Human Colorectal Cancer from the Perspective of Mouse Models
Abstract
1. Introduction
2. Mouse Models of Chemically Induced Colorectal Tumorigenesis
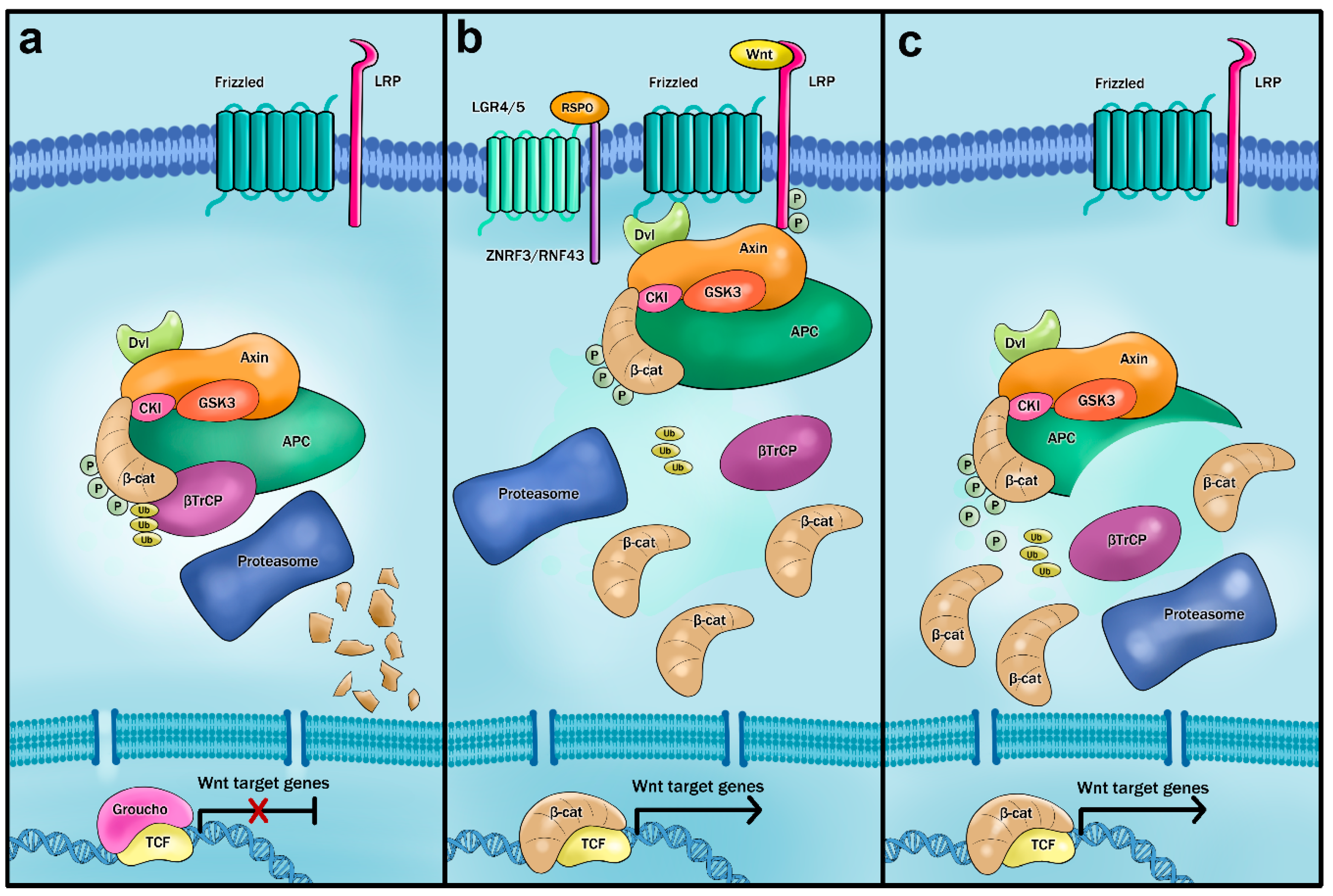
3. Mouse Models of Aberrant Wnt Signaling
3.1. Multiple Intestinal Neoplasia (Min) Mice
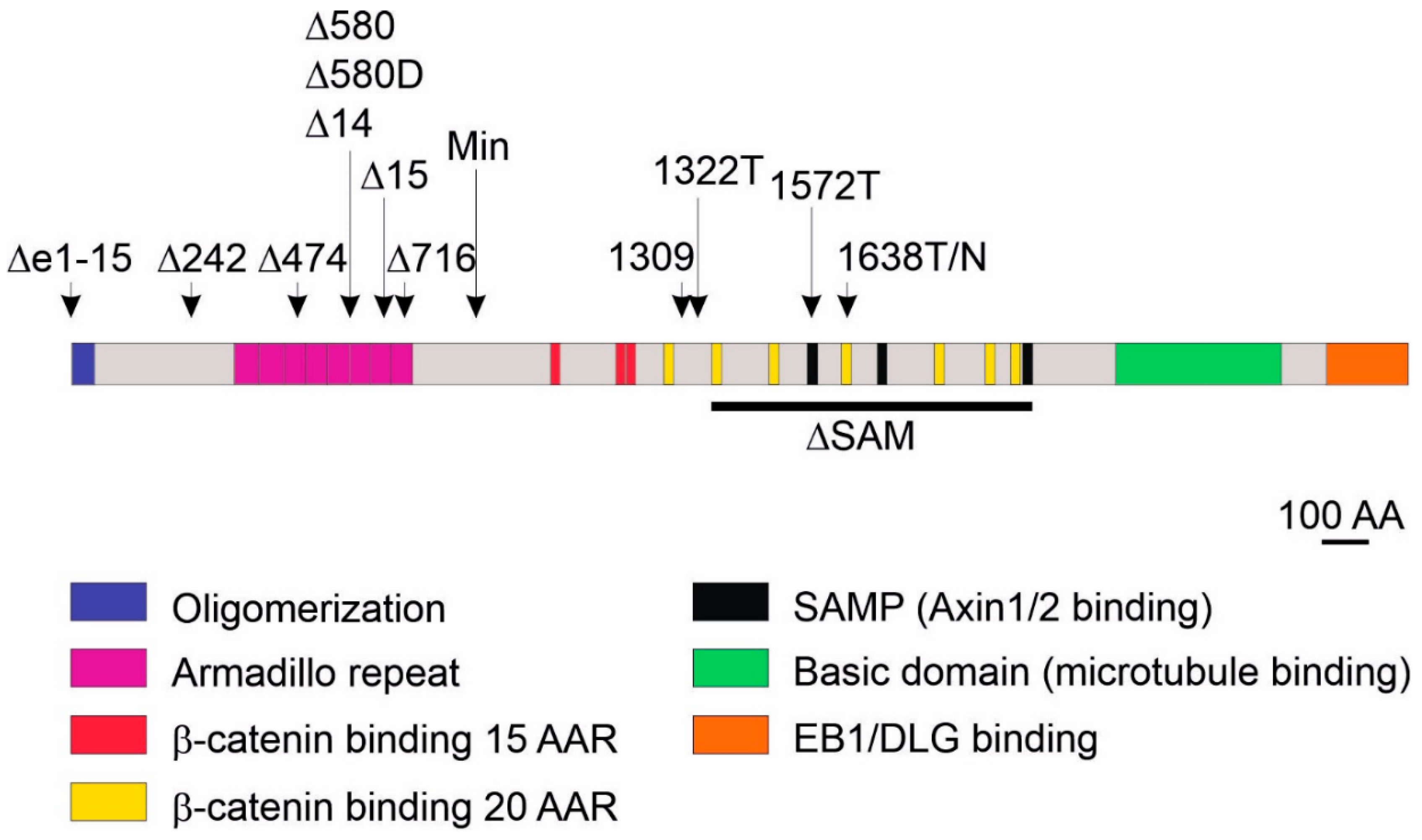
3.2. Models Producing Mutant Apc Variants Longer Than Apc Protein Expressed from the ApcMin Allele
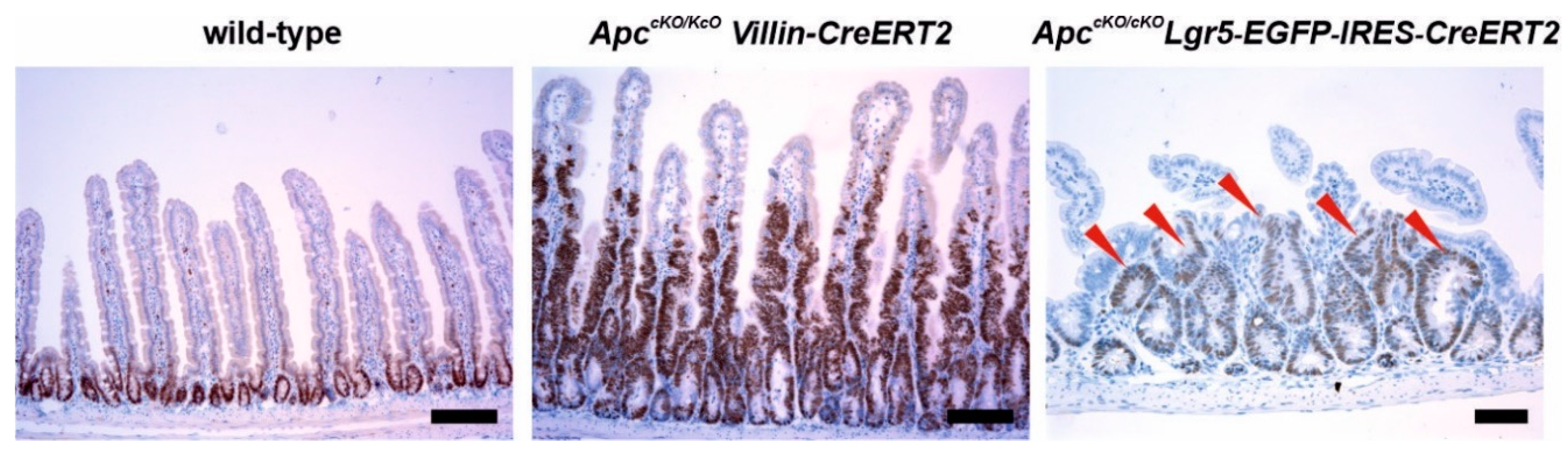
3.3. Models Producing Mutant Apc Variants Shorter Than Apc Protein Expressed from the ApcMin Allele and a Strain with Complete Apc Deletion
3.4. Models Expressing Stabilized β-Catenin
3.5. Alleles Allowing Aberrant (Over) Expression of Wnt Agonists R-Spondins
4. Mouse Models of Inactive Hippo Signaling
5. Mouse Models of p53 Pathway Deficiency
6. Mouse Models of Aberrant Activation of the Epidermal Growth Factor Signaling Pathway
6.1. Mouse Strains Expressing Mutant Epidermal Growth Factor Receptor
6.2. Mouse Models Producing Mutant Kras and Nras
6.3. Mouse Models Harboring Mutant Braf Alleles
7. Mouse Models of Impaired TGFβ Signaling
8. Mouse Models of DNA Mismatch Repair Deficiency
9. Future Perspectives
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Kinzler, K.W.; Vogelstein, B. Lessons from hereditary colorectal cancer. Cell 1996, 87, 159–170. [Google Scholar] [CrossRef]
- Jen, J.; Powell, S.M.; Papadopoulos, N.; Smith, K.J.; Hamilton, S.R.; Vogelstein, B.; Kinzler, K.W. Molecular determinants of dysplasia in colorectal lesions. Cancer Res. 1994, 54, 5523–5526. [Google Scholar] [PubMed]
- Janssen, K.P.; Alberici, P.; Fsihi, H.; Gaspar, C.; Breukel, C.; Franken, P.; Rosty, C.; Abal, M.; El Marjou, F.; Smits, R.; et al. APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology 2006, 131, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; Stern, H.S.; Penner, M.; Hay, K.; Mitri, A.; Bapat, B.V.; Gallinger, S. Somatic APC and K-ras codon 12 mutations in aberrant crypt foci from human colons. Cancer Res. 1994, 54, 5527–5530. [Google Scholar]
- Rodrigues, N.R.; Rowan, A.; Smith, M.E.; Kerr, I.B.; Bodmer, W.F.; Gannon, J.V.; Lane, D.P. p53 mutations in colorectal cancer. Proc. Natl. Acad. Sci. USA 1990, 87, 7555–7559. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Kern, S.E.; Preisinger, A.C.; Leppert, M.; Nakamura, Y.; White, R.; Smits, A.M.; Bos, J.L. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988, 319, 525–532. [Google Scholar] [CrossRef]
- Weinberg, R.A. Oncogenes, antioncogenes, and the molecular bases of multistep carcinogenesis. Cancer Res. 1989, 49, 3713–3721. [Google Scholar]
- Mattar, M.C.; Lough, D.; Pishvaian, M.J.; Charabaty, A. Current management of inflammatory bowel disease and colorectal cancer. Gastrointest. Cancer Res. 2011, 4, 53–61. [Google Scholar]
- Van Der Kraak, L.; Gros, P.; Beauchemin, N. Colitis-associated colon cancer: Is it in your genes? World J. Gastroenterol. 2015, 21, 11688–11699. [Google Scholar] [CrossRef]
- Brentnall, T.A.; Crispin, D.A.; Rabinovitch, P.S.; Haggitt, R.C.; Rubin, C.E.; Stevens, A.C.; Burmer, G.C. Mutations in the p53 gene: An early marker of neoplastic progression in ulcerative colitis. Gastroenterology 1994, 107, 369–378. [Google Scholar] [CrossRef]
- Shenoy, A.K.; Fisher, R.C.; Butterworth, E.A.; Pi, L.; Chang, L.J.; Appelman, H.D.; Chang, M.; Scott, E.W.; Huang, E.H. Transition from colitis to cancer: High Wnt activity sustains the tumor-initiating potential of colon cancer stem cell precursors. Cancer Res. 2012, 72, 5091–5100. [Google Scholar] [CrossRef] [PubMed]
- Yaeger, R.; Shah, M.A.; Miller, V.A.; Kelsen, J.R.; Wang, K.; Heins, Z.J.; Ross, J.S.; He, Y.; Sanford, E.; Yantiss, R.K.; et al. Genomic Alterations Observed in Colitis-Associated Cancers Are Distinct From Those Found in Sporadic Colorectal Cancers and Vary by Type of Inflammatory Bowel Disease. Gastroenterology 2016, 151, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Ullman, T.A.; Itzkowitz, S.H. Intestinal inflammation and cancer. Gastroenterology 2011, 140, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Andreyev, H.J.; Norman, A.R.; Cunningham, D.; Oates, J.R.; Clarke, P.A. Kirsten ras mutations in patients with colorectal cancer: The multicenter “RASCAL” study. J. Natl. Cancer Inst. 1998, 90, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.; Carey, F.A.; Beattie, J.; Wilkie, M.J.; Lightfoot, T.J.; Coxhead, J.; Garner, R.C.; Steele, R.J.; Wolf, C.R. Mutations in APC, Kirsten-ras, and p53—Alternative genetic pathways to colorectal cancer. Proc. Natl. Acad. Sci. USA 2002, 99, 9433–9438. [Google Scholar] [CrossRef] [PubMed]
- Robles, A.I.; Traverso, G.; Zhang, M.; Roberts, N.J.; Khan, M.A.; Joseph, C.; Lauwers, G.Y.; Selaru, F.M.; Popoli, M.; Pittman, M.E.; et al. Whole-Exome Sequencing Analyses of Inflammatory Bowel Disease-Associated Colorectal Cancers. Gastroenterology 2016, 150, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Budinska, E.; Popovici, V.; Tejpar, S.; D’Ario, G.; Lapique, N.; Sikora, K.O.; Di Narzo, A.F.; Yan, P.; Hodgson, J.G.; Weinrich, S.; et al. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J. Pathol. 2013, 231, 63–76. [Google Scholar] [CrossRef]
- Marisa, L.; de Reynies, A.; Duval, A.; Selves, J.; Gaub, M.P.; Vescovo, L.; Etienne-Grimaldi, M.C.; Schiappa, R.; Guenot, D.; Ayadi, M.; et al. Gene expression classification of colon cancer into molecular subtypes: Characterization, validation, and prognostic value. PLoS Med. 2013, 10, e1001453. [Google Scholar] [CrossRef]
- Roepman, P.; Schlicker, A.; Tabernero, J.; Majewski, I.; Tian, S.; Moreno, V.; Snel, M.H.; Chresta, C.M.; Rosenberg, R.; Nitsche, U.; et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int. J. Cancer 2014, 134, 552–562. [Google Scholar] [CrossRef]
- De Sousa, E.M.F.; Wang, X.; Jansen, M.; Fessler, E.; Trinh, A.; de Rooij, L.P.; de Jong, J.H.; de Boer, O.J.; van Leersum, R.; Bijlsma, M.F.; et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat. Med. 2013, 19, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Sadanandam, A.; Lyssiotis, C.A.; Homicsko, K.; Collisson, E.A.; Gibb, W.J.; Wullschleger, S.; Ostos, L.C.; Lannon, W.A.; Grotzinger, C.; Del Rio, M.; et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med. 2013, 19, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Schlicker, A.; Beran, G.; Chresta, C.M.; McWalter, G.; Pritchard, A.; Weston, S.; Runswick, S.; Davenport, S.; Heathcote, K.; Castro, D.A.; et al. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med. Genom. 2012, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Ning, C.; Li, Y.Y.; Wang, Y.; Han, G.C.; Wang, R.X.; Xiao, H.; Li, X.Y.; Hou, C.M.; Ma, Y.F.; Sheng, D.S.; et al. Complement activation promotes colitis-associated carcinogenesis through activating intestinal IL-1β/IL-17A axis. Mucosal Immunol. 2015, 8, 1275–1284. [Google Scholar] [CrossRef] [PubMed]
