Coding and Non-Coding RNA Abnormalities in Bipolar Disorder
Abstract
1. Introduction
2. Methods
2.1. Study Population
2.2. Human Tissue and RNA Processing
2.3. RNAseq Library Preparation, Sequencing and Alignment
2.4. Differential Expression (DE) and Weighted Gene Co-expression Network Analyses (WGCNA)
2.5. Validation and Replication Analyses
2.6. Gene Ontology (GO) term and Gene Set Enrichment Analyses (GSEA)
2.7. Alternative Splicing (AS) Detection
2.8. Detection and Expression Analysis of Circular RNAs
2.9. Code and Data Availability and Data Sharing
3. Results
3.1. Baseline Characteristics
3.2. Coding RNA: Differential Expression, Network and Enrichment Analyses
3.3. Alternative Splicing (AS) Events
3.4. Differentially Expressed Long Non-Coding RNAs in BPD
3.5. Circular RNA Transcripts in Healthy and Affected Brain Tissue
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2015, 386, 743–800. [Google Scholar] [CrossRef]
- Van Heeringen, C.; Marusic, A. Understanding the suicidal brain. Br. J. Psychiatry 2003, 183, 282–284. [Google Scholar] [CrossRef]
- Nock, M.K.; Hwang, I.; Sampson, N.A.; Kessler, R.C. Mental disorders, comorbidity and suicidal behavior: Results from the National Comorbidity Survey Replication. Mol. Psychiatry 2010, 15, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Craddock, N.; Sklar, P. Genetics of bipolar disorder. Lancet 2013, 381, 1654–1662. [Google Scholar] [CrossRef]
- Hou, L.; Bergen, S.E.; Akula, N.; Song, J.; Hultman, C.M.; Landen, M.; Adli, M.; Alda, M.; Ardau, R.; Arias, B.; et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum. Mol. Genet. 2016, 25, 3383–3394. [Google Scholar] [CrossRef] [PubMed]
- Psychiatric GWAS Consortium Bipolar Disorder Working Groupa. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011, 43, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Stahl, E.A.; Breen, G.; Forstner, A.J.; McQuillin, A.; Ripke, S.; Trubetskoy, V.; Mattheisen, M.; Wang, Y.; Coleman, J.R.I.; Gaspar, H.A.; et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 2019, 51, 793–803. [Google Scholar] [CrossRef]
