Abstract
Leaf premature senescence largely determines maize (Zea mays L.) grain yield and quality. A natural recessive premature-senescence mutant was selected from the breeding population, and near-isogenic lines were constructed using Jing24 as the recurrent parent. In the near-isogenic lines, the dominant homozygous material was wild-type (WT), and the recessive material of early leaf senescence was the premature-senescence-type ZmELS5. To identify major genes and regulatory mechanisms involved in leaf senescence, a transcriptome analysis of the ZmELS5 and WT near-isogenic lines (NILs) was performed. A total of 8796 differentially expressed transcripts were identified between ZmELS5 and WT, including 3811 up-regulated and 4985 down-regulated transcripts. By combining gene ontology, Kyoto Encyclopedia of Genes and Genomes, gene set, and transcription factor enrichment analyses, key differentially expressed genes were screened. The senescence regulatory network was predicted based on these key differentially expressed genes, which indicated that the senescence process is mainly regulated by bHLH, WRKY, and AP2/EREBP family transcription factors, leading to the accumulations of jasmonic acid and ethylene. This causes stress responses and reductions in the chlorophyll a/b-binding protein activity level. Then, decreased ATP synthase activity leads to increased photosystem II photodamage, ultimately leading to leaf senescence.
1. Introduction
Plant senescence is the last stage of a developmental program, and it usually starts with leaf yellowing [1]. Leaf senescence determines crop grain yield and biomass formation, which is a highly regulated, well-coordinated, and biologically active process that marks the end of the life cycle of the leaf and, ultimately, the whole plant [2,3]. Delaying plant senescence can effectively prolong photosynthesis and increase the overall crop biomass [4,5,6,7,8]. However, premature senescence causes substantial production losses [9,10]. Leaf senescence is a complex physiological process that comprises of chlorophyll decomposition, photosynthesis termination, protein and nucleic acid degradation, molecular metabolism and nutrient transport decreases, and responses to cell death [11,12,13]. Many kinds of environmental factors can promote leaf senescence, such as drought, nutrient starvation, inhibited pollination, salinity stress, darkness, excessive light intensity, and biotic stresses [14,15,16,17]. The partitioning of sugars to various sinks and the associated signals emerging from the interplay of the photosynthetic output of the plant (source strength), as well as the photoassimilate (sink strength) needs, play key roles in the regulation of senescence [16,18,19,20]. Exploring the regulatory mechanisms underlying plant senescence is essential for crop improvement. However, the corresponding mechanisms in maize are still confusing, especially the onset and precise controlling of the senescence progress.
In recent years, some senescence-associated genes (SAGs) have been identified in various plant species at the transcriptional level [21,22,23]. For instance, almost one fourth of Arabidopsis genes are identified to be associated with senescence, as assessed by transcriptome analyses [24]. Numerous studies have identified many key regulatory molecules, as well as a series of signaling pathways, that are involved in gene expression changes during leaf senescence [25]. These regulatory factors include chromatin-modifying factors (DDM1, DRD1, and HDA9-PWR), transcription factors (NAC, bHLH, JAZ, MYC, and WRKY), post-transcriptional regulatory factors, post-translational regulators (MAPK6, MAPKKK18, ATL31, PUB12/13, PUB44, and SSPP), and metabolic regulators [26,27,28]. Importantly, over 150 Arabidopsis mutants and/or transgenic plants have altered leaf senescence phenotypes [29].
Maize is an important crop for human food and animal feed, as well as an important genetic model plant. Anthesis represents the onset of plant senescence, during which maize leaves start to turn to yellow and gradually die. As leaves senesce, their photosynthetic rates decline. Thus, manipulating the maize leaf senescence process could help achieve high grain yield and quality. In the United States of America (USA) and Canada, maize yield was significantly increased by delaying leaf senescence [30,31]. Similarly, owing to delayed leaf senescence, rice grain yields increased by 24% [32] and 10.3% [33,34], respectively. Recently, several omics analyses have been performed to identify senescence-associated genes [35,36,37,38], miRNAs [5], and proteins [39,40] in maize. Although large amounts of genes have been screened, only ZmSnRK1s and knotted1 were experimentally verified [41,42].
Because of the complicated regulatory networks, it is difficult to identify major senescence-associated genes. Premature senescence mutants provide ideal materials for functionally identifying major senescence-associated genes and corresponding regulatory networks. In our previous study, a premature-senescence mutant, early leaf senescence 5 (ZmELS5), was screened from breeding population. In the present study, we aimed to identify senescence-associated genes using a comparative transcriptome analysis between ZmELS5 and wild type (WT) NILs; to construct the corresponding regulatory networks; and to predict a regulatory model for ZmELS5 in maize leaf senescence.
2. Materials and Methods
2.1. Plant Materials
In our previous study, the premature senescence-mutant ZmELS5 in maize was screened from a breeding population and crossed with the normal Jing24 to create near-isogenic lines (NILs) through five back-crosses. In the NILs, the dominant homozygous material was WT, and the recessive material responsible for the early leaf senescence was the premature-senescence type ZmELS5. In the summer of 2016, the ZmELS5 NILs were planted in the Science and Education Park of Henan Agricultural University (Zhengzhou, China; E113°35′, N34°51′). In the field, the leaves of ZmELS5 NILs started to yellow at V6 stage. At V13 stage, the 11th leaf (ear leaf) of mutants and WT were immediately sampled, frozen in liquid nitrogen, with three biological replicates, and further stored at −80 °C in a refrigerator. Additionally, the candidate gene of ZmELS5 was mapped in a 9.58 Mb chromosomal region at Bin 8.03.
2.2. RNA Extraction
Samples of the 11th leaves of the ZmELS5 and its WT NILs were used to extract total RNA. Total RNAs were extracted using TRIzol reagent (Invitrogen, Waltham, MA, USA) following the manufacturer’s instruction. Six libraries were constructed and sequenced using the Illumina HiSeq 2500 platform (Hangzhou Lianchuan Biotechnology Co. Ltd., Hangzhou, China). The entire original sequence data in fastq format have been uploaded to the NCBI short read archive (accession number: PRJNA555720).
2.3. Transcriptome Analysis
To screen for SAGs, a comparative transcriptome analysis was conducted within ZmELS5 and its WT NILs. The raw data of RNA sequencing (RNA-seq) were quality controlled using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The Q30 ratios of the six sequenced libraries were greater than 93%. The trimmed and low quality (Q < 30) sequencing data were removed by Trimmomatic Software V0.36 [43], and 34.26 Gb sequence data were acquired (Table S1). Then, the clean sequencing data were aligned to the maize B73 RefGen_V4.42 reference genome (http://ensembl.gramene.org/Zea_mays/Info/Index) using HISAT2 V2.1.0 [44]. Sorting and converting the sam file to a bam file was done using SAMtools software V1.9 [45]. The transcripts were assembled using StringTie software V1.3.4 [46], and the count matrix was generated using prepDE.py (https://ccb.jhu.edu/software/stringtie/dl/prepDE.py). The differentially expressed gene (DEG) analysis was performed using DESeq2 software V1.22.2 [47]. A principal component analysis (PCA) was performed using the DESeq2 VST mode. Transcripts with a log2 fold change >2 and a false discovery rate-corrected p-value <0.05 were determined to be differentially expressed. We used the apeglm method for LFC effect-size shrinkage [48], which improved on the previous estimator.