- Linnekamp, J.F.; Hooff, S.R.V.; Prasetyanti, P.R.; Kandimalla, R.; Buikhuisen, J.Y.; Fessler, E.; Ramesh, P.; Lee, K.; Bochove, G.G.W.; de Jong, J.H.; et al. Consensus molecular subtypes of colorectal cancer are recapitulated in in vitro and in vivo models. Cell Death Differ. 2018, 25, 616–633. [Google Scholar] [CrossRef] [PubMed]
- Phesse, T.J.; Durban, V.M.; Sansom, O.J. Defining key concepts of intestinal and epithelial cancer biology through the use of mouse models. Carcinogenesis 2017, 38, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Taketo, M.M.; Edelmann, W. Mouse models of colon cancer. Gastroenterology 2009, 136, 780–798. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lee, H.S.; Choe, G.; Chung, J.H.; Kim, W.H. Clinicopathological characteristics, microsatellite instability, and expression of mucin core proteins and p53 in colorectal mucinous adenocarcinomas in relation to location. Virchows Arch. 2006, 449, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Oshima, H.; Nakayama, M.; Han, T.S.; Naoi, K.; Ju, X.; Maeda, Y.; Robine, S.; Tsuchiya, K.; Sato, T.; Sato, H.; et al. Suppressing TGFβ signaling in regenerating epithelia in an inflammatory microenvironment is sufficient to cause invasive intestinal cancer. Cancer Res. 2015, 75, 766–776. [Google Scholar] [CrossRef]
- Reitmair, A.H.; Redston, M.; Cai, J.C.; Chuang, T.C.; Bjerknes, M.; Cheng, H.; Hay, K.; Gallinger, S.; Bapat, B.; Mak, T.W. Spontaneous intestinal carcinomas and skin neoplasms in Msh2-deficient mice. Cancer Res. 1996, 56, 3842–3849. [Google Scholar] [PubMed]
- Su, L.K.; Kinzler, K.W.; Vogelstein, B.; Preisinger, A.C.; Moser, A.R.; Luongo, C.; Gould, K.A.; Dove, W.F. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 1992, 256, 668–670. [Google Scholar] [CrossRef] [PubMed]
- Sasai, H.; Masaki, M.; Wakitani, K. Suppression of polypogenesis in a new mouse strain with a truncated Apc∆474 by a novel COX-2 inhibitor, JTE-522. Carcinogenesis 2000, 21, 953–958. [Google Scholar] [CrossRef] [PubMed]
- Colnot, S.; Niwa-Kawakita, M.; Hamard, G.; Godard, C.; Le Plenier, S.; Houbron, C.; Romagnolo, B.; Berrebi, D.; Giovannini, M.; Perret, C. Colorectal cancers in a new mouse model of familial adenomatous polyposis: Influence of genetic and environmental modifiers. Lab. Investig. J. Tech. Methods Pathol. 2004, 84, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.; Bazan, V.; Iacopetta, B.; Kerr, D.; Soussi, T.; Gebbia, N. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: Influence of tumor site, type of mutation, and adjuvant treatment. J. Clin. Oncol. 2005, 23, 7518–7528. [Google Scholar] [CrossRef] [PubMed]
- Mercer, K.; Giblett, S.; Green, S.; Lloyd, D.; DaRocha Dias, S.; Plumb, M.; Marais, R.; Pritchard, C. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res. 2005, 65, 11493–11500. [Google Scholar] [CrossRef] [PubMed]
- Samowitz, W.S.; Sweeney, C.; Herrick, J.; Albertsen, H.; Levin, T.R.; Murtaugh, M.A.; Wolff, R.K.; Slattery, M.L. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005, 65, 6063–6069. [Google Scholar] [CrossRef] [PubMed]
- Ito, N.; Hasegawa, R.; Sano, M.; Tamano, S.; Esumi, H.; Takayama, S.; Sugimura, T. A new colon and mammary carcinogen in cooked food, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP). Carcinogenesis 1991, 12, 1503–1506. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, M.; Imai, H.; Sugimura, T.; Nagao, M.; Nakagama, H. Induction of intestinal tumors and lymphomas in C57BL/6N mice by a food-borne carcinogen, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine. Jpn. J. Cancer Res. 2002, 93, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Nakagama, H.; Nakanishi, M.; Ochiai, M. Modeling human colon cancer in rodents using a food-borne carcinogen, PhIP. Cancer Sci. 2005, 96, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Shikata, N.; Mizuoka, H.; Tsubura, A. Colon carcinogenesis in shrews by intrarectal infusion of N-methyl-N-nitrosourea. Cancer Lett. 1996, 110, 105–112. [Google Scholar] [CrossRef]
- Rosenberg, D.W.; Giardina, C.; Tanaka, T. Mouse models for the study of colon carcinogenesis. Carcinogenesis 2009, 30, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Deschner, E.E.; Long, F.C. Colonic neoplasms in mice produced with six injections of 1,2-dimethylhydrazine. Oncology 1977, 34, 255–257. [Google Scholar] [CrossRef] [PubMed]
- Maltzman, T.; Whittington, J.; Driggers, L.; Stephens, J.; Ahnen, D. AOM-induced mouse colon tumors do not express full-length APC protein. Carcinogenesis 1997, 18, 2435–2439. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Nakatsugi, S.; Sugimura, T.; Wakabayashi, K. Frequent mutations of the β-catenin gene in mouse colon tumors induced by azoxymethane. Carcinogenesis 2000, 21, 1117–1120. [Google Scholar] [PubMed]
- Vivona, A.A.; Shpitz, B.; Medline, A.; Bruce, W.R.; Hay, K.; Ward, M.A.; Stern, H.S.; Gallinger, S. K-ras mutations in aberrant crypt foci, adenomas and adenocarcinomas during azoxymethane-induced colon carcinogenesis. Carcinogenesis 1993, 14, 1777–1781. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.S.; Papanikolaou, A.; Sabourin, C.L.; Rosenberg, D.W. Altered expression of cyclin D1 and cyclin-dependent kinase 4 in azoxymethane-induced mouse colon tumorigenesis. Carcinogenesis 1998, 19, 2001–2006. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Huang, X.F. The signal pathways in azoxymethane-induced colon cancer and preventive implications. Cancer Biol. Ther. 2009, 8, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Waaler, J.; Machon, O.; Tumova, L.; Dinh, H.; Korinek, V.; Wilson, S.R.; Paulsen, J.E.; Pedersen, N.M.; Eide, T.J.; Machonova, O.; et al. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res. 2012, 72, 2822–2832. [Google Scholar] [CrossRef] [PubMed]
