.R.; Nevin, P.A.; Ha, P.; Barck, L.E.; Lamadrid-Rose, Y.; Masuno, S.; Hedgecock, D. Sex-ratios and sex-determination in progeny from crosses of surgically sex-reversed freshwater prawns, Macrobrachium rosenbergii. Aquaculture 1992, 105, 201–218. [Google Scholar] [CrossRef]
- Sagi, A. м-Pah Growth, maturation and progeny of sex-reversed. World Aquac. 1990, 21, 87. [Google Scholar]
- Ventura, T.; Aflalo, E.; Weil, S.; Kashkush, K.; Sagi, A. Isolation and characterization of a female-specific DNA marker in the giant freshwater prawn Macrobrachium rosenbergii. Heredity 2011, 107, 456. [Google Scholar] [CrossRef]
- Jiang, X.H.; Qiu, G.F. Female-only sex-linked amplified fragment length polymorphism markers support ZW/ZZ sex determination in the giant freshwater prawn Macrobrachium rosenbergii. Anim. Genet. 2013, 44, 782–785. [Google Scholar] [CrossRef]
- Ma, K.-Y.; Yu, S.-H.; Du, Y.-X.; Feng, S.-Q.; Qiu, L.-J.; Ke, D.-Y.; Luo, M.-Z.; Qiu, G.-F. Construction of a genomic bacterial artificial chromosome (BAC) library for the prawn Macrobrachium rosenbergii and initial analysis of ZW chromosome-derived BAC inserts. Mar. Biotechnol. 2019, 21, 206–216. [Google Scholar] [CrossRef]
- Cao, J.-X.; Yin, G.-L.; Yang, W.-J. Identification of a novel male reproduction-related gene and its regulated expression patterns in the prawn, Macrobrachium rosenbergii. Peptides 2006, 27, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.-X.; Dai, J.-Q.; Dai, Z.-M.; Yin, G.-L.; Yang, W.-J. A male reproduction-related kazal-type peptidase inhibitor gene in the prawn, Macrobrachium rosenbergii: Molecular characterization and expression patterns. Mar. Biotechnol. 2007, 9, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Raviv, S.; Glazer, L.; Sagi, A. Temporal silencing of an androgenic gland-specific insulin-like gene affecting phenotypical gender differences and spermatogenesis. Endocrinology 2008, 150, 1278–1286. [Google Scholar] [CrossRef] [PubMed]
- Phoungpetchara, I.; Tinikul, Y.; Poljaroen, J.; Changklungmoa, N.; Siangcham, T.; Sroyraya, M.; Chotwiwatthanakun, C.; Vanichviriyakit, R.; Hanna, P.J.; Sobhon, P. Expression of the male reproduction-related gene (Mar-Mrr) in the spermatic duct of the giant freshwater prawn, Macrobrachium rosenbergii. Cell Tissue Res. 2012, 348, 609–623. [Google Scholar] [CrossRef]
- Sharabi, O.; Manor, R.; Weil, S.; Aflalo, E.; Lezer, Y.; Levy, T.; Aizen, J.; Ventura, T.; Mather, P.; Khalaila, I. Identification and characterization of an insulin-like receptor involved in crustacean reproduction. Endocrinology 2015, 157, 928–941. [Google Scholar] [CrossRef]
- Liu, X.; Luo, B.Y.; Feng, J.B.; Zhou, L.X.; Ma, K.Y.; Qiu, G.F. Identification and profiling of microRNAs during gonadal development in the giant freshwater prawn Macrobrachium rosenbergii. Sci. Rep. 2019, 9, 2406. [Google Scholar] [CrossRef]
- Jin, S.; Fu, H.; Sun, S.; Jiang, S.; Xiong, Y.; Gong, Y.; Qiao, H.; Zhang, W.; Wu, Y. Integrated analysis of microRNA and mRNA expression profiles during the sex-differentiation sensitive period in oriental river prawn, Macrobrachium nipponense. Sci. Rep. 2017, 7, 12011. [Google Scholar] [CrossRef]
