Transcription Factors That Govern Development and Disease: An Achilles Heel in Cancer
Abstract
:1. Introduction
1.1. Embryonic Development and Cancer: Two Sides of the Same Coin
1.2. Epithelial-to-Mesenchymal Transition: in Development and Cancer
1.3. Cell Migration: Essential for Development and Cancer Progression
2. High Mobility Group Box (HMG)
2.1. HMG Proteins: A Superfamily of Chromatin Remodelers
2.2. Role in Development
2.3. Evolutionary Conservation
2.4. Role in Cancer
3. GATA Transcription Factors
3.1. Role in Development
3.2. Evolutionary Conservation
3.3. Role in Cancer
4. Pax Transcription Factors
4.1. Role in Development
4.2. Evolutionary Conservation
4.3. Role in Cancer
5. bHLH Transcription Factors
5.1. Role in Development
5.2. Evolutionary Conservation
5.3. Role in Cancer
6. Discussion
6.1. Transcription Factors—Crucial Proteins for Development and Homeostasis
6.2. Therapeutic Targeting of Transcription Factor: Need of the Hour
6.3. Natural Resistance Against Cancer: Learning from Life
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| Acute megakaryoblastic leukemia | AMKL |
| Alveolar rhabdomyosarcomas | ARMS |
| basic Helix-loop-Helix | bHLH |
| Bone Morphogenetic Protein | BMP |
| Central nervous system | CNS |
| Chronic myelogenous leukemia | CML |
| Cystein-rich Polycomb-like Proteins | CPP |
| Epithelial-to-mesenchymal transition | EMT |
| Fibroblast Growth Factor | FGF |
| Forkhead box | FOX |
| Hedgehog | HH |
| Hepatocellular carcinoma | HCC |
| Hematopoietic stem cells | HSCs |
| Hematopoietic stem/progenitor cells | HSPC |
| High Mobility Group box | HMG |
| HMG-AT-hook family | HMGA |
| HMG-box family | HMGB |
| HMG-nucleosome binding family | HMGBN |
| Homeodomain | HD |
| Homology-directed repair | HDR |
| Leukemia Inhibitory Factor pseudogene 6 | LIF6 |
| Myelodysplastic syndrome | MDS |
| Myeloproliferative neoplasms | MPN |
| Nephew of atonal 3 | Nato 3 |
| Neurogenin2 | Ngn2 |
| Non homologous end-joining | NHEJ |
| Non-small cell lung carcinomas | NSCLC |
| Nuclear localization signal | NLS |
| Nucleosome-binding domain | NBD |
| Octamer binding transcription factor 4 | Oct-04 |
| Octopeptide | OP |
| Paired box genes | PAX |
| Paired domain | PD |
| Pancreatic ductal adenocarcinoma | PDAC |
| Proliferating cell nuclear antigen | PCNA |
| Renal-cell carcinomas | RCC |
| Specificity proteins | Sp |
| Retinoblastoma | Rb |
| Ten Eleven Translocation | TET |
| Transactivation domain | TD |
| Tumor initiating cells | TIC |
| Thyroglobulin | Ty |
| Thyroid peroxidase | Tpo |
| Transcription Factors | TFs |
References
- Roy, N.; Hebrok, M. Regulation of Cellular Identity in Cancer. Dev. Cell 2015, 35, 674–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knudson, A.G. Mutation and Cancer: Statistical Study of Retinoblastoma. Proc. Natl. Acad. Sci. USA 1971, 68, 820–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knudson, A.G. Cancer genetics through a personal retrospectroscope. Genes Chromosom. Cancer 2003, 38, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.; Zevnik, B.; Anastassiadis, K.; Niwa, H.; Klewe-Nebenius, D.; Chambers, I.; Scholer, H.; Smith, A. Formation of Pluripotent Stem Cells in the Mammalian Embryo Depends on the POU Transcription Factor Oct4. Cell 1998, 95, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Atlasi, Y.; Mowla, S.J.; Ziaee, S.A.; Gokhale, P.J.; Andrews, P.W. OCT4 Spliced Variants Are Differentially Expressed in Human Pluripotent and Nonpluripotent Cells. Stem Cells 2008, 26, 3068–3074. [Google Scholar] [CrossRef]
- Ku, J.-L.; Shin, Y.-K.; Kim, D.-W.; Choi, J.-S.; Hong, S.-H.; Jeon, Y.-K.; Park, J.-H. Establishment and characterization of 13 human colorectal carcinoma cell lines: Mutations of genes and expressions of drug-sensitivity genes and cancer stem cell markers. Carcinogenesis 2010, 31, 1003–1009. [Google Scholar] [CrossRef]
- Liu, A.; Yu, X.; Liu, S. Pluripotency transcription factors and cancer stem cells: Small genes make a big difference. Chin. J. Cancer 2013, 32, 483–487. [Google Scholar] [CrossRef]
- Amini, S.; Fathi, F.; Mobalegi, J.; Sofimajidpour, H.; Ghadimi, T. The expressions of stem cell markers: Oct4, Nanog, Sox2, nucleostemin, Bmi, Zfx, Tcl1, Tbx3, Dppa4, and Esrrb in bladder, colon, and prostate cancer, and certain cancer cell lines. Anat. Cell Boil. 2014, 47, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-J.; Herlyn, M. The emerging roles of Oct4 in tumor-initiating cells. Am. J. Physiol. Physiol. 2015, 309, C709–C718. [Google Scholar] [CrossRef]
- Olmez, I.; Shen, W.; McDonald, H.; Ozpolat, B. Dedifferentiation of patient-derived glioblastoma multiforme cell lines results in a cancer stem cell-like state with mitogen-independent growth. J. Cell. Mol. Med. 2015, 19, 1262–1272. [Google Scholar] [CrossRef]
- Ghosh, D.; Nandi, S.; Bhattacharjee, S. Combination therapy to checkmate Glioblastoma: Clinical challenges and advances. Clin. Transl. Med. 2018, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Borrull, A.; Ghislin, S.; Deshayes, F.; Lauriol, J.; Alcaide-Loridan, C.; Middendorp, S. Nanog and Oct4 overexpression increases motility and transmigration of melanoma cells. J. Cancer Res. Clin. Oncol. 2012, 138, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.M.; Liu, S.; Lu, H.; Zhang, H.; Zhang, P.J.; Gimotty, P.A.; Guerra, M.; Guo, W.; Xu, X. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene 2012, 31, 4898–4911. [Google Scholar] [CrossRef] [Green Version]
- Chiou, S.-H.; Wang, M.-L.; Chou, Y.-T.; Chen, C.-J.; Hong, C.-F.; Hsieh, W.-J.; Chang, H.-T.; Chen, Y.-S.; Lin, T.-W.; Hsu, H.-S.; et al. Coexpression of Oct4 and Nanog Enhances Malignancy in Lung Adenocarcinoma by Inducing Cancer Stem Cell-Like Properties and Epithelial-Mesenchymal Transdifferentiation. Cancer Res. 2010, 70, 10433–10444. [Google Scholar] [CrossRef] [PubMed]
- Cappellen, D.; Schlange, T.; Bauer, M.; Maurer, F.; Hynes, N.E. Novel c-MYC target genes mediate differential effects on cell proliferation and migration. EMBO Rep. 2007, 8, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Khattar, E.; Leow, S.C.; Liu, C.Y.; Müller, J.; Ang, W.X.; Li, Y.; Franzoso, G.; Li, S.; Guccione, E.; et al. Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J. Clin. Investig. 2015, 125, 2109–2122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Dillon, C.P.; Shi, L.Z.; Milasta, S.; Carter, R.; Finkelstein, D.; McCormick, L.L.; Fitzgerald, P.; Chi, H.; Munger, J.; et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 2011, 35, 871–882. [Google Scholar] [CrossRef]
- Beer, S.; Zetterberg, A.; Ihrie, R.A.; McTaggart, R.A.; Yang, Q.; Bradon, N.; Arvanitis, C.; Attardi, L.D.; Feng, S.; Ruebner, B.; et al. Developmental context determines latency of MYC-induced tumorigenesis. PLoS Boil. 2004, 2, e332. [Google Scholar] [CrossRef] [PubMed]
- Felsher, D.W.; Bishop, J. Reversible Tumorigenesis by MYC in Hematopoietic Lineages. Mol. Cell 1999, 4, 199–207. [Google Scholar] [CrossRef]
- Jain, M.; Arvanitis, C.; Chu, K.; Dewey, W.; Leonhardt, E.; Trinh, M.; Sundberg, C.D.; Bishop, J.M.; Felsher, D.W. Sustained Loss of a Neoplastic Phenotype by Brief Inactivation of MYC. Science 2002, 297, 102–104. [Google Scholar] [CrossRef] [PubMed]
- Pelengaris, S.; Khan, M.; I Evan, G. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 2002, 109, 321–334. [Google Scholar] [CrossRef]
- Marinkovic, D.; Marinkovic, T.; Kokai, E.; Barth, T.; Möller, P.; Wirth, T. Identification of novel Myc target genes with a potential role in lymphomagenesis. Nucleic Acids Res. 2004, 32, 5368–5378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hay, E. An Overview of Epithelio-Mesenchymal Transformation. Cells Tissues Organs 1995, 154, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Neilson, E.G. Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Investig. 2003, 112, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- Roche, J. The Epithelial-to-Mesenchymal Transition in Cancer. Cancers 2018, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Vaquer, A.; Viotti, M.; Hadjantonakis, A.-K. Transitions between epithelial and mesenchymal states and the morphogenesis of the early mouse embryo. Cell Adhes. Migr. 2014, 4, 447–457. [Google Scholar] [CrossRef]
- Wang, X.; Kopinke, D.; Lin, J.; McPherson, A.D.; Duncan, R.N.; Otsuna, H.; Moro, E.; Hoshijima, K.; Grunwald, D.J.; Argenton, F.; et al. Wnt signaling regulates postembryonic hypothalamic progenitor differentiation. Dev. Cell 2012, 23, 624–636. [Google Scholar] [CrossRef]
- Duband, J.L.; Thiery, J.P. Appearance and distribution of fibronectin during chick embryo gastrulation and neurulation. Dev. Boil. 1982, 94, 337–350. [Google Scholar] [CrossRef]
- Liem, K.F.; Jessell, T.M.; Briscoe, J. Regulation of the neural patterning activity of sonic hedgehog by secreted BMP inhibitors expressed by notochord and somites. Development 2000, 127, 4855–4866. [Google Scholar]
- Sela-Donenfeld, D.; Kalcheim, C. Localized BMP4–Noggin Interactions Generate the Dynamic Patterning of Noggin Expression in Somites. Dev. Boil. 2002, 246, 311–328. [Google Scholar] [CrossRef]
- Thiery, J.P. Epithelial–mesenchymal transitions in development and pathologies. Curr. Opin. Cell Boil. 2003, 15, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Burstyn-Cohen, T.; Stanleigh, J.; Sela-Donenfeld, D.; Kalcheim, C. Canonical Wnt activity regulates trunk neural crest delamination linking BMP/noggin signaling with G1/S transition. Development 2004, 131, 5327–5339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karafiat, V.; Dvorakova, M.; Pajer, P.; Cermak, V.; Dvorak, M. Melanocyte fate in neural crest is triggered by Myb proteins through activation of c-kit. Cell. Mol. Life Sci. 2007, 64, 2975–2984. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, S.; Glavic, A.; Ruiz, P.; Mayor, R.; Ruiz-Rudolph, P. Posteriorization by FGF, Wnt, and Retinoic Acid Is Required for Neural Crest Induction. Dev. Boil. 2002, 241, 289–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, L.C.; Lohmer, L.L.; Hagedorn, E.J.; Sherwood, D.R. Traversing the basement membrane in vivo: A diversity of strategies. J. Cell Boil. 2014, 204, 291–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tepass, U.; Theres, C.; Knust, E. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell 1990, 61, 787–799. [Google Scholar] [CrossRef]
- Edelman, G.M.; Crossin, K.L. Cell Adhesion Molecules: Implications for a Molecular Histology. Annu. Rev. Biochem. 1991, 60, 155–190. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Boil. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Beaman, J.E.; White, C.R.; Seebacher, F. Evolution of Plasticity: Mechanistic Link between Development and Reversible Acclimation. Trends Ecol. Evol. 2016, 31, 237–249. [Google Scholar] [CrossRef]
- Holtan, S.G.; Creedon, D.J.; Haluska, P.; Markovic, S.N. Cancer and Pregnancy: Parallels in Growth, Invasion, and Immune Modulation and Implications for Cancer Therapeutic Agents. Mayo Clin. Proc. 2009, 84, 985–1000. [Google Scholar] [CrossRef] [Green Version]
- Burger, O.; Baudisch, A.; Vaupel, J.W. Human mortality improvement in evolutionary context. Proc. Natl. Acad. Sci. USA 2012, 109, 18210–18214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, S.; Nakashima, A.; Shima, T.; Ito, M. Th1/Th2/Th17 and Regulatory T-Cell Paradigm in Pregnancy. Am. J. Reprod. Immunol. 2010, 63, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Mor, G. The Unique immunologic and microbial aspects of pregnancy. Placenta 2017, 57, 226. [Google Scholar] [CrossRef]
- Sakaguchi, S. Regulatory T cells: Key controllers of immunologic self-tolerance. Cell 2000, 101, 455–458. [Google Scholar] [CrossRef]
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336. [Google Scholar] [CrossRef]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef]