- Bissahoyo, A.; Pearsall, R.S.; Hanlon, K.; Amann, V.; Hicks, D.; Godfrey, V.L.; Threadgill, D.W. Azoxymethane is a genetic background-dependent colorectal tumor initiator and promoter in mice: Effects of dose, route, and diet. Toxicol. Sci. 2005, 88, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Greten, F.R.; Eckmann, L.; Greten, T.F.; Park, J.M.; Li, Z.W.; Egan, L.J.; Kagnoff, M.F.; Karin, M. IKKβ links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 2004, 118, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Neufert, C.; Becker, C.; Neurath, M.F. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc. 2007, 2, 1998–2004. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kohno, H.; Suzuki, R.; Yamada, Y.; Sugie, S.; Mori, H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 2003, 94, 965–973. [Google Scholar] [CrossRef] [PubMed]
- De Robertis, M.; Massi, E.; Poeta, M.L.; Carotti, S.; Morini, S.; Cecchetelli, L.; Signori, E.; Fazio, V.M. The AOM/DSS murine model for the study of colon carcinogenesis: From pathways to diagnosis and therapy studies. J. Carcinog. 2011, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Taketo, M.M. Adenomatous polyposis coli (APC): A multi-functional tumor suppressor gene. J. Cell Sci. 2007, 120, 3327–3335. [Google Scholar] [CrossRef] [PubMed]
- Valenta, T.; Hausmann, G.; Basler, K. The many faces and functions of β-catenin. EMBO J. 2012, 31, 2714–2736. [Google Scholar] [CrossRef] [PubMed]
- Kimelman, D.; Xu, W. β-catenin destruction complex: Insights and questions from a structural perspective. Oncogene 2006, 25, 7482–7491. [Google Scholar] [CrossRef]
- Stamos, J.L.; Weis, W.I. The β-catenin destruction complex. Cold Spring Harb. Perspect. Biol. 2013, 5, a007898. [Google Scholar] [CrossRef]
- Saito-Diaz, K.; Chen, T.W.; Wang, X.; Thorne, C.A.; Wallace, H.A.; Page-McCaw, A.; Lee, E. The way Wnt works: Components and mechanism. Growth Factors 2013, 31, 1–31. [Google Scholar] [CrossRef]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef]
- Shtutman, M.; Zhurinsky, J.; Simcha, I.; Albanese, C.; D’Amico, M.; Pestell, R.; Ben-Ze’ev, A. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 5522–5527. [Google Scholar] [CrossRef] [PubMed]
- Wielenga, V.J.; Smits, R.; Korinek, V.; Smit, L.; Kielman, M.; Fodde, R.; Clevers, H.; Pals, S.T. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am. J. Pathol. 1999, 154, 515–523. [Google Scholar] [CrossRef]
- Coppede, F.; Lopomo, A.; Spisni, R.; Migliore, L. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of colorectal cancer. World J. Gastroenterol. 2014, 20, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Segditsas, S.; Tomlinson, I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 2006, 25, 7531–7537. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Ikeda, S.; Fujimori, M.; Kodama, S.; Nakahara, M.; Okajima, M.; Asahara, T. Frequent alterations in the Wnt signaling pathway in colorectal cancer with microsatellite instability. Genes Chromosomes Cancer 2002, 33, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, S.M.; Fearon, E.R. AXIN1 and AXIN2 variants in gastrointestinal cancers. Cancer Lett. 2014, 355, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nishisho, I.; Nakamura, Y.; Miyoshi, Y.; Miki, Y.; Ando, H.; Horii, A.; Koyama, K.; Utsunomiya, J.; Baba, S.; Hedge, P. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science 1991, 253, 665–669. [Google Scholar] [CrossRef] [PubMed]
- Groden, J.; Thliveris, A.; Samowitz, W.; Carlson, M.; Gelbert, L.; Albertsen, H.; Joslyn, G.; Stevens, J.; Spirio, L.; Robertson, M.; et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 1991, 66, 589–600. [Google Scholar] [CrossRef]
- Galiatsatos, P.; Foulkes, W.D. Familial adenomatous polyposis. Am. J. Gastroenterol. 2006, 101, 385–398. [Google Scholar] [CrossRef] [PubMed]
- Rubinfeld, B.; Albert, I.; Porfiri, E.; Fiol, C.; Munemitsu, S.; Polakis, P. Binding of GSK3β to the APC-β-catenin complex and regulation of complex assembly. Science 1996, 272, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Behrens, J.; Jerchow, B.A.; Wurtele, M.; Grimm, J.; Asbrand, C.; Wirtz, R.; Kuhl, M.; Wedlich, D.; Birchmeier, W. Functional interaction of an axin homolog, conductin, with β-catenin, APC, and GSK3β. Science 1998, 280, 596–599. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, Y.; Nagase, H.; Ando, H.; Horii, A.; Ichii, S.; Nakatsuru, S.; Aoki, T.; Miki, Y.; Mori, T.; Nakamura, Y. Somatic mutations of the APC gene in colorectal tumors: Mutation cluster region in the APC gene. Hum. Mol. Genet. 1992, 1, 229–233. [Google Scholar] [PubMed]
- Miyaki, M.; Konishi, M.; Kikuchi-Yanoshita, R.; Enomoto, M.; Igari, T.; Tanaka, K.; Muraoka, M.; Takahashi, H.; Amada, Y.; Fukayama, M.; et al. Characteristics of somatic mutation of the adenomatous polyposis coli gene in colorectal tumors. Cancer Res. 1994, 54, 3011–3020. [Google Scholar] [PubMed]
- Hayashi, S.; Rubinfeld, B.; Souza, B.; Polakis, P.; Wieschaus, E.; Levine, A.J. A Drosophila homolog of the tumor suppressor gene adenomatous polyposis coli down-regulates β-catenin but its zygotic expression is not essential for the regulation of Armadillo. Proc. Natl. Acad. Sci. USA 1997, 94, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Moser, A.R.; Pitot, H.C.; Dove, W.F. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 1990, 247, 322–324. [Google Scholar] [CrossRef]
- Moser, A.R.; Mattes, E.M.; Dove, W.F.; Lindstrom, M.J.; Haag, J.D.; Gould, M.N. ApcMin, a mutation in the murine Apc gene, predisposes to mammary carcinomas and focal alveolar hyperplasias. Proc. Natl. Acad. Sci. USA 1993, 90, 8977–8981. [Google Scholar] [CrossRef]
- Tomita, H.; Yamada, Y.; Oyama, T.; Hata, K.; Hirose, Y.; Hara, A.; Kunisada, T.; Sugiyama, Y.; Adachi, Y.; Linhart, H.; et al. Development of gastric tumors in ApcMin/+ mice by the activation of the β-catenin/Tcf signaling pathway. Cancer Res. 2007, 67, 4079–4087. [Google Scholar] [CrossRef]
- Svendsen, C.; Alexander, J.; Knutsen, H.K.; Husoy, T. The min mouse on FVB background: Susceptibility to spontaneous and carcinogen-induced intestinal tumourigenesis. Anticancer Res. 2011, 31, 785–788. [Google Scholar]
- Sodring, M.; Gunnes, G.; Paulsen, J.E. Spontaneous initiation, promotion and progression of colorectal cancer in the novel A/J Min/+ mouse. Int. J. Cancer 2016, 138, 1936–1946. [Google Scholar] [CrossRef]