- Tian, C.; Li, Z.; Dong, Z.; Huang, Y.; Du, T.; Chen, H.; Jiang, D.; Deng, S.; Zhang, Y.; Wanida, S.; et al. Transcriptome Analysis of Male and Female Mature Gonads of Silver Sillago (Sillago sihama). Genes 2019, 10, 129. [Google Scholar] [CrossRef]
- Fan, Z.; You, F.; Wang, L.; Weng, S.; Wu, Z.; Hu, J.; Zou, Y.; Tan, X.; Zhang, P. Gonadal transcriptome analysis of male and female olive flounder (Paralichthys olivaceus). BioMed Res. Int. 2014, 2014, 291067. [Google Scholar] [CrossRef]
- Yu, L.; Xu, D.; Ye, H.; Yue, H.; Ooka, S.; Kondo, H.; Yazawa, R.; Takeuchi, Y. Gonadal Transcriptome Analysis of Pacific Abalone Haliotis discus discus: Identification of Genes Involved in Germ Cell Development. Mar. Biotechnol. 2018, 20, 467–480. [Google Scholar] [CrossRef]
- Peng, J.; Wei, P.; Zhang, B.; Zhao, Y.; Zeng, D.; Chen, X.; Li, M.; Chen, X. Gonadal transcriptomic analysis and differentially expressed genes in the testis and ovary of the Pacific white shrimp (Litopenaeus vannamei). BMC Genom. 2015, 16, 1006. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-J.; Zhou, Y.-L.; Wang, Z.-W.; Li, G.-H.; Jin, F.-P.; Cui, L.-L.; Gao, H.-T.; Li, X.-P.; Zhou, L.; Gui, J.-F. Comparative Transcriptome Analysis Reveals Differentially Expressed Genes and Signaling Pathways Between Male and Female Red-Tail Catfish (Mystus wyckioides). Mar. Biotechnol. 2019, 21, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Y.; Gong, Q.; Lai, J.; Song, M.; Du, J.; Deng, X. Gonadal transcriptome sequencing of the critically endangered Acipenser dabryanus to discover candidate sex-related genes. PeerJ 2018, 6, e5389. [Google Scholar] [CrossRef] [PubMed]
- Levy, T.; Rosen, O.; Eilam, B.; Azulay, D.; Aflalo, E.D.; Manor, R.; Shechter, A.; Sagi, A. A Single Injection of Hypertrophied Androgenic Gland Cells Produces All-Female Aquaculture. Mar. Biotechnol. 2016, 18, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ai, C.; Kong, L. CGPS: A machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. J. Genet. Genom. 2018, 45, 489–504. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef]
- Zhou, S.-M.; Tao, Z.; Shen, C.; Qian, D.; Wang, C.-L.; Zhou, Q.-C.; Jin, S. β-actin gene expression is variable among individuals and not suitable for normalizing mRNA levels in Portunus trituberculatus. Fish Shellfish. Immunol. 2018, 81, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Kuchipudi, S.V.; Tellabati, M.; Nelli, R.K.; White, G.A.; Perez, B.B.; Sebastian, S.; Slomka, M.J.; Brookes, S.M.; Brown, I.H.; Dunham, S.P. 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol. J. 2012, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sagi, A.; Ra’anan, Z.; Cohen, D.; Wax, Y. Production of Macrobrachium rosenbergii in monosex populations: Yield characteristics under intensive monoculture conditions in cages. Aquaculture 1986, 51, 265–275. [Google Scholar] [CrossRef]
- Cao, J.; Wu, L.; Jin, M.; Li, T.; Hui, K.; Ren, Q. Transcriptome profiling of the Macrobrachium rosenbergii lymphoid organ under the white spot syndrome virus challenge. Fish Shellfish. Immunol. 2017, 67, 27–39. [Google Scholar] [CrossRef]