- Kataoka, M.; Matoba, N.; Sawada, T.; Kazuno, A.A.; Ishiwata, M.; Fujii, K.; Matsuo, K.; Takata, A.; Kato, T. Exome sequencing for bipolar disorder points to roles of de novo loss-of-function and protein-altering mutations. Mol. Psychiatry 2016, 21, 885–893. [Google Scholar] [CrossRef]
- Cruceanu, C.; Ambalavanan, A.; Spiegelman, D.; Gauthier, J.; Lafrenière, R.G.; Dion, P.A.; Alda, M.; Turecki, G.; Rouleau, G.A. Family-based exome-sequencing approach identifies rare susceptibility variants for lithium-responsive bipolar disorder. Genome 2013, 56, 634–640. [Google Scholar] [CrossRef][Green Version]
- Goes, F.S.; Pirooznia, M.; Parla, J.S.; Kramer, M.; Ghiban, E.; Mavruk, S.; Chen, Y.C.; Monson, E.T.; Willour, V.L.; Karchin, R.; et al. Exome Sequencing of Familial Bipolar Disorder. JAMA Psychiatry 2016, 73, 590–597. [Google Scholar] [CrossRef]
- Middleton, F.A.; Pato, C.N.; Gentile, K.L.; McGann, L.; Brown, A.M.; Trauzzi, M.; Diab, H.; Morley, C.P.; Medeiros, H.; Macedo, A.; et al. Gene expression analysis of peripheral blood leukocytes from discordant sib-pairs with schizophrenia and bipolar disorder reveals points of convergence between genetic and functional genomic approaches. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2005, 136, 12–25. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gandal, M.J.; Haney, J.R.; Parikshak, N.N.; Leppa, V.; Ramaswami, G.; Hartl, C.; Schork, A.J.; Appadurai, V.; Buil, A.; Werge, T.M.; et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 2018, 359, 693–697. [Google Scholar] [CrossRef] [PubMed]
- Akula, N.; Barb, J.; Jiang, X.; Wendland, J.R.; Choi, K.H.; Sen, S.K.; Hou, L.; Chen, D.T.W.; Laje, G.; Johnson, K.; et al. RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms and GTPase binding in bipolar disorder. Mol. Psychiatry 2014, 19, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Kohen, R.; Dobra, A.; Tracy, J.H.; Haugen, E. Transcriptome profiling of human hippocampus dentate gyrus granule cells in mental illness. Transl. Psychiatry 2014, 4, e366. [Google Scholar] [CrossRef] [PubMed]