- Cooper, H.S.; Chang, W.C.; Coudry, R.; Gary, M.A.; Everley, L.; Spittle, C.S.; Wang, H.; Litwin, S.; Clapper, M.L. Generation of a unique strain of multiple intestinal neoplasia (Apc+/Min-FCCC) mice with significantly increased numbers of colorectal adenomas. Mol. Carcinog. 2005, 44, 31–41. [Google Scholar] [CrossRef]
- Bashir, O.; FitzGerald, A.J.; Goodlad, R.A. Both suboptimal and elevated vitamin intake increase intestinal neoplasia and alter crypt fission in the ApcMin/+ mouse. Carcinogenesis 2004, 25, 1507–1515. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lawrance, A.K.; Deng, L.; Brody, L.C.; Finnell, R.H.; Shane, B.; Rozen, R. Genetic and nutritional deficiencies in folate metabolism influence tumorigenicity in Apcmin/+ mice. J. Nutr. Biochem. 2007, 18, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Mutanen, M.; Pajari, A.M.; Oikarinen, S.I. Beef induces and rye bran prevents the formation of intestinal polyps in ApcMin mice: Relation to β-catenin and PKC isozymes. Carcinogenesis 2000, 21, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Lamprecht, S.A.; Shinozaki, H.; Fan, K.; Yang, W.; Newmark, H.L.; Kopelovich, L.; Edelmann, W.; Jin, B.; Gravaghi, C.; et al. Dietary calcium and cholecalciferol modulate cyclin D1 expression, apoptosis, and tumorigenesis in intestine of adenomatous polyposis coli1638N/+ mice. J. Nutr. 2008, 138, 1658–1663. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kwong, L.N.; Dove, W.F. APC and its modifiers in colon cancer. Adv. Exp. Med. Biol. 2009, 656, 85–106. [Google Scholar] [PubMed]
- Lamlum, H.; Ilyas, M.; Rowan, A.; Clark, S.; Johnson, V.; Bell, J.; Frayling, I.; Efstathiou, J.; Pack, K.; Payne, S.; et al. The type of somatic mutation at APC in familial adenomatous polyposis is determined by the site of the germline mutation: A new facet to Knudson’s ‘two-hit’ hypothesis. Nat. Med. 1999, 5, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Sieber, O.M.; Heinimann, K.; Gorman, P.; Lamlum, H.; Crabtree, M.; Simpson, C.A.; Davies, D.; Neale, K.; Hodgson, S.V.; Roylance, R.R.; et al. Analysis of chromosomal instability in human colorectal adenomas with two mutational hits at APC. Proc. Natl. Acad. Sci. USA 2002, 99, 16910–16915. [Google Scholar] [CrossRef]
- Lewis, A.; Segditsas, S.; Deheragoda, M.; Pollard, P.; Jeffery, R.; Nye, E.; Lockstone, H.; Davis, H.; Clark, S.; Stamp, G.; et al. Severe polyposis in Apc1322T mice is associated with submaximal Wnt signalling and increased expression of the stem cell marker Lgr5. Gut 2010, 59, 1680–1686. [Google Scholar] [CrossRef]
- Pollard, P.; Deheragoda, M.; Segditsas, S.; Lewis, A.; Rowan, A.; Howarth, K.; Willis, L.; Nye, E.; McCart, A.; Mandir, N.; et al. The Apc1322T mouse develops severe polyposis associated with submaximal nuclear β-catenin expression. Gastroenterology 2009, 136, 2204–2213. [Google Scholar] [CrossRef]
- Bakker, E.R.; Hoekstra, E.; Franken, P.F.; Helvensteijn, W.; van Deurzen, C.H.; van Veelen, W.; Kuipers, E.J.; Smits, R. β-Catenin signaling dosage dictates tissue-specific tumor predisposition in Apc-driven cancer. Oncogene 2013, 32, 4579–4585. [Google Scholar] [CrossRef]
- Quesada, C.F.; Kimata, H.; Mori, M.; Nishimura, M.; Tsuneyoshi, T.; Baba, S. Piroxicam and acarbose as chemopreventive agents for spontaneous intestinal adenomas in APC gene 1309 knockout mice. Jpn. J. Cancer Res. 1998, 89, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Niho, N.; Takahashi, M.; Kitamura, T.; Shoji, Y.; Itoh, M.; Noda, T.; Sugimura, T.; Wakabayashi, K. Concomitant suppression of hyperlipidemia and intestinal polyp formation in Apc-deficient mice by peroxisome proliferator-activated receptor ligands. Cancer Res. 2003, 63, 6090–6095. [Google Scholar] [PubMed]
- Deka, J.; Kuhlmann, J.; Muller, O. A domain within the tumor suppressor protein APC shows very similar biochemical properties as the microtubule-associated protein tau. Eur. J. Biochem. 1998, 253, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.; Davis, H.; Deheragoda, M.; Pollard, P.; Nye, E.; Jeffery, R.; Segditsas, S.; East, P.; Poulsom, R.; Stamp, G.; et al. The C-terminus of Apc does not influence intestinal adenoma development or progression. J. Pathol. 2012, 226, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Fodde, R.; Edelmann, W.; Yang, K.; van Leeuwen, C.; Carlson, C.; Renault, B.; Breukel, C.; Alt, E.; Lipkin, M.; Khan, P.M.; et al. A targeted chain-termination mutation in the mouse Apc gene results in multiple intestinal tumors. Proc. Natl. Acad. Sci. USA 1994, 91, 8969–8973. [Google Scholar] [CrossRef] [PubMed]
- Smits, R.; van der Houven van Oordt, W.; Luz, A.; Zurcher, C.; Jagmohan-Changur, S.; Breukel, C.; Khan, P.M.; Fodde, R. Apc1638N: A mouse model for familial adenomatous polyposis-associated desmoid tumors and cutaneous cysts. Gastroenterology 1998, 114, 275–283. [Google Scholar] [CrossRef]
- Caspari, R.; Olschwang, S.; Friedl, W.; Mandl, M.; Boisson, C.; Boker, T.; Augustin, A.; Kadmon, M.; Moslein, G.; Thomas, G.; et al. Familial adenomatous polyposis: Desmoid tumours and lack of ophthalmic lesions (CHRPE) associated with APC mutations beyond codon 1444. Hum. Mol. Genet. 1995, 4, 337–340. [Google Scholar] [CrossRef]
- Davies, D.R.; Armstrong, J.G.; Thakker, N.; Horner, K.; Guy, S.P.; Clancy, T.; Sloan, P.; Blair, V.; Dodd, C.; Warnes, T.W.; et al. Severe Gardner syndrome in families with mutations restricted to a specific region of the APC gene. Am. J. Hum. Genet. 1995, 57, 1151–1158. [Google Scholar]
- Ikenoue, T.; Yamaguchi, K.; Komura, M.; Imoto, S.; Yamaguchi, R.; Shimizu, E.; Kasuya, S.; Shibuya, T.; Hatakeyama, S.; Miyano, S.; et al. Attenuated familial adenomatous polyposis with desmoids caused by an APC mutation. Hum. Genome Var. 2015, 2, 15011. [Google Scholar] [CrossRef]
- Wang, T.; Onouchi, T.; Yamada, N.O.; Matsuda, S.; Senda, T. A disturbance of intestinal epithelial cell population and kinetics in APC1638T mice. Med. Mol. Morphol. 2017, 50, 94–102. [Google Scholar] [CrossRef]
- Smits, R.; Kielman, M.F.; Breukel, C.; Zurcher, C.; Neufeld, K.; Jagmohan-Changur, S.; Hofland, N.; van Dijk, J.; White, R.; Edelmann, W.; et al. Apc1638T: A mouse model delineating critical domains of the adenomatous polyposis coli protein involved in tumorigenesis and development. Genes Dev. 1999, 13, 1309–1321. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, Y.S.; Dabdoub, A.; Smallwood, P.M.; Williams, J.; Woods, C.; Kelley, M.W.; Jiang, L.; Tasman, W.; Zhang, K.; et al. Vascular development in the retina and inner ear: Control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell 2004, 116, 883–895. [Google Scholar] [CrossRef]
