Cloning, Characterization and Functional Analysis of the LtuPTOX Gene, a Homologue of Arabidopsis thaliana IMMUTANS Derived from Liriodendron tulipifera
Abstract
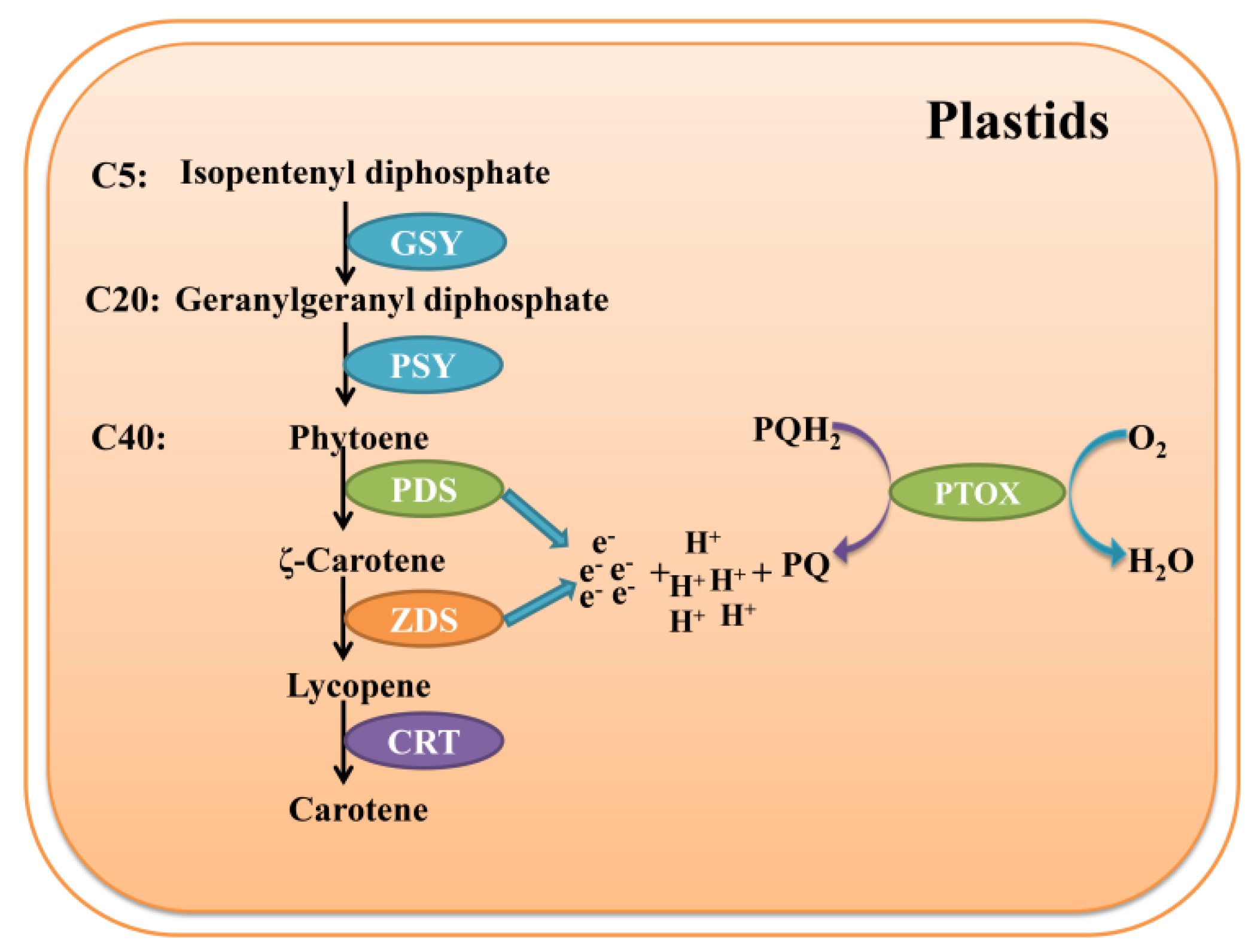
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Identification and Isolation of the Full-Length LcPTOX Gene in L. tulipifera
2.3. Structural and Functional Identification of LtuPTOX by Bioinformatic Approaches
2.4. Transient Expression and Subcellular Localization of the LtuPTOX Gene
2.5. Expression Pattern of the LtuPTOX Gene in Various Tissues/Organs
2.6. Expression Levels of the LtuPTOX Gene within the Orange Petal Band at Different Stages
2.7. Quantification of Chlorophyll and Carotenoid Contents in the Orange Petal Band
3. Results
3.1. Full-Length cDNA Isolation of the LtuPTOX Gene
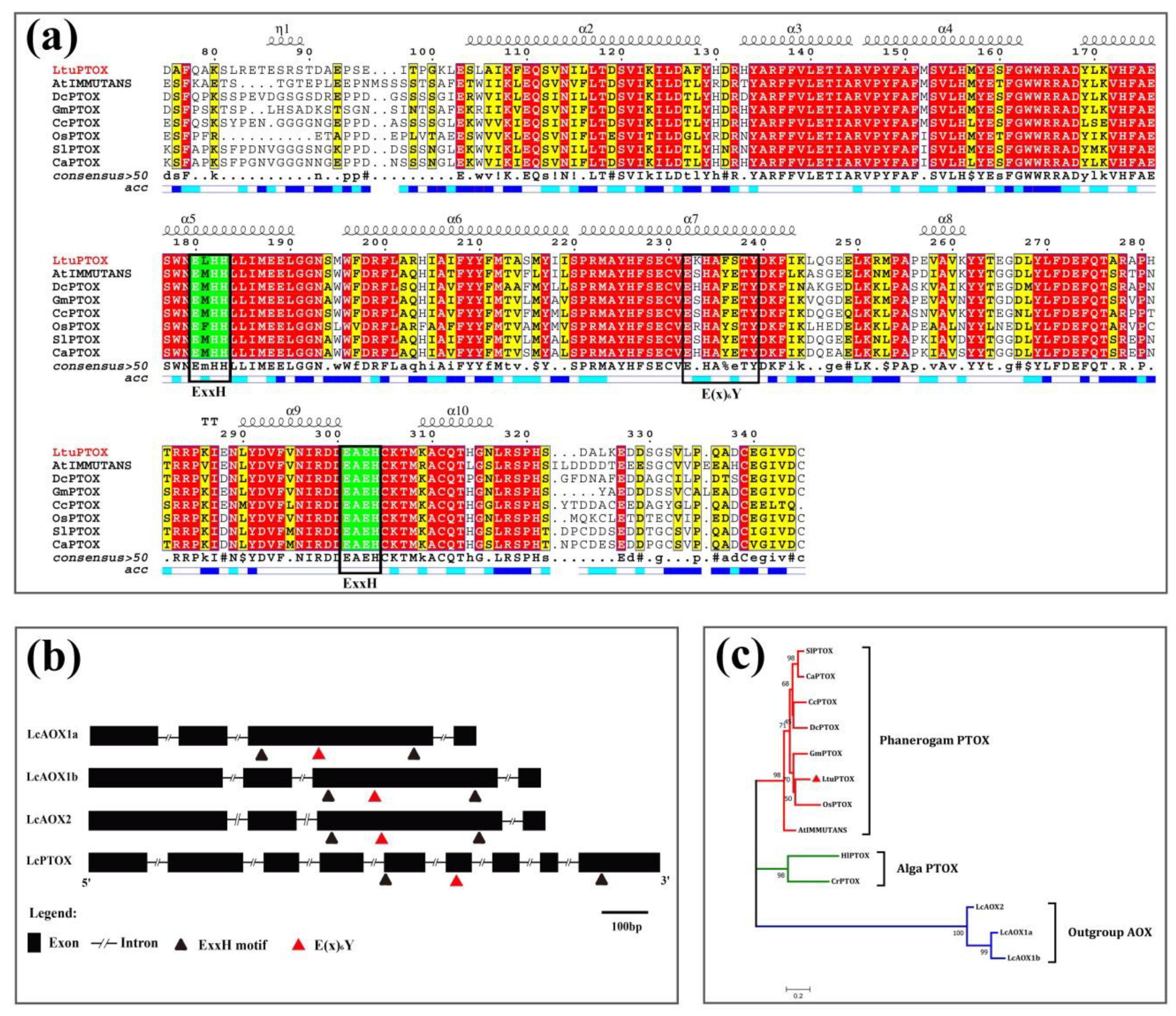
3.2. In Silico Analysis of the Function and Structure of LtuPTOX Gene
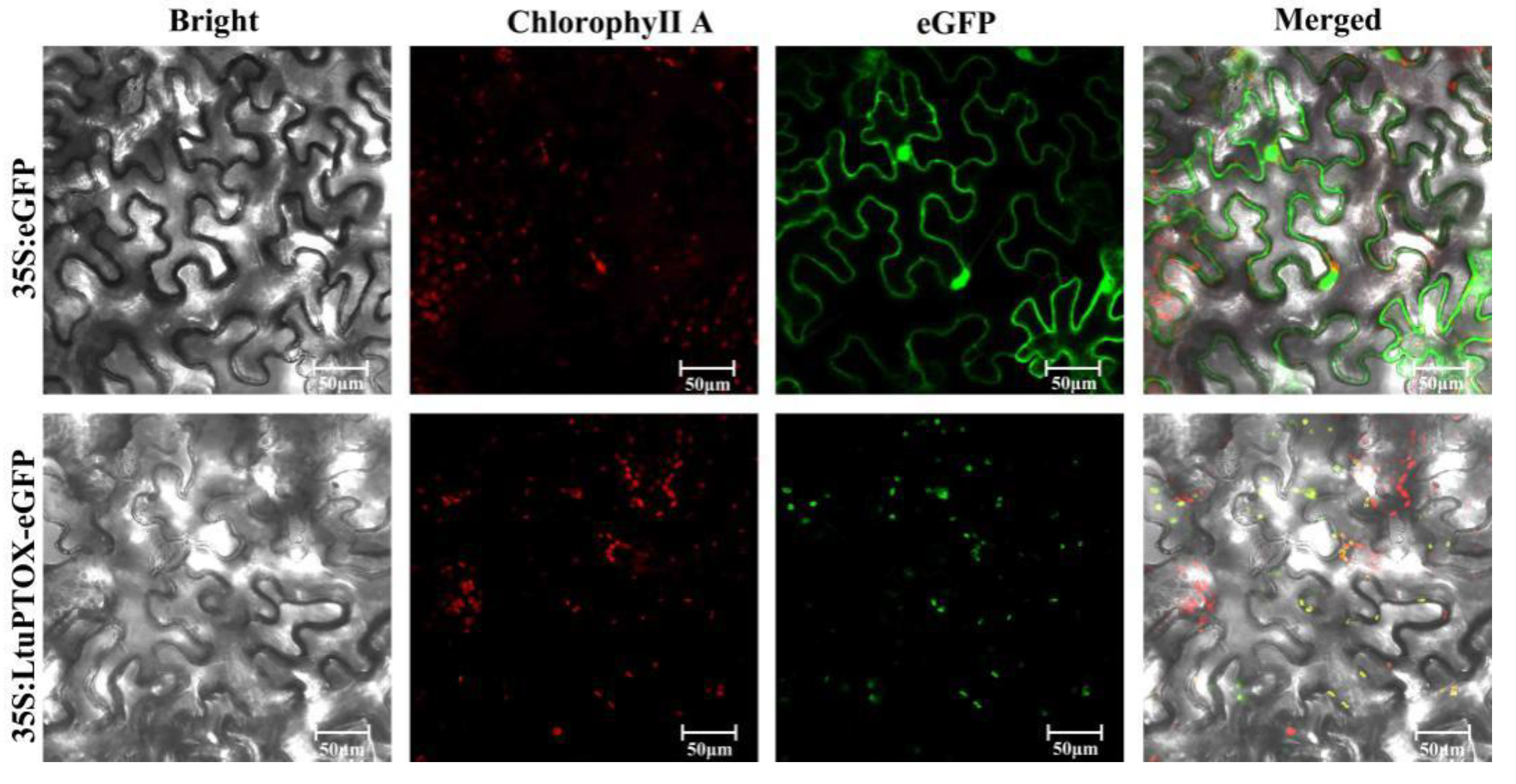
3.3. Transient Transformation and Subcellular Localization of LtuPTOX
3.4. Expression Pattern of the LtuPTOX Gene in Various Tissues/Organs
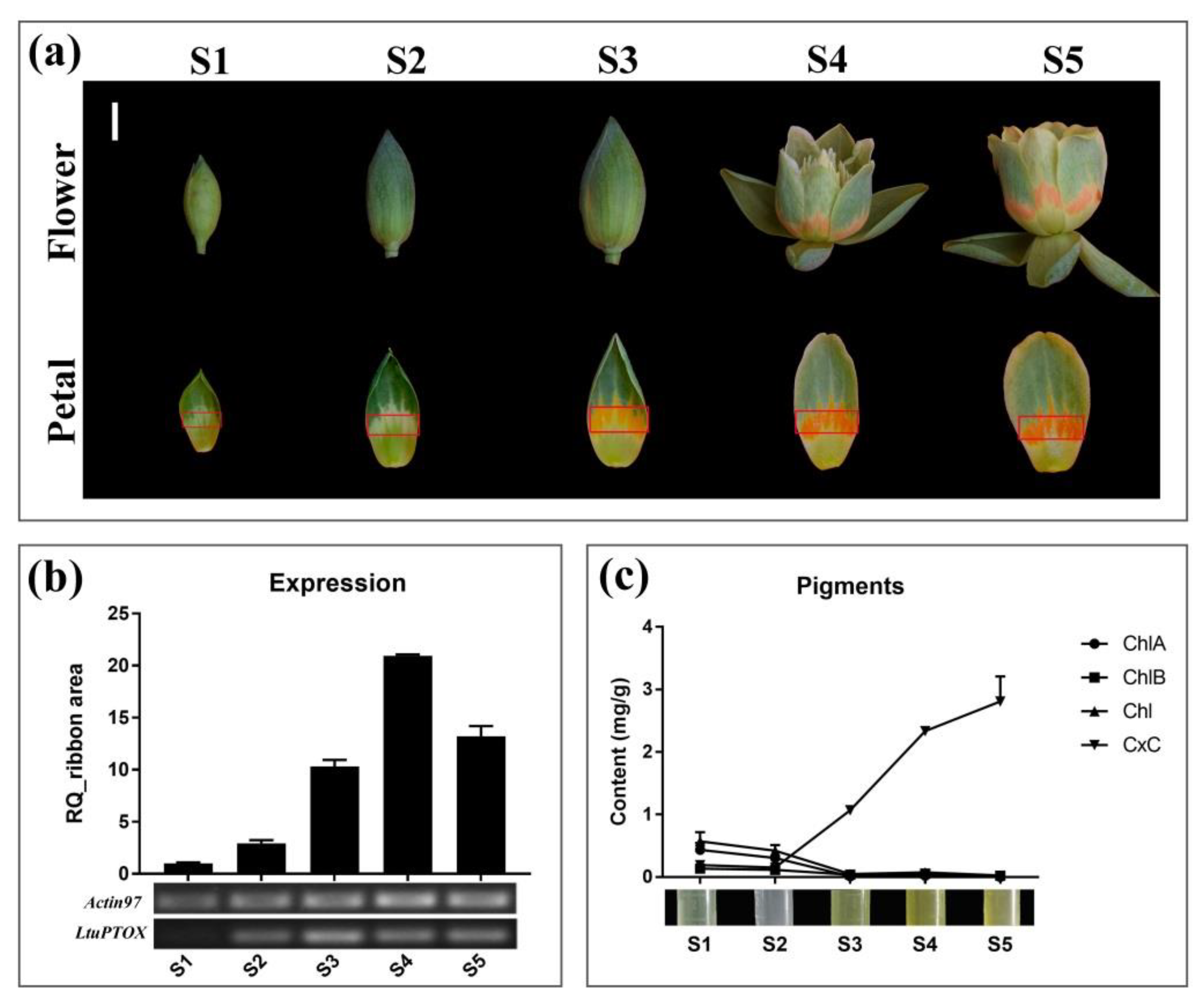
3.5. LtuPTOX Involved in Carotenoid Metabolism and Orange-Band Formation on the Tepal
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Primer Name | Primer Sequence | TM/°C |
|---|---|---|
| nG31435-F | AACCCAGGAAGACCATCTCC | 57 |
| nG31435-R | GGAGCTCTTGCTGTTTGAA | 54 |
| 5GSP1 | CAAATAATCAGCTCTTCTCCAC | 55.6 |
| 5GSP2 | CCTCCACGATAACTTCTTCCTCC | 61 |
| 3GSP1 | GTTTGACCGTTTTCTTGCTC | 54.6 |
| 3GSP2 | GGAGAAGAATTGAAAAGAATGCCTG | 58.1 |
| LtuPTOX-F | TACTCAGCAACGAATAAACC | 52.2 |
| LtuPTOX-R | TTGGATAAATATGGACCCCT | 52.9 |