- Cruceanu, C.; Tan, P.P.C.; Rogic, S.; Lopez, J.P.; Torres-Platas, S.G.; Gigek, C.O.; Alda, M.; Rouleau, G.A.; Pavlidis, P.; Turecki, G. Transcriptome Sequencing of the Anterior Cingulate in Bipolar Disorder: Dysregulation of G Protein-Coupled Receptors. Am. J. Psychiatry 2015, 172, 1131–1140. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, Y.; Webster, M.J.; Lee, D. Differential activation of immune/inflammatory response-related co-expression modules in the hippocampus across the major psychiatric disorders. Mol. Psychiatry 2016, 21, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Pacifico, R.; Davis, R.L. Transcriptome sequencing implicates dorsal striatum-specific gene network, immune response and energy metabolism pathways in bipolar disorder. Mol. Psychiatry 2016, 22, 441–449. [Google Scholar] [CrossRef]
- Hu, J.; Xu, J.; Pang, L.; Zhao, H.; Li, F.; Deng, Y.; Liu, L.; Lan, Y.; Zhang, X.; Zhao, T.; et al. Systematically characterizing dysfunctional long intergenic non-coding RNAs in multiple brain regions of major psychosis. Oncotarget 2016, 7, 71087–71098. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and Functions of Long Noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef]
- Nagano, T.; Fraser, P. No-Nonsense Functions for Long Noncoding RNAs. Cell 2011, 145, 178–181. [Google Scholar] [CrossRef]
- Brettschneider, J.; Del Tredici, K.; Toledo, J.B.; Robinson, J.L.; Irwin, D.J.; Grossman, M.; Suh, E.; Van Deerlin, V.M.; Wood, E.M.; Baek, Y.; et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann. Neurol. 2013, 74, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Ziats, M.N.; Rennert, O.M. Aberrant expression of long noncoding RNAs in autistic brain. J. Mol. Neurosci. 2013, 49, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; St Laurent, G., III; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, Y.; Nakagawa, S.; Hirose, T.; Okano, H.J.; Takao, M.; Shibata, S.; Suyama, S.; Kuwako, K.I.; Imai, T.; Murayama, S.; et al. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 2013, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Soreq, L.; Guffanti, A.; Salomonis, N.; Simchovitz, A.; Israel, Z.; Bergman, H.; Soreq, H. Long Non-Coding RNA and Alternative Splicing Modulations in Parkinson’s Leukocytes Identified by RNA Sequencing. PLoS Comput. Biol. 2014, 10, e1003517. [Google Scholar] [CrossRef] [PubMed]
- Ganzola, R.; Duchesne, S. Voxel-based morphometry meta-analysis of gray and white matter finds significant areas of differences in bipolar patients from healthy controls. Bipolar Disord. 2017, 386, 743–783. [Google Scholar] [CrossRef] [PubMed]
- Vargas, C.; López-Jaramillo, C.; Vieta, E. A systematic literature review of resting state network—Functional MRI in bipolar disorder. J. Affect. Disord. 2013, 150, 727–735. [Google Scholar] [CrossRef]
- Özerdem, A.; Ceylan, D.; Can, G. Neurobiology of Risk for Bipolar Disorder. Curr. Treat. Options Psychiatry 2016, 3, 315–329. [Google Scholar] [CrossRef]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary Sequence-Mediated Exon Circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef]
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.H.; Bindereif, A. Exon Circularization Requires Canonical Splice Signals. Cell Rep. 2015, 10, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef]
- Mercer, T.R.; Qureshi, I.A.; Gokhan, S.; Dinger, M.E.; Li, G.; Mattick, J.S.; Mehler, M.F. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.D.; Diaz, A.; Nellore, A.; Delgado, R.N.; Park, K.Y.; Gonzales-Roybal, G.; Oldham, M.C.; Song, J.S.; Lim, D.A. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 2013, 12, 616–628. [Google Scholar] [CrossRef]
- Liu, S.J.; Nowakowski, T.J.; Pollen, A.A.; Lui, J.H.; Horlbeck, M.A.; Attenello, F.J.; He, D.; Weissman, J.S.; Kriegstein, A.R.; Diaz, A.A.; et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016, 17, 67. [Google Scholar] [CrossRef]