- Gaspar, C.; Franken, P.; Molenaar, L.; Breukel, C.; van der Valk, M.; Smits, R.; Fodde, R. A targeted constitutive mutation in the APC tumor suppressor gene underlies mammary but not intestinal tumorigenesis. PLoS Genet. 2009, 5, e1000547. [Google Scholar] [CrossRef] [PubMed]
- Crist, R.C.; Roth, J.J.; Baran, A.A.; McEntee, B.J.; Siracusa, L.D.; Buchberg, A.M. The armadillo repeat domain of Apc suppresses intestinal tumorigenesis. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2010, 21, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Oshima, M.; Oshima, H.; Kitagawa, K.; Kobayashi, M.; Itakura, C.; Taketo, M. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc. Natl. Acad. Sci. USA 1995, 92, 4482–4486. [Google Scholar] [CrossRef] [PubMed]
- Shibata, H.; Toyama, K.; Shioya, H.; Ito, M.; Hirota, M.; Hasegawa, S.; Matsumoto, H.; Takano, H.; Akiyama, T.; Toyoshima, K.; et al. Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science 1997, 278, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Kuraguchi, M.; Wang, X.P.; Bronson, R.T.; Rothenberg, R.; Ohene-Baah, N.Y.; Lund, J.J.; Kucherlapati, M.; Maas, R.L.; Kucherlapati, R. Adenomatous polyposis coli (APC) is required for normal development of skin and thymus. PLoS Genet. 2006, 2, e146. [Google Scholar] [CrossRef]
- Colnot, S.; Decaens, T.; Niwa-Kawakita, M.; Godard, C.; Hamard, G.; Kahn, A.; Giovannini, M.; Perret, C. Liver-targeted disruption of Apc in mice activates β-catenin signaling and leads to hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 2004, 101, 17216–17221. [Google Scholar] [CrossRef]
- El Marjou, F.; Janssen, K.P.; Chang, B.H.; Li, M.; Hindie, V.; Chan, L.; Louvard, D.; Chambon, P.; Metzger, D.; Robine, S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 2004, 39, 186–193. [Google Scholar] [CrossRef]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Horazna, M.; Janeckova, L.; Svec, J.; Babosova, O.; Hrckulak, D.; Vojtechova, M.; Galuskova, K.; Sloncova, E.; Kolar, M.; Strnad, H.; et al. Msx1 loss suppresses formation of the ectopic crypts developed in the Apc-deficient small intestinal epithelium. Sci. Rep. 2019, 9, 1629. [Google Scholar] [CrossRef] [PubMed]
- Robanus-Maandag, E.C.; Koelink, P.J.; Breukel, C.; Salvatori, D.C.; Jagmohan-Changur, S.C.; Bosch, C.A.; Verspaget, H.W.; Devilee, P.; Fodde, R.; Smits, R. A new conditional Apc-mutant mouse model for colorectal cancer. Carcinogenesis 2010, 31, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.F.; Carter, A.M.; Kostova, K.K.; Woodruff, J.F.; Crowley, D.; Bronson, R.T.; Haigis, K.M.; Jacks, T. Complete deletion of Apc results in severe polyposis in mice. Oncogene 2010, 29, 1857–1864. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Wang, Y.M.; Broaddus, R.; Sun, L.H.; Xue, F.X.; Zhang, W. Exon 3 mutations of CTNNB1 drive tumorigenesis: A review. Oncotarget 2018, 9, 5492–5508. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Jeong, S. Mutation Hotspots in the β-Catenin Gene: Lessons from the Human Cancer Genome Databases. Mol. Cells 2019, 42, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Harada, N.; Tamai, Y.; Ishikawa, T.; Sauer, B.; Takaku, K.; Oshima, M.; Taketo, M.M. Intestinal polyposis in mice with a dominant stable mutation of the β-catenin gene. EMBO J. 1999, 18, 5931–5942. [Google Scholar] [CrossRef] [PubMed]
- Kriz, V.; Korinek, V. Wnt, RSPO and Hippo Signalling in the Intestine and Intestinal Stem Cells. Genes 2018, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Seshagiri, S.; Stawiski, E.W.; Durinck, S.; Modrusan, Z.; Storm, E.E.; Conboy, C.B.; Chaudhuri, S.; Guan, Y.; Janakiraman, V.; Jaiswal, B.S.; et al. Recurrent R-spondin fusions in colon cancer. Nature 2012, 488, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Ogawa, R.; Yoshida, H.; Taniguchi, H.; Kojima, M.; Saito, Y.; Sekine, S. EIF3E-RSPO2 and PIEZO1-RSPO2 fusions in colorectal traditional serrated adenoma. Histopathology 2019. [Google Scholar] [CrossRef]
- Hilkens, J.; Timmer, N.C.; Boer, M.; Ikink, G.J.; Schewe, M.; Sacchetti, A.; Koppens, M.A.J.; Song, J.Y.; Bakker, E.R.M. RSPO3 expands intestinal stem cell and niche compartments and drives tumorigenesis. Gut 2017, 66, 1095–1105. [Google Scholar] [CrossRef]
- Han, T.; Schatoff, E.M.; Murphy, C.; Zafra, M.P.; Wilkinson, J.E.; Elemento, O.; Dow, L.E. R-Spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat. Commun. 2017, 8, 15945. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Tumaneng, K.; Guan, K.L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Boil. 2011, 13, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.X.; Meng, Z.; Plouffe, S.W.; Guan, K.L. Hippo pathway regulation of gastrointestinal tissues. Annu. Rev. Physiol. 2015, 77, 201–227. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shi, S.; Guo, Z.; Zhang, X.; Han, S.; Yang, A.; Wen, W.; Zhu, Q. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLoS ONE 2013, 8, e65539. [Google Scholar] [CrossRef]
- Yuen, H.F.; McCrudden, C.M.; Huang, Y.H.; Tham, J.M.; Zhang, X.; Zeng, Q.; Zhang, S.D.; Hong, W. TAZ expression as a prognostic indicator in colorectal cancer. PLoS ONE 2013, 8, e54211. [Google Scholar] [CrossRef] [PubMed]
- Avruch, J.; Zhou, D.; Bardeesy, N. YAP oncogene overexpression supercharges colon cancer proliferation. Cell Cycle 2012, 11, 1090–1096. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Gwak, J.W.; Shin, Y.C.; Moon, D.; Ahn, J.; Sol, H.W.; Kim, S.; Kim, G.; Shin, H.M.; Lee, K.H.; et al. Expression of Hippo pathway genes and their clinical significance in colon adenocarcinoma. Oncol. Lett. 2018, 15, 4926–4936. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, X.; Yu, T.; Yuan, L.; Dai, J.; Wang, W.; Chen, G.; Jiao, C.; Zhou, W.; Huang, Q.; et al. REGγ Controls Hippo Signaling and Reciprocal NF-κB-YAP Regulation to Promote Colon Cancer. Clin. Cancer Res. 2018, 24, 2015–2025. [Google Scholar] [CrossRef]
- Camargo, F.D.; Gokhale, S.; Johnnidis, J.B.; Fu, D.; Bell, G.W.; Jaenisch, R.; Brummelkamp, T.R. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Boil. CB 2007, 17, 2054–2060. [Google Scholar] [CrossRef]
- Barry, E.R.; Morikawa, T.; Butler, B.L.; Shrestha, K.; de la Rosa, R.; Yan, K.S.; Fuchs, C.S.; Magness, S.T.; Smits, R.; Ogino, S.; et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature 2013, 493, 106–110. [Google Scholar] [CrossRef]
