An SNP-Based Genetic Map and QTL Mapping for Growth Traits in the Red-Spotted Grouper (Epinephelus akaara)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mapping Family and DNA Isolation
2.2. Restriction Site-Associated DNA Sequencing
2.3. Sequence Data Analysis and Genotyping
2.4. Genetic Map Construction
2.5. QTL Mapping Analysis and Gene Annotation
2.6. Network Analysis Between the Candidate Genes
3. Results
3.1. Genotyping by Sequencing
3.2. Construction of the Genetic Map
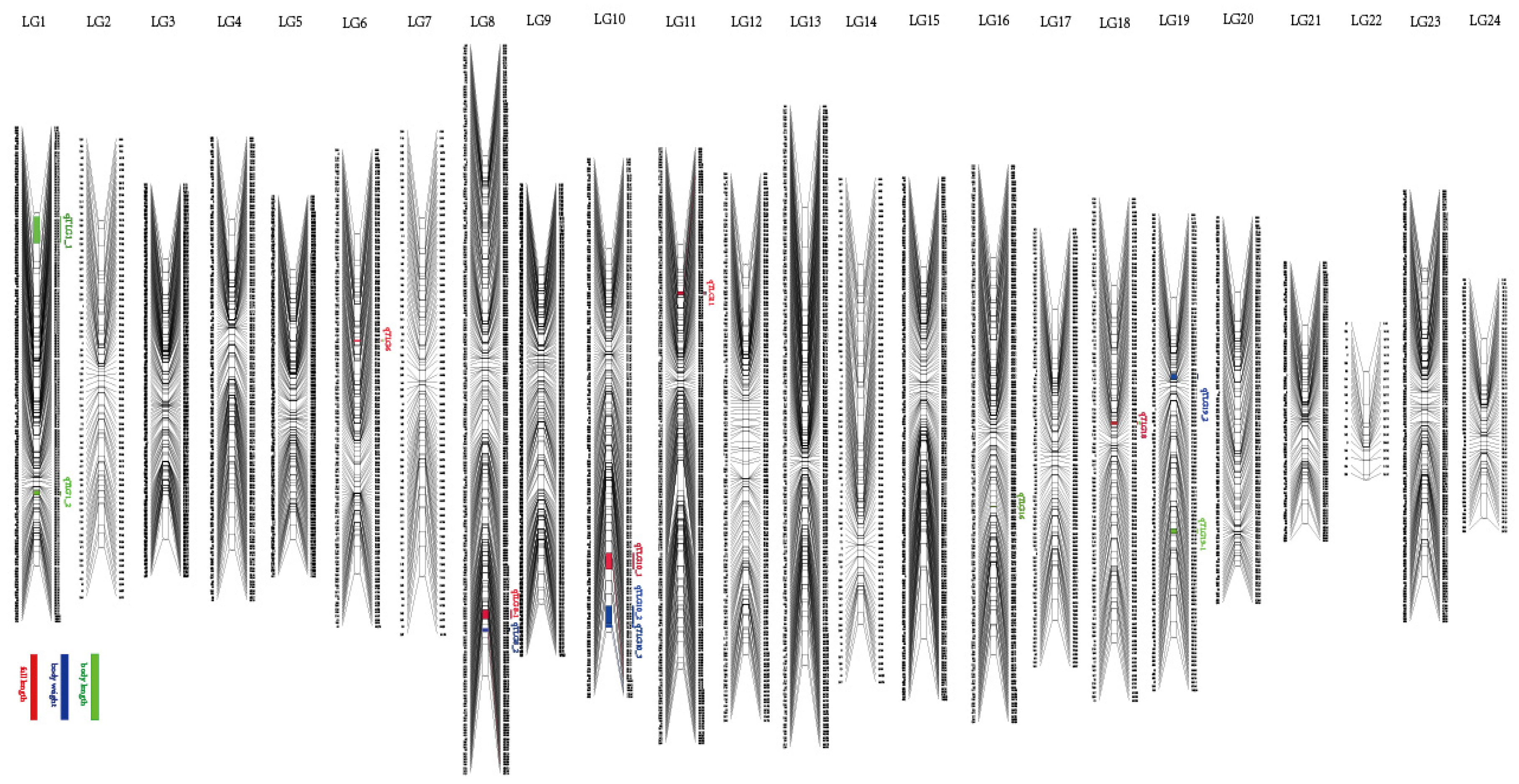
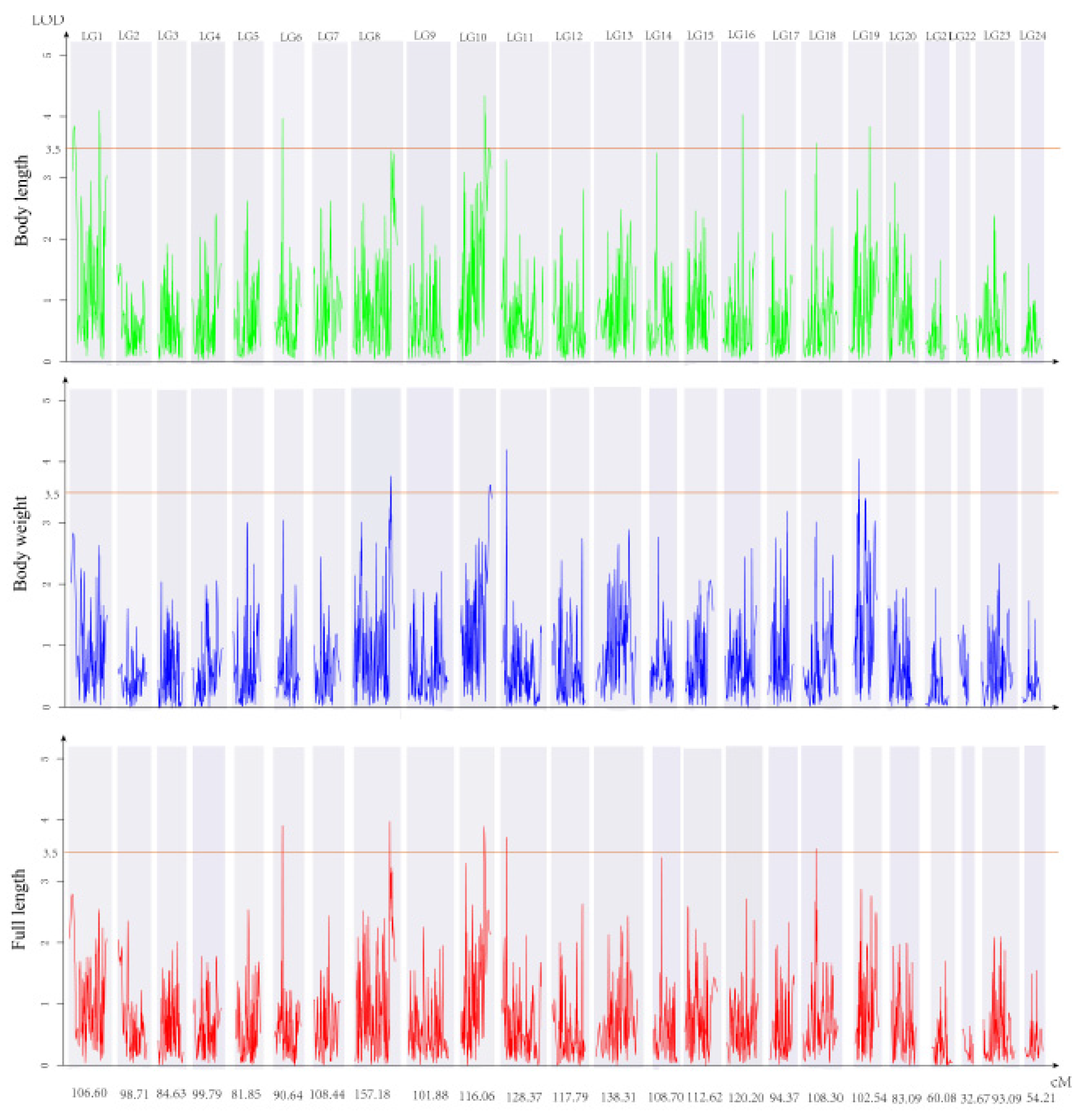
3.3. QTL Identification and Related Genes
3.4. Network Analysis Between the Candidate Genes
4. Discussion
4.1. Molecular Marker Development and Utilization of the Genetic Map and QTL Analysis
4.2. High-Density SNP Genetic Map Construction in the Red-Spotted Grouper Based on RAD-Seq
4.3. QTL Analysis Based on the High-Density SNP Genetic Map
4.4. Protein Network Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Kayano, Y.; Yao, S.; Yamamoto, S.; Nakagawa, H. Effects of feeding frequency on the growth and body constituents of young red-spotted grouper, Epinephelus akaara. Aquaculture 1993, 110, 271–278. [Google Scholar] [CrossRef]
- Liao, I.C.; Su, H.M.; Chang, E.Y. Techniques in finfish larviculture in Taiwan. Aquaculture 2001, 200, 1–31. [Google Scholar] [CrossRef]
- Andrews, K.R.; Good, J.M.; Miller, M.R.; Luikart, G.; Hohenlohe, P.A. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet. 2016, 17, 81. [Google Scholar] [CrossRef]
- Robledo, D.; Palaiokostas, C.; Bargelloni, L.; Martínez, P.; Houston, R. Applications of genotyping by sequencing in aquaculture breeding and genetics. Rev. Aquac. 2018, 10, 670–682. [Google Scholar] [CrossRef]
- Lien, S.; Gidskehaug, L.; Moen, T.; Hayes, B.J.; Berg, P.R.; Davidson, W.S.; Omholt, S.W.; Kent, M.P. A dense SNP-based linkage map for Atlantic salmon (Salmo salar) reveals extended chromosome homeologies and striking differences in sex-specific recombination patterns. BMC Genom. 2011, 12, 615. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Pang, R.; Niu, Y.; Gao, F.; Zhao, Y.; Zhang, J.; Sun, J.; Shao, C.; Liao, X.; Wang, L. Construction of high-density genetic linkage maps and mapping of growth-related quantitative trail loci in the Japanese flounder (Paralichthys olivaceus). PLoS ONE 2012, 7, e50404. [Google Scholar] [CrossRef] [PubMed]
- Palti, Y.; Gao, G.; Liu, S.; Kent, M.; Lien, S.; Miller, M.; Rexroad III, C.; Moen, T. The development and characterization of a 57 K single nucleotide polymorphism array for rainbow trout. Mol. Ecol. Resour. 2015, 15, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Palti, Y.; Vallejo, R.L.; Gao, G.; Liu, S.; Hernandez, A.G.; Rexroad, C.E., III; Wiens, G.D. Detection and validation of QTL affecting bacterial cold water disease resistance in rainbow trout using restriction-site associated DNA sequencing. PLoS ONE 2015, 10, e0138435. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.-F.; Xiong, L.-W.; Han, Z.-K.; Liu, Z.-Q.; Feng, J.-B.; Wu, X.-G.; Yan, Y.-L.; Shen, H.; Huang, L.; Chen, L. A second generation SNP and SSR integrated linkage map and QTL mapping for the Chinese mitten crab Eriocheir sinensis. Sci. Rep. 2017, 7, 39826. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Ming, Y.; Shao, F.; Jian, J.; Zhang, Y.; Peng, Z. Restriction site-associated DNA sequencing for SNP discovery and high-density genetic map construction in southern catfish (Silurus meridionalis). R. Soc. Open Sci. 2018, 5, 172054. [Google Scholar] [CrossRef]