- Rybak-Wolf, A.; Stottmeister, C.; Glažar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Venø, M.T.; Hansen, T.B.; Venø, S.T.; Clausen, B.H.; Grebing, M.; Finsen, B.; Holm, I.E.; Kjems, J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015, 16, 245. [Google Scholar] [CrossRef]
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610. [Google Scholar] [CrossRef]
- Chen, W.; Schuman, E. Circular RNAs in Brain and Other Tissues: A Functional Enigma. Trends Neurosci. 2016, 39, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yuan, J.; Chen, R. NONCODEv4: Annotation of Noncoding RNAs with Emphasis on Long Noncoding RNAs. Methods Mol. Biol. 2016, 1402, 243–254. [Google Scholar] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef]
- Fleige, S.; Pfaffl, M.W. RNA integrity and the effect on the real-time qRT-PCR performance. Mol. Asp. Med. 2006, 27, 126–139. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From reads to genes to pathways: Differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Research 2016, 5, 1438. [Google Scholar]
- Vitting-Seerup, K.; Porse, B.T.; Sandelin, A.; Waage, J. spliceR: An R package for classification of alternative splicing and prediction of coding potential from RNA-seq data. BMC Bioinform. 2014, 15, 81. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Falcon, S.; Gentleman, R. Using GOstats to test gene lists for GO term association. Bioinformatics 2007, 23, 257–258. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bosnjak, M.; Skunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Heilbronner, U.; Degenhardt, F.; Adli, M.; Akiyama, K.; Akula, N.; Ardau, R.; Arias, B.; Backlund, L.; Banzato, C.E.; et al. Genetic variants associated with response to lithium treatment in bipolar disorder: A genome-wide association study. Lancet 2016, 387, 1085–1093. [Google Scholar] [CrossRef]
- Chen, C.H.; Suckling, J.; Lennox, B.R.; Ooi, C.; Bullmore, E. A quantitative meta-analysis of fMRI studies in bipolar disorder. Bipolar Disord. 2011, 13. [Google Scholar] [CrossRef]