| p1302-LtuPTOX-TF | actcttgaccatggtagatctATGACTACAAGATCAACATCTCTCTCTTC | 60.6 |
| p1302-LtuPTOX-TR | aagttcttctcctttactagtTTCTTGTTTCCTTTCATGAGGGG | 62.4 |
| 35S primer | GACGCACAATCCCACTATCC | 56 |
| LtuPTOX-qF | CTGTTCTGCACATGTACGAGA | 57.8 |
| LtuPTOX-qR | GCAAGAAAACGGTCAAACCAC | 57.9 |
| Actin97-qF | TTCCCGTTCAGCAGTGGTCGTGGTCG | 58 |
| Actin97-qR | TGGTCGCACAACTGGTATCG | 58 |

References
- Waser, N.M.; Price, M.V. Pollinator Choice and Stabilizing Selection for Flower Color in Delphinium nelsonii. Evolution 1981, 35, 376–390. [Google Scholar] [CrossRef] [PubMed]
- Menze, R.; Shmida, A. The ecology of flower colours and the natural colour vision of insect pollinators: The Israeli flora as a study case. Biol. Rev. 1993, 68, 81–120. [Google Scholar] [CrossRef]
- Briscoe, A.D.; Chittka, L. The evolution of color vision in insects. Annu. Rev. Entomol. 2001, 46, 471–510. [Google Scholar] [CrossRef] [PubMed]
- Vickery, R.K. Speciation in Mimulus, or, can a simple flower color mutant lead to species divergence. Great Basin Nat. 1995, 55, 177–180. [Google Scholar] [CrossRef]
- Zhang, H.F.; Wei, Q.C.; Li, C.Z.; Jiang, C.M.; Zhang, H.C. Comparative Proteomic Analysis Provides Insights into the Regulation of Flower Bud Differentiation in Crocus Sativus L. J. Food Biochem. 2016, 40, 567–582. [Google Scholar] [CrossRef]
- Yue, Y.; Liu, J.; Shi, T.; Chen, M.; Li, Y.; Du, J.; Jiang, H.; Yang, X.; Hu, H.; Wang, L. Integrating Transcriptomic and GC-MS Metabolomic Analysis to Characterize Color and Aroma Formation during Tepal Development in Lycoris longituba. Plants 2019, 8, 53. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Rodriguezconcepcion, M.; Avalos, J.; Bonet, M.L. A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Progress Lipid Res. 2018, 70, 62–93. [Google Scholar] [CrossRef]