The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using the maize profile database (org. Zeamays; e.g., sqlite) in the clusterProfiler software V3.10.1 [49], and AnnotationHub (V2.14.5) R package [50]. The enrichment analyses used the ensemble database to convert the gene number (maizegbdId) to the corresponding Entrez ID.
The gene set enrichment analysis (GSEA) analysis used Cluster Profiler’s [51] gseGO command. GSEA uses all the transcript data sets expressed in the sequenced materials. The FTP database based on the Ensembl genome (ftp://ftp.ensemblgenomes.org/pub/current/plants/tsv/zea_mays/) was used to convert transcription IDs into Entrez IDs using a Python 3 script. For the GSEA, gseGO parameters were set as repeat 1000 times, nPerm = 1000, minGSSize = 15, and maxGSSize = 500 [52]. In addition, the online tool MCENet (http://bioinformatics.cau.edu.cn/MCENet/index.php) was used for the transcription factor enrichment analysis of the gene sets [53]. Based on the transcription factors corresponding to the expressed genes, the transcription factors corresponding to the DEGs were selected for the enrichment analysis.
2.4. Quantitative Reverse Transcription PCR (RT-qPCR) Validation
The gene expressions, which correlated with premature senescence, were verified by RT-qPCR. RT-qPCR primers (http://primer3.ut.ee/) are listed in Table S2. The relative expression levels of DEGs identified in the transcriptome analyses were measured using the PrimeScript™ RT reagent kit with gDNA Eraser (Perfect Real Time) and the SYBR® Premix EX Taq™ II (Tli RNaseH Plus) Kit (TaKaRa, Dalian, China). The RT-qPCR was performed using the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The Actin gene was used as the endogenous control [54]. The relative expressions of RNAs and targets were calculated using the 2−ΔΔCt method [55].
2.5. Determination of Chlorophyll Content
In total, 0.1 g leaf was soaked in 80% acetone for 24 h. The chlorophyll content was determined using a spectrophotometer (UV-3200PC, Shanghai MAPADA Instruments Co. Ltd., Shanghai, China) according to the method by Porra [56]. Three biological replicates were performed per material, with three technical replicates per biological replicate.
2.6. Putative Senescence Regulatory Network
The senescence regulatory network was predicted using the online tool STRING V11 (https://string-db.org/) [57] by the GO and GSEA enrichment analyses for these identified important DEGs.
3. Results
3.1. Characterization of Maize Premature-Senescence Mutant ZmELS5
From 2016 to 2019, ZmELS5 and its WT NILs were planted in different environments (Zhengzhou, China, E113°35′, N34°51′; Sanya, China, E108°92′, N18°44′). In the field, the ZmELS5 NIL plants began to senesce at the V6 stage, as indicated by their leaves gradually turning yellow from the bottom up, and by the V13 stage, the 12th leaves margin of the ZmELS5’s started to yellow (Figure 1A). Furthermore, the 11th leaves (ear leaves) of ZmELS5 NIL plants showed significant senescence phenotypes (Figure 1B). However, all the leaves of the WT NIL plants were green. The 11th leaves (ear leaves) were sampled as shown in Figure 1B, and the chlorophyll content was measured as shown in Figure 2. During this period, the chlorophyll content in ZmELS5 was significantly lower than that in WT. Chlorophyll a and chlorophyll b were significantly lower in mutants than in WT. This indicates that, at this stage, the mutant has started to senesce. There were significant differences in the metabolic activities between the ZmELS5 and WT NILs.
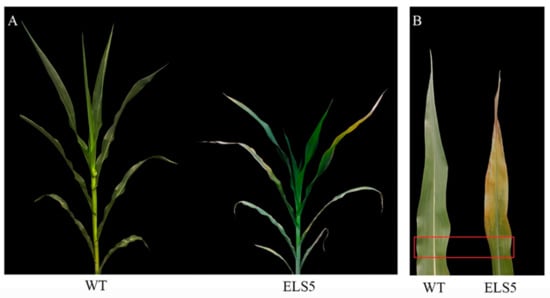
Figure 1.
Phenotyping of wild-type (WT) and ZmELS5 near-isogenic lines (NILs) at V13 stage: (A) Whole plant of WT and ZmELS5 NILs at V13 stage; (B) the 11th leaf phenotype of the WT and ZmELS5 NILs at V13 stage.
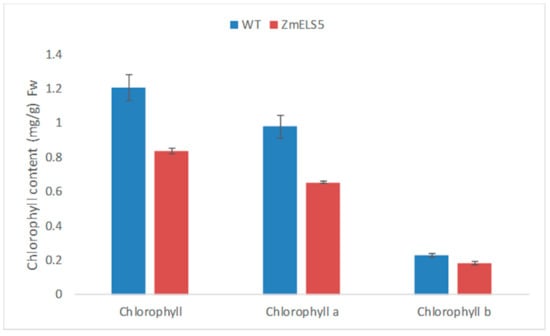
Figure 2.
Chlorophyll contents in the leaves of WT and ZmELS5 NILs.
3.2. Differentially Expressed Gene Identification
For the comparative transcriptome analysis, six samples (three replicates each for the WT and ZmELS5 NILs) were used to construct cDNA libraries and for RNA-seq, and 34.26 Gb raw data were obtained. After low quality reads were filtered, the clean reads were then aligned with the maize B73 genome (RefGen_V4.42). Approximately 85% of the clean reads were unique and could be mapped (Table 1).

Table 1.
Overview of mapping quality in RNA-seq data.
Between WT and ZmELS5 NILs, 8796 (log2 fold change >2.0, count >10, 10.2% of 85,861 total) significant differentially expressed transcripts were identified, including 4985 down-regulated transcripts and 3811 up-regulated transcripts. Among them, 5428 differentially expressed transcripts were annotated, 2292 transcripts (1808 genes) were up-regulated, and 3136 transcripts (2746 genes) were down-regulated (Figure 3). Of the DEGs, the up-regulated genes Zm00001d016441, Zm00001d036370, Zm00001d024767, Zm00001d045251, and Zm00001d014993, and the down-regulated genes Zm00001d048702, Zm00001d010821, Zm00001d026405, Zm00001d018460, and Zm00001d004855, showed the most significant differences. These genes may play important roles in ZmELS5 NILs’ premature leaf senescence.