- Song, J.; Li, Q.; Yu, Y.; Wan, S.; Han, L.; Du, S. Mapping genetic loci for quantitative traits of golden shell color, mineral element contents, and growth-related traits in Pacific oyster (Crassostrea gigas). Mar. Biotechnol. 2018, 20, 666–675. [Google Scholar] [CrossRef] [PubMed]
- Ashton, D.T.; Ritchie, P.A.; Wellenreuther, M. High-density linkage map and QTLs for growth in snapper (Chrysophrys auratus). G3 Genes Genomes Genet. 2019, 9, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xie, N.; Shen, Y.; Ye, B.; Yue, G.H.; Feng, X. Constructing High-Density Genetic Maps and Developing Sexing Markers in Northern Snakehead (Channa argus). Mar. Biotechnol. 2019, 21, 348–358. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Shu, L.; Li, S.; Chen, J.; Luo, J.; Lu, J.; Mu, Q.; Bai, J.; Xia, Q.; Chen, Q. Construction of high-density genetic linkage maps for orange-spotted grouper Epinephelus coioides using multiplexed shotgun genotyping. BMC Genet. 2013, 14, 113. [Google Scholar] [CrossRef]
- Yu, H.; You, X.; Li, J.; Liu, H.; Meng, Z.; Xiao, L.; Zhang, H.; Lin, H.-R.; Zhang, Y.; Shi, Q. Genome-wide mapping of growth-related quantitative trait loci in orange-spotted grouper (Epinephelus coioides) using double digest restriction-site associated DNA sequencing (ddRADseq). Int. J. Mol. Sci. 2016, 17, 501. [Google Scholar] [CrossRef]
- Watanabe, M.; Shimizu, T.; Kamarudin, A.S.B.; Kuniyoshi, H.; Ohara, K.; Takagi, M.; Umino, T. Ten novel polymorphic microsatellite loci of Red-spotted grouper (Epinephelus akaara) revealed from full-sib progeny and unrelated individuals. Conserv. Genet. Resour. 2011, 3, 613–616. [Google Scholar] [CrossRef]
- Dor, L.; Shirak, A.; Gorshkov, S.; Band, M.R.; Korol, A.; Ronin, Y.; Curzon, A.; Hulata, G.; Seroussi, E.; Ron, M. Construction of a microsatellites-based linkage map for the white grouper (Epinephelus aeneus). G3 Genes Genomes Genet. 2014, 4, 1455–1464. [Google Scholar] [CrossRef]
- Liu, Q.; Sakamoto, T.; Kubota, S.; Okamoto, N.; Yamashita, H.; Takagi, M.; Shigenobu, Y.; Sugaya, T.; Nakamura, Y.; Sano, M. A genetic linkage map of kelp grouper (Epinephelus bruneus) based on microsatellite markers. Aquaculture 2013, 414, 63–81. [Google Scholar] [CrossRef]
- Ge, H.; Lin, K.; Shen, M.; Wu, S.; Wang, Y.; Zhang, Z.; Wang, Z.; Zhang, Y.; Huang, Z.; Zhou, C. De novo assembly of a chromosome-level reference genome of red-spotted grouper (Epinephelus akaara) using nanopore sequencing and Hi-C. Mol. Ecol. Resour. 2019, 19, 1322–1332. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, 362–368. [Google Scholar] [CrossRef]
- Liang, W.; Sun, F. Competing Endogenous RNA Network Analysis Reveals Pivotal ceRNAs in Adrenocortical Carcinoma. Front. Endocrinol. 2019, 10, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Khan, Z.A.; Labala, R.K.; Mondal, G.; Devi, H.S.; Rajiv, C.; Yumnamcha, T.; Devi, S.D.; Bharali, R.; Thorat, S.S.; Chattoraj, A. The daily expression profile of Neuropeptides (gnih, gnrh3, kiss1 and kiss2): A study of possible interaction in the brain of Zebrafish (Danio rerio). J. Endocrinol. Reprod. 2017, 20, 46–54. [Google Scholar]