- Carol, P.; Kuntz, M. A plastid terminal oxidase comes to light: Implications for carotenoid biosynthesis and chlororespiration. Trends Plant Sci. 2001, 6, 31–36. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Welsch, R.; Beyer, P.; Hugueney, P.; Kleinig, H.; Von Lintig, J. Regulation and activation of phytoene synthase, a key enzyme in carotenoid biosynthesis, during photomorphogenesis. Planta 2000, 211, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Vallabhaneni, R.; Yu, J.; Rocheford, T.; Wurtzel, E.T. The maize phytoene synthase gene family: Overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol. 2008, 147, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi, M.; Gilbert, M.; Laboure, A.M.; Kuntz, M. Dual role of the plastid terminal oxidase in tomato. Plant Physiol. 2007, 145, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Foudree, A.; Putarjunan, A.; Kambakam, S.; Nolan, T.; Fussell, J.; Pogorelko, G.; Rodermel, S. The mechanism of variegation in immutans provides insight into chloroplast biogenesis. Front. Plant Sci. 2012, 3, 260. [Google Scholar] [CrossRef] [PubMed]
- Welsch, R.; Maass, D.; Voegel, T.; Dellapenna, D.; Beyer, P. Transcription factor RAP2.2 and its interacting partner SINAT2: Stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 2007, 145, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Diner, B.; Mauzerall, D. Feedback controlling oxygen production in a cross-reaction between two photosystems in photosynthesis. Biochim. Biophys. Acta 1973, 305, 329–352. [Google Scholar] [CrossRef]