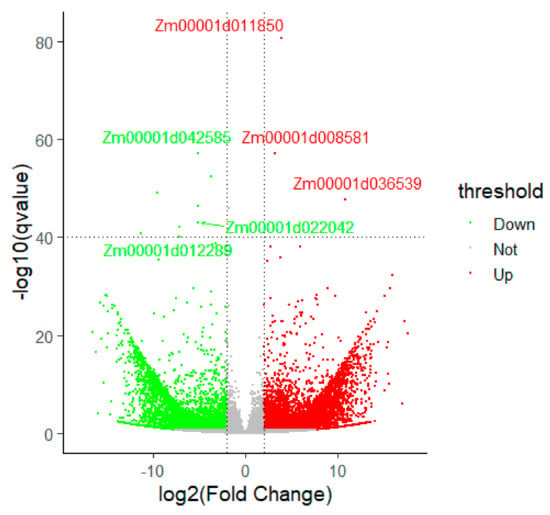
Figure 3.
Functional classification of differentially expressed genes. Red dots indicate up-regulated genes and green dots indicate down-regulated genes under significant levels of Padj <0.05 and Log2 fold change >2.
Based on the Annotation Hub database, these DEGs were enriched to different classes and further analyzed. In the GO analysis, the identified DEGs were classified into three classes: Molecular function, biological process, and cellular component-related genes (Figure 4A). The molecular function-related genes were all enriched in the chlorophyll-binding group. Most of the biological process-related genes were involved in photosynthesis, precursor metabolite generation, and abiotic stimulus responses (Figure 4A and Figure 5, and Figure S1). The other DEGs were mainly enriched in the synthesis of thylakoid, thylakoid part, thylakoid membrane, chloroplast, photosystem (PS) I, and photosynthetic membrane (Figure 4A and Figure 6).
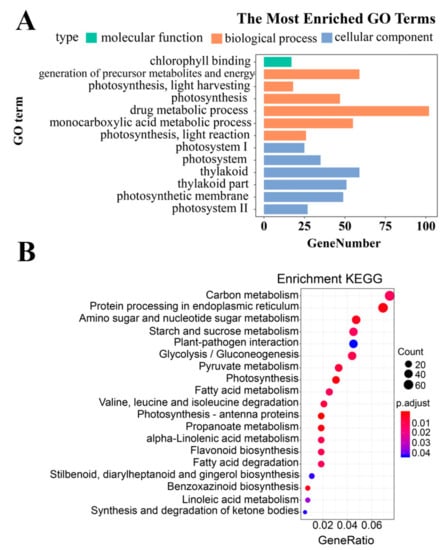
Figure 4.
(A) Gene ontology (GO) and (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed genes.
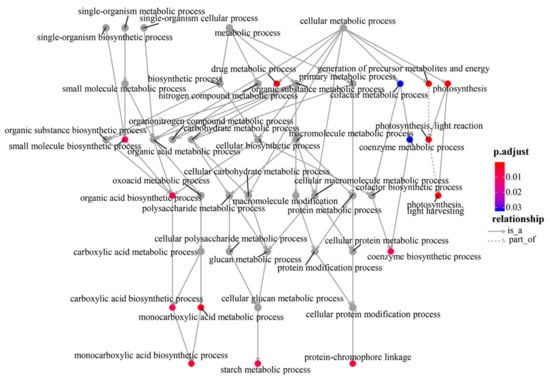
Figure 5.
Gene and biological process connect network.
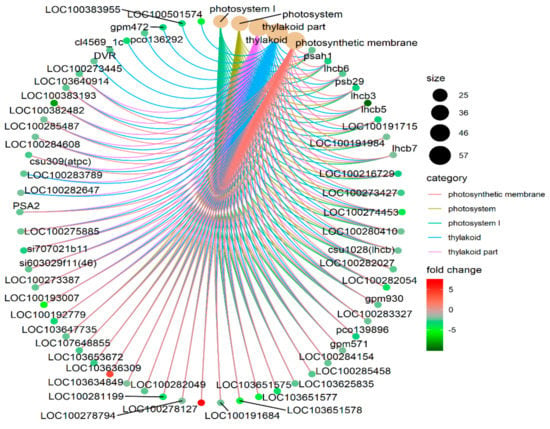
Figure 6.
Different cellular components connect network.
The KEGG analysis indicated that the DEGs were mainly enriched in carbon metabolism, starch and sucrose, amino sugar and nucleotide sugar metabolism, protein processing in endoplasmic reticulum, photosynthesis, and photosynthesis-antenna proteins (Figure 4B).
The GSEA was performed on all the tested genes in WT and ZmELS5 NILs (Figure 7 and Figure 8). The genes up-regulated during the leaf senescence of ZmELS5 NILs were mainly distributed in cell membranes, vacuoles, and other cell components, and mainly involved in the regulation of stress response. The down-regulated genes were mainly distributed in thylakoid, chloroplast, light system, and other related cell components, and they were mainly involved in the regulation of photosynthesis and light response (Figure 7). The enrichment of GSEA cell components revealed two core networks. One is the network of photosynthesis and the other is the vacuole-related network (Figure 8). The GSEA enrichment results indicated that most of the up-regulated genes were located in vacuoles and tonoplasts. There were 16 genes co-expressed in the vacuolar core module of the GSEA co-expression analysis. The functions of the genes enriched in the vacuoles mainly included histone H4, β-galactosidase 15, hydrogen exchanger 4, and tonoplast intrinsic protein 3.
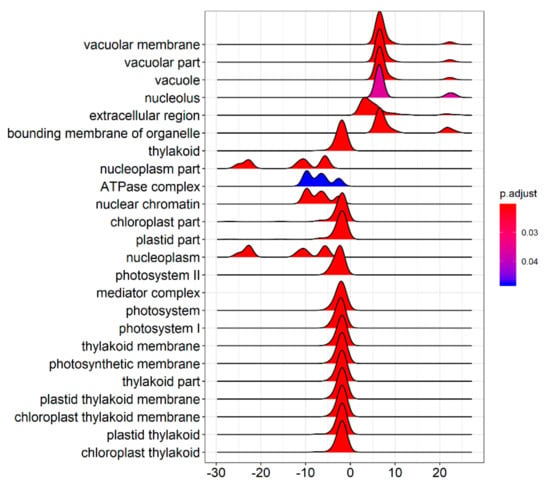
Figure 7.
Gene set enrichment analysis of cell component distribution.
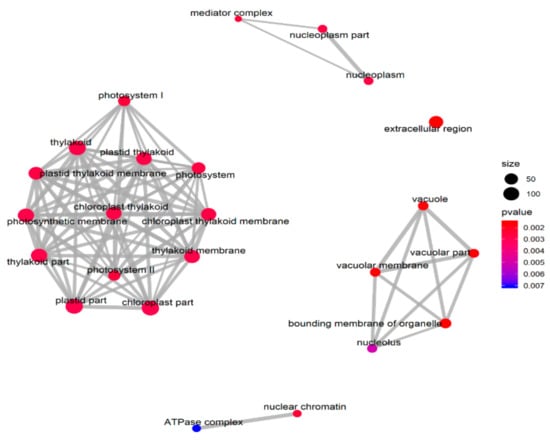
Figure 8.
Network of roles of cellular components enriched in gene sets.