- Khattri, R.; Cox, T.; Yasayko, S.A.; Ramsdell, F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 2003, 4, 337–342. [Google Scholar] [CrossRef]
- Fontenot, J.D.; Rasmussen, J.P.; Williams, L.M.; Dooley, J.L.; Farr, A.G.; Rudensky, A.Y. Regulatory T Cell Lineage Specification by the Forkhead Transcription Factor Foxp3. Immunity 2005, 22, 329–341. [Google Scholar] [CrossRef]
- Wan, Y.Y.; Flavell, R.A. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc. Natl. Acad. Sci. USA 2005, 102, 5126–5131. [Google Scholar] [CrossRef] [Green Version]
- Tremellen, K.P.; Jasper, M.J.; Robertson, S.A. Primary unexplained infertility is associated with reduced expression of the T-regulatory cell transcription factor Foxp3 in endometrial tissue. Mol. Hum. Reprod. 2006, 12, 301–308. [Google Scholar] [Green Version]
- Wang, L.; Liu, R.; Li, W.; Chen, C.; Katoh, H.; Chen, G.-Y.; McNally, B.; Lin, L.; Zhou, P.; Zuo, T.; et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell 2009, 16, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Ladoire, S.; Mignot, G.; Apetoh, L.; Ghiringhelli, F. Human FOXP3 and cancer. Oncogene 2010, 29, 4121–4129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Calisto, J.; Araya, C.; Marchant, L.; Riaz, C.F.; Mayor, R. Essential role of non-canonical Wnt signalling in neural crest migration. Development 2005, 132, 2587–2597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giles, R.H.; Van Es, J.H.; Clevers, H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta (BBA) Bioenergy 2003, 1653, 1–24. [Google Scholar] [CrossRef]
- Polakis, P. The many ways of Wnt in cancer. Curr. Opin. Genet. Dev. 2007, 17, 45–51. [Google Scholar] [CrossRef]
- Bell, S.M.; Schreiner, C.M.; Goetz, J.A.; Robbins, D.J.; Scott, W.J. Shh signaling in limb bud ectoderm: Potential role in teratogen-induced postaxial ectrodactyly. Dev. Dyn. 2005, 233, 313–325. [Google Scholar] [CrossRef]
- Bruner, H.C.; Derksen, P.W.B. Loss of E-Cadherin-Dependent Cell-Cell Adhesion and the Development and Progression of Cancer. Cold Spring Harb. Perspect. Biol. 2018, 10, a029330. [Google Scholar] [CrossRef]
- Lee, Y.C.; Baath, J.A.; Bastle, R.M.; Bhattacharjee, S.; Cantoria, M.J.; Dornan, M.; Gamero-Estevez, E.; Ford, L.; Halova, L.; Kernan, J.; et al. Impact of Detergents on Membrane Protein Complex Isolation. J. Proteome Res. 2018, 17, 348–358. [Google Scholar] [CrossRef]
- Monks, A.; Scudiero, D.; Skehan, P.; Shoemaker, R.; Paull, K.; Vistica, D.; Hose, C.; Langley, J.; Cronise, P.; Vaigro-Wolff, A.; et al. Feasibility of a High-Flux Anticancer Drug Screen Using a Diverse Panel of Cultured Human Tumor Cell Lines. J. Natl. Cancer Inst. 1991, 83, 757–766. [Google Scholar] [CrossRef]
- El-Deiry, W.S.; Kern, S.E.; Pietenpol, J.A.; Kinzler, K.W.; Vogelstein, B. Definition of a consensus binding site for p53. Nat. Genet. 1992, 1, 45–49. [Google Scholar] [CrossRef]
- Kautiainen, T.L.; A Jones, P. DNA methyltransferase levels in tumorigenic and nontumorigenic cells in culture. J. Boil. Chem. 1986, 261, 1594–1598. [Google Scholar]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Dawlaty, M.M.; Breiling, A.; Le, T.; Raddatz, G.; Barrasa, M.I.; Cheng, A.W.; Gao, Q.; Powell, B.E.; Li, Z.; Xu, M.; et al. Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development. Dev. Cell 2013, 24, 310–323. [Google Scholar] [CrossRef] [PubMed]
- Dawlaty, M.M.; Breiling, A.; Le, T.; Barrasa, M.I.; Raddatz, G.; Gao, Q.; Powell, B.E.; Cheng, A.W.; Faull, K.F.; Lyko, F.; et al. Loss of Tet enzymes compromises proper differentiation of embryonic stem cells. Dev. Cell 2014, 29, 102–111. [Google Scholar] [CrossRef]
- An, J.; González-Avalos, E.; Chawla, A.; Jeong, M.; López-Moyado, I.F.; Li, W.; Goodell, M.A.; Chavez, L.; Ko, M.; Rao, A. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat. Commun. 2015, 6, 10071. [Google Scholar] [CrossRef] [Green Version]
- Cimmino, L.; Dawlaty, M.M.; Ndiaye-Lobry, D.; Yap, Y.S.; Bakogianni, S.; Yu, Y.T.; Bhattacharyya, S.; Shaknovich, R.; Geng, H.M.; Lobry, C.; et al. TET1 is a tumor suppressor of hematopoietic malignancy. Nat. Immunol. 2015, 16, 653. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, K.; Shen, Q.; Han, Y.; Gu, Y.; Li, X.; Zhao, D.; Liu, Y.; Wang, C.; Zhang, X.; et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 2015, 525, 389–393. [Google Scholar] [CrossRef] [Green Version]
- Gaidzik, V.I.; Paschka, P.; Späth, D.; Habdank, M.; Köhne, C.-H.; Germing, U.; Von Lilienfeld-Toal, M.; Held, G.; Horst, H.-A.; Haase, D.; et al. TET2 Mutations in Acute Myeloid Leukemia (AML): Results from a Comprehensive Genetic and Clinical Analysis of the AML Study Group. J. Clin. Oncol. 2012, 30, 1350–1357. [Google Scholar] [CrossRef]
- Boumber, Y.A.; Kondo, Y.; Chen, X.; Shen, L.; Gharibyan, V.; Konishi, K.; Estey, E.; Kantarjian, H.; Garcia-Manero, G.; Issa, J.J. RIL, a LIM Gene on 5q31, Is Silenced by Methylation in Cancer and Sensitizes Cancer Cells to Apoptos.pdf. Cancer Res. 2007, 67, 1997–2005. [Google Scholar] [CrossRef]
- Liu, Z.; Zhan, Y.; Tu, Y.; Chen, K.; Liu, Z.; Wu, C. PDZ and LIM domain protein 1(PDLIM1)/CLP36 promotes breast cancer cell migration, invasion and metastasis through interaction with alpha-actinin. Oncogene 2015, 34, 1300–1311. [Google Scholar] [CrossRef]
- Tremblay, M.; Sanchez-Ferras, O.; Bouchard, M. GATA transcription factors in development and disease. Development 2018, 145, dev164384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, J.; A Travers, A. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001, 26, 167–174. [Google Scholar] [CrossRef]
- Goodwin, G.H.; Sanders, C.; Johns, E.W. A New Group of Chromatin-Associated Proteins with a High Content of Acidic and Basic Amino Acids. JBIC J. Boil. Inorg. Chem. 1973, 38, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.E.; Agresti, A. HMG proteins: Dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005, 15, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Asin-Cayuela, J.; Gustafsson, C.M. Mitochondrial transcription and its regulation in mammalian cells. Trends Biochem. Sci. 2007, 32, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Clayton, D.A.; Shadel, G.S. Initiation and Beyond: Multiple Functions of the Human Mitochondrial Transcription Machinery. Mol. Cell 2006, 24, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Štros, M. HMGB proteins: Interactions with DNA and chromatin. Biochim. Biophys. Acta (BBA) Bioenergy 2010, 1799, 101–113. [Google Scholar] [CrossRef]
- Furusawa, T.; Cherukuri, S. Developmental function of HMGN proteins. Biochim. Biophys. Acta (BBA) Bioenergy 2010, 1799, 69–73. [Google Scholar] [CrossRef] [Green Version]
- Reeves, R. Structure and Function of the HMGI(Y) Family of Architectural Transcription Factors. Environ. Health Perspect. 2000, 108, 803. [Google Scholar] [CrossRef]
- Reeves, R. Molecular biology of HMGA proteins: Hubs of nuclear function. Gene 2001, 277, 63–81. [Google Scholar] [CrossRef]
- Tkachuk, D.C.; Kohler, S.; Cleary, M.L. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 1992, 71, 691–700. [Google Scholar] [CrossRef]
- Maher, J.F.; Nathans, D. Multivalent DNA-binding properties of the HMG-1 proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 6716–6720. [Google Scholar] [CrossRef] [PubMed]
- Duguet, M.; de Recondo, A.M. A deoxyribonucleic acid unwinding protein isolated from regenerating rat liver. Physical and functional properties. J. Biol. Chem. 1978, 253, 1660–1666. [Google Scholar] [PubMed]
- Bustin, M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 2001, 26, 152–153. [Google Scholar] [CrossRef]
- Körner, U.; Bustin, M.; Scheer, U.; Hock, R. Developmental role of HMGN proteins in Xenopus laevis. Mech. Dev. 2003, 120, 1177–1192. [Google Scholar] [CrossRef]
- Furusawa, T.; Lim, J.H.; Catez, F.; Birger, Y.; Mackem, S.; Bustin, M. Down-regulation of nucleosomal binding protein HMGN1 expression during embryogenesis modulates Sox9 expression in chondrocytes. Mol. Cell. Biol. 2006, 26, 592–604. [Google Scholar] [CrossRef]
- Birger, Y.; West, K.L.; Postnikov, Y.V.; Lim, J.; Furusawa, T.; Wagner, J.P.; Laufer, C.S.; Kraemer, K.H.; Bustin, M. Chromosomal protein HMGN1 enhances the rate of DNA repair in chromatin. EMBO J. 2003, 22, 1665–1675. [Google Scholar] [CrossRef] [Green Version]
- Birger, Y.; Davis, J.; Furusawa, T.; Rand, E.; Piatigorsky, J.; Bustin, M. A role for chromosomal protein HMGN1 in corneal maturation. Differ. Res. Biol. Divers. 2006, 74, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Birger, Y.; Catez, F.; Furusawa, T.; Lim, J.-H.; Prymakowska-Bosak, M.; West, K.L.; Postnikov, Y.V.; Haines, D.C.; Bustin, M. Increased tumorigenicity and sensitivity to ionizing radiation upon loss of chromosomal protein HMGN1. Cancer Res. 2005, 65, 6711–6718. [Google Scholar] [CrossRef]
- Chieffi, P.; Battista, S.; Barchi, M.; Di Agostino, S.; Pierantoni, G.M.; Fedele, M.; Chiariotti, L.; Tramontano, D.; Fusco, A. HMGA1 and HMGA2 protein expression in mouse spermatogenesis. Oncogene 2002, 21, 3644–3650. [Google Scholar] [CrossRef]
- Fedele, M.; Fidanza, V.; Battista, S.; Pentimalli, F.; Klein-Szanto, A.J.; Visone, R.; De Martino, I.; Curcio, A.; Morisco, C.; Del Vecchio, L.; et al. Haploinsufficiency of the Hmga1 Gene Causes Cardiac Hypertrophy and Myelo-Lymphoproliferative Disorders in Mice. Cancer Res. 2006, 66, 2536–2543. [Google Scholar] [CrossRef] [PubMed]
- Foti, D.; Chiefari, E.; Fedele, M.; Iuliano, R.; Brunetti, L.; Paonessa, F.; Manfioletti, G.; Barbetti, F.; Brunetti, A.; Croce, C.M.; et al. Lack of the architectural factor HMGA1 causes insulin resistance and diabetes in humans and mice. Nat. Med. 2005, 11, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Benson, K.F.; Ashar, H.R.; Chada, K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 1995, 376, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Chada, K. In vivo modulation of Hmgic reduces obesity. Nat. Genet. 2000, 24, 377–380. [Google Scholar] [CrossRef]
- Itou, J.; Taniguchi, N.; Oishi, I.; Kawakami, H.; Lotz, M.; Kawakami, Y. HMGB factors are required for posterior digit development through integrating signaling pathway activities. Dev. Dyn. 2011, 240, 1151–1162. [Google Scholar] [CrossRef] [Green Version]
- Abraham, A.B.; Bronstein, R.; I Chen, E.; Koller, A.; Ronfani, L.; Maletic-Savatic, M.; E Tsirka, S. Members of the high mobility group B protein family are dynamically expressed in embryonic neural stem cells. Proteome Sci. 2013, 11, 18. [Google Scholar] [CrossRef]
- Nemeth, M.J.; Kirby, M.R.; Bodine, D.M. Hmgb3 regulates the balance between hematopoietic stem cell self-renewal and differentiation. Proc. Natl. Acad. Sci. USA 2006, 103, 13783–13788. [Google Scholar] [CrossRef] [Green Version]
- Nemeth, M.J.; Cline, A.P.; Anderson, S.M.; Garrett-Beal, L.J.; Bodine, D.M. Hmgb3 deficiency deregulates proliferation and differentiation of common lymphoid and myeloid progenitors. Blood 2005, 105, 627–634. [Google Scholar] [CrossRef] [Green Version]
- Ronfani, L.; Ferraguti, M.; Croci, L.; Ovitt, C.E.; Scholer, H.R.; Consalez, G.G.; Bianchi, M.E. Reduced fertility and spermatogenesis defects in mice lacking chromosomal protein Hmgb2. Development 2001, 128, 1265–1273. [Google Scholar]
- Taniguchi, N.; Carames, B.; Lotz, M. 217 Chromatin Protein Hmgb2 Regulates Articular Cartilage Surface Maintenance Via Beta-Catenin Pathways. Osteoarthr. Cartil. 2009, 17, S123. [Google Scholar] [CrossRef]