- Bennoun, P. Evidence for a respiratory chain in the chloroplast. Proc. Natl. Acad. Sci. USA 1982, 79, 4352–4356. [Google Scholar] [CrossRef]
- Wu, D.; Wright, D.A.; Wetzel, C.M. The immutans variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 1999, 11, 43–55. [Google Scholar] [CrossRef]
- Carol, P.; Stevenson, D.; Bisanz, C.; Breitenbach, J.; Sandmann, G.; Mache, R.; Coupland, G.; Kuntz, M. Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 1999, 11, 57–68. [Google Scholar] [CrossRef]
- Wetzel, C.M.; Jiang, C.; Meehan, L.J. Nuclear-organelle interactions: The immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J. 1994, 6, 161–175. [Google Scholar] [CrossRef]
- Sun, X.; Wen, T. Physiological roles of plastid terminal oxidase in plant stress responses. J. Biosci. 2011, 36, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Atteia, A.; Van Lis, R.; Van Hellemond, J.J.; Tielens, A.G.M.; Martin, W.; Henze, K. Identification of prokaryotic homologues indicates an endosymbiotic origin for the alternative oxidases of mitochondria (AOX) and chloroplasts (PTOX). Gene 2004, 330, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Li, H. Interspecies evolutionary divergence in Liriodendron, evidence from the nucleotide variations of LcDHN-like gene. BMC Evol. Biol. 2018, 18, 195. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hao, Z.; Guang, X. Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat. Plants 2019, 5, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Li, M.; Zhao, F.; Zha, H.; Yang, L.; Lu, Y.; Wang, G.; Shi, J.; Chen, J. Floral Nectary Morphology and Proteomic Analysis of Nectar of Liriodendron tulipifera Linn. Front. Plant Sci. 2016, 7, 826. [Google Scholar] [CrossRef] [PubMed]