There were 14 co-expressed genes in the light and action core modules of GO and GSEA (Table 2). The functions of these genes were mainly involved with the light-harvesting complex, which consists of chlorophylls a and b and the chlorophyll a/b-binding protein. These genes were significantly down-regulated in the ZmELS5 NILs.

Table 2.
Differentially expressed genes screened by GO analysis and gene set enrichment analysis (GSEA) in ZmELS5 NILs leaf senescence.
3.3. Characterizing the Expression of Transcription Factor Genes in Leaf Senescence
Transcription factors play critical roles in the onset of leaf senescence. Of the identified 3170 DEGs during maize leaves senescence, 170 transcription factors were identified that belonged to 17 transcription factor families, including WRKY, bHLH, MYB, NAC, and ERF (Table S3). In particular, the WRKY, NAC, MYB, bHLH, and HD-ZIP transcription factor families had significantly differential expressions that were induced by premature senescence. Most of these transcription factor families have been identified as important leaf senescence regulators in Arabidopsis.
3.4. Verifying Differential Gene Expression Profiles Using RT-qPCR
DEGs involved in multiple core pathways during leaf senescence were selected for RT-qPCR validation. These genes mainly encoded chlorophyll a/b-binding protein, light harvesting complex, PSI subunit O, heat shock proteins, and WRKY transcription factors. The genes were significantly differentially expressed between WT and ZmELS5 NILs, as assessed by RT-qPCR (Figure 9). Of them, chlorophyll a/b-binding protein, light harvesting complex a/b-binding protein, and PSI subunit O-related genes were significantly down-regulated, while heat-shock protein and WRKY transcription factor-related genes were up-regulated. The RT-qPCR results also revealed that the expression levels of the selected genes were consistent with those determined using RNA-seq, and the two showed a strong correlation. The expression pattern obtained by RT-qPCR was strongly correlated with the RNA-seq results (R2 = 0.8981), indicating that the RNA-seq data are reliable (Figure S2).
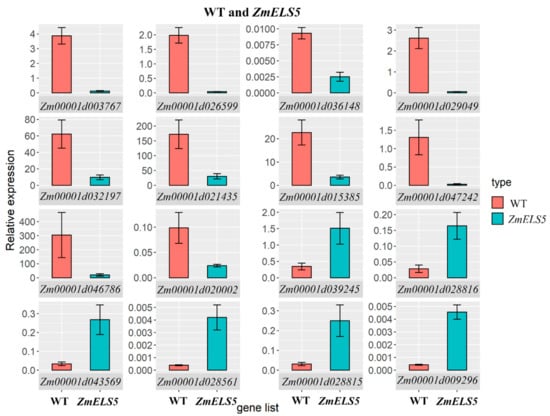
Figure 9.
Comparison of expression of WT and ZmELS5 NILs.
4. Discussion
4.1. Key Genes in ZmELS5 Leaf Premature Senescence
Leaf senescence is accompanied by a change in leaf color, which reflects chlorophyll loss, and ends with the death or abscission of the leaf. In the cells of senescing leaves, a highly ordered disassembly and degradation of cellular components occurs [58]. During leaf senescence, the chlorophyll is gradually degraded with the thylakoid membrane converting to a colorless decomposition product that is stored in the vacuole through a multi-step pathway. The order in which the various components in the leaves senesce is as follows: Chlorophyll, chlorophyll-binding protein, PSII, and PSI. In the present study, the leaves of ZmELS5 NILs gradually senesced from the bottom. DEGs were identified using a comparative transcriptome analysis. The results revealed that the genes involved in photosynthesis-related and stress responses, and the cellular components of the thylakoid, thylakoid membrane, chloroplast, PSI, and photosynthetic membrane, were all down-regulated, which is consistent with chlorophyll decomposition and decreasing photosynthesis. However, the genes mainly encoding protein degradation-related enzymes, transcription factors, and stress-responsive proteins were up-regulated. They may be involved in the onset of leaf senescence and senescence-induced metabolisms. Of them, the homologue of Zm00001d016441 in Arabidopsis encodes a kind of tyrosine aminotransferase that is involved in stress tolerance [59]. The gene Zm00001d036370 encodes chitinase, which is expressed in plants in response to abiotic stresses and during developmental processes [60]. The gene Zm00001d024767 encodes aspartic proteinase A1, and its homologue in Arabidopsis is involved in several physiological processes in plants, including protein processing, senescence, and stress responses [61]. The gene Zm00001d014993 encodes luminal-binding protein 2 that has a role in the regulation of tobacco stress-induced programmed cell death [62]. The gene Zm00001d049217 is a homologue of Arabidopsis SAG12, which regulates senescence processes in a Gibberellic acid-dependent pathway [63]. The genes Zm00001d043025, Zm00001d003412, and Zm00001d012427 encode WRKY33, P-type R2R3 Myb protein, and ZAT11 transcription factor, respectively, which respond to abiotic stresses in Arabidopsis [64,65,66]. Additionally, several pathogenesis-related genes were significantly up-regulated, such as Zm00001d009296 (ZmPRms), Zm00001d018738 (prp4), and Zm00001d023811 (PR10). In maize, ZmPRms possibly to play an important role in the defense of aflatoxin infection [67]; Prp4 is involved in responding to the infection by Aspergillus flavus [68], and PR1 family genes were associated with Fusarium graminearum infection [69], which is the primary pathogen causing stalk rot in maize [70]. This indicates that pathogenesis-related genes (PR proteins) play crucial roles in plants’ defense system and leaf senescence.
In our previous study, the candidate gene of ZmELS5 mutant senescence was mapped to the chromosomal region Chr8: 86,964,414-92,548,025. In this region, the several differently expressed genes were distributed, such as Zm00001d009945, Zm00001d009910, Zm00001d009903, Zm00001d009928, and Zm00001d009936. Zm00001d009910 encoded a NADH protein. Zm00001d009928 encoding photosystem II reaction center PsbP family protein was highly down-regulated. Zm00001d00009936 encoded a Basic endochitinase A protein.