- Taniguchi, N.; Yoshida, K.; Ito, T.; Tsuda, M.; Mishima, Y.; Furumatsu, T.; Ronfani, L.; Abeyama, K.; Kawahara, K.-I.; Komiya, S.; et al. Stage-Specific Secretion of HMGB1 in Cartilage Regulates Endochondral Ossification. Mol. Cell. Boil. 2007, 27, 5650–5663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniguchi, N.; Caramés, B.; Ronfani, L.; Ulmer, U.; Komiya, S.; Bianchi, M.E.; Lotz, M. Aging-related loss of the chromatin protein HMGB2 in articular cartilage is linked to reduced cellularity and osteoarthritis. Proc. Natl. Acad. Sci. USA 2009, 106, 1181–1186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, J.-H.; Catez, F.; Birger, Y.; West, K.L.; Prymakowska-Bosak, M.; Postnikov, Y.V.; Bustin, M. Chromosomal Protein HMGN1 Modulates Histone H3 Phosphorylation. Mol. Cell 2004, 15, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Arce-Cerezo, A.; Garcia, M.; Rodríguez-Nuevo, A.; Crosa-Bonell, M.; Enguix, N.; Pero, A.; Muñoz, S.; Roca, C.; Ramos, D.; Franckhauser, S.; et al. HMGA1 overexpression in adipose tissue impairs adipogenesis and prevents diet-induced obesity and insulin resistance. Sci. Rep. 2015, 5, 14487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, S.N.; Cope, L.; Poh, W.; Belton, A.; Roy, S.; Talbot, C.C.; Sukumar, S.; Huso, D.L.; Resar, L.M.S. HMGA1: A Master Regulator of Tumor Progression in Triple-Negative Breast Cancer Cells. PLoS ONE 2013, 8, e63419. [Google Scholar] [CrossRef] [PubMed]
- Pallante, P.; Sepe, R.; Puca, F.; Fusco, A. High Mobility Group A Proteins as Tumor Markers. Front. Med. 2015, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Chen, R.; Zhang, Q.; Hou, W.; Wu, S.; Cao, L.; Huang, J.; Yu, Y.; Fan, X.G.; Yan, Z.; et al. HMGB1 in health and disease. Mol. Asp. Med. 2014, 40, 1–116. [Google Scholar] [CrossRef] [Green Version]
- Simon, M.C. Gotta have GATA. Nat Genet. 1995, 11, 9–11. [Google Scholar] [CrossRef]
- Weiss, M.J.; Orkin, S.H. GATA transcription factors: Key regulators of hematopoiesis. Exp. Hematol. 1995, 23, 99–107. [Google Scholar]
- Zheng, R.; Blobel, G.A. GATA Transcription Factors and Cancer. Genes Cancer 2010, 1, 1178–1188. [Google Scholar] [CrossRef] [Green Version]
- Grass, J.A.; Boyer, M.E.; Pal, S.; Wu, J.; Weiss, M.J.; Bresnick, E.H. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. USA 2003, 100, 8811–8816. [Google Scholar] [CrossRef]
- Zhang, S.-J.; Ma, L.-Y.; Huang, Q.-H.; Li, G.; Gu, B.-W.; Gao, X.-D.; Shi, J.-Y.; Wang, Y.-Y.; Gao, L.; Cai, X.; et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 2076–2081. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Bravo, V.; Carceles-Cordon, M.; Hoshida, Y.; Cordon-Cardo, C.; Galsky, M.D.; Domingo-Domenech, J. The role of GATA2 in lethal prostate cancer aggressiveness. Nat. Rev. Urol. 2017, 14, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Hu, Y.; Tian, Y.; Fan, Z.; Wang, K.; Li, H.; Zhou, Q.; Zeng, G.; Hu, X.; Yu, L.; et al. Lung cancer deficient in the tumor suppressor GATA4 is sensitive to TGFBR1 inhibition. Nat. Commun. 2019, 10, 1665. [Google Scholar] [CrossRef] [PubMed]
- Ho, I.C.; Vorhees, P.; Marin, N.; Oakley, B.K.; Tsai, S.F.; Orkin, S.H.; Leiden, J.M. Human GATA-3: A lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. 1991, 10, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Usary, J.; Llaca, V.; Karaca, G.; Presswala, S.; Karaca, M.; He, X.; Langerød, A.; Kåresen, R.; Oh, D.S.; Dressler, L.G.; et al. Mutation of GATA3 in human breast tumors. Oncogene 2004, 23, 7669–7678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heineke, J.; Auger-Messier, M.; Xu, J.; Oka, T.; Sargent, M.A.; York, A.; Klevitsky, R.; Vaikunth, S.; Duncan, S.A.; Aronow, B.J.; et al. Cardiomyocyte GATA4 functions as a stress-responsive regulator of angiogenesis in the murine heart. J. Clin. Investig. 2007, 117, 3198–3210. [Google Scholar] [CrossRef] [Green Version]
- Beuling, E.; Baffour–Awuah, N.Y.A.; Stapleton, K.A.; Aronson, B.E.; Noah, T.K.; Shroyer, N.F.; Duncan, S.A.; Fleet, J.C.; Krasinski, S.D. GATA factors regulate proliferation, differentiation, and gene expression in small intestine of mature mice. Gastroenterol. 2011, 140, 1219–1229. [Google Scholar] [CrossRef]
- Akiyama, Y.; Watkins, N.; Suzuki, H.; Jair, K.-W.; Van Engeland, M.; Esteller, M.; Sakai, H.; Ren, C.-Y.; Yuasa, Y.; Herman, J.G.; et al. GATA-4 and GATA-5 Transcription Factor Genes and Potential Downstream Antitumor Target Genes Are Epigenetically Silenced in Colorectal and Gastric Cancer. Mol. Cell. Boil. 2003, 23, 8429–8439. [Google Scholar] [CrossRef]
- Gong, Y.; Zhang, L.; Zhang, A.; Chen, X.; Gao, P.; Zeng, Q. GATA4 inhibits cell differentiation and proliferation in pancreatic cancer. PLoS ONE 2018, 13, e0202449. [Google Scholar] [CrossRef]
- Ip, H.S.; Morrisey, E.E.; Tang, Z.; Parmacek, M.S. GATA-4 Activates Transcription Via Two Novel Domains That Are Conserved within the GATA-4/5/6 Subfamily. J. Boil. Chem. 1997, 272, 8515–8524. [Google Scholar] [Green Version]
- Laforest, B.; Nemer, M. GATA5 interacts with GATA4 and GATA6 in outflow tract development. Dev. Boil. 2011, 358, 368–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.; Zhu, M.; Zhang, R.; Wang, Q.; Li, W.; Dong, X.; Chen, Y.; Lu, Y.; Liu, K.; Lin, B.; et al. GATA5 inhibits hepatocellular carcinoma cells malignant behaviours by blocking expression of reprogramming genes. J. Cell. Mol. Med. 2019, 23, 2536–2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepore, J.J.; Mericko, P.A.; Cheng, L.; Lu, M.M.; Morrisey, E.E.; Parmacek, M.S. GATA-6 regulates semaphorin 3C and is required in cardiac neural crest for cardiovascular morphogenesis. J. Clin. Investig. 2006, 116, 929–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Yuan, L.; Goss, A.M.; Wang, T.; Yang, J.; Lepore, J.J.; Zhou, D.; Schwartz, R.J.; Patel, V.; Cohen, E.D.; et al. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Dev. Cell 2010, 18, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Kamnasaran, D.; Qian, B.; Hawkins, C.; Stanford, W.L.; Guha, A. GATA6 is an astrocytoma tumor suppressor gene identified by gene trapping of mouse glioma model. Proc. Natl. Acad. Sci. USA 2007, 104, 8053–8058. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, P.; Carrillo-de Santa Pau, E.; Cox, T.; Sainz, B., Jr.; Dusetti, N.; Greenhalf, W.; Rinaldi, L.; Costello, E.; Ghaneh, P.; Malats, N.; et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut 2017, 66, 1665–1676. [Google Scholar] [CrossRef]
- Shen, W.; Niu, N.; Lawson, B.; Qi, L.; Zhang, J.; Li, T.; Zhang, H.; Liu, J. GATA6: A new predictor for prognosis in ovarian cancer. Hum. Pathol. 2019, 86, 163–169. [Google Scholar] [CrossRef]
- Peters, H.; Doll, U.; Niessing, J. Differential expression of the chicken Pax-1 and Pax-9 Gene: In situ hybridization and immunohistochemical analysis. Dev. Dyn. 1995, 203, 1–16. [Google Scholar] [CrossRef]
- Lai, H.-C.; Lin, Y.-W.; Huang, T.H.; Yan, P.; Huang, R.-L.; Wang, H.-C.; Liu, J.; Chan, M.W.; Chu, T.-Y.; Sun, C.-A.; et al. Identification of novel DNA methylation markers in cervical cancer. Int. J. Cancer 2008, 123, 161–167. [Google Scholar] [CrossRef]
- Imgrund, M.; Gröne, E.; Gröne, H.-J.; Kretzler, M.; Holzman, L.; Schlöndorff, D.; Rothenpieler, U.W. Re-expression of the developmental gene Pax-2 during experimental acute tubular necrosis in mice1. Kidney Int. 1999, 56, 1423–1431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeshima, A.; Nojima, Y.; Kojima, I. Involvement of Pax-2 in the Action of Activin A on Tubular Cell Regeneration. J. Am. Soc. Nephrol. 2002, 13, 2850–2859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doberstein, K.; Pfeilschifter, J.; Gutwein, P. The transcription factor PAX2 regulates ADAM10 expression in renal cell carcinoma. Carcinogenesis 2011, 32, 1713–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Chen, Y.; Liang, J.; Shi, B.; Wu, G.; Zhang, Y.; Wang, D.; Li, R.; Yi, X.; Zhang, H.; et al. Hypomethylation-linked activation of PAX2 mediates tamoxifen-stimulated endometrial carcinogenesis. Nature 2005, 438, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Koblar, S.A.; Murphy, M.; Barrett, G.L.; Underhill, A.; Gros, P.; Bartlett, P.F. Pax3 regulates neurogenesis in neural crest-derived precursor cells. J. Neurosci. Res. 1999, 56, 518–530. [Google Scholar] [CrossRef]
- Lang, D.; Lu, M.M.; Huang, L.; Engleka, K.A.; Zhang, M.; Chu, E.Y.; Lipner, S.; Skoultchi, A.; Millar, S.E.; Epstein, J.A. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature 2005, 433, 884–887. [Google Scholar] [CrossRef]
- Galili, N.; Davis, R.J.; Fredericks, W.J.; Mukhopadhyay, S.; Rauscher, F.J.; Emanuel, B.S.; Rovera, G.; Barr, F.G. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 1993, 5, 230–235. [Google Scholar] [CrossRef]
- Bennicelli, J.L.; Edwards, R.H.; Barr, F.G. Mechanism for transcriptional gain of function resulting from chromosomal translocation in alveolar rhabdomyosarcoma. Proc. Natl. Acad. Sci. USA 1996, 93, 5455–5459. [Google Scholar] [CrossRef]
- Takeuchi, H.; Morton, D.L.; Kuo, C.; Turner, R.R.; Elashoff, D.; Elashoff, R.; Taback, B.; Fujimoto, A.; Hoon, D.S. Prognostic significance of molecular upstaging of paraffin-embedded sentinel lymph nodes in melanoma patients. J. Clin. Oncol. 2004, 22, 2671–2680. [Google Scholar] [CrossRef]
- Brun, T.; Franklin, I.; St-Onge, L.; Biason-Lauber, A.; Schoenle, E.J.; Wollheim, C.B.; Gauthier, B.R. The diabetes-linked transcription factor PAX4 promotes {beta}-cell proliferation and survival in rat and human islets. J. Cell Biol. 2004, 167, 1123–1135. [Google Scholar] [CrossRef]
- Brun, T.; Duhamel, D.L.; Hu He, K.H.; Wollheim, C.B.; Gauthier, B.R. The transcription factor PAX4 acts as a survival gene in INS-1E insulinoma cells. Oncogene 2007, 26, 4261–4271. [Google Scholar] [CrossRef] [PubMed]
- Sanz, E.; Alvarez-Mon, M.; Martinez, A.C.; de la Hera, A. Human cord blood CD34+Pax-5+ B-cell progenitors: Single-cell analyses of their gene expression profiles. Blood 2003, 101, 3424–3430. [Google Scholar] [CrossRef] [PubMed]
- Krenacs, L.; Himmelmann, A.W.; Quintanilla-Martinez, L.; Fest, T.; Riva, A.; Wellmann, A.; Bagdi, E.; Kehrl, J.H.; Jaffe, E.S.; Raffeld, M. Transcription Factor B-Cell–Specific Activator Protein (BSAP) Is Differentially Expressed in B Cells. Blood 1998, 1308–1316. [Google Scholar]
- Liu, W.; Li, X.; Chu, E.S.; Go, M.Y.; Xu, L.; Zhao, G.; Li, L.; Dai, N.; Si, J.; Tao, Q.; et al. Paired box gene 5 is a novel tumor suppressor in hepatocellular carcinoma through interaction with p53 signaling pathway. Hepatology 2011, 53, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Vidal, L.J.; Perry, J.K.; Vouyovitch, C.M.; Pandey, V.; Brunet-Dunand, S.E.; Mertani, H.C.; Liu, D.X.; Lobie, P.E. PAX5alpha enhances the epithelial behavior of human mammary carcinoma cells. Mol. Cancer Res. MCR 2010, 8, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Chi, N.; Epstein, J.A. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002, 18, 41–47. [Google Scholar] [CrossRef]
- Mascarenhas, J.B.; Young, K.P.; Littlejohn, E.L.; Yoo, B.K.; Salgia, R.; Lang, D. PAX6 Is Expressed in Pancreatic Cancer and Actively Participates in Cancer Progression through Activation of the MET Tyrosine Kinase Receptor Gene. J. Boil. Chem. 2009, 284, 27524–27532. [Google Scholar] [CrossRef] [Green Version]
- Mayes, D.A.; Hu, Y.; Teng, Y.; Siegel, E.; Wu, X.; Panda, K.; Tan, F.; Yung, W.A.; Zhou, Y.-H. PAX6 Suppresses the Invasiveness of Glioblastoma Cells and the Expression of the Matrix Metalloproteinase-2 Gene. Cancer Res. 2006, 66, 9809–9817. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.H.; Hu, Y.; Mayes, D.; Siegel, E.; Kim, J.G.; Mathews, M.S.; Hsu, N.; Eskander, D.; Yu, O.; Tromberg, B.J.; et al. PAX6 suppression of glioma angiogenesis and the expression of vascular endothelial growth factor A. J. Neuro-Oncol. 2010, 96, 191–200. [Google Scholar] [CrossRef]
- Buckingham, M.; Relaix, F. The Role ofPaxGenes in the Development of Tissues and Organs: Pax3 and Pax7 Regulate Muscle Progenitor Cell Functions. Annu. Rev. Cell Dev. Boil. 2007, 23, 645–673. [Google Scholar] [CrossRef]