- Zhang, B.; Zhuang, Z.; Wang, X.; Huang, H.; Fu, Q.; Yan, Q. Dual RNA-Seq reveals the role of a transcriptional regulator gene in pathogen-host interactions between Pseudomonas plecoglossicida and Epinephelus coioides. Fish. Shellfish Immunol. 2019, 87, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Puente-Marin, S.; Nombela, I.; Chico, V.; Ciordia, S.; Mena, M.C.; Perez, L.G.; Coll, J.; Ortega-Villaizan, M.d.M. Potential Role of Rainbow Trout Erythrocytes as Mediators in the Immune Response Induced by a DNA Vaccine in Fish. Vaccines 2019, 7, 60. [Google Scholar] [CrossRef]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef]
- Catchen, J.M.; Amores, A.; Hohenlohe, P.; Cresko, W.; Postlethwait, J.H. Stacks: Building and genotyping loci de novo from short-read sequences. G3 Genes Genomes Genet. 2011, 1, 171–182. [Google Scholar] [CrossRef]
- Voorrips, R. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Rastas, P.; Paulin, L.; Hanski, I.; Lehtonen, R.; Auvinen, P. Lep-MAP: Fast and accurate linkage map construction for large SNP datasets. Bioinformatics 2013, 29, 3128–3134. [Google Scholar] [CrossRef]
- Luciano Da Costa, E.S.; Wang, S.; Zeng, Z.-B. Composite interval mapping and multiple interval mapping: Procedures and guidelines for using Windows QTL Cartographer. In Quantitative Trait Loci (QTL); Springer: Berlin, Germany, 2012; pp. 75–119. [Google Scholar]
- Wang, W.; Huang, S.; Liu, Y.; Fang, Z.; Yang, L.; Hua, W.; Yuan, S.; Liu, S.; Sun, J.; Zhuang, M. Construction and analysis of a high-density genetic linkage map in cabbage (Brassica oleracea L. var. capitata). BMC Genom. 2012, 13, 523. [Google Scholar] [CrossRef]
- Van Ooijen, J.W. Accuracy of mapping quantitative trait loci in autogamous species. Theor. Appl. Genet. 1992, 84, 803–811. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Zhang, H.; Liufu, Y. Karyotypes of Epinephelus coioides and Epinephelus adaara. J. Zhanjiang Ocean. Univ. 2004, 24, 4–8. [Google Scholar]
- Wang, B.; Jia, J.; Yang, G.; Qin, J.; Zhang, C.; Zhang, Q.; Sun, C.; Li, W. In vitro effects of somatostatin on the growth hormone-insulin-like growth factor axis in orange-spotted grouper (Epinephelus coioides). Gen. Comp. Endocrinol. 2016, 237, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, K.; Dufour, S. Phylogenetic evolution of the neuroendocrine control of growth hormone: Contribution from teleosts. Cybium 2004, 28, 181–198. [Google Scholar]
- Yunker, W.K.; Smith, S.; Graves, C.; Davis, P.J.; Unniappan, S.; Rivier, J.E.; Peter, R.E.; Chang, J.P. Endogenous hypothalamic somatostatins differentially regulate growth hormone secretion from goldfish pituitary somatotropes in vitro. Endocrinology 2003, 144, 4031–4041. [Google Scholar] [CrossRef] [PubMed]
- Murray, R.D.; Kim, K.; Ren, S.-G.; Chelly, M.; Umehara, Y.; Melmed, S. Central and peripheral actions of somatostatin on the growth hormone–IGF-I axis. J. Clin. Investig. 2004, 114, 349–356. [Google Scholar] [CrossRef]
- Li, W.; Chu, M.; Lin, H.; Wong, A. In Interaction of pituitary adenylate cyclase-activating polypeptide with gonadotropin-releasing hormone, dopamine, and somatostatin in regulating growth hormone gene expression in grass carp pituitary cells. In Proceedings of the 4th International Symposium on Fish Endocrinology, Seattle, WA, USA, 31 July–3 August 2000. [Google Scholar]