- Zhou, D.; Zhang, Y.; Wu, H.; Barry, E.; Yin, Y.; Lawrence, E.; Dawson, D.; Willis, J.E.; Markowitz, S.D.; Camargo, F.D.; et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc. Natl. Acad. Sci. USA 2011, 108, E1312–E1320. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Zhang, N.; Zheng, Y.; de Wilde, R.F.; Maitra, A.; Pan, D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010, 24, 2383–2388. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.; Vousden, K.H. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 2014, 25, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Oliner, J.D.; Pietenpol, J.A.; Thiagalingam, S.; Gyuris, J.; Kinzler, K.W.; Vogelstein, B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 1993, 362, 857–860. [Google Scholar] [CrossRef] [PubMed]
- Amaral, J.D.; Xavier, J.M.; Steer, C.J.; Rodrigues, C.M. The role of p53 in apoptosis. Discov. Med. 2010, 9, 145–152. [Google Scholar] [PubMed]
- Abukhdeir, A.M.; Park, B.H. P21 and p27: Roles in carcinogenesis and drug resistance. Expert Rev. Mol. Med. 2008, 10, e19. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.J.; Preisinger, A.C.; Jessup, J.M.; Paraskeva, C.; Markowitz, S.; Willson, J.K.; Hamilton, S.; Vogelstein, B. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 1990, 50, 7717–7722. [Google Scholar] [PubMed]
- Hainaut, P.; Hollstein, M. p53 and human cancer: The first ten thousand mutations. Adv. Cancer Res. 2000, 77, 81–137. [Google Scholar]
- Lopez, I.; Oliveira, L.P.; Tucci, P.; Alvarez-Valin, F.; Coudry, R.A.; Marin, M. Different mutation profiles associated to P53 accumulation in colorectal cancer. Gene 2012, 499, 81–87. [Google Scholar] [CrossRef]
- Li, X.L.; Zhou, J.; Chen, Z.R.; Chng, W.J. P53 mutations in colorectal cancer—Molecular pathogenesis and pharmacological reactivation. World J. Gastroenterol. 2015, 21, 84–93. [Google Scholar] [CrossRef]
- Ogino, S.; Nosho, K.; Shima, K.; Baba, Y.; Irahara, N.; Kirkner, G.J.; Hazra, A.; De Vivo, I.; Giovannucci, E.L.; Meyerhardt, J.A.; et al. p21 expression in colon cancer and modifying effects of patient age and body mass index on prognosis. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2513–2521. [Google Scholar] [CrossRef]
- Ogino, S.; Kawasaki, T.; Kirkner, G.J.; Ogawa, A.; Dorfman, I.; Loda, M.; Fuchs, C.S. Down-regulation of p21 (CDKN1A/CIP1) is inversely associated with microsatellite instability and CpG island methylator phenotype (CIMP) in colorectal cancer. J. Pathol. 2006, 210, 147–154. [Google Scholar] [CrossRef]
- Jacks, T.; Remington, L.; Williams, B.O.; Schmitt, E.M.; Halachmi, S.; Bronson, R.T.; Weinberg, R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Boil. CB 1994, 4, 1–7. [Google Scholar] [CrossRef]
- Lang, G.A.; Iwakuma, T.; Suh, Y.A.; Liu, G.; Rao, V.A.; Parant, J.M.; Valentin-Vega, Y.A.; Terzian, T.; Caldwell, L.C.; Strong, L.C.; et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004, 119, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Halberg, R.B.; Katzung, D.S.; Hoff, P.D.; Moser, A.R.; Cole, C.E.; Lubet, R.A.; Donehower, L.A.; Jacoby, R.F.; Dove, W.F. Tumorigenesis in the multiple intestinal neoplasia mouse: Redundancy of negative regulators and specificity of modifiers. Proc. Natl. Acad. Sci. USA 2000, 97, 3461–3466. [Google Scholar] [CrossRef] [PubMed]
- Funabashi, H.; Uchida, K.; Kado, S.; Matsuoka, Y.; Ohwaki, M. Establishment of a Tcrb and Trp53 genes deficient mouse strain as an animal model for spontaneous colorectal cancer. Exp. Anim. 2001, 50, 41–47. [Google Scholar] [CrossRef]
- Cooks, T.; Pateras, I.S.; Tarcic, O.; Solomon, H.; Schetter, A.J.; Wilder, S.; Lozano, G.; Pikarsky, E.; Forshew, T.; Rosenfeld, N.; et al. Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell 2013, 23, 634–646. [Google Scholar] [CrossRef]
- Chang, W.C.; Coudry, R.A.; Clapper, M.L.; Zhang, X.; Williams, K.L.; Spittle, C.S.; Li, T.; Cooper, H.S. Loss of p53 enhances the induction of colitis-associated neoplasia by dextran sulfate sodium. Carcinogenesis 2007, 28, 2375–2381. [Google Scholar] [CrossRef]
- Vyas, M.; Yang, X.; Zhang, X. Gastric Hamartomatous Polyps-Review and Update. Clin. Med. Insights Gastroenterol. 2016, 9, 3–10. [Google Scholar] [CrossRef]
- Karuman, P.; Gozani, O.; Odze, R.D.; Zhou, X.C.; Zhu, H.; Shaw, R.; Brien, T.P.; Bozzuto, C.D.; Ooi, D.; Cantley, L.C.; et al. The Peutz-Jegher gene product LKB1 is a mediator of p53-dependent cell death. Mol. Cell 2001, 7, 1307–1319. [Google Scholar] [CrossRef]
- Tiainen, M.; Vaahtomeri, K.; Ylikorkala, A.; Makela, T.P. Growth arrest by the LKB1 tumor suppressor: Induction of p21(WAF1/CIP1). Hum. Mol. Genet. 2002, 11, 1497–1504. [Google Scholar] [CrossRef] [PubMed]
- Tiainen, M.; Ylikorkala, A.; Makela, T.P. Growth suppression by Lkb1 is mediated by a G1 cell cycle arrest. Proc. Natl. Acad. Sci. USA 1999, 96, 9248–9251. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, H.; Nakau, M.; Ishikawa, T.O.; Seldin, M.F.; Oshima, M.; Taketo, M.M. Gastrointestinal hamartomatous polyposis in Lkb1 heterozygous knockout mice. Cancer Res. 2002, 62, 2261–2266. [Google Scholar] [PubMed]
- Wei, C.J.; Amos, C.I.; Stephens, L.C.; Campos, I.; Deng, J.M.; Behringer, R.R.; Rashid, A.; Frazier, M.L. Mutation of Lkb1 and p53 genes exert a cooperative effect on tumorigenesis. Cancer Res. 2005, 65, 11297–11303. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Zhang, P.; Harper, J.W.; Elledge, S.J.; Leder, P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 1995, 82, 675–684. [Google Scholar] [CrossRef]
- Brugarolas, J.; Chandrasekaran, C.; Gordon, J.I.; Beach, D.; Jacks, T.; Hannon, G.J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 1995, 377, 552–557. [Google Scholar] [CrossRef]
- Martin-Caballero, J.; Flores, J.M.; Garcia-Palencia, P.; Serrano, M. Tumor susceptibility of p21Waf1/Cip1-deficient mice. Cancer Res. 2001, 61, 6234–6238. [Google Scholar] [PubMed]
- Poole, A.J.; Heap, D.; Carroll, R.E.; Tyner, A.L. Tumor suppressor functions for the Cdk inhibitor p21 in the mouse colon. Oncogene 2004, 23, 8128–8134. [Google Scholar] [CrossRef][Green Version]
- Jackson, R.J.; Engelman, R.W.; Coppola, D.; Cantor, A.B.; Wharton, W.; Pledger, W.J. p21Cip1 nullizygosity increases tumor metastasis in irradiated mice. Cancer Res. 2003, 63, 3021–3025. [Google Scholar]