- Zannini, M.; Francis-Lang, H.; Plachov, D.; Di Lauro, R. Pax-8, a paired domain-containing protein, binds to a sequence overlapping the recognition site of a homeodomain and activates transcription from two thyroid-specific promoters. Mol. Cell. Boil. 1992, 12, 4230–4241. [Google Scholar] [CrossRef] [PubMed]
- Tong, G.-X.; Yu, W.M.; Beaubier, N.T.; Weeden, E.M.; Hamele-Bena, D.; Mansukhani, M.M.; O’Toole, K.M. Expression of PAX8 in normal and neoplastic renal tissues: An immunohistochemical study. Mod. Pathol. 2009, 22, 1218–1227. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; Campbell, H.G.; Wiles, A.K.; Eccles, M.R.; Reddel, R.R.; Braithwaite, A.W.; Royds, J.A. PAX8 Regulates Telomerase Reverse Transcriptase and Telomerase RNA Component in Glioma. Cancer Res. 2008, 68, 5724–5732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suda, N.; Ogawa, T.; Kojima, T.; Saito, C.; Moriyama, K. Non-syndromic oligodontia with a novel mutation of PAX9. J. Dent. Res. 2011, 90, 382–386. [Google Scholar] [CrossRef]
- Kendall, J.; Liu, Q.; Bakleh, A.; Krasnitz, A.; Nguyen, K.C.Q.; Lakshmi, B.; Gerald, W.L.; Powers, S.; Mu, D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 16663–16668. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.C.; Sharma, M.; Lee, Y.H.; Lee, N.H.; Kim, S.Y.; Yun, J.S.; Nam, S.Y.; Hwang, P.H.; Jhee, E.C.; Yi, H.K. Pax9 mediated cell survival in oral squamous carcinoma cell enhanced by c-myb. Cell Biochem. Funct. 2008, 26, 892–899. [Google Scholar] [CrossRef]
- Huang, C.; Chan, J.A.; Schuurmans, C. Proneural bHLH Genes in Development and Disease. Curr. Top Dev. Biol. 2014, 110, 75–127. [Google Scholar] [CrossRef]
- Bialek, P.; Kern, B.; Yang, X.; Schrock, M.; Sosic, D.; Hong, N.; Wu, H.; Yu, K.; Ornitz, D.M.; Olson, E.N.; et al. A twist code determines the onset of osteoblast differentiation. Dev. Cell 2004, 6, 423–435. [Google Scholar] [CrossRef]
- Yang, J.; A Mani, S.; Donaher, J.L.; Ramaswamy, S.; Itzykson, R.A.; Come, C.; Savagner, P.; Gitelman, I.; Richardson, A.; A Weinberg, R. Twist, a Master Regulator of Morphogenesis, Plays an Essential Role in Tumor Metastasis. Cell 2004, 117, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Chang, A.T.; Liu, Y.; Ayyanathan, K.; Benner, C.; Jiang, Y.; Prokop, J.W.; Paz, H.; Wang, D.; Li, H.-R.; Fu, X.-D.; et al. An evolutionarily conserved DNA architecture determines target specificity of the TWIST family bHLH transcription factors. Genes Dev. 2015, 29, 603–616. [Google Scholar] [CrossRef] [Green Version]
- Tsai, J.H.; Yang, J. Epithelial–mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013, 27, 2192–2206. [Google Scholar] [CrossRef] [PubMed]
- Hurlin, P.J. Control of Vertebrate Development by MYC. Cold Spring Harb. Perspect. Med. 2013, 3, a014332. [Google Scholar] [CrossRef] [PubMed]
- Witkiewicz, A.K.; McMillan, E.A.; Balaji, U.; Baek, G.; Lin, W.-C.; Mansour, J.; Mollaee, M.; Wagner, K.-U.; Koduru, P.; Yopp, A.; et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015, 6, 6744. [Google Scholar] [CrossRef] [PubMed]
- Chesi, M.; Robbiani, D.F.; Sebag, M.; Chng, W.J.; Affer, M.; Tiedemann, R.; Valdez, R.; Palmer, S.E.; Haas, S.S.; Stewart, A.K.; et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell 2008, 13, 167–180. [Google Scholar] [CrossRef]
- Bermingham, N.A. Math1: An Essential Gene for the Generation of Inner Ear Hair Cells. Science 1999, 284, 1837–1841. [Google Scholar] [CrossRef]
- Price, S.D.; Shope, C.; Himes, D.; Liu, M.; Pereira, F.A.; Chu, M.-J.; Eatock, R.A.; Brownell, W.E.; Lysakowski, A.; Tsai, M.-J. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 2000, 14, 2839–2854. [Google Scholar]
- Boulay, G.; Sandoval, G.J.; Riggi, N.; Iyer, S.; Buisson, R.; Naigles, B.; Awad, M.E.; Rengarajan, S.; Volorio, A.; McBride, M.J.; et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171, 163–178. [Google Scholar] [CrossRef]
- Khan, S.; Stott, S.R.; Chabrat, A.; Truckenbrodt, A.M.; Spencer-Dene, B.; Nave, K.-A.; Guillemot, F.; Levesque, M.; Ang, S.-L. Survival of a Novel Subset of Midbrain Dopaminergic Neurons Projecting to the Lateral Septum Is Dependent on NeuroD Proteins. J. Neurosci. 2017, 37, 2305–2316. [Google Scholar] [CrossRef] [Green Version]
- Laclé, M.M.; Van Diest, P.J.; Goldschmeding, R.; Van Der Wall, E.; Nguyen, T.Q. Expression of Connective Tissue Growth Factor in Male Breast Cancer: Clinicopathologic Correlations and Prognostic Value. PLoS ONE 2015, 10, e0118957. [Google Scholar] [CrossRef]
- Barnes, R.M.; Firulli, B.A.; Conway, S.J.; Vincentz, J.W.; Firulli, A.B. Analysis of the Hand1 cell lineage reveals novel contributions to cardiovascular, neural crest, extra-embryonic, and lateral mesoderm derivatives. Dev. Dyn. 2010, 239, 3086–3097. [Google Scholar] [CrossRef] [Green Version]
- Barnes, R.M.; Firulli, B.A.; VanDusen, N.J.; Morikawa, Y.; Conway, S.J.; Cserjesi, P.; Vincentz, J.W.; Firulli, A.B. Hand2 loss-of-function in Hand1-expressing cells reveals distinct roles in epicardial and coronary vessel development. Circ. Res. 2011, 108, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Martı́nez-Espinoza, A.D.; Garcı́a-Pedrajas, M.D.; Gold, S.E. The Ustilaginales as Plant Pests and Model Systems. Fungal Genet. Boil. 2002, 35, 1–20. [Google Scholar]
- Kato, N.; Iwase, A.; Ishida, C.; Nagai, T.; Mori, M.; Bayasula; Nakamura, T.; Osuka, S.; Ganiyeva, U.; Qin, Y.; et al. Upregulation of Fibroblast Growth Factors Caused by Heart and Neural Crest Derivatives Expressed 2 Suppression in Endometriotic Cells: A Possible Therapeutic Target in Endometriosis. Reprod. Sci. 2019, 26, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Anderson, D.J. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell 2002, 109, 61–73. [Google Scholar] [CrossRef]
- Brena, R.M.; Morrison, C.; Liyanarachchi, S.; Jarjoura, D.; Davuluri, R.V.; Otterson, G.A.; Reisman, D.; Glaros, S.; Rush, L.J.; Plass, C. Aberrant DNA Methylation of OLIG1, a Novel Prognostic Factor in Non-Small Cell Lung Cancer. PLoS Med. 2007, 4, e108. [Google Scholar] [CrossRef]
- Kosty, J.; Lu, F.; Kupp, R.; Mehta, S.; Lu, Q.R. Harnessing OLIG2 function in tumorigenicity and plasticity to target malignant gliomas. Cell Cycle 2017, 16, 1654–1660. [Google Scholar] [CrossRef] [Green Version]
- MacLean, H.E.; Kronenberg, H.M. Expression of Stra13 during mouse endochondral bone development. Gene Expr. Patterns 2004, 4, 633–636. [Google Scholar] [CrossRef]
- Huang, K.-L.; Mashl, R.J.; Wu, Y.; Ritter, D.I.; Wang, J.; Oh, C.; Paczkowska, M.; Wyczalkowski, M.A.; Oak, N.; Scott, A.D.; et al. Pathogenic Germline Variants in 10,389 Adult Cancers. Cell 2018, 173, 355–370. [Google Scholar] [CrossRef]
- Bhawal, U.K.; Sato, F.; Arakawa, Y.; Fujimoto, K.; Kawamoto, T.; Tanimoto, K.; Ito, Y.; Sasahira, T.; Sakurai, T.; Kobayashi, M.; et al. Basic helix-loop-helix transcription factor DEC1 negatively regulates cyclin D1. J. Pathol. 2011, 224, 420–429. [Google Scholar] [CrossRef]
- Sasamoto, T.; Fujimoto, K.; Kanawa, M.; Kimura, J.; Takeuchi, J.; Harada, N.; Goto, N.; Kawamoto, T.; Noshiro, M.; Suardita, K.; et al. DEC2 is a negative regulator for the proliferation and differentiation of chondrocyte lineage-committed mesenchymal stem cells. Int. J. Mol. Med. 2016, 38, 876–884. [Google Scholar] [CrossRef]
- Liu, J.; Uygur, B.; Zhang, Z.; Shao, L.; Romero, D.; Vary, C.; Ding, Q.; Wu, W.-S. Slug inhibits proliferation of human prostate cancer cells via downregulation of cyclin D1 expression. Prostate 2010, 70, 1768–1777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blanpain, C.; Lowry, W.E.; Pasolli, H.A.; Fuchs, E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genome Res. 2006, 20, 3022–3035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichijo, R.; Iizuka, Y.; Kubo, H.; Toyoshima, F. Essential roles of Tbx3 in embryonic skin development during epidermal stratification. Genes Cells 2017, 22, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Kageyama, R. Hes1 regulates embryonic stem cell differentiation by suppressing Notch signaling. Genes Cells 2010, 15, 689–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imayoshi, I.; Ishidate, F.; Kageyama, R. Real-time imaging of bHLH transcription factors reveals their dynamic control in the multipotency and fate choice of neural stem cells. Front. Cell. Neurosci. 2015, 9, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baek, J.H.; Hatakeyama, J.; Sakamoto, S.; Ohtsuka, T.; Kageyama, R. Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development 2006, 133, 2467–2476. [Google Scholar] [CrossRef] [Green Version]
- Carré, A.; Rachdi, L.; Tron, E.; Richard, B.; Castanet, M.; Schlumberger, M.; Bidart, J.-M.; Szinnai, G.; Polak, M. Hes1 Is Required for Appropriate Morphogenesis and Differentiation during Mouse Thyroid Gland Development. PLoS ONE 2011, 6, e16752. [Google Scholar] [CrossRef]
- Huang, Q.; Raya, A.; DeJesus, P.; Chao, S.-H.; Quon, K.C.; Caldwell, J.S.; Chanda, S.K.; Izpisua-Belmonte, J.C.; Schultz, P.G. Identification of p53 regulators by genome-wide functional analysis. Proc. Natl. Acad. Sci. USA 2004, 101, 3456–3461. [Google Scholar] [CrossRef] [Green Version]
- Fischer, A.; Schumacher, N.; Maier, M.; Sendtner, M.; Gessler, M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genome Res. 2004, 18, 901–911. [Google Scholar] [CrossRef]
- Benito-Gonzalez, A.; Doetzlhofer, A. Hey1 and Hey2 control the spatial and temporal pattern of mammalian auditory hair cell differentiation downstream of Hedgehog signaling. J. Neurosci. 2014, 34, 12865–12876. [Google Scholar] [CrossRef]
- Yin, X.; Zeng, Z.; Xing, J.; Zhang, A.; Jiang, W.; Wang, W.; Sun, H.; Ni, L. Hey1 functions as a positive regulator of odontogenic differentiation in odontoblastlineage cells. Int. J. Mol. Med. 2018, 41, 331–339. [Google Scholar] [PubMed]
- Laudet, V.; Stéhelin, D.; Clevers, H. Ancestry and diversity of the HMG box superfamily. Nucleic Acids Res. 1993, 21, 2493–2501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Štros, M.; Launholt, D.; Grasser, K.D. The HMG-box: A versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 2007, 64, 2590–2606. [Google Scholar] [CrossRef] [PubMed]
- Sessa, L.; Bianchi, M.E. The evolution of High Mobility Group Box (HMGB) chromatin proteins in multicellular animals. Gene 2007, 387, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.; Felsenfeld, G. The erythroid-specific transcription factor eryf1: A new finger protein. Cell 1989, 58, 877–885. [Google Scholar] [CrossRef]
- Katsumura, K.R.; Bresnick, E.H. GATA Factor Mechanisms Group; the GATA Factor Mechanisms Group the GATA factor revolution in hematology. Blood 2017, 129, 2092–2102. [Google Scholar] [CrossRef]
- Lentjes, M.H.; Niessen, H.E.; Akiyama, Y.; De Bruïne, A.P.; Melotte, V.; Van Engeland, M. The emerging role of GATA transcription factors in development and disease. Expert Rev. Mol. Med. 2016, 18, e3. [Google Scholar] [CrossRef]
- Simon, M.C.; Pevny, L.; Wiles, M.V.; Keller, G.; Costantini, F.; Orkin, S.H. Rescue of erythroid development in gene targeted GATA–1−mouse embryonic stem cells. Nat. Genet. 1992, 1, 92–98. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Browne, C.P.; Cunniff, K.; Goff, S.C.; Orkin, S.H. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. USA 1996, 93, 12355–12358. [Google Scholar] [CrossRef]
- Hosoya, T.; Maillard, I.; Engel, J.D. From the cradle to the grave: Activities of GATA-3 throughout T-cell development and differentiation. Immunol. Rev. 2010, 238, 110–125. [Google Scholar] [CrossRef]