- Händel, M.; Schulz, S.; Stanarius, A.; Schreff, M.; Erdtmann-Vourliotis, M.; Schmidt, H.; Wolf, G.; Höllt, V. Selective targeting of somatostatin receptor 3 to neuronal cilia. Neuroscience 1999, 89, 909–926. [Google Scholar] [CrossRef]
- Rescan, P.-Y.; Montfort, J.; Rallière, C.; Le Cam, A.; Esquerré, D.; Hugot, K. Dynamic gene expression in fish muscle during recovery growth induced by a fasting-refeeding schedule. BMC Genom. 2007, 8, 438. [Google Scholar] [CrossRef]
- Podgorniak, T.; Brockmann, S.; Konstantinidis, I.; Fernandes, J.M. Differences in the fast muscle methylome provide insight into sex-specific epigenetic regulation of growth in Nile tilapia during early stages of domestication. Epigenetics 2019, 14, 818–836. [Google Scholar] [CrossRef]
- Andersen, S.M.; Waagbø, R.; Espe, M. Functional amino acids in fish health and welfare. Front. Biosci. 2016, 8, 143–169. [Google Scholar]
- Chatchaiphan, S.; Srisapoome, P.; Kim, J.-H.; Devlin, R.H.; Na-Nakorn, U. De novo transcriptome characterization and growth-related gene expression profiling of diploid and triploid bighead catfish (Clarias macrocephalus Günther, 1864). Mar. Biotechnol. 2017, 19, 36–48. [Google Scholar] [CrossRef]
- Kaushik, G.; Xia, Y.; Pfau, J.C.; Thomas, M.A. Dysregulation of autism-associated synaptic proteins by psychoactive pharmaceuticals at environmental concentrations. Neurosci. Lett. 2017, 661, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Sazhenova, E.; Nikitina, T.; Skryabin, N.; Minaycheva, L.; Ivanova, T.; Nemtseva, T.; Yuriev, S.Y.; Evtushenko, I.; Lebedev, I. Epigenetic status of imprinted genes in placenta during recurrent pregnancy loss. Russ. J. Genet. 2017, 53, 376–387. [Google Scholar] [CrossRef]
- Lam, S.W.; van IJzendoorn, D.G.; Cleton-Jansen, A.-M.; Szuhai, K.; Bovée, J.V. Molecular pathology of bone tumors. J. Mol. Diagn. 2018, 18, 30329–30333. [Google Scholar] [CrossRef] [PubMed]
- Migdalska-Richards, A.; Mill, J. Epigenetic studies of schizophrenia: Current status and future directions. Curr. Opin. Behav. Sci. 2019, 25, 102–110. [Google Scholar] [CrossRef]
- Kolshus, E.; Dalton, V.; Ryan, K.; McLoughlin, D. When less is more–microRNAs and psychiatric disorders. Acta Psychiatr. Scand. 2014, 129, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Nadeau, N.J.; Minvielle, F.; Ito, S.; Inoue-Murayama, M.; Gourichon, D.; Follett, S.A.; Burke, T.; Mundy, N.I. Characterization of Japanese quail yellow as a genomic deletion upstream of the avian homolog of the mammalian ASIP (agouti) gene. Genetics 2008, 178, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Zopf, C.; Quinn, K.; Zeidman, J.; Maheshri, N. Cell-cycle dependence of transcription dominates noise in gene expression. PLoS Comput. Biol. 2013, 9, e1003161. [Google Scholar] [CrossRef]
- Yan, J.; Liao, K.; Wang, T.; Mai, K.; Xu, W.; Ai, Q. Dietary lipid levels influence lipid deposition in the liver of large yellow croaker (Larimichthys crocea) by regulating lipoprotein receptors, fatty acid uptake and triacylglycerol synthesis and catabolism at the transcriptional level. PLoS ONE 2015, 10, e0129937. [Google Scholar] [CrossRef]
- Endres, L.; Fasullo, M.; Rose, R. tRNA modification and cancer: Potential for therapeutic prevention and intervention. Future Med. Chem. 2019, 11, 885–900. [Google Scholar] [CrossRef]