- Yang, W.C.; Mathew, J.; Velcich, A.; Edelmann, W.; Kucherlapati, R.; Lipkin, M.; Yang, K.; Augenlicht, L.H. Targeted inactivation of the p21WAF1/cip1 gene enhances Apc-initiated tumor formation and the tumor-promoting activity of a Western-style high-risk diet by altering cell maturation in the intestinal mucosal. Cancer Res. 2001, 61, 565–569. [Google Scholar]
- Zirbes, T.K.; Baldus, S.E.; Moenig, S.P.; Nolden, S.; Kunze, D.; Shafizadeh, S.T.; Schneider, P.M.; Thiele, J.; Hoelscher, A.H.; Dienes, H.P. Prognostic impact of p21/waf1/cip1 in colorectal cancer. Int. J. Cancer 2000, 89, 14–18. [Google Scholar] [CrossRef]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef]
- Tuveson, D.A.; Shaw, A.T.; Willis, N.A.; Silver, D.P.; Jackson, E.L.; Chang, S.; Mercer, K.L.; Grochow, R.; Hock, H.; Crowley, D.; et al. Endogenous oncogenic K-rasG12D stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell 2004, 5, 375–387. [Google Scholar] [CrossRef]
- Johnson, L.; Mercer, K.; Greenbaum, D.; Bronson, R.T.; Crowley, D.; Tuveson, D.A.; Jacks, T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 2001, 410, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, N.; Minamoto, T.; Ochiai, A.; Onda, M.; Esumi, H. Frequent and characteristic K-ras activation and absence of p53 protein accumulation in aberrant crypt foci of the colon. Gastroenterology 1995, 108, 434–440. [Google Scholar] [CrossRef]
- Roerink, S.F.; Sasaki, N.; Lee-Six, H.; Young, M.D.; Alexandrov, L.B.; Behjati, S.; Mitchell, T.J.; Grossmann, S.; Lightfoot, H.; Egan, D.A.; et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 2018, 556, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Kavuri, S.M.; Jain, N.; Galimi, F.; Cottino, F.; Leto, S.M.; Migliardi, G.; Searleman, A.C.; Shen, W.; Monsey, J.; Trusolino, L.; et al. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov. 2015, 5, 832–841. [Google Scholar] [CrossRef]
- Dougherty, U.; Cerasi, D.; Taylor, I.; Kocherginsky, M.; Tekin, U.; Badal, S.; Aluri, L.; Sehdev, A.; Cerda, S.; Mustafi, R.; et al. Epidermal growth factor receptor is required for colonic tumor promotion by dietary fat in the azoxymethane/dextran sulfate sodium model: Roles of transforming growth factor-α and PTGS2. Clin. Cancer Res. 2009, 15, 6780–6789. [Google Scholar] [CrossRef] [PubMed]
- Dube, P.E.; Yan, F.; Punit, S.; Girish, N.; McElroy, S.J.; Washington, M.K.; Polk, D.B. Epidermal growth factor receptor inhibits colitis-associated cancer in mice. J. Clin. Investig. 2012, 122, 2780–2792. [Google Scholar] [CrossRef]
- Roberts, R.B.; Min, L.; Washington, M.K.; Olsen, S.J.; Settle, S.H.; Coffey, R.J.; Threadgill, D.W. Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 1521–1526. [Google Scholar] [CrossRef]
- Nagahara, H.; Mimori, K.; Ohta, M.; Utsunomiya, T.; Inoue, H.; Barnard, G.F.; Ohira, M.; Hirakawa, K.; Mori, M. Somatic mutations of epidermal growth factor receptor in colorectal carcinoma. Clin. Cancer Res. 2005, 11, 1368–1371. [Google Scholar] [CrossRef] [PubMed]
- Moroni, M.; Veronese, S.; Benvenuti, S.; Marrapese, G.; Sartore-Bianchi, A.; Di Nicolantonio, F.; Gambacorta, M.; Siena, S.; Bardelli, A. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: A cohort study. Lancet Oncol. 2005, 6, 279–286. [Google Scholar] [CrossRef]
- Brink, M.; de Goeij, A.F.; Weijenberg, M.P.; Roemen, G.M.; Lentjes, M.H.; Pachen, M.M.; Smits, K.M.; de Bruine, A.P.; Goldbohm, R.A.; van den Brandt, P.A. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis 2003, 24, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Haigis, K.M.; Kendall, K.R.; Wang, Y.; Cheung, A.; Haigis, M.C.; Glickman, J.N.; Niwa-Kawakita, M.; Sweet-Cordero, A.; Sebolt-Leopold, J.; Shannon, K.M.; et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 2008, 40, 600–608. [Google Scholar] [CrossRef]
- Guerra, C.; Mijimolle, N.; Dhawahir, A.; Dubus, P.; Barradas, M.; Serrano, M.; Campuzano, V.; Barbacid, M. Tumor induction by an endogenous K-ras oncogene is highly dependent on cellular context. Cancer Cell 2003, 4, 111–120. [Google Scholar] [CrossRef]
- Ireland, H.; Kemp, R.; Houghton, C.; Howard, L.; Clarke, A.R.; Sansom, O.J.; Winton, D.J. Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: Effect of loss of β-catenin. Gastroenterology 2004, 126, 1236–1246. [Google Scholar] [CrossRef]
- Luo, F.; Brooks, D.G.; Ye, H.; Hamoudi, R.; Poulogiannis, G.; Patek, C.E.; Winton, D.J.; Arends, M.J. Mutated K-rasAsp12 promotes tumourigenesis in ApcMin mice more in the large than the small intestines, with synergistic effects between K-ras and Wnt pathways. Int. J. Exp. Pathol. 2009, 90, 558–574. [Google Scholar] [CrossRef]
- Hung, K.E.; Maricevich, M.A.; Richard, L.G.; Chen, W.Y.; Richardson, M.P.; Kunin, A.; Bronson, R.T.; Mahmood, U.; Kucherlapati, R. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc. Natl. Acad. Sci. USA 2010, 107, 1565–1570. [Google Scholar] [CrossRef]
- Poulin, E.J.; Bera, A.K.; Lu, J.; Lin, Y.J.; Strasser, S.D.; Paulo, J.A.; Huang, T.Q.; Morales, C.; Yan, W.; Cook, J.; et al. Tissue-Specific Oncogenic Activity of KRASA146T. Cancer Discov. 2019. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Rajagopalan, H.; Bardelli, A.; Lengauer, C.; Kinzler, K.W.; Vogelstein, B.; Velculescu, V.E. Tumorigenesis—RAF/RAS oncogenes and mismatch-repair status. Nature 2002, 418, 934. [Google Scholar] [CrossRef] [PubMed]
- Dankort, D.; Filenova, E.; Collado, M.; Serrano, M.; Jones, K.; McMahon, M. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007, 21, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Rad, R.; Cadinanos, J.; Rad, L.; Varela, I.; Strong, A.; Kriegl, L.; Constantino-Casas, F.; Eser, S.; Hieber, M.; Seidler, B.; et al. A Genetic Progression Model of BrafV600E-Induced Intestinal Tumorigenesis Reveals Targets for Therapeutic Intervention. Cancer Cell 2013, 24, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Kang, B.; Petkovich, D.A.; Bhandari, Y.R.; In, J.; Stein-O’Brien, G.; Kong, X.; Xie, W.; Zachos, N.; Maegawa, S.; et al. Aging-like Spontaneous Epigenetic Silencing Facilitates Wnt Activation, Stemness, and BrafV600)-Induced Tumorigenesis. Cancer Cell 2019, 35, 315–328.e6. [Google Scholar] [CrossRef] [PubMed]
- Biemer-Huttmann, A.E.; Walsh, M.D.; McGuckin, M.A.; Simms, L.A.; Young, J.; Leggett, B.A.; Jass, J.R. Mucin core protein expression in colorectal cancers with high levels of microsatellite instability indicates a novel pathway of morphogenesis. Clin. Cancer Res. 2000, 6, 1909–1916. [Google Scholar]