- Frelin, C.; Herrington, R.; Janmohamed, S.; Barbara, M.; Tran, G.; Paige, C.J.; Benveniste, P.; Zúñiga-Pflücker, J.-C.; Souabni, A.; Busslinger, M.; et al. GATA-3 regulates the self-renewal of long-term hematopoietic stem cells. Nat. Immunol. 2013, 14, 1037–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fitch, S.R.; Kimber, G.M.; Wilson, N.K.; Parker, A.; Mirshekar-Syahkal, B.; Göttgens, B.; Medvinsky, A.; Dzierzak, E.; Ottersbach, K. Signaling from the sympathetic nervous system regulates hematopoietic stem cell emergence during embryogenesis. Cell Stem Cell 2012, 11, 554–566. [Google Scholar] [CrossRef] [PubMed]
- Yamak, A.; Latinkić, B.V.; Dali, R.; Temsah, R.; Nemer, M. Cyclin D2 is a GATA4 cofactor in cardiogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 1415–1420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, R.; Watt, A.J.; Li, J.; Luebke-Wheeler, J.; Morrisey, E.E.; Duncan, S.A. GATA6 Is Essential for Embryonic Development of the Liver but Dispensable for Early Heart Formation. Mol. Cell. Boil. 2005, 25, 2622–2631. [Google Scholar] [CrossRef] [Green Version]
- Scazzocchio, C. The fungal GATA factors. Curr. Opin. Microbiol. 2000, 3, 126–131. [Google Scholar] [CrossRef]
- Lowry, J.A.; Atchley, W.R. Molecular Evolution of the GATA Family of Transcription Factors: Conservation Within the DNA-Binding Domain. J. Mol. Evol. 2000, 50, 103–115. [Google Scholar] [CrossRef]
- He, C.; Cheng, H.; Zhou, R. GATA family of transcription factors of vertebrates: Phylogenetics and chromosomal synteny. J. Biosci. 2007, 32, 1273–1280. [Google Scholar] [CrossRef]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA Family of Transcription Factors in Arabidopsis and Rice1. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef]
- Arguello-Astorga, G.; Herrera-Estrella, L. Evolution of Light-Regulated Plant Promoters. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 525–555. [Google Scholar] [CrossRef]
- Shimizu, R.; Ohneda, K.; Engel, J.D.; Trainor, C.D.; Yamamoto, M. Transgenic rescue of GATA-1-deficient mice with GATA-1 lacking a FOG-1 association site phenocopies patients with X-linked thrombocytopenia. Blood 2004, 103, 2560–2567. [Google Scholar] [CrossRef]
- Chang, A.N.; Cantor, A.B.; Fujiwara, Y.; Lodish, M.B.; Droho, S.; Crispino, J.D.; Orkin, S.H. GATA-factor dependence of the multitype zinc-finger protein FOG-1 for its essential role in megakaryopoiesis. Proc. Natl. Acad. Sci. USA 2002, 99, 9237–9242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, M.S.; Hancock, D.C.; Molina-Arcas, M.; Steckel, M.; East, P.; Diefenbacher, M.; Armenteros-Monterroso, E.; Lassailly, F.; Matthews, N.; Nye, E.; et al. The GATA2 Transcriptional Network Is Requisite for RAS Oncogene-Driven Non-Small Cell Lung Cancer. Cell 2012, 149, 642–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kouros-Mehr, H.; Bechis, S.K.; Slorach, E.M.; Littlepage, L.E.; Egeblad, M.; Ewald, A.J.; Pai, S.-Y.; Ho, I.-C.; Werb, Z. GATA-3 links tumor differentiation and dissemination in a luminal breast cancer model. Cancer Cell 2008, 13, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Dydensborg, A.B.; Rose, A.A.N.; Wilson, B.J.; Grote, D.; Paquet, M.; Giguère, V.; Siegel, P.M.; Bouchard, M. GATA3 inhibits breast cancer growth and pulmonary breast cancer metastasis. Oncogene 2009, 28, 2634–2642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, J.; Lin, J.H.; Brenot, A.; Kim, J.-W.; Provot, S.; Werb, Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat. Cell Biol. 2013, 15, 201–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, B.; Luo, M.; Lakkur, S.; Lucito, R.; Iacobuzio-Donahue, C.A. Frequent genomic copy number gain and overexpression of GATA-6 in pancreatic carcinoma. Cancer Boil. Ther. 2008, 7, 1593–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shureiqi, I.; Zuo, X.; Broaddus, R.; Wu, Y.; Guan, B.; Morris, J.S.; Lippman, S.M. The transcription factor GATA-6 is overexpressed in vivo and contributes to silencing 15-LOX-1 in vitro in human colon cancer. FASEB J. 2007, 21, 743–753. [Google Scholar] [CrossRef]
- Lang, D.; Powell, S.K.; Plummer, R.S.; Young, K.P.; Ruggeri, B.A. PAX genes: Roles in development, pathophysiology, and cancer. Biochem. Pharmacol. 2007, 73, 1–14. [Google Scholar] [CrossRef]
- Buckingham, M. Skeletal muscle progenitor cells and the role of Pax genes. C. R. Boil. 2007, 330, 530–533. [Google Scholar] [CrossRef]
- Vorobyov, E.; Horst, J. Getting the proto-Pax by the tail. J. Mol. Evol. 2006, 63, 153–164. [Google Scholar] [CrossRef]
- Eberhard, D.; Jiménez, G.; Heavey, B.; Busslinger, M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000, 19, 2292–2303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apuzzo, S.; Gros, P. Cooperative Interactions between the Two DNA Binding Domains of Pax3: Helix 2 of the Paired Domain Is in the Proximity of the Amino Terminus of the Homeodomain. Biochemistry 2007, 46, 2984–2993. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Fang, W.-H.; Krupinski, J.; Kumar, S.; Slevin, M.; Kumar, P. Pax genes in embryogenesis and oncogenesis. J. Cell. Mol. Med. 2008, 12, 2281–2294. [Google Scholar] [CrossRef] [PubMed]
- Robichaud, G.A.; Nardini, M.; Laflamme, M.; Čuperlović-Culf, M.; Ouellette, R.J. Human Pax-5 C-terminal Isoforms Possess Distinct Transactivation Properties and Are Differentially Modulated in Normal and Malignant B Cells. J. Boil. Chem. 2004, 279, 49956–49963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robson, E.J.D.; He, S.-J.; Eccles, M.R. A PANorama of PAX genes in cancer and development. Nat. Rev. Cancer 2006, 6, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Tong, G.-X.; Melamed, J.; Mansukhani, M.; Memeo, L.; Hernandez, O.; Deng, F.-M.; Chiriboga, L.; Waisman, J. PAX2: A reliable marker for nephrogenic adenoma. Mod. Pathol. 2006, 19, 356–363. [Google Scholar] [CrossRef]
- Tong, G.X.; Chiriboga, L.; Hamele-Bena, D.; Borczuk, A.C. Expression of PAX2 in papillary serous carcinoma of the ovary: Immunohistochemical evidence of fallopian tube or secondary Mullerian system origin? Mod. Pathol. 2007, 20, 856–863. [Google Scholar] [CrossRef]
- Silberstein, G.B.; Dressler, G.R.; Van Horn, K. Expression of the PAX2 oncogene in human breast cancer and its role in progesterone-dependent mammary growth. Oncogene 2002, 21, 1009–1016. [Google Scholar] [CrossRef] [Green Version]
- Hanna, J.; Markoulaki, S.; Schorderet, P.; Carey, B.W.; Beard, C.; Wernig, M.; Creyghton, M.P.; Steine, E.J.; Cassady, J.P.; Foreman, R.; et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 2008, 133, 250–264. [Google Scholar] [CrossRef]
- Simpson, T.I.; Price, D.J. Pax6; A pleiotropic player in development. BioEssays 2002, 24, 1041–1051. [Google Scholar] [CrossRef]
- Pichaud, F.; Desplan, C. Pax genes and eye organogenesis. Curr. Opin. Genet. Dev. 2002, 12, 430–434. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, C.T.; Chen, J.; Pankratz, M.T.; Xi, J.; Li, J.; Yang, Y.; LaVaute, T.M.; Li, X.-J.; Ayala, M.; et al. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 2010, 7, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Sansom, S.N.; Griffiths, D.S.; Faedo, A.; Kleinjan, D.-J.; Ruan, Y.; Smith, J.; Van Heyningen, V.; Rubenstein, J.L.; Livesey, F.J. The Level of the Transcription Factor Pax6 Is Essential for Controlling the Balance between Neural Stem Cell Self-Renewal and Neurogenesis. PLoS Genet. 2009, 5, e1000511. [Google Scholar] [CrossRef] [PubMed]
- Sivak, J.M.; Mohan, R.; Rinehart, W.B.; Xu, P.-X.; Maas, R.L.; Fini, M.E. Pax-6 expression and activity are induced in the reepithelializing cornea and control activity of the transcriptional promoter for matrix metalloproteinase gelatinase B. Dev. Boil. 2000, 222, 41–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, J.; Lowes, C.; Collinson, J.M. Cytoskeletal and cell adhesion defects in wounded and Pax6+/- corneal epithelia. Investig. Ophthalmol. Vis. Sci. 2010, 51, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Ramaesh, T.; Ramaesh, K.; Martin Collinson, J.; Chanas, S.A.; Dhillon, B.; West, J.D. Developmental and cellular factors underlying corneal epithelial dysgenesis in the Pax6+/− mouse model of aniridia. Exp. Eye Res. 2005, 81, 224–235. [Google Scholar] [CrossRef]
- Relaix, F.; Montarras, D.; Zaffran, S.; Gayraud-Morel, B.; Rocancourt, D.; Tajbakhsh, S.; Mansouri, A.; Cumano, A.; Buckingham, M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 2006, 172, 91–102. [Google Scholar] [CrossRef]
- De Felice, M.; Di Lauro, R. Minireview: Intrinsic and Extrinsic Factors in Thyroid Gland Development: An Update. Endocrinology 2011, 152, 2948–2956. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.; Friedman, S.; Lin, R.-Y. Thyroid stem cells: Lessons from normal development and thyroid cancer. Endocr.-Relat. Cancer 2008, 15, 51–58. [Google Scholar] [CrossRef]
- Little, M.H.; Bertram, J.F. Is there such a thing as a renal stem cell? J. Am. Soc. Nephrol. JASN 2009, 20, 2112–2117. [Google Scholar] [CrossRef]
- Li, C.G.; E Nyman, J.; Braithwaite, A.W.; Eccles, M.R. PAX8 promotes tumor cell growth by transcriptionally regulating E2F1 and stabilizing RB protein. Oncogene 2011, 30, 4824–4834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazet, F.; Hutt, J.A.; Millard, J.; Shimeld, S.M. Pax gene expression in the developing central nervous system of Ciona intestinalis. Gene Expr. Patterns 2003, 3, 743–745. [Google Scholar] [CrossRef]
- Sagasser, S.; Hadrys, T.; DeSalle, R.; Fischer, N.; Schierwater, B. The Trichoplax PaxB Gene: A Putative Proto-PaxA/B/C Gene Predating the Origin of Nerve and Sensory Cells. Mol. Boil. Evol. 2005, 22, 1569–1578. [Google Scholar] [Green Version]
- Putnam, N.H.; Butts, T.; Ferrier, D.E.K.; Furlong, R.F.; Hellsten, U.; Kawashima, T.; Robinson-Rechavi, M.; Shoguchi, E.; Terry, A.; Yu, J.-K.; et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 2008, 453, 1064–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dehal, P.; Boore, J.L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Boil. 2005, 3, e314. [Google Scholar] [CrossRef] [PubMed]
- Goode, D.K.; Elgar, G. ThePAX258gene subfamily: A comparative perspective. Dev. Dyn. 2009, 238, 2951–2974. [Google Scholar] [CrossRef]
- Bassham, S.; Cañestro, C.; Postlethwait, J.H. Evolution of developmental roles of Pax2/5/8 paralogs after independent duplication in urochordate and vertebrate lineages. BMC Boil. 2008, 6, 35. [Google Scholar] [CrossRef]
- Paixão-Côrtes, V.R.; Salzano, F.M.; Bortolini, M.C. Evolutionary History of Chordate PAX Genes: Dynamics of Change in a Complex Gene Family. PLoS ONE 2013, 8, e73560. [Google Scholar] [CrossRef]
- Miller, D.J.; Hayward, D.C.; Reece-Hoyes, J.S.; Scholten, I.; Catmull, J.; Gehring, W.J.; Callaerts, P.; Larsen, J.E.; Ball, E.E. Pax gene diversity in the basal cnidarian Acropora millepora (Cnidaria, Anthozoa): Implications for the evolution of the Pax gene family. Proc. Natl. Acad. Sci. USA 2000, 97, 4475–4480. [Google Scholar] [CrossRef]
- Galliot, B.; De Vargas, C.; Miller, D. Evolution of homeobox genes: Q 50 Paired-like genes founded the Paired class. Dev. Genes Evol. 1999, 209, 186–197. [Google Scholar] [CrossRef]
- Hoshiyama, D.; Suga, H.; Iwabe, N.; Koyanagi, M.; Nikoh, N.; Kuma, K.-I.; Matsuda, F.; Honjo, T.; Miyata, T. Sponge Pax cDNA Related to Pax-2/5/8 and Ancient Gene Duplications in the Pax Family. J. Mol. Evol. 1998, 47, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Balczarek, K.A.; Lai, Z.C.; Kumar, S. Evolution of functional diversification of the paired box (Pax) DNA-binding domains. Mol. Boil. Evol. 1997, 14, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, M.; Remppis, A.; Fredericks, W.J.; Rauscher, F.J.; Schäfer, B.W. Induction of apoptosis in rhabdomyosarcoma cells through down-regulation of PAX proteins. Proc. Natl. Acad. Sci. USA 1996, 93, 13164–13169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gnarra, J.R.; Dressler, G.R. Expression of Pax-2 in human renal cell carcinoma and growth inhibition by antisense oligonucleotides. Cancer Res. 1995, 55, 4092–4098. [Google Scholar]