4.2. Transcription Factors Involved in the Regulation of ZmELS5 Leaf Senescence
Transcription factors play critical roles in responding to different environmental factors and to the onset of many biological progresses. To date, several transcription factor families have been identified as taking part in the regulation of leaf senescence progress, such as NAC, WRKY, MYB, C2H2 zinc-finger, bZIP, and AP2/EREBP [71]. In Arabidopsis, the WRKY family of transcription factors plays an important role in regulating plant leaf senescence by a jasmonic acid (JA)-dependent pathway or chrlorophyll degradation [17,72,73,74], such as WRKY53 [74] and WRKY6 [75]. In global transcriptome profiling, more than 30 NAC genes showed enhanced expression levels during natural leaf senescence in Arabidopsis, highlighting their importance in plant senescence regulation [21,76], such as ORESARA1 [77] and Oresara1 sister 1 (ORS1) [78]. The bHLH subgroup IIId factors (bHLH03, bHLH13, bHLH14, and bHLH17) bind to the promoter of SAG29 and repress its expression to attenuate MYC2/MYC3/MYC4/MYC5-activated JA-induced leaf senescence [79,80]. The Arabidopsis MYBS3 plays a critical role in the developmentally-regulated and dark-induced leaf senescence by repressing transcription [81]. In addition, heat shock proteins are involved in the degradation of protein components during leaf senescence [82], such as heat shock protein (Hsp) 70 and Hsp90. In maize, several TF families have been identified to be associated with leaf senescence by RNA-seq profiling, such as NAC, MYB, bHLH, AP2/EREBP, C2h2, and bZIP families [83]. However, only NAC007 has been experimentally verified [38]. In the transcription factor enrichment analysis of the present study, these transcription factor family members were determined to be associated with the premature leaf senescence of ZmELS5 NILs. Transcription factors, such as WRKY, TCP, MYC, and NAC, usually take part in leaf senescence regulation through a JA signaling-dependent pathway. Chlorophyll breakdown is a feature of JA-induced, as well as natural, leaf senescence, and the JA-responsive enzyme chlorophyllase is the main enzyme responsible for chlorophyll breakdown [84].
4.3. Predicted Regulatory Networks of Maize Leaf Senescence
The onset of leaf senescence depends on developmental stages and environmental signals. Once leaf senescence begins, concerted cellular events are triggered, such as rapid chlorophyll degradation, increased membrane ion leakage, and, eventually, the decline of photochemical biosynthesis [85]. We predicted the possible senescence regulatory networks using important DEGs and transcription factors (Figure 10 and Figure 11). The premature senescence of the ZmELS5 NILs may be regulated by a variety of signals, ultimately leading to leaf senescence. In the JA-regulated senescence signaling pathway in Arabidopsis, the bHLH transcription factor negatively regulates the senescence gene SAG29 [86]. Signal pathways for leaf senescence and plant defense responses may overlap [14,87]. The upstream bHLH86 transcription factor of the senescence regulatory network negatively regulates stress-responsive proteins. The homeodomain-leucine zipper transcription factor hb12 up-regulates expression during senescence initiation and positively regulates stress-responsive proteins. Stress-responsive proteins, such as Hsp90, luminal-binding protein, and chitinase 2, are up-regulated. The stress-responsive proteins positively regulate the chlorophyll a/b-binding proteins. The expression levels of chlorophyll a/b-binding protein, as well as Cab48, Lhcb3, gpm571, and Zm00001d011285, gradually decrease as the senescence process extends. The corresponding chlorophyll content gradually decreases, leading to the senescence of phenotypic leaves. The ndh gene is involved in the protection of chloroplasts from photooxidative stress in mature senescent leaves [88]. The down-regulated expression of ndhE and ndhK reduces the inhibition of photooxidative stress on chloroplasts in senescent leaves and aggravates the degradation of chloroplast proteins. The decreased ATP synthase activity leads to increased PSII photodamage [89]. Starch synthase4 is expressed in a specific region of the thylakoid and restricts the formation of starch granules in these areas of the chloroplast. The down-regulation of starch synthase4 leads to the accumulation of polymers as large as starch granules in this region of the thylakoid, which affects photosynthetic reactions [90], ultimately leading to leaf senescence. The WRKY53 transcription factor activates SAG12 from upstream. This eventually leads to the accumulation of JA, a decreased chlorophyll content, decreased PSII activity, and the promotion of senescence [17]. The accumulation of JA causes a stress response. The overexpression of ERF1 induces defense responses in ethylene or JA mutants, as well as an induction of many other ethylene- and JA-reactive genes [91].
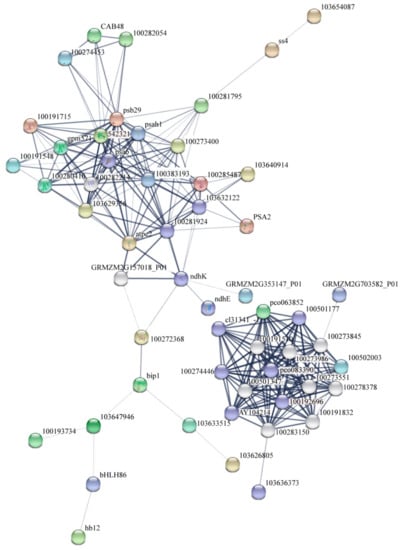
Figure 10.
The string network of proteins (encoded by the differentially expressed genes (DEGs)) involved in senescence signaling pathways.
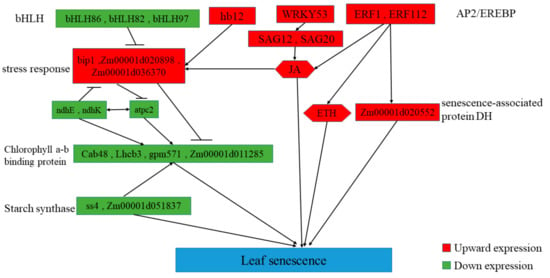
Figure 11.
Predicting the regulatory process of premature senescence of mutant leaves.
4.4. SAG Mutant Identification and the Dissection of Related Regulatory Mechanisms
Leaf senescence is a complex process that is regulated by intricate gene networks in plants [25,92]. Uncovering the related gene regulatory networks is critical for crop improvement. A comparative omics analysis and bioinformatics analysis will be helpful for SAG screening and constructing the related regulatory networks. However, the SAG functions still need to be experimentally verified, as well as the identities of key SAGs. Leaf senescence-related mutants can provide materials for identifying key SAGs and the corresponding regulatory mechanisms. In the present work, we conducted a comparative transcriptome analysis between the premature-senescence mutant ZmELS5 and its WT NILs. This transcriptome analysis found important DEGs and pathways involved in the process of premature leaf senescence, and, in combination with previous studies, can aid in predicting possible regulatory networks for premature senescence. This provides a theoretical basis for further gene cloning and functional validation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/11/944/s1: Figure S1: Different molecular function connect network; Figure S2: RNA-seq and RT-qPCR; Table S1: The detailed information of raw reads from different samples; Table S2: RT-qPCR primer information; Table S3: Results of transcription factor enrichment analysis; Table S4: Significant differences in DEGs, and DEGs involved in important pathways.
Author Contributions
J.T. and Z.Z. conceived and designed the experiments. M.C., X.S., and Y.L. performed the experiments. M.C. and Z.G. analyzed the data. M.C. and Z.Z. wrote the article. Z.Z. and J.T. revised the article.
Funding
This research was supported by the fund from the Henan Provincial Higher Education Key Research Project (20A210003) and the 2019 Henan Agricultural University Science and Technology Innovation Fund (KJCX2019A01).
Acknowledgments
The authors thank Lesley Benyon, from Liwen Bianji, Edanz Group, China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Conflicts of Interest
Authors declare no conflict of interest.