- Demuth, P.; Join, F.S. Carotenoid flower pigments in Liodendron and Magnolia. Bull. Torrey Bot. Club 1978, 105, 65. [Google Scholar] [CrossRef]
- Liang, H.Y.; Carlson, J.E.; Leebens-Mack, J.H.; Wall, P.K.; Mueller, L.A.; Buzgo, M.; Landherr, L.L.; Hu, Y.; DiLoreto, D.S.; Ilut, D.C.; et al. An EST database for Liriodendron tulipifera L. floral buds: The first EST resource for functional and comparative genomics in Liriodendron. Tree Genet. Genomes 2008, 4, 419–433. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 1949, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.J.; Je, S.M.; Cheng, H.C.; Seo, S.M.; Park, J.H.; Baek, S.G.; Khaine, I.; Lee, T.; Jang, J.; Li, Y.; et al. Night Light-Adaptation Strategies for Photosynthetic Apparatus in Yellow-Poplar (Liriodendron tulipifera L.) Exposed to Artificial Night Lighting. Forests 2018, 9, 74. [Google Scholar] [CrossRef]
- Suzuki, T.; Hashimoto, T.; Yabu, Y.; Majiwa, P.A.; Ohshima, S.; Suzuki, M.; Lu, S.; Hato, M.; Kido, Y.; Sakamoto, K.; et al. Alternative oxidase (AOX) genes of African trypanosomes: Phylogeny and evolution of AOX and plastid terminal oxidase families. J. Eukaryot. Microbiol. 2005, 52, 374–381. [Google Scholar] [CrossRef]
- Nakamura, K.; Sakamoto, K.; Kido, Y.; Fujimoto, Y.; Suzuki, T.; Suzuki, M.; Yabu, Y.; Ohta, N.; Tsuda, A.; Onuma, M.; et al. Mutational analysis of the Trypanosoma vivax alternative oxidase: The E(X)6Y motif is conserved in both mitochondrial alternative oxidase and plastid terminal oxidase and is indispensable for enzyme activity. Biochem. Biophys. Res. Commun. 2005, 334, 593–600. [Google Scholar] [CrossRef]