- Nguyen Thanh, H.; Zhao, L.; Liu, Q. De novo transcriptome sequencing analysis and comparison of differentially expressed genes (DEGs) in Macrobrachium rosenbergii in China. PLoS ONE 2014, 9, e109656. [Google Scholar] [CrossRef]
- Jung, H.; Yoon, B.-H.; Kim, W.-J.; Kim, D.-W.; Hurwood, D.; Lyons, R.; Salin, K.; Kim, H.-S.; Baek, I.; Chand, V. Optimizing hybrid de novo transcriptome assembly and extending genomic resources for giant freshwater prawns (Macrobrachium rosenbergii): The identification of genes and markers associated with reproduction. Int. J. Mol. Sci. 2016, 17, 690. [Google Scholar] [CrossRef]
- Bose, U.; Kruangkum, T.; Wang, T.; Zhao, M.; Ventura, T.; Mitu, S.A.; Hodson, M.P.; Shaw, P.N.; Sobhon, P.; Cummins, S.F. Biomolecular changes that occur in the antennal gland of the giant freshwater prawn (Machrobrachium rosenbergii). PLoS ONE 2017, 12, e0177064. [Google Scholar] [CrossRef]
- Huntriss, J.; Lu, J.; Hemmings, K.; Bayne, R.; Anderson, R.; Rutherford, A.; Balen, A.; Elder, K.; Picton, H.M. Isolation and expression of the human gametocyte-specific factor 1 gene (GTSF1) in fetal ovary, oocytes, and preimplantation embryos. J. Assist. Reprod. Genet. 2017, 34, 23–31. [Google Scholar] [CrossRef]
- Takemoto, N.; Yoshimura, T.; Miyazaki, S.; Tashiro, F.; Miyazaki, J.-I. Gtsf1l and Gtsf2 are specifically expressed in gonocytes and spermatids but are not essential for spermatogenesis. PLoS ONE 2016, 11, e0150390. [Google Scholar] [CrossRef]
- Jiang, D.N.; Yang, H.H.; Li, M.H.; Shi, H.J.; Zhang, X.B.; Wang, D.S. gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 2016, 83, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.; Chen, J.; Tan, D.; Yang, J.; Sun, L.; Wei, J.; Conte, M.A.; Kocher, T.D.; Wang, D. Transcriptome display during tilapia sex determination and differentiation as revealed by RNA-Seq analysis. BMC Genom. 2018, 19, 363. [Google Scholar] [CrossRef] [PubMed]
- Neill, J.D.; Duck, L.W.; Sellers, J.C.; Musgrove, L.C. A gonadotropin-releasing hormone (GnRH) receptor specific for GnRH II in primates. Biochem. Biophys. Res. Commun. 2001, 282, 1012–1018. [Google Scholar] [CrossRef] [PubMed]
- Millar, R.; Lowe, S.; Conklin, D.; Pawson, A.; Maudsley, S.; Troskie, B.; Ott, T.; Millar, M.; Lincoln, G.; Sellar, R.; et al. A novel mammalian receptor for the evolutionarily conserved type II GnRH. Proc. Natl. Acad. Sci. USA 2001, 98, 9636–9641. [Google Scholar] [CrossRef]
- Desaulniers, A.T.; Cederberg, R.A.; Lents, C.A.; White, B.R. Expression and Role of Gonadotropin-Releasing Hormone 2 and Its Receptor in Mammals. Front. Endocrinol. 2017, 8, 269. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Rosen, O.; Sagi, A. Timing sexual differentiation: Full functional sex reversal achieved through silencing of a single insulin-like gene in the prawn, Macrobrachium rosenbergii. Biol. Reprod. 2012, 86, 91–96. [Google Scholar] [CrossRef]
- Fu, C.; Zeng, Q.; Li, F.; Wang, H.; Sun, J.; Wang, H. Comparative Transcriptome Analysis Reveals Related Regulatory Mechanisms of Androgenic Gland in Eriocheir sinensis. BioMed. Res. Int. 2017, 2017, 4956216. [Google Scholar] [CrossRef]