- Takahashi, K.; Mitoma, J.; Hosono, M.; Shiozaki, K.; Sato, C.; Yamaguchi, K.; Kitajima, K.; Higashi, H.; Nitta, K.; Shima, H. Sialidase NEU4 hydrolyzes polysialic acids of neural cell adhesion molecules and negatively regulates neurite formation by hippocampal neurons. J. Biol. Chem. 2012, 287, 14816–14826. [Google Scholar] [CrossRef]
- Liu, Z.J.; Cordes, J. DNA marker technologies and their applications in aquaculture genetics. Aquaculture 2004, 238, 1–37. [Google Scholar] [CrossRef]
- Joshi, R.; Arnyasi, M.; Lien, S.; Gjøen, H.M.; Alvarez, A.T.; Kent, M. Development and validation of 58K SNP-array and high-density linkage map in Nile tilapia (O. niloticus). Front. Genet. 2018, 9, 472. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.Y.; Robledo, D.; Lowe, N.R.; Bekaert, M.; Taggart, J.B.; Bron, J.E.; Houston, R.D. Construction and annotation of a high density SNP linkage map of the Atlantic salmon (Salmo salar) genome. G3 Genes Genomes Genet. 2016, 6, 2173–2179. [Google Scholar] [CrossRef] [PubMed]
- Ren, P.; Peng, W.; You, W.; Huang, Z.; Guo, Q.; Chen, N.; He, P.; Ke, J.; Gwo, J.-C.; Ke, C. Genetic mapping and quantitative trait loci analysis of growth-related traits in the small abalone Haliotis diversicolor using restriction-site-associated DNA sequencing. Aquaculture 2016, 454, 163–170. [Google Scholar] [CrossRef]
- Zhang, G.; Zhang, X.; Ye, H.; Jiang, S.; Yu, H.; Li, J.; Shi, Q.; Chen, G.; Zhou, Z.; Luo, J. Construction of high-density genetic linkage maps and QTL mapping in the golden pompano. Aquaculture 2018, 482, 90–95. [Google Scholar] [CrossRef]
- Palaiokostas, C.; Ferraresso, S.; Franch, R.; Houston, R.D.; Bargelloni, L. Genomic prediction of resistance to pasteurellosis in gilthead sea bream (Sparus aurata) using 2b-RAD sequencing. G3 Genes Genomes Genet. 2016, 6, 3693–3700. [Google Scholar] [CrossRef]
- Liu, H.; Fu, B.; Pang, M.; Feng, X.; Yu, X.; Tong, J. A high-density genetic linkage map and QTL fine mapping for body weight in crucian carp (Carassius auratus) using 2b-RAD sequencing. G3 Genes Genomes Genet. 2017, 7, 2473–2487. [Google Scholar] [CrossRef]
- Dou, J.; Li, X.; Fu, Q.; Jiao, W.; Li, Y.; Li, T.; Wang, Y.; Hu, X.; Wang, S.; Bao, Z. Evaluation of the 2b-RAD method for genomic selection in scallop breeding. Sci. Rep. 2016, 6, 19244. [Google Scholar] [CrossRef]
- Kong, S.; Ke, Q.; Chen, L.; Zhou, Z.; Pu, F.; Zhao, J.; Bai, H.; Peng, W.; Xu, P. Constructing a High-Density Genetic Linkage Map for Large Yellow Croaker (Larimichthys crocea) and Mapping Resistance Trait Against Ciliate Parasite Cryptocaryon irritans. Mar. Biotechnol. 2019, 21, 262–275. [Google Scholar] [CrossRef]
- Jones, D.B.; Jerry, D.R.; Khatkar, M.S.; Raadsma, H.W.; Zenger, K.R. A high-density SNP genetic linkage map for the silver-lipped pearl oyster, Pinctada maxima: A valuable resource for gene localisation and marker-assisted selection. BMC Genom. 2013, 14, 810. [Google Scholar] [CrossRef]
- Wang, L.; Wan, Z.Y.; Bai, B.; Huang, S.Q.; Chua, E.; Lee, M.; Pang, H.Y.; Wen, Y.F.; Liu, P.; Liu, F. Construction of a high-density linkage map and fine mapping of QTL for growth in Asian seabass. Sci. Rep. 2015, 5, 16358. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Xu, J.; Zhang, Y.; Feng, J.; Dong, C.; Jiang, L.; Feng, J.; Chen, B.; Gong, Y.; Chen, L. An ultra-high density linkage map and QTL mapping for sex and growth-related traits of common carp (Cyprinus carpio). Sci. Rep. 2016, 6, 26693. [Google Scholar] [CrossRef] [PubMed]