- Baena-Gonzalez, E.; Allahverdiyeva, Y.; Svab, Z.; Maliga, P.; Josse, E.M.; Kuntz, M.; Maenpaa, P.; Aro, E.M. Deletion of the tobacco plastid psbA gene triggers an upregulation of the thylakoid-associated NAD(P)H dehydrogenase complex and the plastid terminal oxidase (PTOX). Plant J. 2003, 35, 704–716. [Google Scholar] [CrossRef]
- Houille-Vernes, L.; Rappaport, F.; Wollman, F.A.; Alric, J.; Johnson, X. Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. Proc. Natl. Acad. Sci. USA 2011, 108, 20820–20825. [Google Scholar] [CrossRef]
- Ahmad, N.; Michoux, F.; Nixon, P.J. Investigating the production of foreign membrane proteins in tobacco chloroplasts: Expression of an algal plastid terminal oxidase. PLoS ONE 2012, 7, e41722. [Google Scholar] [CrossRef][Green Version]
- Josse, E.M.; Simkin, A.J.; Gaffe, J.; Laboure, A.M.; Kuntz, M.; Carol, P. A plastid terminal oxidase associated with carotenoid desaturation during chromoplast differentiation. Plant Physiol. 2000, 123, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sommerfeld, M.; Hu, Q. Occurrence and environmental stress responses of two plastid terminal oxidases in Haematococcus pluvialis (Chlorophyceae). Planta 2009, 230, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Laureau, C.; De Paepe, R.; Latouche, G.; Moreno-Chacon, M.; Finazzi, G.; Kuntz, M.; Cornic, G.; Streb, P. Plastid terminal oxidase (PTOX) has the potential to act as a safety valve for excess excitation energy in the alpine plant species Ranunculus glacialis L. Plant Cell Environ. 2013, 36, 1296–1310. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, G.; Bartley, G.E.; Scolnik, P.A. Regulation of carotenoid biosynthesis during tomato development. Plant Cell 1993, 5, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, P.M.; Umbach, A.L.; Wilce, J.A. Prokaryotic origins for the mitochondrial alternative oxidase and plastid terminal oxidase nuclear genes. FEBS Lett. 2003, 555, 425–430. [Google Scholar] [CrossRef]
- McDonald, A.E.; Vanlerberghe, G.C. Origins, evolutionary history, and taxonomic distribution of alternative oxidase and plastoquinol terminal oxidase. Comp. Biochem. Physiol. Part D Genomics Proteomics 2006, 1, 357–364. [Google Scholar] [CrossRef]
- Fu, A.; Liu, H.; Yu, F.; Kambakam, S.; Luan, S.; Rodermel, S. Alternative oxidases (AOX1a and AOX2) can functionally substitute for plastid terminal oxidase in Arabidopsis chloroplasts. Plant Cell 2012, 24, 1579–1595. [Google Scholar] [CrossRef]
- Kuntz, M. Plastid terminal oxidase and its biological significance. Planta 2004, 218, 896–899. [Google Scholar] [CrossRef]
- Cournac, L.; Redding, K.; Ravenel, J. Electron flow between photosystem II and oxygen in chloroplasts of photosystem Ideficient algae is mediated by a quinol oxidase involved in chlororespiration. J. Biol. Chem. 2000, 275, 17256–17262. [Google Scholar] [CrossRef]
- Sun, X.; Yang, C.Q.; Wen, T.; Zeng, F.C.; Wang, Q.; Yang, W.Y.; Lin, H.H. Water stress enhances expression of genes encoding plastid terminal oxidase and key components of chlororespiration and alternative respiration in soybean seedlings. Z. Naturforsch C 2014, 69, 300–308. [Google Scholar] [CrossRef]
- Qian, Y.; Wang, D.; Aigen, F. Molecular features and physiological roles of plant plastid terminal oxidase. Plant Physiol. J. 2016, 52, 1710–1720. [Google Scholar] [CrossRef]