- Von Philipsborn, A.C.; Jörchel, S.; Tirian, L.; Demir, E.; Morita, T.; Stern, D.L.; Dickson, B.J. Cellular and behavioral functions of fruitless isoforms in Drosophila courtship. Curr. Biol. 2014, 24, 242–251. [Google Scholar] [CrossRef]
- Liu, Y.; Hui, M.; Cui, Z.; Luo, D.; Song, C.; Li, Y.; Liu, L. Comparative transcriptome analysis reveals sex-biased gene expression in juvenile Chinese mitten crab Eriocheir sinensis. PLoS ONE 2015, 10, e0133068. [Google Scholar] [CrossRef]
- Forstbauer, L.M.; Brockschmidt, F.F.; Moskvina, V.; Herold, C.; Redler, S.; Herzog, A.; Hillmer, A.M.; Meesters, C.; Heilmann, S.; Albert, F. Genome-wide pooling approach identifies SPATA5 as a new susceptibility locus for alopecia areata. Eur. J. Hum. Genet. 2012, 20, 326. [Google Scholar] [CrossRef]
- Ge, L.-Q.; Xia, T.; Huang, B.; Song, Q.-S.; Zhang, H.-W.; Stanley, D.; Yang, G.-Q.; Wu, J.-C. Suppressing male spermatogenesis-associated protein 5-like gene expression reduces vitellogenin gene expression and fecundity in Nilaparvata lugens Stål. Sci. Rep. 2016, 6, 28111. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Wang, Y.-L.; Setsuie, R.; Sekiguchi, S.; Sakurai, M.; Sato, Y.; Lee, W.-W.; Ishii, Y.; Kyuwa, S.; Noda, M. Developmental regulation of ubiquitin C-terminal hydrolase isozyme expression during spermatogenesis in mice. Biol. Reprod. 2004, 71, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J. The new function of two ubiquitin C-terminal hydrolase isozymes as reciprocal modulators of germ cell apoptosis. Exp. Anim. 2007, 56, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.T.; Petryk, A.; Marqués, G.; Jarcho, M.; Parvy, J.-P.; Dauphin-Villemant, C.; O’Connor, M.B.; Gilbert, L.I. Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2002, 99, 11043–11048. [Google Scholar] [CrossRef]
- Ranson, H.; Claudianos, C.; Ortelli, F.; Abgrall, C.; Hemingway, J.; Sharakhova, M.V.; Unger, M.F.; Collins, F.H.; Feyereisen, R. Evolution of supergene families associated with insecticide resistance. Science 2002, 298, 179–181. [Google Scholar] [CrossRef]
- Chávez, V.M.; Marqués, G.; Delbecque, J.P.; Kobayashi, K.; Hollingsworth, M.; Burr, J.; Natzle, J.E.; O’Connor, M.B. The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development 2000, 127, 4115–4126. [Google Scholar]
- Zhu, J.; Chen, Y.; Zhao, Q.; Tang, S.; Huang, J.; Shen, X. Expression profile of several genes on ecdysteroidogenic pathway related to diapause in pupal stage of Bombyx mori bivoltine strain. Gene 2019, 707, 109–116. [Google Scholar] [CrossRef]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schütz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835. [Google Scholar] [CrossRef]
- Uhlenhaut, N.H.; Jakob, S.; Anlag, K.; Eisenberger, T.; Sekido, R.; Kress, J.; Treier, A.C.; Klugmann, C.; Klasen, C.; Holter, N.I.; et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 2009, 139, 1130–1142. [Google Scholar] [CrossRef]
- Pisarska, M.D.; Barlow, G.; Kuo, F.T. Minireview: Roles of the forkhead transcription factor FOXL2 in granulosa cell biology and pathology. Endocrinology 2011, 152, 1199–1208. [Google Scholar] [CrossRef]