- Hubert, S.; Hedgecock, D. Linkage maps of microsatellite DNA markers for the Pacific oyster Crassostrea gigas. Genetics 2004, 168, 351–362. [Google Scholar] [CrossRef]
- Haldane, J. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 1922, 12, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Kong, A.; Gudbjartsson, D.F.; Sainz, J.; Jonsdottir, G.M.; Gudjonsson, S.A.; Richardsson, B.; Sigurdardottir, S.; Barnard, J.; Hallbeck, B.; Masson, G. A high-resolution recombination map of the human genome. Nat. Genet. 2002, 31, 241. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, H.; Pan, Z.; Chang, G.; Wang, H.; Wu, N.; Ding, H.; Yu, X. Construction of a high-density genetic linkage map and QTL mapping for growth traits in Pseudobagrus ussuriensis. Aquaculture 2019, 511, 734213. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, X.; Chen, X.; Xu, T.; Wang, M.; Qin, Q.; Zhong, L.; Jiang, H.; Zhu, X.; Liu, H. Construction of a High-density Linkage Map and QTL Fine Mapping for Growth and Sex Related Traits in Channel Catfish (Ictalurus punctatus). Front. Genet. 2019, 10, 251. [Google Scholar] [CrossRef]
- Qiu, C.; Han, Z.; Li, W.; Ye, K.; Xie, Y.; Wang, Z. A high-density genetic linkage map and QTL mapping for growth and sex of yellow drum (Nibea albiflora). Sci. Rep. 2018, 8, 17271. [Google Scholar] [CrossRef]
- Sun, C.; Niu, Y.; Ye, X.; Dong, J.; Hu, W.; Zeng, Q.; Chen, Z.; Tian, Y.; Zhang, J.; Lu, M. Construction of a high-density linkage map and mapping of sex determination and growth-related loci in the mandarin fish (Siniperca chuatsi). BMC Genom. 2017, 18, 446. [Google Scholar] [CrossRef]
- Uchino, T.; Hosoda, E.; Nakamura, Y.; Yasuike, M.; Mekuchi, M.; Sekino, M.; Fujiwara, A.; Sugaya, T.; Tanaka, Y.; Kumon, K. Genotyping-by-sequencing for construction of a new genetic linkage map and QTL analysis of growth-related traits in Pacific bluefin tuna. Aquac. Res. 2018, 49, 1293–1301. [Google Scholar] [CrossRef]
- Guo, J.; Li, C.; Teng, T.; Shen, F.; Chen, Y.; Wang, Y.; Pan, C.; Ling, Q. Construction of the first high-density genetic linkage map of pikeperch (Sander lucioperca) using specific length amplified fragment (SLAF) sequencing and QTL analysis of growth-related traits. Aquaculture 2018, 497, 299–305. [Google Scholar] [CrossRef]
- Wan, S.-M.; Liu, H.; Zhao, B.-W.; Nie, C.-H.; Wang, W.-M.; Gao, Z.-X. Construction of a high-density linkage map and fine mapping of QTLs for growth and gonad related traits in blunt snout bream. Sci. Rep. 2017, 7, 46509. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Li, J.; You, X.; Mu, Y.; Ding, Y.; Mao, K.; Bian, C.; Mu, P.; Shi, Q.; Chen, X. Construction of the high-density genetic linkage map and chromosome map of large yellow croaker (Larimichthys crocea). Int. J. Mol. Sci. 2015, 16, 26237–26248. [Google Scholar] [CrossRef] [PubMed]
- Andriantahina, F.; Liu, X.; Huang, H. Genetic map construction and quantitative trait locus (QTL) detection of growth-related traits in Litopenaeus vannamei for selective breeding applications. PLoS ONE 2013, 8, e75206. [Google Scholar] [CrossRef]
- Simon, L.M.; Robin, E.D. Relationship of cytochrome oxidase activity to vertebrate total and organ oxygen consumption. Int. J. Biochem. 1971, 2, 569–573. [Google Scholar] [CrossRef]