- DelVecchio, G.; Fossati, P.; Boyer, P.; Brambilla, P.; Falkai, P.; Gruber, O.; Hietala, J.; Lawrie, S.M.; Martinot, J.L.; McIntosh, A.M.; et al. Common and distinct neural correlates of emotional processing in Bipolar Disorder and Major Depressive Disorder: A voxel-based meta-analysis of functional magnetic resonance imaging studies. Eur. Neuropsychopharmacol. 2012, 22, 100–113. [Google Scholar] [CrossRef]
- Dell’Osso, L.; Del Grande, C.; Gesi, C.; Carmassi, C.; Musetti, L. A new look at an old drug: Neuroprotective effects and therapeutic potentials of lithium salts. Neuropsychiatr. Dis. Treat. 2016, 12, 1687–1703. [Google Scholar]
- Chiu, C.T.; Wang, Z.; Hunsberger, J.G.; Chuang, D.M. Therapeutic potential of mood stabilizers lithium and valproic acid: Beyond bipolar disorder. Pharmacol. Rev. 2013, 65, 105–142. [Google Scholar] [CrossRef]
- Tsai, L.K.; Wang, Z.; Munasinghe, J.; Leng, Y.; Leeds, P.; Chuang, D.M. Mesenchymal stem cells primed with valproate and lithium robustly migrate to infarcted regions and facilitate recovery in a stroke model. Stroke 2011, 42, 2932–2939. [Google Scholar] [CrossRef]
- Ludwig, B.; Dwivedi, Y. Dissecting bipolar disorder complexity through epigenomic approach. Mol. Psychiatry 2016, 21, 1490–1498. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mo, Y.; Gong, Z.; Yang, X.; Yang, M.; Zhang, S.; Xiong, F.; Xiang, B.; Zhou, M.; Liao, Q.; et al. Circular RNAs in human cancer. Mol. Cancer 2017, 16, 25. [Google Scholar] [CrossRef]
- Dines, M.; Lamprecht, R. The Role of Ephs and Ephrins in Memory Formation. Int. J. Neuropsychopharmacol. 2016, 19. [Google Scholar] [CrossRef] [PubMed]
- Dines, M.; Lamprecht, R. EphrinA4 mimetic peptide targeted to EphA binding site impairs the formation of long-term fear memory in lateral amygdala. Transl. Psychiatry 2014, 4, e450. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Attwood, B.K.; Bourgognon, J.M.; Patel, S.; Mucha, M.; Schiavon, E.; Skrzypiec, A.E.; Young, K.W.; Shiosaka, S.; Korostynski, M.; Piechota, M.; et al. Neuropsin cleaves EphB2 in the amygdala to control anxiety. Nature 2011, 473, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Boyd, A.W.; Bartlett, P.F.; Lackmann, M. Therapeutic targeting of EPH receptors and their ligands. Nat. Rev. Drug Discov. 2014, 13, 39–62. [Google Scholar] [CrossRef]
- Lamminmaki, U.; Nikolov, D.; Himanen, J. Eph Receptors as Drug Targets: Single-Chain Antibodies and Beyond. Curr. Drug Targets 2015, 16, 1021–1030. [Google Scholar] [CrossRef]
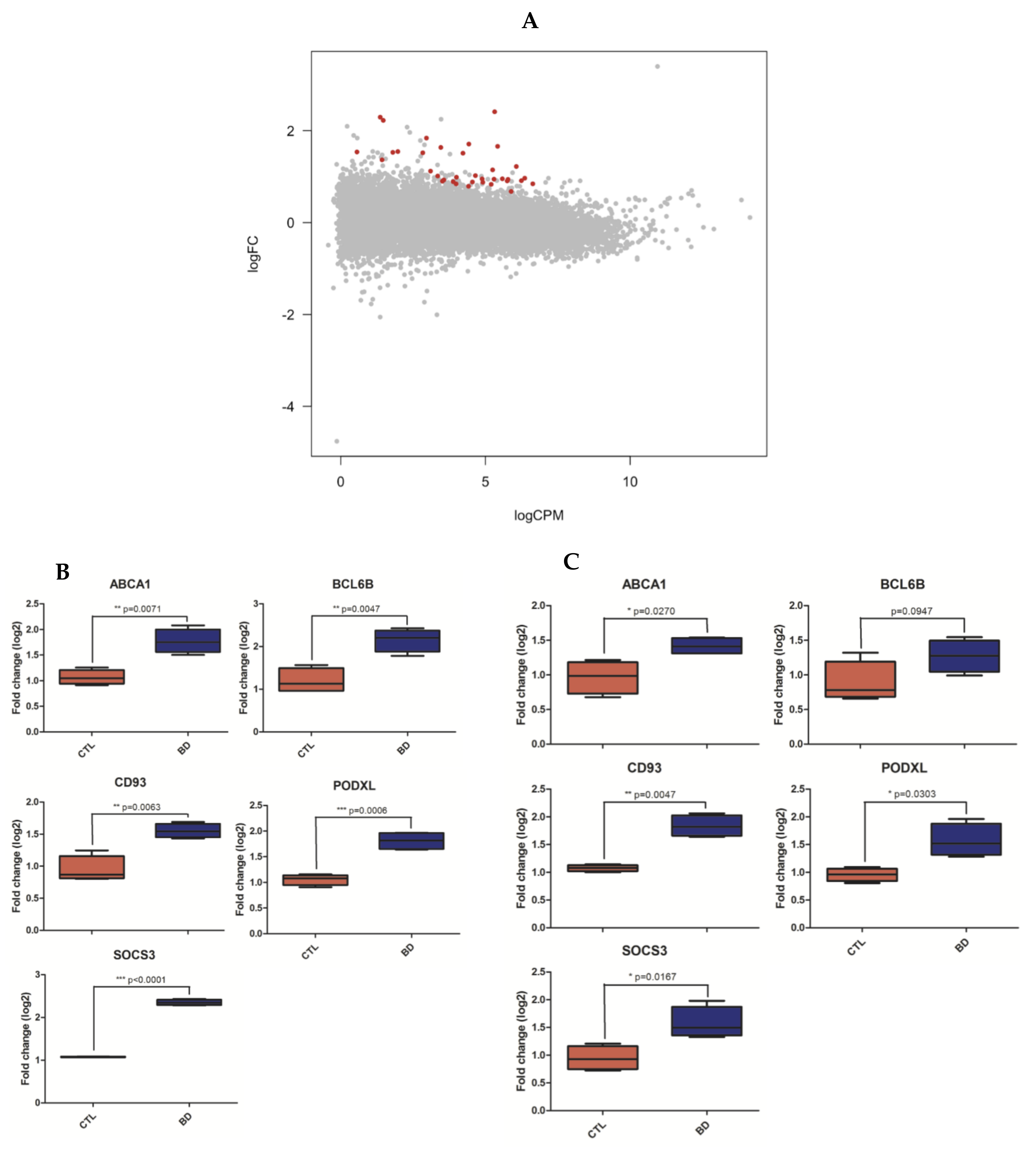
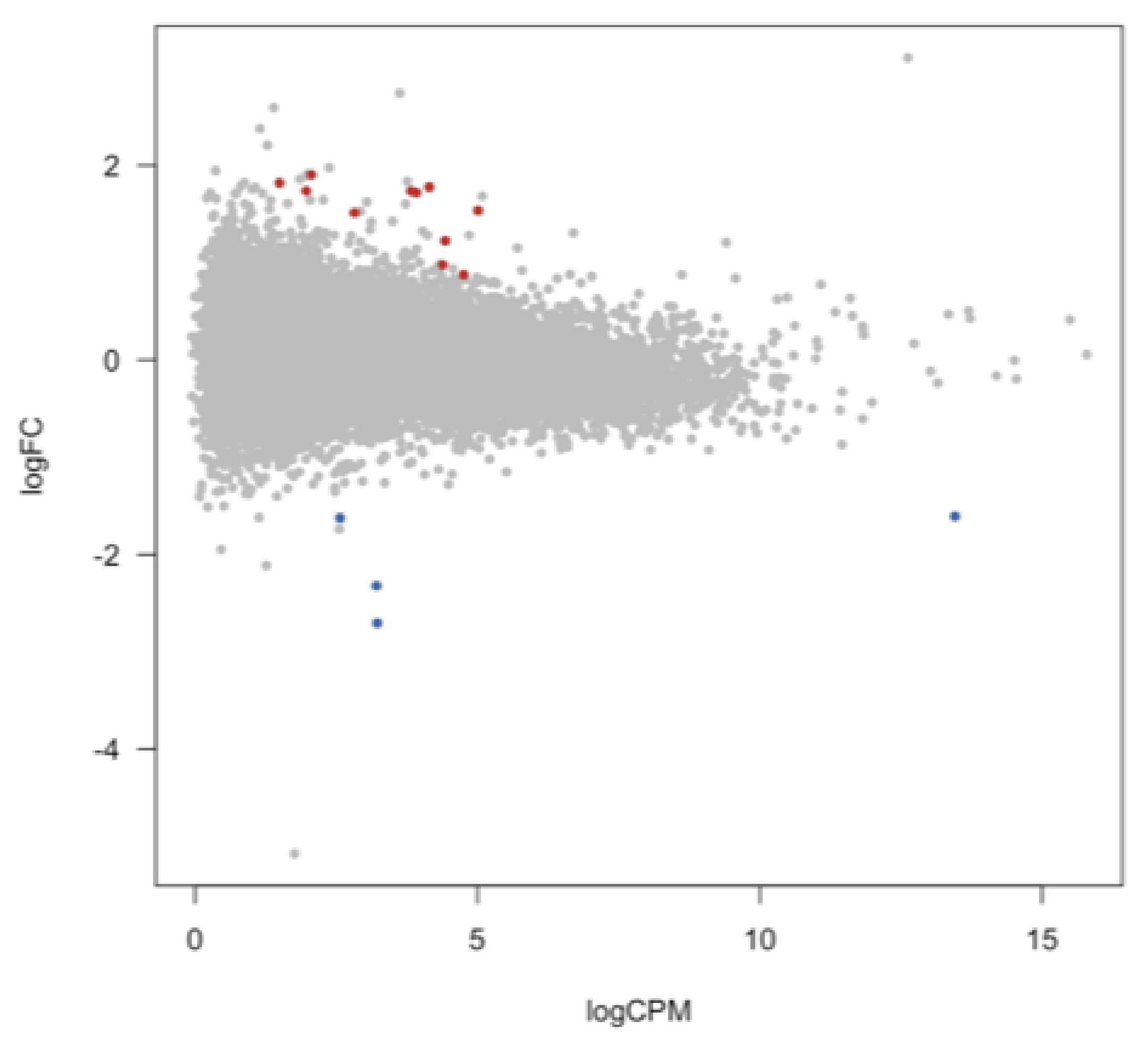
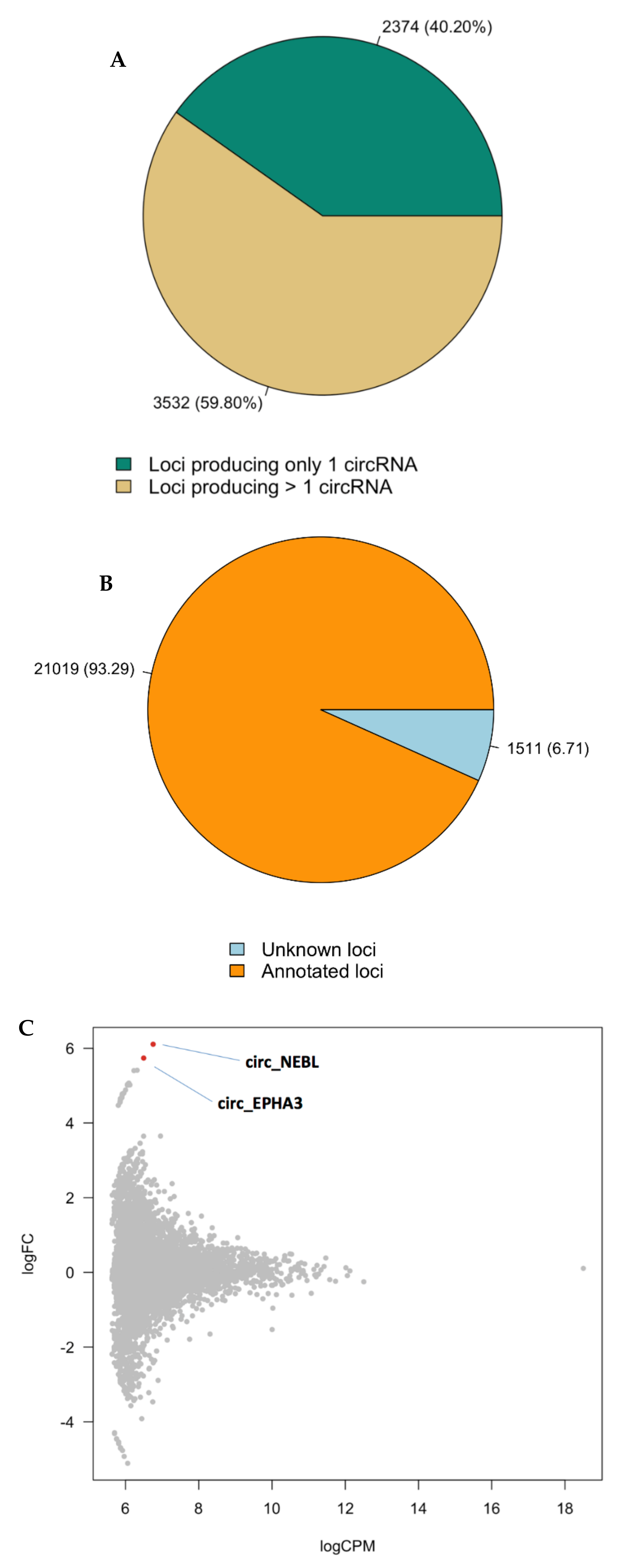
| Cohort | Discovery (N = 8) | Replication (N = 8) | ||
|---|---|---|---|---|
| Controls (N = 4) | Bipolar Disorder (N = 4) | Controls (N = 4) | Bipolar Disorder (N = 4) | |
| PMI | 6 h 55 | 6h 25 | 6 h 47 | 5 h 21 |
| PMI range | 6 h 15–7 h 15 | 4 h 50–8 h 00 | 5 h 05–8 h 10 | 4 h 35–6 h 40 |
| pH | 6.41 | 6.39 | 6.53 | 6.48 |
| pH range | 6.23–6.76 | 6.26–6.53 | 6.47–6.58 | 6.38–6.70 |
| Sex | 1 F, 3 M | 1 F, 3 M | 3 F, 1 M | 4 M |
| Mean age | 65.7 | 72.5 | 70 | 73.5 |
| Age range | 49–88 | 70–79 | 57–93 | 68–81 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luykx, J.J.; Giuliani, F.; Giuliani, G.; Veldink, J. Coding and Non-Coding RNA Abnormalities in Bipolar Disorder. Genes 2019, 10, 946. https://doi.org/10.3390/genes10110946
Luykx JJ, Giuliani F, Giuliani G, Veldink J. Coding and Non-Coding RNA Abnormalities in Bipolar Disorder. Genes. 2019; 10(11):946. https://doi.org/10.3390/genes10110946
Chicago/Turabian StyleLuykx, Jurjen J., Fabrizio Giuliani, Giuliano Giuliani, and Jan Veldink. 2019. "Coding and Non-Coding RNA Abnormalities in Bipolar Disorder" Genes 10, no. 11: 946. https://doi.org/10.3390/genes10110946
APA StyleLuykx, J. J., Giuliani, F., Giuliani, G., & Veldink, J. (2019). Coding and Non-Coding RNA Abnormalities in Bipolar Disorder. Genes, 10(11), 946. https://doi.org/10.3390/genes10110946