References
- Woo, H.R.; Masclaux-Daubresse, C.; Lim, P.O. Plant senescence: How plants know when and how to die. J. Exp. Bot. 2018, 69, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, J.; Gregersen, P.L.; Krupinska, K. Identification of predominant genes involved in regulation and execution of senescence-associated nitrogen remobilization in flag leaves of field grown barley. J. Exp. Bot. 2014, 65, 3963–3973. [Google Scholar] [CrossRef] [PubMed]
- Kohl, S.; Hollmann, J.; Blattner, F.R.; Radchuk, V.; Andersch, F.; Steuernagel, B.; Schmutzer, T.; Scholz, U.; Krupinska, K.; Weber, H.; et al. A putative role for amino acid permeases in sink-source communication of barley tissues uncovered by RNA-seq. BMC Plant Biol. 2012, 12, 154. [Google Scholar]
- Ra, R. Selectable traits to increase crop photosynthesis and yield of grain crops. J. Exp. Bot. 2000, 51, 447–458. [Google Scholar]
- Wu, X.; Ding, D.; Shi, C.; Xue, Y.; Zhang, Z.; Tang, G.; Tang, J. microRNA-dependent gene regulatory networks in maize leaf senescence. BMC Plant Biol. 2016, 16, 73. [Google Scholar] [CrossRef] [PubMed]
- Robson, P.; Mos, M.; Clifton-Brown, J.; Donnison, I. Phenotypic Variation in Senescence in Miscanthus: Towards Optimising Biomass Quality and Quantity. BioEnergy Res. 2012, 5, 95–105. [Google Scholar] [CrossRef]
- Gregersen, P.L.; Culetic, A.; Boschian, L.; Krupinska, K. Plant senescence and crop productivity. Plant Mol. Biol. 2013, 82, 603–622. [Google Scholar] [CrossRef]
- Khan, M.; Rozhon, W.; Poppenberger, B. The role of hormones in the aging of plants—A mini-review. Gerontology 2014, 60, 49–55. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Li, B.; Tong, Y.; Yang, X.; Li, Z. Comparative study on physiological traits related with grain filling and photosynthesis of flag leaf in early aging and normal near--isogenic lines of common wheat. Acta Agron. Sin. 2006, 32, 1642–1648. [Google Scholar]
- Pleuel, H.; Ojanpera, K.; Danielsson, H.; Sild, E.; Gelang, J.; Wallin, G.; Skärby, L.; Sellden, G.; Fl, P. Effects of Ozone on Leaf Senescence in Spring Wheat-Possible Consequences for Grain Yield; Food and Agriculture Organization: Rome, Italy, 1997; Volume 37. [Google Scholar]
- Koyama, T. The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front. Plant Sci. 2014, 5, 650. [Google Scholar] [CrossRef]
- Sarwat, M.; Naqvi, A.R.; Ahmad, P.; Ashraf, M.; Akram, N.A. Phytohormones and microRNAs as sensors and regulators of leaf senescence: Assigning macro roles to small molecules. Biotechnol. Adv. 2013, 31, 1153–1171. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, B.; Du, M.; Eneji, A.E.; Wang, B.; Duan, L.; Li, Z.; Tian, X. Mechanism of phytohormone involvement in feedback regulation of cotton leaf senescence induced by potassium deficiency. J. Exp. Bot. 2012, 63, 5887–5901. [Google Scholar] [CrossRef] [PubMed]
- Quirino, B.F.; Noh, Y.-S.; Himelblau, E.; Amasino, R.M. Molecular aspects of leaf senescence. Trends Plant Sci. 2000, 5, 278–282. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Gil Nam, H. Leaf Senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Thomas, H. Senescence, ageing and death of the whole plant. New Phytol. 2013, 197, 696–711. [Google Scholar] [CrossRef]
- Schippers, J.H.M. Transcriptional networks in leaf senescence. Curr. Opin. Plant Biol. 2015, 27, 77–83. [Google Scholar] [CrossRef]
- Lee, E.A.; Tollenaar, M. Physiological Basis of Successful Breeding Strategies for Maize Grain Yield. Crop Sci. 2007, 47, S202–S215. [Google Scholar] [CrossRef]
- Wingler, A.; Masclaux-Daubresse, C.; Fischer, A.M. Sugars, senescence, and ageing in plants and heterotrophic organisms. J. Exp. Bot. 2009, 60, 1063–1066. [Google Scholar] [CrossRef]
- Thomas, H.; Ougham, H. The stay-green trait. J. Exp. Bot. 2014, 65, 3889–3900. [Google Scholar] [CrossRef]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.-S.; Penfold, C.A.; Jenkins, D. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Earl, S.; Harrison, E.; Mathas, E.; Navabpour, S.; Page, T.; Pink, D. The molecular analysis of leaf senescence—A genomics approach. Plant Biotechnol. J. 2003, 1, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.-Y.; Hu, W.-J.; Luo, H.; Xia, Y.; Zhao, Y.; Wang, L.-D.; Zhang, L.-M.; Luo, J.-C.; Jing, H.-C. Transcriptome profiling of developmental leaf senescence in sorghum (Sorghum bicolor). Plant Mol. Biol. 2016, 92, 555–580. [Google Scholar] [CrossRef] [PubMed]
- Zentgraf, U.; Doll, J.; Riester, L. Live and Let Die: The Core Circadian Oscillator Coordinates Plant Life History and Pilots Leaf Senescence. Mol. Plant 2018, 11, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, J.H.; Lyu, J.I.; Woo, H.R.; Lim, P.O. New insights into the regulation of leaf senescence in Arabidopsis. J. Exp. Bot. 2018, 69, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Woo, H.R.; Nam, H.G. Toward Systems Understanding of Leaf Senescence: An Integrated Multi-Omics Perspective on Leaf Senescence Research. Mol. Plant 2016, 9, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Podzimska-Sroka, D.; O’Shea, C.; Gregersen, P.L.; Skriver, K. NAC Transcription Factors in Senescence: From Molecular Structure to Function in Crops. Plants 2015, 4, 412–448. [Google Scholar] [CrossRef]
- Woo, H.R.; Kim, H.J.; Nam, H.G.; Lim, P.O. Plant leaf senescence and death - regulation by multiple layers of control and implications for aging in general. J. Cell Sci. 2013, 126, 4823–4833. [Google Scholar] [CrossRef]
- Li, Z.H.; Zhao, Y.; Liu, X.C.; Peng, J.Y.; Guo, H.W.; Luo, J.C. LSD 2.0: An update of the leaf senescence database. Nucleic Acids Res. 2014, 42, D1200–D1205. [Google Scholar] [CrossRef]
- Duvick, D.N. Genetic contributions to yield gains of US hybrid maize, 1930 to 1980. In Genetic Contributions to Yield Gains of Five Major Crop Plants; CSSA Special Publication Number 7; CSSA: Fitchburg, WI, USA; ASA: Madison, WI, USA, 1984; pp. 15–47. [Google Scholar]