- Luu, V.-D.; Boysen, G.; Struckmann, K.; Casagrande, S.; Von Teichman, A.; Wild, P.J.; Sulser, T.; Schraml, P.; Moch, H. Loss of VHL and Hypoxia Provokes PAX2 Up-Regulation in Clear Cell Renal Cell Carcinoma. Clin. Cancer Res. 2009, 15, 3297–3304. [Google Scholar] [CrossRef] [Green Version]
- Buttiglieri, S.; Deregibus, M.C.; Bravo, S.; Cassoni, P.; Chiarle, R.; Bussolati, B.; Camussi, G. Role of Pax2 in apoptosis resistance and proinvasive phenotype of Kaposi’s sarcoma cells. J. Biol. Chem. 2004, 279, 4136–4143. [Google Scholar] [CrossRef]
- Relaix, F.; Polimeni, M.; Rocancourt, D.; Ponzetto, C.; Schäfer, B.W.; Buckingham, M. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genome Res. 2003, 17, 2950–2965. [Google Scholar] [CrossRef]
- Miyamoto, T.; Kakizawa, T.; Ichikawa, K.; Nishio, S.; Kajikawa, S.; Hashizume, K. Expression of Dominant Negative Form of PAX4 in Human Insulinoma. Biochem. Biophys. Res. Commun. 2001, 282, 34–40. [Google Scholar] [CrossRef]
- Chang, J.Y.; Hu, Y.; Siegel, E.; Stanley, L.; Zhou, Y.-H. PAX6 increases glioma cell susceptibility to detachment and oxidative stress. J. Neuro-Oncol. 2007, 84, 9–19. [Google Scholar] [CrossRef]
- Kroll, T.G.; Sarraf, P.; Pecciarini, L.; Chen, C.J.; Mueller, E.; Spiegelman, B.M.; A Fletcher, J. PAX8-PPARgamma1 fusion oncogene in human thyroid carcinoma [corrected]. Science 2000, 289, 1357–1360. [Google Scholar] [CrossRef]
- Li, C.G.; Eccles, M.R. PAX Genes in Cancer; Friends or Foes? Front. Genet. 2012, 3, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertrand, N.; Castro, D.S.; Guillemot, F. Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 2002, 3, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Murre, C.; Massari, M.E. Helix-Loop-Helix Proteins: Regulators of Transcription in Eucaryotic Organisms. Mol. Cell. Boil. 2000, 20, 429–440. [Google Scholar] [Green Version]
- Jones, S. An overview of the basic helix-loop-helix proteins. Genome Boil. 2004, 5, 226. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Morgan, M. Breast cancer progression with a Twist. Nat. Med. 2004, 10, 777–778. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef]
- Dennis, D.J.; Han, S.; Schuurmans, C. bHLH transcription factors in neural development, disease, and reprogramming. Brain Res. 2019, 1705, 48–65. [Google Scholar] [CrossRef]
- Fode, C.; Gradwohl, G.; Morin, X.; Dierich, A.; LeMeur, M.; Goridis, C.; Guillemot, F. The bHLH Protein NEUROGENIN 2 Is a Determination Factor for Epibranchial Placode–Derived Sensory Neurons. Neuron 1998, 20, 483–494. [Google Scholar] [CrossRef]
- Guillemot, F.; Lo, L.-C.; Johnson, J.E.; Auerbach, A.; Anderson, D.J.; Joyner, A.L. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 1993, 75, 463–476. [Google Scholar] [CrossRef]
- Scardigli, R.; Schuurmans, C.; Gradwohl, G.; Guillemot, F. Crossregulation between Neurogenin2 and Pathways Specifying Neuronal Identity in the Spinal Cord. Neuron 2001, 31, 203–217. [Google Scholar] [CrossRef] [Green Version]
- Casarosa, S.; Fode, C.; Guillemot, F. Mash1 regulates neurogenesis in the ventral telencephalon. Development 1999, 126, 525–534. [Google Scholar] [PubMed]
- Horton, S.; Meredith, A.; Richardson, J.A.; Johnson, J.E. Correct Coordination of Neuronal Differentiation Events in Ventral Forebrain Requires the bHLH Factor MASH1. Mol. Cell. Neurosci. 1999, 14, 355–369. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Sommer, L.; Cserjesi, P.; Anderson, D.J. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J. Neurosci. 1997, 17, 3644–3652. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.; Schuurmans, C.; Britz, O.; Guillemot, F. Neural bHLH Genes Control the Neuronal versus Glial Fate Decision in Cortical Progenitors. Neuron 2001, 29, 401–413. [Google Scholar] [CrossRef] [Green Version]
- Tomita, K.; Moriyoshi, K.; Nakanishi, S.; Guillemot, F.; Kageyama, R. Mammalian achaete–scute and atonal homologs regulate neuronal versus glial fate determination in the central nervous system. EMBO J. 2000, 19, 5460–5472. [Google Scholar] [CrossRef]
- Caiazzo, M.; Dell’Anno, M.T.; Dvoretskova, E.; Lazarevic, D.; Taverna, S.; Leo, D.; Sotnikova, T.D.; Menegon, A.; Roncaglia, P.; Colciago, G.; et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 2011, 476, 224–227. [Google Scholar] [CrossRef]
- Pfisterer, U.; Kirkeby, A.; Torper, O.; Wood, J.; Nelander, J.; Dufour, A.; Björklund, A.; Lindvall, O.; Jakobsson, J.; Parmar, M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. USA 2011, 108, 10343–10348. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.P.; Yang, N.; Vierbuchen, T.; Ostermeier, A.; Fuentes, D.R.; Yang, T.Q.; Citri, A.; Sebastiano, V.; Marro, S.; Südhof, T.C.; et al. Induction of human neuronal cells by defined transcription factors. Nature 2011, 476, 220–223. [Google Scholar] [CrossRef]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef] [Green Version]
- Mazzoni, E.; Mahony, S.; Closser, M.; A Morrison, C.; Nedelec, S.; Williams, D.J.; An, D.; Gifford, D.K.; Wichterle, H. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 2013, 16, 1219–1227. [Google Scholar] [CrossRef] [Green Version]
- Schwab, M.H.; Bartholomae, A.; Heimrich, B.; Feldmeyer, D.; Druffel-Augustin, S.; Goebbels, S.; Naya, F.J.; Zhao, S.; Frotscher, M.; Tsai, M.-J.; et al. Neuronal Basic Helix-Loop-Helix Proteins (NEX and BETA2/Neuro D) Regulate Terminal Granule Cell Differentiation in the Hippocampus. J. Neurosci. 2000, 20, 3714–3724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takebayashi, H.; Nabeshima, Y.; Yoshida, S.; Chisaka, O.; Ikenaka, K.; Nabeshima, Y.-I. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr. Boil. 2002, 12, 1157–1163. [Google Scholar] [CrossRef]
- Lu, Q.R.; Cai, L.; Rowitch, D.; Cepko, C.L.; Stiles, C.D. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat. Neurosci. 2001, 4, 973–974. [Google Scholar] [CrossRef] [PubMed]
- Hartenstein, V.; Takashima, S.; Hartenstein, P.; Asanad, S.; Asanad, K. bHLH proneural genes as cell fate determinants of entero-endocrine cells, an evolutionarily conserved lineage sharing a common root with sensory neurons. Dev. Boil. 2017, 431, 36–47. [Google Scholar] [CrossRef]
- VanDussen, K.L.; Samuelson, L.C. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev. Boil. 2010, 346, 215–223. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; De Faria, J.P.; Andrew, P.; Nitarska, J.; Richardson, W.D. Phosphorylation regulates OLIG2 cofactor choice and the motor neuron-oligodendrocyte fate switch. Neuron 2011, 69, 918–929. [Google Scholar] [CrossRef]
- Huang, H.-P.; Tsai, M.-J.; Naya, F.J.; Qiu, Y.; Mutoh, H.; DeMayo, F.J.; Leiter, A.B. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev. 1997, 11, 2323–2334. [Google Scholar] [Green Version]
- Risebro, C.A.; Smart, N.; Dupays, L.; Breckenridge, R.; Mohun, T.J.; Riley, P.R. Hand1 regulates cardiomyocyte proliferation versus differentiation in the developing heart. Development 2006, 133, 4595–4606. [Google Scholar] [CrossRef] [Green Version]
- Quarto, N.; Shailendra, S.; Meyer, N.P.; Menon, S.; Renda, A.; Longaker, M.T. Twist1-Haploinsufficiency Selectively Enhances the Osteoskeletal Capacity of Mesoderm-Derived Parietal Bone Through Downregulation of Fgf23. Front. Physiol. 2018, 9, 1426. [Google Scholar] [CrossRef]
- Bildsoe, H.; Loebel, D.A.; Jones, V.J.; Chen, Y.-T.; Behringer, R.R.; Tam, P.P. Requirement for Twist1 in frontonasal and skull vault development in the mouse embryo. Dev. Boil. 2009, 331, 176–188. [Google Scholar] [CrossRef] [Green Version]
- Segev, E.; Halachmi, N.; Salzberg, A.; Ben-Arie, N. Nato3 is an evolutionarily conserved bHLH transcription factor expressed in the CNS of Drosophila and mouse. Mech. Dev. 2001, 106, 197–202. [Google Scholar] [CrossRef]
- Han, Z.; Yi, P.; Li, X.; Olson, E.N. Hand, an evolutionarily conserved bHLH transcription factor required for Drosophila cardiogenesis and hematopoiesis. Development 2006, 133, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Ledent, V.; Vervoort, M. The Basic Helix-Loop-Helix Protein Family: Comparative Genomics and Phylogenetic Analysis. Genome Res. 2001, 11, 754–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Goossens, J.; Mertens, J.; Goossens, A. Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 2017, 68, 1333–1347. [Google Scholar] [CrossRef]
- Zhu, Z.; Chen, G.; Guo, X.; Yin, W.; Yu, X.; Hu, J.; Hu, Z. Overexpression of SlPRE2, an atypical bHLH transcription factor, affects plant morphology and fruit pigment accumulation in tomato. Sci. Rep. 2017, 7, 5786. [Google Scholar] [CrossRef]
- Morgenstern, B.; Atchley, W.R. Evolution of bHLH transcription factors: Modular evolution by domain shuffling? Mol. Biol. Evol. 1999, 16, 1654–1663. [Google Scholar] [CrossRef]
- Li, X.; Duan, X.; Jiang, H.; Sun, Y.; Tang, Y.; Yuan, Z.; Guo, J.; Liang, W.; Chen, L.; Yin, J.; et al. Genome-Wide Analysis of Basic/Helix-Loop-Helix Transcription Factor Family in Rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Robertson, D.L.; Oliver, S.G.; Bornberg-Bauer, E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep. 2004, 5, 274–279. [Google Scholar] [CrossRef]
- Simionato, E.; Ledent, V.; Richards, G.; Thomas-Chollier, M.; Kerner, P.; Coornaert, D.; Degnan, B.M.; Vervoort, M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: Insights from comparative genomics. BMC Evol. Boil. 2007, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Paulet, L.; Galstyan, A.; Roig-Villanova, I.; Martínez-García, J.F.; Bilbao-Castro, J.R.; Robertson, D.L. Genome-Wide Classification and Evolutionary Analysis of the bHLH Family of Transcription Factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol. 2010, 153, 1398–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pires, N.; Dolan, L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mo. Biol. Evol. 2010, 27, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Schild, C.; Wirth, M.; Reichert, M.; Schmid, R.M.; Saur, D.; Schneider, G. PI3K signaling maintains c-myc expression to regulate transcription of E2F1 in pancreatic cancer cells. Mol. Carcinog. 2009, 48, 1149–1158. [Google Scholar] [CrossRef]
- Palomero, T.; Lim, W.K.; Odom, D.T.; Sulis, M.L.; Real, P.J.; Margolin, A.; Barnes, K.C.; O’Neil, J.; Neuberg, D.; Weng, A.P.; et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA 2006, 103, 18261–18266. [Google Scholar] [CrossRef]
- He, T.-C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; Da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a Target of the APC Pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef]
- Gysin, S.; Salt, M.; Young, A.; McCormick, F. Therapeutic Strategies for Targeting Ras Proteins. Genes Cancer 2011, 2, 359–372. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.M.; Harris, A.W.; Pinkert, C.A.; Corcoran, L.M.; Alexander, W.S.; Cory, S.; Palmiter, R.D.; Brinster, R.L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985, 318, 533–538. [Google Scholar] [CrossRef]
- Belandia, B.; Powell, S.M.; García-Pedrero, J.M.; Walker, M.M.; Bevan, C.L.; Parker, M.G. Hey1, a Mediator of Notch Signaling, Is an Androgen Receptor Corepressor. Mol. Cell. Boil. 2005, 25, 1425–1436. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Sato, F.; Yamada, T.; Bhawal, U.K.; Kawamoto, T.; Fujimoto, K.; Noshiro, M.; Seino, H.; Morohashi, S.; Hakamada, K.; et al. The BHLH transcription factor DEC1 plays an important role in the epithelial-mesenchymal transition of pancreatic cancer. Int. J. Oncol. 2012, 41, 1337–1346. [Google Scholar] [CrossRef] [Green Version]