- Goolish, E.M.; Adelman, I.R. Tissue-specific cytochrome oxidase activity in largemouth bass: The metabolic costs of feeding and growth. Physiol. Zool. 1987, 60, 454–464. [Google Scholar] [CrossRef]

| Linkage Group | Consensus | Female | Male | |||
|---|---|---|---|---|---|---|
| Marker | Size (cM) | Marker | Size (cM) | Marker | Size (cM) | |
| LG1 | 184 | 106.60 | 146 | 100.12 | 118 | 126.50 |
| LG2 | 75 | 98.71 | 57 | 70.92 | 75 | 106.19 |
| LG3 | 193 | 84.63 | 193 | 131.33 | 20 | 40.38 |
| LG4 | 145 | 99.79 | 126 | 95.30 | 74 | 109.50 |
| LG5 | 171 | 81.85 | 157 | 112.50 | 29 | 44.82 |
| LG6 | 140 | 90.64 | 140 | 99.33 | 13 | 32.17 |
| LG7 | 73 | 108.44 | 70 | 92.97 | 43 | 73.68 |
| LG8 | 251 | 157.18 | 231 | 160.84 | 40 | 36.32 |
| LG9 | 240 | 101.88 | 213 | 131.38 | 27 | 44.35 |
| LG10 | 174 | 116.06 | 164 | 118.29 | 65 | 90.05 |
| LG11 | 214 | 128.37 | 200 | 177.05 | 82 | 79.71 |
| LG12 | 155 | 17.79 | 155 | 130.54 | 63 | 75.75 |
| LG13 | 193 | 138.31 | 164 | 92.35 | 85 | 119.22 |
| LG14 | 80 | 108.70 | 80 | 95.26 | 80 | 103.62 |
| LG15 | 179 | 112.62 | 169 | 128.81 | 58 | 74.76 |
| LG16 | 174 | 120.20 | 166 | 118.02 | 75 | 90.29 |
| LG17 | 109 | 94.37 | 90 | 78.62 | 16 | 35.54 |
| LG18 | 118 | 108.30 | 118 | 100.84 | 29 | 42.07 |
| LG19 | 118 | 102.54 | 118 | 101.20 | 75 | 87.26 |
| LG20 | 114 | 83.09 | 114 | 94.58 | 66 | 85.05 |
| LG21 | 98 | 60.08 | 98 | 64.59 | 17 | 69.82 |
| LG22 | 20 | 32.67 | 20 | 45.69 | 7 | 22.78 |
| LG23 | 154 | 93.09 | 105 | 70.01 | 37 | 60.82 |
| LG24 | 63 | 54.21 | 57 | 54.02 | 26 | 21.32 |
| Total | 3435 | 2300.12 | 3151 | 2464.56 | 1220 | 1671.97 |
| Average | 143 | 95.84 | 131 | 102.69 | 51 | 69.67 |
| Trait | QTL | SNP ID | Genetic Position (cM) | Logarithm of Odds (LOD) | Explained Phenotype (%) |
|---|---|---|---|---|---|
| FL | qTLG6 _FL | 21190 | 32.24_32.59 | 3.92 | 11.8 |
| 449098 | |||||
| FL | qTLG8_1 | 169102 | 137.04_137.43 | 3.98 | 11.9 |
| 597906 | |||||
| 633637 | |||||
| FL | qTLG10_FL | 858434 | 93.08_96.14 | 3.9 | 11.7 |
| 17495 | |||||
| FL | qTLG11_FL | 755146 | 14.33_15.33 | 3.72 | 11.2 |
| FL | qTLG18_FL | 343730 | 41.39_42.10 | 3.52 | 10.7 |
| BL | qTLG1_1 | 741153 | 3_9.12 | 3.83 | 11.5 |
| 931102 | |||||
| BL | qTLG1_2 | 110627 | 84.0_85.29 | 4.08 | 12.2 |
| 365461 | |||||
| BL | qTLG6_BL | 21190 | 32.24_32.59 | 3.95 | 11.9 |
| 449098 | |||||
| BL | qTLG10_BL | 858434 | 93.08_96.14 | 4.32 | 12.9 |
| 17495 | |||||
| BL | qTLG16 | 341254 | 75.1_75.36 | 4.01 | 12 |
| 444247 | |||||
| BL | qTLG18_BL | 343730 | 41.39_4210 | 3.54 | 10.7 |
| BL | qTLG19_1 | 978465 | 69.82_70.81 | 3.82 | 11.5 |
| BW | qTLG8_2 | 695031 | 141.58_143.58 | 3.76 | 11.3 |
| BW | qTLG10_2 | 819257 | 109.22_113.22 | 3.63 | 11 |
| BW | qTLG10_3 | 689551 | 113.47_114.47 | 3.55 | 10.7 |
| BW | qTLG11-BW | 755146 | 14.33_15.33 | 4.21 | 12.6 |
| BW | qTLG19_2 | 196799 | 23.84_24.57 | 4.06 | 12.2 |
| 964460 |
| QTL | Related Traits | SNP ID | Loci | Gene ID | Gene Description | Reference |
|---|---|---|---|---|---|---|
| qTLG6 | FL and BL | 21190 | Chr4 | Eco_gene_10017022 | mRNA, disks large-associated protein 4-like (LOC108900233) | |
| qTLG6 | FL and BL | 449098 | Chr4 | Eco_gene_10020133 | mRNA, somatostatin-3B-like (somatostatin3) | [38] |
| Eco_gene_10020134 | mRNA, spermidine synthase (srm) | [41,42] | ||||
| qTLG8_1 | FL | 633637 | Chr6 | Eco_gene_10016761 | mRNA, gamma-aminobutyric acid receptor subunit alpha-6 (gabra6) | [43] |