- Tollenaar, M.; Wu, J. Yield improvement in temperate maize is attributable to greater stress tolerance. Crop Sci. 1999, 39, 1597–1604. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S.-S. Translational researches on leaf senescence for enhancing plant productivity and quality. J. Exp. Bot. 2014, 65, 3901–3913. [Google Scholar] [CrossRef]
- Jehanzeb, M.; Zheng, X.; Miao, Y. The role of the S40 gene family in leaf senescence. Int. J. Mol. Sci. 2017, 18, 2152. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [PubMed]
- Sekhon, R.S.; Childs, K.L.; Santoro, N.; Foster, C.E.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Transcriptional and metabolic analysis of senescence induced by preventing pollination in maize. Plant Physiol. 2012, 159, 1730–1744. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Geng, M.; Shen, P.; Chen, X.; Li, Y.; Wen, X. Effect of post-silking drought stress on the expression profiles of genes involved in carbon and nitrogen metabolism during leaf senescence in maize (Zea mays L.). Plant Physiol. Biochem. PPB 2019, 135, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Li, M.; Tian, L.; Wang, S.; Wu, L.; Ku, L.; Zhang, J.; Song, X.; Liu, H.; Chen, Y. Global transcriptome analysis of the maize (Zea mays L.) inbred line 08LF during leaf senescence initiated by pollination-prevention. PLoS ONE 2017, 12, e0185838. [Google Scholar] [CrossRef]
- Zhang, W.Y.; Xu, Y.C.; Li, W.L.; Yang, L.; Yue, X.; Zhang, X.S.; Zhao, X.Y. Transcriptional analyses of natural leaf senescence in maize. PLoS ONE 2014, 9, e115617. [Google Scholar] [CrossRef]
- Wu, L.; Wang, S.; Tian, L.; Wu, L.; Li, M.; Zhang, J.; Li, P.; Zhang, W.; Chen, Y. Comparative proteomic analysis of the maize responses to early leaf senescence induced by preventing pollination. J. Proteom. 2018, 177, 75–87. [Google Scholar] [CrossRef]
- Wei, S.; Wang, X.; Zhang, J.; Liu, P.; Zhao, B.; Li, G.; Dong, S. The role of nitrogen in leaf senescence of summer maize and analysis of underlying mechanisms using comparative proteomics. Plant Sci. Int. J. Exp. Plant Biol. 2015, 233, 72–81. [Google Scholar] [CrossRef]
- Wang, J.; Guan, H.; Dong, R.; Liu, C.; Liu, Q.; Liu, T.; Wang, L.; He, C. Overexpression of maize sucrose non-fermenting-1-related protein kinase 1 genes, ZmSnRK1s, causes alteration in carbon metabolism and leaf senescence in Arabidopsis thaliana. Gene 2019, 691, 34–44. [Google Scholar] [CrossRef]
- Luo, K.; Deng, W.; Xiao, Y.; Zheng, X.; Li, Y.; Pei, Y. Leaf senescence is delayed in tobacco plants expressing the maize knotted1 gene under the control of a wound-inducible promoter. Plant Cell Rep. 2006, 25, 1246–1254. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef]
- Yu, G.C.; Wang, L.G.; Han, Y.Y.; He, Q.Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Morgan, M.; Carlson, M.; Tenenbaum, D.; Arora, S. AnnotationHub: Client to access AnnotationHub resources. R Package Version 2017, 2. Available online: https://rdrr.io/bioc/AnnotationHub/ (accessed on 19 November 2019).
- Yu, G. clusterProfiler: An universal enrichment tool for functional and comparative study. bioRxiv 2018, 256784. [Google Scholar] [CrossRef]
- Yu, C.-P.; Chen, S.C.-C.; Chang, Y.-M.; Liu, W.-Y.; Lin, H.-H.; Lin, J.-J.; Chen, H.J.; Lu, Y.-J.; Wu, Y.-H.; Lu, M.-Y.J. Transcriptome dynamics of developing maize leaves and genomewide prediction of cis elements and their cognate transcription factors. Proc. Natl. Acad. Sci. USA 2015, 112, E2477–E2486. [Google Scholar] [CrossRef]
- Tian, T.; You, Q.; Yan, H.Y.; Xu, W.Y.; Su, Z. MCENet: A database for maize conditional co-expression network and network characterization collaborated with multi-dimensional omics levels. J. Genet. Genom. 2018, 45, 351–360. [Google Scholar] [CrossRef]
- Wang, W.; Dai, Y.; Wang, M.; Yang, W.; Zhao, D. Transcriptome Dynamics of Double Recessive Mutant, o2o2o16o16, Reveals the Transcriptional Mechanisms in the Increase of Its Lysine and Tryptophan Content in Maize. Genes 2019, 10, 316. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Porra, R.J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 2002, 73, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018, 47, D607–D613. [Google Scholar] [CrossRef]
- Yoshida, S. Molecular regulation of leaf senescence. Curr. Opin. Plant Biol. 2003, 6, 79–84. [Google Scholar] [CrossRef]
- Hollander-Czytko, H.; Grabowski, J.; Sandorf, I.; Weckermann, K.; Weiler, E.W. Tocopherol content and activities of tyrosine aminotransferase and cystine lyase in Arabidopsis under stress conditions. J. Plant Physiol. 2005, 162, 767–770. [Google Scholar] [CrossRef]
- Kesari, P.; Patil, D.N.; Kumar, P.; Tomar, S.; Sharma, A.K.; Kumar, P. Structural and functional evolution of chitinase-like proteins from plants. Proteomics 2015, 15, 1693–1705. [Google Scholar] [CrossRef]
- Mazorra-Manzano, M.A.; Tanaka, T.; Dee, D.R.; Yada, R.Y. Structure-function characterization of the recombinant aspartic proteinase A1 from Arabidopsis thaliana. Phytochemistry 2010, 71, 515–523. [Google Scholar] [CrossRef]
- Xu, H.; Xu, W.; Xi, H.; Ma, W.; He, Z.; Ma, M. The ER luminal binding protein (BiP) alleviates Cd(2+)-induced programmed cell death through endoplasmic reticulum stress-cell death signaling pathway in tobacco cells. J. Plant Physiol. 2013, 170, 1434–1441. [Google Scholar] [CrossRef]
- John, C.F.; Morris, K.; Jordan, B.R.; Thomas, B.; A-H-Mackerness, S. Ultraviolet-B exposure leads to up-regulation of senescence-associated genes in Arabidopsis thaliana. J. Exp. Bot. 2001, 52, 1367–1373. [Google Scholar] [CrossRef]
- Jiang, Y.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.M.; An, J.; Han, H.J.; Kim, S.H.; Lim, C.O.; Yun, D.J.; Chung, W.S. ZAT11, a zinc finger transcription factor, is a negative regulator of nickel ion tolerance in Arabidopsis. Plant Cell Rep. 2014, 33, 2015–2021. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.; Rajasekaran, K.; Sickler, C.; Lebar, M.; Musungu, B.M.; Fakhoury, A.M.; Payne, G.A.; Geisler, M.; Carter-Wientjes, C.; Wei, Q.; et al. The Pathogenesis-Related Maize Seed (PRms) Gene Plays a Role in Resistance to Aspergillus flavus Infection and Aflatoxin Contamination. Front. Plant Sci. 2017, 8, 1758. [Google Scholar] [CrossRef]