- Kambakam, S.; Bhattacharjee, U.; Petrich, J.; Rodermel, S. PTOX Mediates Novel Pathways of Electron Transport in Etioplasts of Arabidopsis. Mol. Plant 2016, 9, 1240–1259. [Google Scholar] [CrossRef] [PubMed]

| AAGP Gene NO. 1 | RNA-seq Dataset | Gene ID 2 | Annotation |
|---|---|---|---|
| gnl|Liriodendron|b4_c3586 | Liriodend_newGene_31435 | Lchi07087 | PTOX/IMMUTANS |
| gnl|Liriodendron|b4_c15469 | |||
| gnl|Liriodendron|b4_c23296 | |||
| gnl|Liriodendron|b4_c24190 | |||
| gnl|Liriodendron|b4_s124823 | |||
| gnl|Liriodendron|b4_rep_c78233 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Z.; Zong, Y.; Liu, H.; Tu, Z.; Li, H. Cloning, Characterization and Functional Analysis of the LtuPTOX Gene, a Homologue of Arabidopsis thaliana IMMUTANS Derived from Liriodendron tulipifera. Genes 2019, 10, 878. https://doi.org/10.3390/genes10110878
Hao Z, Zong Y, Liu H, Tu Z, Li H. Cloning, Characterization and Functional Analysis of the LtuPTOX Gene, a Homologue of Arabidopsis thaliana IMMUTANS Derived from Liriodendron tulipifera. Genes. 2019; 10(11):878. https://doi.org/10.3390/genes10110878
Chicago/Turabian StyleHao, Ziyuan, Yaxian Zong, Huanhuan Liu, Zhonghua Tu, and Huogen Li. 2019. "Cloning, Characterization and Functional Analysis of the LtuPTOX Gene, a Homologue of Arabidopsis thaliana IMMUTANS Derived from Liriodendron tulipifera" Genes 10, no. 11: 878. https://doi.org/10.3390/genes10110878
APA StyleHao, Z., Zong, Y., Liu, H., Tu, Z., & Li, H. (2019). Cloning, Characterization and Functional Analysis of the LtuPTOX Gene, a Homologue of Arabidopsis thaliana IMMUTANS Derived from Liriodendron tulipifera. Genes, 10(11), 878. https://doi.org/10.3390/genes10110878