- Patel, D.; Chaudhary, J. Increased expression of bHLH Transcription Factor E2A (TCF3) in prostate cancer promotes proliferation and confers resistance to doxorubicin induced apoptosis. Biochem. Biophys. Res. Commun. 2012, 422, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Holmquist-Mengelbier, L.; Fredlund, E.; Lofstedt, T.; Noguera, R.; Navarro, S.; Nilsson, H.; Pietras, A.; Vallon-Christersson, J.; Borg, A.; Gradin, K.; et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell 2006, 10, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Villarino, N.; Signaevskaia, L.; Van Niekerk, J.; Medal, R.; Kim, H.; Lahmy, R.; Scully, K.; Pinkerton, A.; Kim, S.; Lowy, A.; et al. A screen for inducers of bHLH activity identifies pitavastatin as a regulator of p21, Rb phosphorylation and E2F target gene expression in pancreatic cancer. Oncotarget 2017, 8, 53154–53167. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Ravasi, T.; Suzuki, H.; Cannistraci, C.V.; Katayama, S.; Bajic, V.B.; Tan, K.; Akalin, A.; Schmeier, S.; Kanamori-Katayama, M.; Bertin, N.; et al. An Atlas of Combinatorial Transcriptional Regulation in Mouse and Man. Cell 2010, 140, 744–752. [Google Scholar] [CrossRef] [Green Version]
- Chou, S.-J.; Tole, S. Lhx2, an evolutionarily conserved, multifunctional regulator of forebrain development. Brain Res. 2019, 1705, 1–14. [Google Scholar] [CrossRef]
- Matthews, J.M.; Lester, K.; Joseph, S.; Curtis, D.J. LIM-domain-only proteins in cancer. Nat. Rev. Cancer 2013, 13, 111–122. [Google Scholar] [CrossRef]
- Safe, S.; Abbruzzese, J.; Abdelrahim, M.; Hedrick, E. Specificity Protein Transcription Factors and Cancer: Opportunities for Drug Development. Cancer Prev. Res. 2018, 11, 371–382. [Google Scholar] [CrossRef]
- Gilding, L.N.; Somervaille, T.C.P. The Diverse Consequences of FOXC1 Deregulation in Cancer. Cancers 2019, 11, 184. [Google Scholar] [CrossRef]
- Gartel, A.L. FOXM1 in Cancer: Interactions and Vulnerabilities. Cancer Res. 2017, 77, 3135–3139. [Google Scholar] [CrossRef]
- Golson, M.L.; Kaestner, K.H. Fox transcription factors: From development to disease. Development 2016, 143, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Hannenhalli, S.; Kaestner, K.H. The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 2009, 10, 233–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moens, C.B.; Selleri, L. Hox cofactors in vertebrate development. Dev. Boil. 2006, 291, 193–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatlekar, S.; Addya, S.; Salunek, M.; Orr, C.R.; Surrey, S.; McKenzie, S.; Fields, J.Z.; Boman, B.M. Identification of a developmental gene expression signature, including HOX genes, for the normal human colonic crypt stem cell niche: Overexpression of the signature parallels stem cell overpopulation during colon tumorigenesis. Stem Cells Dev. 2014, 23, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Feng, F.Y.; Brenner, J.C.; Hussain, M.; Chinnaiyan, A.M. Molecular pathways: Targeting ETS gene fusions in cancer. Clin. Cancer Res. 2014, 20, 4442–4448. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, G.; Chung, T.; Bejerano, G. Changes in the enhancer landscape during early placental development uncover a trophoblast invasion gene-enhancer network. Placenta 2016, 37, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Sizemore, G.M.; Pitarresi, J.R.; Balakrishnan, S.; Ostrowski, M.C. The ETS family of oncogenic transcription factors in solid tumours. Nat. Rev. Cancer 2017, 17, 337–351. [Google Scholar] [CrossRef]
- Ahmad, N.; Kumar, R. Steroid hormone receptors in cancer development: A target for cancer therapeutics. Cancer Lett. 2011, 300, 1–9. [Google Scholar] [CrossRef]
- Mendell, A.L.; MacLusky, N.J. Neurosteroid Metabolites of Gonadal Steroid Hormones in Neuroprotection: Implications for Sex Differences in Neurodegenerative Disease. Front. Mol. Neurosci. 2018, 11, 359. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.-U.; Maeda, T. POK/ZBTB proteins: An emerging family of proteins that regulate lymphoid development and function. Immunol. Rev. 2012, 247, 107–119. [Google Scholar] [CrossRef]
- Romano, K.A.; Campo, A.M.-D.; Kasahara, K.; Chittim, C.L.; Vivas, E.I.; Amador-Noguez, D.; Balskus, E.P.; Rey, F.E. Metabolic, Epigenetic, and Transgenerational Effects of Gut Bacterial Choline Consumption. Cell Host Microbe 2017, 22, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Diamantopoulos, P.T.; Kotsianidis, I.; Symeonidis, A.; Pappa, V.; Galanopoulos, A.; Gogos, D.; Karakatsanis, S.; Papadaki, H.; Palla, A.; Hatzimichael, E.; et al. Chronic myelomonocytic leukemia treated with 5-azacytidine—Results from the Hellenic 5-Azacytidine Registry: Proposal of a new risk stratification system. Leuk. Lymphoma 2018, 60, 1721–1730. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Higgins, P.J. Drugging the undruggable: Transcription therapy for cancer. Biochim. Biophys. Acta 2013, 1835, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jin, G.; Zhou, X. Modeling the relationship of epigenetic modifications to transcription factor binding. Nucleic Acids Res. 2015, 43, 3873–3885. [Google Scholar] [CrossRef] [Green Version]
- Freed-Pastor, W.A.; Prives, C. Mutant p53: One name, many proteins. Genes Dev. 2012, 26, 1268–1286. [Google Scholar] [CrossRef]
- Baliou, S.; Adamaki, M.; Kyriakopoulos, A.M.; Spandidos, D.A.; Panayiotidis, M.; Christodoulou, I.; Zoumpourlis, V. CRISPR therapeutic tools for complex genetic disorders and cancer (Review). Int. J. Oncol. 2018, 53, 443–468. [Google Scholar] [CrossRef]
- Ghosh, D.; Venkataramani, P.; Nandi, S.; Bhattacharjee, S. CRISPR-Cas9 a boon or bane: The bumpy road ahead to cancer therapeutics. Cancer Cell Int. 2019, 19, 12. [Google Scholar] [CrossRef]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.-W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef]
- Friedland, A.E.; Tzur, Y.B.; Esvelt, K.M.; Colaiácovo, M.P.; Church, G.M.; Calarco, J.A. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 2013, 10, 741–743. [Google Scholar] [CrossRef] [Green Version]
- Shachaf, C.M.; Kopelman, A.M.; Arvanitis, C.; Karlsson, Å.; Beer, S.; Mandl, S.; Bachmann, M.H.; Borowsky, A.D.; Ruebner, B.; Cardiff, R.D.; et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 2004, 431, 1112–1117. [Google Scholar] [CrossRef]
- Polstein, L.R.; Gersbach, C.A. A light-inducible CRISPR/Cas9 system for control of endogenous gene activation. Nat. Methods 2015, 11, 198–200. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Guernet, A.; Mungamuri, S.K.; Cartier, D.; Sachidanandam, R.; Jayaprakash, A.; Adriouch, S.; Vezain, M.; Charbonnier, F.; Rohkin, G.; Coutant, S.; et al. CRISPR-Barcoding for Intratumor Genetic Heterogeneity Modeling and Functional Analysis of Oncogenic Driver Mutations. Mol. Cell 2016, 63, 526–538. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharjee, S.; Nandi, S. Choices have consequences: The nexus between DNA repair pathways and genomic instability in cancer. Clin. Transl. Med. 2016, 5, 45. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Nandi, S. DNA damage response and cancer therapeutics through the lens of the Fanconi Anemia DNA repair pathway. Cell Commun. Signal. 2017, 15, 41. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Nandi, S. Rare Genetic Diseases with Defects in DNA Repair: Opportunities and Challenges in Orphan Drug Development for Targeted Cancer Therapy. Cancers 2018, 10, 298. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Nandi, S. Synthetic lethality in DNA repair network: A novel avenue in targeted cancer therapy and combination therapeutics. IUBMB Life 2017, 69, 929–937. [Google Scholar] [CrossRef] [Green Version]
- Philipsen, S. A new twist to the GATA switch. Blood 2013, 122, 3391–3392. [Google Scholar] [CrossRef] [Green Version]
- Smith, A.N.; Miller, L.-A.; Radice, G.; Ashery-Padan, R.; Lang, R.A. Stage-dependent modes of Pax6-Sox2 epistasis regulate lens development and eye morphogenesis. Development 2009, 136, 3377. [Google Scholar] [CrossRef]
- Ali, F.; Hindley, C.; McDowell, G.; Deibler, R.; Jones, A.; Kirschner, M.; Guillemot, F.; Philpott, A. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 2011, 138, 4267–4277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.-C.; Kim, S.M.; Choi, J.E.; Kim, C.H.; Duong, H.-Q.; Han, S.I.; Kang, H.S. Sodium salicylate switches glucose depletion-induced necrosis to autophagy and inhibits high mobility group box protein 1 release in A549 lung adenocarcinoma cells. Oncol. Rep. 2008, 19, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Ohmori, H.; Fujii, K.; Moriwaka, Y.; Sasahira, T.; Kurihara, M.; Tatsumoto, N.; Sasaki, T.; Yamashita, Y.; Kuniyasu, H. HMGB1 attenuates anti-metastatic defence of the liver in colorectal cancer. Eur. J. Cancer 2010, 46, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Jothi, M.; Mal, M.; Keller, C.; Mal, A.K. Small molecule inhibition of PAX3-FOXO1 through AKT activation suppresses malignant phenotypes of alveolar rhabdomyosarcoma. Mol. Cancer Ther. 2013, 12, 2663–2674. [Google Scholar] [CrossRef]
- Wang, H.; Hammoudeh, D.I.; Follis, A.V.; Reese, B.E.; Lazo, J.S.; Metallo, S.J.; Prochownik, E.V. Improved low molecular weight Myc-Max inhibitors. Mol. Cancer Ther. 2007, 6, 2399–2408. [Google Scholar] [CrossRef]
- Carabet, L.A.; Rennie, P.S.; Cherkasov, A. Therapeutic Inhibition of Myc in Cancer. Structural Bases and Computer-Aided Drug Discovery Approaches. Int. J. Mol. Sci. 2018, 20, 120. [Google Scholar] [CrossRef]
- Wang, X.-N.; Su, X.-X.; Cheng, S.-Q.; Sun, Z.-Y.; Huang, Z.-S.; Ou, T.-M. MYC modulators in cancer: A patent review. Expert Opin. Ther. Patents 2019, 29, 353–367. [Google Scholar] [CrossRef]
- Peto, R.; Roe, F.J.; Lee, P.N.; Levy, L.; Clack, J. Cancer and ageing in mice and men. Br. J. Cancer 1975, 32, 411–426. [Google Scholar] [CrossRef]
- Nunney, L. Lineage selection and the evolution of multistage carcinogenesis. Proc. R. Soc. B Boil. Sci. 1999, 266, 493–498. [Google Scholar] [CrossRef] [Green Version]
- Caulin, A.F.; Maley, C.C. Peto’s Paradox: evolution’s prescription for cancer prevention. Trends Ecol. Evol. 2011, 26, 175–182. [Google Scholar] [CrossRef]
- Gaughran, S.J.; Evlyn, P.; Stearns, S.C. How elephants beat cancer. eLife 2016. [Google Scholar] [CrossRef] [PubMed]
- Abegglen, L.M.; Caulin, A.F.; Chan, A.; Lee, K.; Robinson, R.; Campbell, M.S.; Kiso, W.K.; Schmitt, D.L.; Waddell, P.J.; Bhaskara, S.; et al. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 2015, 314, 1850–1860. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, J.M.; Sulak, M.; Chigurupati, S.; Lynch, V.J. A Zombie LIF Gene in Elephants Is Upregulated by TP53 to Induce Apoptosis in Response to DNA Damage. Cell Rep. 2018, 24, 1765–1776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haupt, S.; Haupt, Y. P53 at the start of the 21st century: Lessons from elephants. F1000Research 2017, 6, 2041. [Google Scholar] [CrossRef] [PubMed]
- Nagy, J.D.; Victor, E.M.; Cropper, J.H. Why don’t all whales have cancer? A novel hypothesis resolving Peto’s paradox. Integr. Comp. Boil. 2007, 47, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Martineau, D.; Lemberger, K.; Dallaire, A.; Labelle, P.; Lipscomb, T.P.; Michel, P.; Mikaelian, I. Cancer in wildlife, a case study: Beluga from the St. Lawrence estuary, Quebec, Canada. Environ. Health Perspect. 2002, 110, 285–292. [Google Scholar] [CrossRef]
- George, J.C.; Bada, J.; Zeh, J.; Scott, L.; Brown, S.E.; O’Hara, T.; Suydam, R. Age and growth estimates of bowhead whales (Balaena mysticetus) via aspartic acid racemization. Can. J. Zool. 1999, 77, 571–580. [Google Scholar] [CrossRef]
- Keane, M.; Semeiks, J.; Webb, A.E.; Li, Y.I.; Quesada, V.; Craig, T.; Madsen, L.B.; Van Dam, S.; Brawand, D.; Marques, P.I.; et al. Insights into the Evolution of Longevity from the Bowhead Whale Genome. Cell Rep. 2015, 10, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Gladyshev, V.N. Molecular signatures of longevity: Insights from cross-species comparative studies. Semin. Cell Dev. Boil. 2017, 70, 190–203. [Google Scholar] [CrossRef]