- Hawkins, L.K.; Warburton, M.L.; Tang, J.D.; Tomashek, J.; Alves Oliveira, D.; Ogunola, O.F.; Smith, J.S.; Williams, W.P. Survey of Candidate Genes for Maize Resistance to Infection by Aspergillus flavus and/or Aflatoxin Contamination. Toxins 2018, 10, 61. [Google Scholar] [CrossRef]
- Yuan, G.; He, X.; Li, H.; Xiang, K.; Liu, L.; Zou, C.; Lin, H.; Zhang, Z.; Pan, G. Transcriptomic responses in resistant and susceptible maize infected with Fusarium graminearum. Crop J. 2019, 2214–5141. [Google Scholar] [CrossRef]
- Yang, Q.; Balint-Kurti, P.; Xu, M. Quantitative Disease Resistance: Dissection and Adoption in Maize. Mol. Plant 2017, 10, 402–413. [Google Scholar] [CrossRef]
- Balazadeh, S.; Riaño-Pachón, D.M.; Mueller-Roeber, B. Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 2008, 10, 63–75. [Google Scholar] [CrossRef]
- Wei, L.; Guo, Y. Transcriptome, transcription factors and transcriptional regulation of leaf senescence. J. Bioinform. Comp. Genom. 2014, 1, 1. [Google Scholar]
- Rushton, D.L.; Tripathi, P.; Rabara, R.C.; Lin, J.; Ringler, P.; Boken, A.K.; Langum, T.J.; Smidt, L.; Boomsma, D.D.; Emme, N.J. WRKY transcription factors: Key components in abscisic acid signalling. Plant Biotechnol. J. 2012, 10, 2–11. [Google Scholar] [CrossRef]
- Miao, Y.; Laun, T.; Zimmermann, P.; Zentgraf, U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004, 55, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Z.; Wang, X.; Wang, J.; Fan, K.; Li, Z.; Lin, W. DELLA proteins negatively regulate dark-induced senescence and chlorophyll degradation in Arabidopsis through interaction with the transcription factor WRKY6. Plant Cell Rep. 2018, 37, 981–992. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.L.; Holm, P.B. Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol. J. 2007, 5, 192–206. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Woo, H.R.; Kim, J.; Lim, P.O.; Lee, I.C.; Choi, S.H.; Hwang, D.; Nam, H.G. Trifurcate Feed-Forward Regulation of Age-Dependent Cell Death Involving miR164 in Arabidopsis. Science 2009, 323, 1053–1057. [Google Scholar] [CrossRef]
- Balazadeh, S.; Kwasniewski, M.; Caldana, C.; Mehrnia, M.; Zanor, M.I.; Xue, G.-P.; Mueller-Roeber, B. ORS1, an H2O2-Responsive NAC Transcription Factor, Controls Senescence in Arabidopsis thaliana. Mol. Plant 2011, 4, 346–360. [Google Scholar] [CrossRef]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Song, S.; Xie, D. Regulation of Jasmonate-Induced Leaf Senescence by Antagonism between bHLH Subgroup IIIe and IIId Factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef]
- Song, S.; Huang, H.; Wang, J.; Liu, B.; Qi, T.; Xie, D. MYC5 is Involved in Jasmonate-Regulated Plant Growth, Leaf Senescence and Defense Responses. Plant Cell Physiol. 2017, 58, 1752–1763. [Google Scholar] [CrossRef]
- Huang, C.-K.; Lo, P.-C.; Huang, L.-F.; Wu, S.-J.; Yeh, C.-H.; Lu, C.-A. A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant Mol. Biol. 2015, 88, 269–286. [Google Scholar] [CrossRef]
- Moschen, S.; Bengoa Luoni, S.; Di Rienzo, J.A.; Caro, M.D.P.; Tohge, T.; Watanabe, M.; Hollmann, J.; González, S.; Rivarola, M.; García-García, F.; et al. Integrating transcriptomic and metabolomic analysis to understand natural leaf senescence in sunflower. Plant Biotechnol. J. 2016, 14, 719–734. [Google Scholar] [CrossRef]
- Bengoa Luoni, S.; Astigueta, F.H.; Nicosia, S.; Moschen, S.; Fernandez, P.; Heinz, R. Transcription Factors Associated with Leaf Senescence in Crops. Plants 2019, 8, 411. [Google Scholar] [CrossRef]
- Ullah, A.; Akbar, A.; Yang, X. Jasmonic Acid (JA)-Mediated Signaling in Leaf Senescence. In Senescence Signalling and Control in Plants; Sarwat, M., Tuteja, N., Eds.; Academic Press: Cambridge, MA, USA, 2019; Chapter 7; pp. 111–123. [Google Scholar]
- Zhang, Y.; Wang, Y.; Wei, H.; Li, N.; Tian, W.; Chong, K.; Wang, L. Circadian Evening Complex Represses Jasmonate-Induced Leaf Senescence in Arabidopsis. Mol. Plant 2018, 11, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, Y.; Han, X.; Wang, H.; Pan, J.; Yu, D. Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. J. Exp. Bot. 2017, 68, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Provart, N.J.; Glazebrook, J.; Katagiri, F.; Chang, H.-S.; Eulgem, T.; Mauch, F.; Luan, S.; Zou, G.; Whitham, S.A.; et al. Expression Profile Matrix of Arabidopsis Transcription Factor Genes Suggests Their Putative Functions in Response to Environmental Stresses. Plant Cell 2002, 14, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Martín, M.; Casano, L.M.; Sabater, B. Identification of the Product of ndhA Gene as a Thylakoid Protein Synthesized in Response to Photooxidative Treatment. Plant Cell Physiol. 1996, 37, 293–298. [Google Scholar] [CrossRef]
- Davis, G.A.; Kanazawa, A.; Schöttler, M.A.; Kohzuma, K.; Froehlich, J.E.; Rutherford, A.W.; Satoh-Cruz, M.; Minhas, D.; Tietz, S.; Dhingra, A.; et al. Limitations to photosynthesis by proton motive force-induced photosystem II photodamage. ELife 2016, 5, e16921. [Google Scholar] [CrossRef]
- Gámez-Arjona, F.M.; Raynaud, S.; Ragel, P.; Mérida, Á. Starch synthase 4 is located in the thylakoid membrane and interacts with plastoglobule-associated proteins in Arabidopsis. Plant J. 2014, 80, 305–316. [Google Scholar] [CrossRef]
- Trobacher, C.P. Ethylene and programmed cell death in plants. Botany 2009, 87, 757–769. [Google Scholar] [CrossRef]
- Grosskinsky, D.K.; Syaifullah, S.J.; Roitsch, T. Integration of multi-omics techniques and physiological phenotyping within a holistic phenomics approach to study senescence in model and crop plants. J. Exp. Bot. 2018, 69, 825–844. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).