- Picard, D. Chaperoning steroid hormone action. Trends Endocrinol. Metab. TEM 2006, 17, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Kohno, S.; Katsu, Y.; Urushitani, H.; Ohta, Y.; Iguchi, T.; Guillette, L.J., Jr. Potential contributions of heat shock proteins to temperature-dependent sex determination in the American alligator. Sex. Dev. 2010, 4, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Kurata, S.; Fujimoto, H.; Hoshi, M. Haploid specific activations of protamine 1 and hsc70t genes in mouse spermatogenesis. Biochim. Biophys. Acta 1993, 1174, 274–278. [Google Scholar] [CrossRef]
- Dai, Z.-M.; Zhu, X.-J.; Yang, W.-J. Full-length normalization subtractive hybridization: A novel method for generating differentially expressed cDNAs. Mol. Biotechnol. 2009, 43, 257. [Google Scholar] [CrossRef] [PubMed]
- Sroyraya, M.; Hanna, P.J.; Changklungmoa, N.; Senarai, T.; Siangcham, T.; Tinikul, Y.; Sobhon, P. Expression of the male reproduction-related gene in spermatic ducts of the blue swimming crab, Portunus pelagicus, and transfer of modified protein to the sperm acrosome. Microsc. Res. Tech. 2013, 76, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.M.; Mathialagan, N.; Duffy, J.Y.; Smith, G.W. Regulation and regulatory role of proteinase inhibitors. Crit. Rev. Eukaryot. Gene Expr. 1995, 5, 385–436. [Google Scholar] [CrossRef] [PubMed]
- Coleman, S.; DrÄhn, B.; Petersen, G.; Stolorov, J.; Kraus, K. A Drosophila male accessory gland protein that is a member of the serpin superfamily of proteinase inhibitors is transferred to females during mating. Insect Biochem. Mol. Biol. 1995, 25, 203–207. [Google Scholar] [CrossRef]
- Li, Y.; Qian, Y.-Q.; Ma, W.-M.; Yang, W.-J. Inhibition mechanism and the effects of structure on activity of male reproduction-related peptidase inhibitor Kazal-type (MRPINK) of Macrobrachium rosenbergii. Mar. Biotechnol. 2009, 11, 252–259. [Google Scholar] [CrossRef]
- Qian, Y.Q.; Li, Y.; Yang, F.; Yu, Y.Q.; Yang, J.S.; Yang, W.J. Two Kazal-type protease inhibitors from Macrobrachium nipponense and Eriocheir sinensis: Comparative analysis of structure and activities. Fish Shellfish. Immunol. 2012, 32, 446–458. [Google Scholar] [CrossRef]
- Li, M.; Yang, H.; Zhao, J.; Fang, L.; Shi, H.; Li, M.; Sun, Y.; Zhang, X.; Jiang, D.; Zhou, L.; et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 2014, 197, 591–599. [Google Scholar] [CrossRef]
- Li, M.; Sun, Y.; Zhao, J.; Shi, H.; Zeng, S.; Ye, K.; Jiang, D.; Zhou, L.; Sun, L.; Tao, W. A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile tilapia, Oreochromis niloticus. PLoS Genet. 2015, 11, e1005678. [Google Scholar] [CrossRef] [PubMed]
- See, L.M.; Hassan, R.; Tan, S.G.; Bhassu, S. Novel polymorphic microsatellite DNA markers from Malaysian giant freshwater prawn, Macrobrachium rosenbergii. Genetika 2011, 47, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Jin, M.; Ren, Q. Transcriptome analysis of Macrobrachium rosenbergii intestines under the white spot syndrome virus and poly (I:C) challenges. PLoS ONE 2018, 13, e0204626. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).