| Eco_gene_10016762 | mRNA, gamma-aminobutyric acid receptor subunit alpha-2-like (gabra5) | [44] | ||||
| qTLG10 | FL and BL | 858434 | Chr21 | Eco_gene_10016634 | mRNA, decay activator protein ZFP36-like (zfp36) | [45] |
| Eco_gene_10016635 | mRNA, leucine-rich repeat and fibronectin type III domain-containing protein 1-like protein (LOC111216932) | |||||
| qTLG10 | FL and BL | 17495 | Chr21 | Eco_gene_10021594 | mRNA, leucine rich repeat and fibronectin type III domain containing 1 (lrfn1) | [46] |
| qTLG18 | FL and BL | 343730 | Chr15 | Eco_gene_10003697 | mRNA, delta-like canonical Notch ligand 1 (grid1)) | [47] |
| qTLG1_1 | BL | 741153 | Chr9 | Eco_gene_10012245 | mRNA, RALY heterogeneous nuclear ribonucleoprotein (raly) | [48] |
| Eco_gene_10012246 | mRNA, cytochrome c1, heme protein, mitochondrial-like (cyc1) | [49] | ||||
| qTLG1_1 | BL | 931102 | Chr9 | Eco_gene_10014886 | mRNA, serine/threonine-protein kinase WNK2-like (LOC108886535) | |
| qTLG1_2 | BL | 365461 | Chr9 | Eco_gene_10010099 | mRNA, low-density lipoprotein receptor-related protein 1-like (lrp1) | [50] |
| qTLG10_3 | BW | 689551 | Chr21 | Eco_gene_10016615 | mRNA, alkB homolog 8, tRNA methyltransferase (alkbh8) | [51] |
| Eco_gene_10016616 | mRNA, kelch repeat and BTB domain containing 3 (kbtbd3) | |||||
| qTLG19_2 | BW | 96400 | Chr21 | Eco_gene_10000580 | mRNA, sialidase-4-like (neu4) | [40,52] |
| qTLG16 | BL | 444247 | Chr19 | Eco_gene_10010609 | mRNA, protein kinase C gamma type-like (LOC114557331) |
| GO Term | Term ID | Description | p-Value |
| Biological process | GO:0006122 | Mitochondrial electron transport, ubiquinol to cytochrome c | 9.3 × 10−4 |
| GO:0046034 | ATP metabolic process | 5.0 × 10−3 | |
| GO:0009167 | Purine ribonucleoside monophosphate metabolic process | 5.7 × 10−3 | |
| Molecular function | GO:0009055 | Electron transfer activity | 7.3 × 10−4 |
| Cellular component | GO:0070469 | Respirasome | 6.2 × 10−3 |
| KEGG pathway ID | Description | p-value | |
| dre00190 | Oxidative phosphorylation | 7.19 × 10−14 | |
| dre04260 | Cardiac muscle contraction | 2.14 × 10−13 | |
| dre01100 | Metabolic pathways | 4.09 × 10−8 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Jiang, S.; Zheng, L.; Xiao, L.; Zhang, X.; Wang, D.; Li, S.; Shi, Q.; Wu, S.; Lin, H.; et al. An SNP-Based Genetic Map and QTL Mapping for Growth Traits in the Red-Spotted Grouper (Epinephelus akaara). Genes 2019, 10, 793. https://doi.org/10.3390/genes10100793
Wang X, Jiang S, Zheng L, Xiao L, Zhang X, Wang D, Li S, Shi Q, Wu S, Lin H, et al. An SNP-Based Genetic Map and QTL Mapping for Growth Traits in the Red-Spotted Grouper (Epinephelus akaara). Genes. 2019; 10(10):793. https://doi.org/10.3390/genes10100793
Chicago/Turabian StyleWang, Xiang, Shoujia Jiang, Leyun Zheng, Ling Xiao, Xinhui Zhang, Dengdong Wang, Shuisheng Li, Qiong Shi, Shuiqing Wu, Haoran Lin, and et al. 2019. "An SNP-Based Genetic Map and QTL Mapping for Growth Traits in the Red-Spotted Grouper (Epinephelus akaara)" Genes 10, no. 10: 793. https://doi.org/10.3390/genes10100793
APA StyleWang, X., Jiang, S., Zheng, L., Xiao, L., Zhang, X., Wang, D., Li, S., Shi, Q., Wu, S., Lin, H., You, X., & Zhang, Y. (2019). An SNP-Based Genetic Map and QTL Mapping for Growth Traits in the Red-Spotted Grouper (Epinephelus akaara). Genes, 10(10), 793. https://doi.org/10.3390/genes10100793