- Smith, E.F.; Townsend, C.O. A Plant-Tumor of Bacterial Origin. Science 1907, 25, 671–673. [Google Scholar] [CrossRef] [Green Version]
- Sharp, W.R.; Gunckel, J.E. Physiological Comparisons of Pith Callus with Crown-Gall and Genetic Tumors of Nicotiana glauca, N. langsdorffii, and N. glauca-langsdorffii Grown in Vitro. I. Tumor Induction and Proliferation. Plant Physiol. 1969, 44, 1069–1072. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H.; Kasahara, H.; Ueda, N.; Kojima, M.; Takei, K.; Hishiyama, S.; Asami, T.; Okada, K.; Kamiya, Y.; Yamaya, T.; et al. Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc. Natl. Acad. Sci. USA 2005, 102, 9972–9977. [Google Scholar] [CrossRef] [PubMed]
- Gutzat, R.; Borghi, L.; Gruissem, W. Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends Plant Sci. 2012, 17, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.-J.; Hanley-Bowdoin, L. A Geminivirus Replication Protein Interacts with a Protein Kinase and a Motor Protein That Display Different Expression Patterns during Plant Development and Infection. Plant Cell 2002, 14, 1817–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
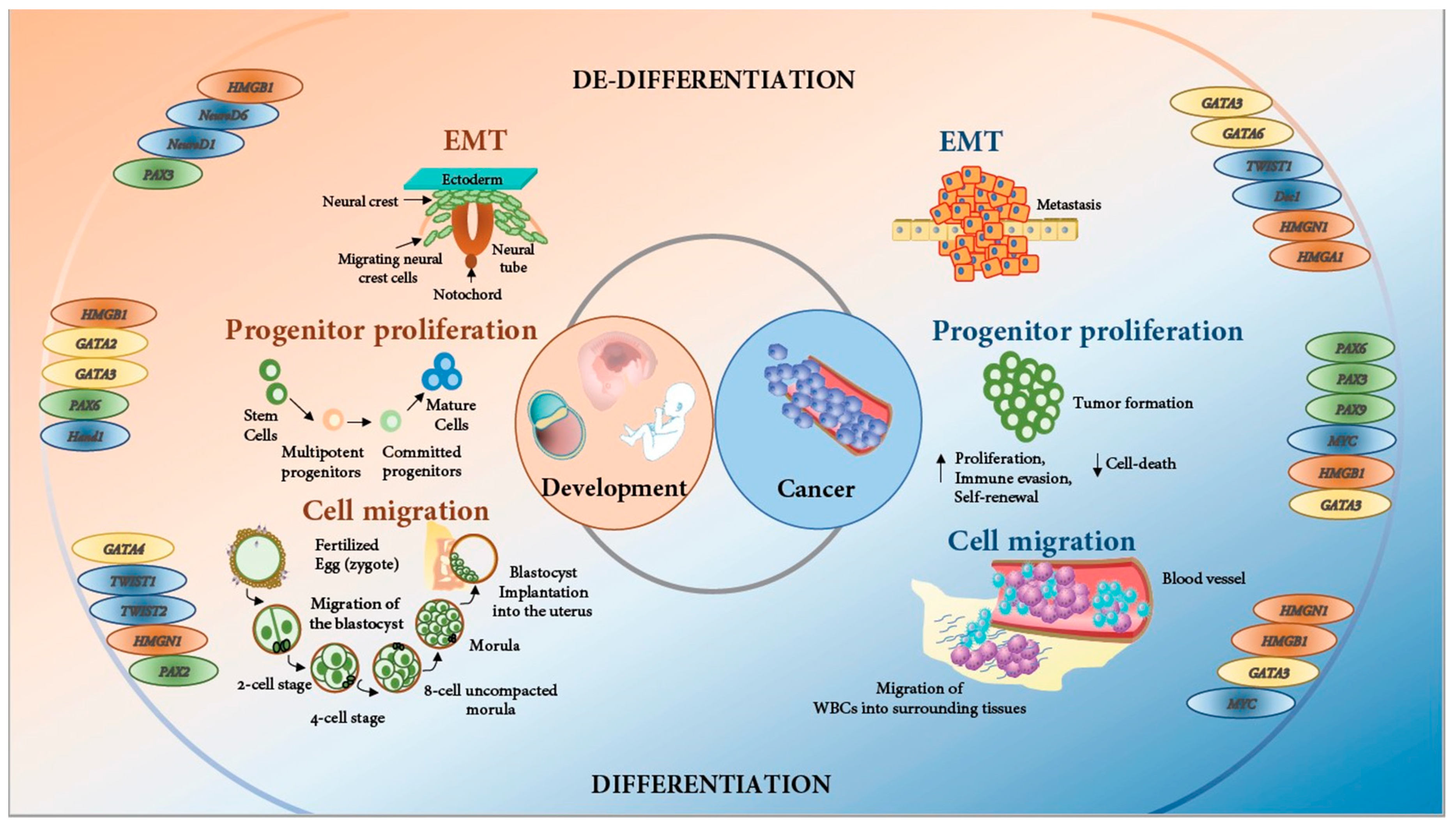
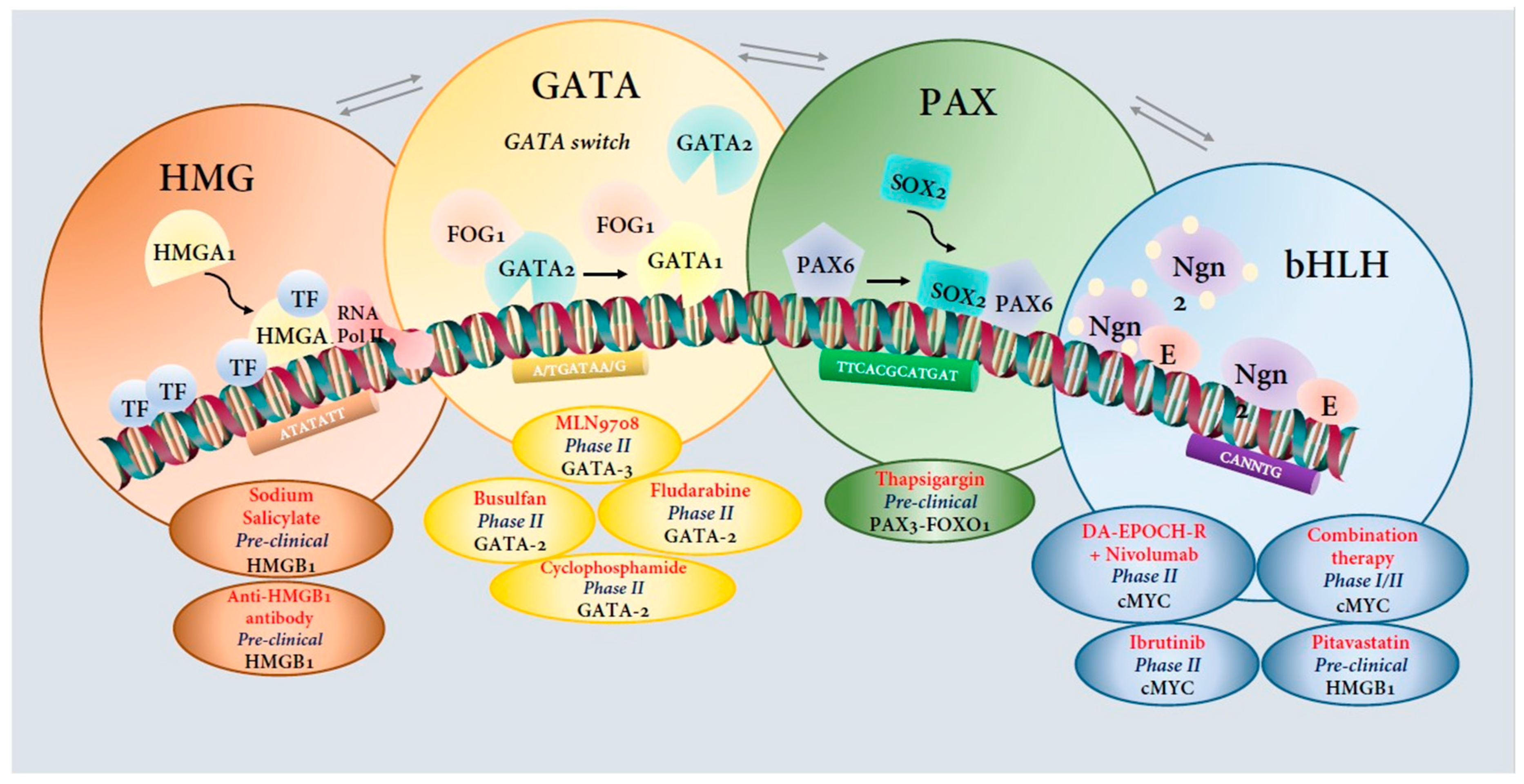
| Transcription Factor Family | Subtype | Role in Development | Role in Cancer |
|---|---|---|---|
| High Mobility Group Proteins (HMG) | HMGN1 | Corneal epithelium development and maintenance [87,88,89] | Regulates transcription of proto-oncogenes and pro-metastatic genes like c-fos, BCL3, N-cadherin, JunB and c-Jun [103] |
| HMGA1 | Regulator of adipogenesis [104], stem cell state [105] and lymphohematopoietic differentiation; crucial for normal sperm production in mouse | Overexpressed in colon, breast and invasive ovarian carcinomas, pancreatic and non-small cell lung adenocarcinomas [106] | |
| HMGA2 | Neural crest cells specification in Xenopus (essential for animal growth) [93]; governs the exit of embryonic stem cells from pluripotent ground state; cell proliferation and distal epithelium differentiation during embryonic lung development | Overexpressed in pancreatic and non-small cell lung adenocarcinomas [106] | |
| HMGB1 | Neural stem cell proliferation, differentiation, and maintenance [96] | Overexpressed in pancreatic (PDAC), gastric, colon, hepatocellular, and non-small cell lung adenocarcinomas [107] | |
| GATA | GATA1 | Development of erythrocytes, megakaryocytes, mast cells, and eosinophils [108,109] | Mutations in GATA1 in Down syndrome patients associated with DS-AMKL [110] |
| GATA2 | Hematopoiesis [111] | Mediates Kras-driven tumorigenesis in NSCLC; GATA2 mutated in a subset of human CML [112,113,114] | |
| GATA3 | T-cell lymphopoiesis, self-renewal, and differentiation of long-term HSCs [115] | Tumor suppressor and strong prognostic marker in breast cancer [116] | |
| GATA4 | Cardiac angiogenesis and bile homoeostasis [117,118] | Downregulated in gastric, lung, ovarian, colorectal, esophageal, glioblastoma, and large B-cell lymphoma [114,119,120] | |
| GATA5 | Cardiac development [121,122] | Downregulated in gastric, lung, ovarian, colorectal, esophageal, glioblastoma, and large B-cell lymphoma [119,123] | |
| GATA6 | Hepatic and cardiac development [124,125] | Tumor suppressor in astrocytoma; overexpressed in colon and pancreatic cancer [126,127,128] | |
| PAX | PAX1 | Maturation of thymocytes [129] | Hypermethylated in cervical cancer [130] |
| PAX2 | Prevention of tubular cells from apoptosis post-injury [131,132] | Overexpressed in ovarian, renal cell, and bladder carcinomas. Regulates ERBB2 expression in breast cancer [133,134] | |
| PAX3 | Early neurogenesis; regulation of sensory neuron generation from precursor cells. Maintenance of undifferentiated state of muscle stem cells [135,136] | PAX3-FKHR fusion protein acts as an oncogene in alveolar rhabdomyosarcomas. Overexpressed in primary melanomas [137,138,139] | |
| PAX4 | Protection of pancreaticβ-cells from apoptosis [140] | Upregulated in human insulinomas [141] | |
| PAX5 | B lymphopoiesis [142] | Tumor suppressor in hepatocellular carcinomas; overexpressed in B-cell neoplasms; good prognostic marker in breast cancer [143,144,145] | |
| PAX6 | Eye organogenesis and neural stem cell self-renewal, neuroectoderm cell fate determination [146] | Oncogenic role in pancreatic adenocarcinoma and glioblastoma [147,148,149] | |
| PAX7 | Proliferation and maintenance of postnatal and muscle satellite cells [150] | PAX7-FKHR fusion protein acts as an oncogene in alveolar rhabdomyosarcomas [137] | |
| PAX8 | Thyroglobulin regulation; maintenance of thyroid progenitor cells [151,152] | Oncogenic role in renal, ovarian, lung, and thyroid cancers and certain glioblastoma subtypes [153] | |
| PAX9 | Development of permanent teeth [154] | Oncogenic role in lung cancer and oral squamous cell carcinomas [155,156] | |
| bHLH | TWIST 1 | Osteogenesis and craniofacial development [157,158] | Induces EMT; activated during tumor progression [159,160,161] |
| TWIST 2 | Osteogenesis and bone proliferation [157] | Induces EMT; activated during tumor progression [159] | |
| MYC | Skeletal development, osteogenesis, stem and progenitor cell maintenance and self-renewal, organogenesis [162] | Oncogenic role in various cancer signaling pathways; tumor maintenance; copy number variations observed in pancreatic ductal adenocarcinoma [163,164] | |
| ATOH1 | Differentiation of granule cells of the cerebellum and inner ear hair cells [165] | Tumor suppressor; silenced in most colorectal cancers; induces differentiation of gastric cancer stem cells; drives metastasis of medulloblastoma; lineage-dependency oncogene in Merkel cell carcinoma. | |
| NEUROD1 | Differentiation of inner ear sensory neurons, cerebellum, and the hippocampus [166] | Survival and migration of neuroendocrine lung carcinomas; cell motility and tumor formation of neuroblastoma; in cooperation with Otx2, controls Group 3 medulloblastoma active enhancer landscape [167] | |
| NEUROD2 | Formation of corpus callosum, essential for communication between the two cerebral hemispheres [168] | Tumor suppressor and prognostic biomarker in Glioblastoma; copy number gains of NEUROD2 in male breast cancer (prognostic value) [169] | |
| HAND1 | Proliferation, differentiation, and morphogenesis of embryonic ventricle cardiomyocytes [170,171] | Downregulated in medulloblastoma; facilitates proliferation and metastasis in gastrointestinal stromal tumor; silenced in over 90% of human primary colorectal tumors. Methylation of HAND1 associated with poor survival in gastric cancer; involved in thyroid carcinogenesis [172] | |
| HAND2 | Proliferation, differentiation, and morphogenesis of embryonic ventricle cardiomyocytes [170,171] | Tumor suppressor in endometroid endometrial carcinoma. HAND2 suppression upregulates Fgfs in endometriosis [173]. | |
| OLIG1 | Oligodendrocyte differentiation in the neocortex [174] | Aberrant DNA methylation in non-small cell lung cancer [175] | |
| OLIG2 | Oligodendrocyte differentiation in the spinal cord [174] | Universally expressed in gliomas [176] | |
| DEC1 | Embryonic endochondral bone development [177]; upregulated in growth plate cartilage and chondrocytes; cartilage terminal differentiation; blocks myogenesis in bovine cells [178] | Critical in cell cycle regulation and cell death in breast and oral cancer; DEC1 induces EMT in pancreatic cancer [179] | |
| DEC2 | Proliferation and differentiation of chondrocytes; neuronal differentiation; adipogenesis. Negative regulator of proliferation and differentiation of chondrocyte-lineage committed mesenchymal stem cells [180] | Critical in cell cycle regulation and cell death in breast and oral cancer [181] | |
| HES1 | Cell fate determination and epidermal development [182]; epidermal development [183]; heterogenous ES cell differentiation [184]; proneural gene expression and neuronal differentiation [185]; brain morphogenesis [186]; development of the arterial pole of the heart; thyroid gland development [187]. | Deregulated in several cancers and positively regulate levels of the tumor suppressor gene p53 [188] | |
| HEY1 & HEY2 | Embryonic vascular development [189]; maintenance of neural precursor cells; spatial-temporal pattern of mammalian auditory hair cell differentiation [190]. HEY1 is involved in odontogenic/osteogenic differentiation and cardiac development [191]. | Deregulated in several cancers and positively regulates levels of the tumor suppressor gene p53 [188] |
| Molecular Target | Candidate Drug | Condition or Disease | Stage of Testing | Other Targets & Disease Conditions | Direct or Nonselective Inhibition | Reference/ClinicalTrial.gov Identifier |
|---|---|---|---|---|---|---|
| HMGB1 | Sodium salicylate | Lung adenocarcinoma | Preclinical | Targets—Mitogen-activated protein kinases (MAPK), Caspase 3, NF-κb, p38 kinase, AP-1 Disease—Acute Myeloid Leukemia | Nonselective | [352] |
| HMGB1 | Anti-HMGB1 antibody | Colorectal cancer | Preclinical | Disease—Stroke, Epilepsy, Neudegenerative diseases, neuropathic pain | Direct | [353] |
| GATA-3 | MLN9708 | Lymphoma | Phase II | Targets—p38 kinase, Janus Kinase (JNK), NF-κb Disease—Breast Cancer | Nonselective | NCT02158975 |
| GATA-2 | Busulfan, Fludarabine, Busulfan and Cyclophosphamide | Myelodysplastic Syndromes | Phase II | Disease—Chronic Myelogenous Leukemia, Lymphomas | Chemo-therapy | NCT01861106 |
| Pax3-Foxo1 | Thapsigargin | Alveolar Rhabdomyosarcoma | Preclinical | Targets—Sarco/endoplasmic reticulum Ca2+ ATPase (SERCA), Nicotinic acetylcholine receptors | Nonselective | [354] |
| bHLH | Pitavastatin | Pancreatic cancer | Preclinical | Targets—3-hydroxy-3-methyl glutaryl coenzyme A reductase Disease—Hypercholestrolemia and dyslipidemia | Nonselective | [313] |
| Reverse the association between Myc and its obligate bHLH heterodimerization partner, Max | 10058-F4 | Promyelocytic leukemia | Preclinical | Targets—MYCN, Myc/Max dimerization Disease—MYCN-amplified neuroblastoma, Acute Myeloid Leukemia | Direct | [355] |
| Myc | Mycro1, Mycro2 and Mycro3 | Leukemia | Preclinical | Targets—Myc/Max dimerization | Direct | [314,356,357] |
| MYC | Lenalidomide and Combination chemotherapy | B-cell lymphoma | Phase I/II | Targets—CRL4 E3 Ubiquitin ligase Disease—Multiple Myeloma | Nonselective | NCT02213913 |
| MYC | Ibrutinib | Gastrooesophageal Cancer | Phase II | Targets—Bruton’s tyrosine kinase (BTK), CD20 Disease—B-cell cancers such as mantle cell lymphoma, chronic lymphocytic leukemia and Waldenstrom’s macroglobulinemia | Nonselective | NCT02884453 |
| MYC | DA-EPOCH-R followed by Nivolumab | B-cell lymphoma | Phase II | Targets (Nivolumab)—PD-L1 Disease—Squamous non-small cell lung cancer, renal-cell carcinoma, small cell lung cancer | Nonselective | NCT03620578 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huilgol, D.; Venkataramani, P.; Nandi, S.; Bhattacharjee, S. Transcription Factors That Govern Development and Disease: An Achilles Heel in Cancer. Genes 2019, 10, 794. https://doi.org/10.3390/genes10100794
Huilgol D, Venkataramani P, Nandi S, Bhattacharjee S. Transcription Factors That Govern Development and Disease: An Achilles Heel in Cancer. Genes. 2019; 10(10):794. https://doi.org/10.3390/genes10100794
Chicago/Turabian StyleHuilgol, Dhananjay, Prabhadevi Venkataramani, Saikat Nandi, and Sonali Bhattacharjee. 2019. "Transcription Factors That Govern Development and Disease: An Achilles Heel in Cancer" Genes 10, no. 10: 794. https://doi.org/10.3390/genes10100794
APA StyleHuilgol, D., Venkataramani, P., Nandi, S., & Bhattacharjee, S. (2019). Transcription Factors That Govern Development and Disease: An Achilles Heel in Cancer. Genes, 10(10), 794. https://doi.org/10.3390/genes10100794