- Walsh, M.D.; Clendenning, M.; Williamson, E.; Pearson, S.A.; Walters, R.J.; Nagler, B.; Packenas, D.; Win, A.K.; Hopper, J.L.; Jenkins, M.A.; et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Mod. Pathol. 2013, 26, 1642–1656. [Google Scholar] [CrossRef]
- Winterford, C.M.; Walsh, M.D.; Leggett, B.A.; Jass, J.R. Ultrastructural localization of epithelial mucin core proteins in colorectal tissues. J. Histochem. Cytochem. 1999, 47, 1063–1074. [Google Scholar] [CrossRef] [PubMed]
- Velcich, A.; Yang, W.; Heyer, J.; Fragale, A.; Nicholas, C.; Viani, S.; Kucherlapati, R.; Lipkin, M.; Yang, K.; Augenlicht, L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 2002, 295, 1726–1729. [Google Scholar] [CrossRef] [PubMed]
- Stambolic, V.; Suzuki, A.; de la Pompa, J.L.; Brothers, G.M.; Mirtsos, C.; Sasaki, T.; Ruland, J.; Penninger, J.M.; Siderovski, D.P.; Mak, T.W. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 1998, 95, 29–39. [Google Scholar] [CrossRef]
- Velho, S.; Oliveira, C.; Ferreira, A.; Ferreira, A.C.; Suriano, G.; Schwartz, S., Jr.; Duval, A.; Carneiro, F.; Machado, J.C.; Hamelin, R.; et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur. J. Cancer 2005, 41, 1649–1654. [Google Scholar] [CrossRef] [PubMed]
- Samuels, Y.; Waldman, T. Oncogenic mutations of PIK3CA in human cancers. Curr. Top. Microbiol. Immunol. 2010, 347, 21–41. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, C.B.; Phillips, W.A. Mouse Models for Exploring the Biological Consequences and Clinical Significance of PIK3CA Mutations. Biomolecules 2019, 9, 158. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Arnold, C.N.; Niedzwiecki, D.; Carethers, J.M.; Dowell, J.M.; Wasserman, L.; Compton, C.; Mayer, R.J.; Bertagnolli, M.M.; Boland, C.R. Frequent inactivation of PTEN by promoter hypermethylation in microsatellite instability-high sporadic colorectal cancers. Cancer Res. 2004, 64, 3014–3021. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.; Danielsen, S.A.; Ahlquist, T.; Merok, M.A.; Agesen, T.H.; Vatn, M.H.; Mala, T.; Sjo, O.H.; Bakka, A.; Moberg, I.; et al. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS ONE 2010, 5, e13978. [Google Scholar] [CrossRef] [PubMed]
- Carpten, J.D.; Faber, A.L.; Horn, C.; Donoho, G.P.; Briggs, S.L.; Robbins, C.M.; Hostetter, G.; Boguslawski, S.; Moses, T.Y.; Savage, S.; et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007, 448, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Bleeker, F.E.; Felicioni, L.; Buttitta, F.; Lamba, S.; Cardone, L.; Rodolfo, M.; Scarpa, A.; Leenstra, S.; Frattini, M.; Barbareschi, M.; et al. AKT1E17K in human solid tumours. Oncogene 2008, 27, 5648–5650. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Pasche, B. TGF-β signaling alterations and susceptibility to colorectal cancer. Hum. Mol. Genet. 2007, 16, R14–R20. [Google Scholar] [CrossRef]
- Meulmeester, E.; Ten Dijke, P. The dynamic roles of TGF-β in cancer. J. Pathol. 2011, 223, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Lampropoulos, P.; Zizi-Sermpetzoglou, A.; Rizos, S.; Kostakis, A.; Nikiteas, N.; Papavassiliou, A.G. TGF-β signalling in colon carcinogenesis. Cancer Lett. 2012, 314, 1–7. [Google Scholar] [CrossRef]
- Xie, W.; Rimm, D.L.; Lin, Y.; Shih, W.J.; Reiss, M. Loss of Smad signaling in human colorectal cancer is associated with advanced disease and poor prognosis. Cancer J. 2003, 9, 302–312. [Google Scholar] [CrossRef]
- Fleming, N.I.; Jorissen, R.N.; Mouradov, D.; Christie, M.; Sakthianandeswaren, A.; Palmieri, M.; Day, F.; Li, S.; Tsui, C.; Lipton, L.; et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013, 73, 725–735. [Google Scholar] [CrossRef]
- Miyaki, M.; Iijima, T.; Konishi, M.; Sakai, K.; Ishii, A.; Yasuno, M.; Hishima, T.; Koike, M.; Shitara, N.; Iwama, T.; et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene 1999, 18, 3098–3103. [Google Scholar] [CrossRef]
- Takagi, Y.; Kohmura, H.; Futamura, M.; Kida, H.; Tanemura, H.; Shimokawa, K.; Saji, S. Somatic alterations of the DPC4 gene in human colorectal cancers in vivo. Gastroenterology 1996, 111, 1369–1372. [Google Scholar] [CrossRef]
- Howe, J.R.; Roth, S.; Ringold, J.C.; Summers, R.W.; Jarvinen, H.J.; Sistonen, P.; Tomlinson, I.P.; Houlston, R.S.; Bevan, S.; Mitros, F.A.; et al. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 1998, 280, 1086–1088. [Google Scholar] [CrossRef]
- Calon, A.; Lonardo, E.; Berenguer-Llergo, A.; Espinet, E.; Hernando-Momblona, X.; Iglesias, M.; Sevillano, M.; Palomo-Ponce, S.; Tauriello, D.V.; Byrom, D.; et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet. 2015, 47, 320–329. [Google Scholar] [CrossRef]
- Nakagawa, H.; Liyanarachchi, S.; Davuluri, R.V.; Auer, H.; Martin, E.W., Jr.; de la Chapelle, A.; Frankel, W.L. Role of cancer-associated stromal fibroblasts in metastatic colon cancer to the liver and their expression profiles. Oncogene 2004, 23, 7366–7377. [Google Scholar] [CrossRef]
- Calon, A.; Espinet, E.; Palomo-Ponce, S.; Tauriello, D.V.; Iglesias, M.; Cespedes, M.V.; Sevillano, M.; Nadal, C.; Jung, P.; Zhang, X.H.; et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 2012, 22, 571–584. [Google Scholar] [CrossRef]
- Kulkarni, A.B.; Huh, C.G.; Becker, D.; Geiser, A.; Lyght, M.; Flanders, K.C.; Roberts, A.B.; Sporn, M.B.; Ward, J.M.; Karlsson, S. Transforming Growth Factor-β-1 Null Mutation in Mice Causes Excessive Inflammatory Response and Early Death. Proc. Natl. Acad. Sci. USA 1993, 90, 770–774. [Google Scholar] [CrossRef]
- Shull, M.M.; Ormsby, I.; Kier, A.B.; Pawlowski, S.; Diebold, R.J.; Yin, M.Y.; Allen, R.; Sidman, C.; Proetzel, G.; Calvin, D.; et al. Targeted Disruption of the Mouse Transforming Growth Factor-β-1 Gene Results in Multifocal Inflammatory Disease. Nature 1992, 359, 693–699. [Google Scholar] [CrossRef]
- Engle, S.J.; Hoying, J.B.; Boivin, G.P.; Ormsby, I.; Gartside, P.S.; Doetschman, T. Transforming growth factor β1 suppresses nonmetastatic colon cancer at an early stage of tumorigenesis. Cancer Res. 1999, 59, 3379–3386. [Google Scholar]
- Takaku, K.; Miyoshi, H.; Matsunaga, A.; Oshima, M.; Sasaki, N.; Taketo, M.M. Gastric and duodenal polyps in Smad4 (Dpc4) knockout mice. Cancer Res. 1999, 59, 6113–6117. [Google Scholar]
- Zhu, Y.; Richardson, J.A.; Parada, L.F.; Graff, J.M. Smad3 mutant mice develop metastatic colorectal cancer. Cell 1998, 94, 703–714. [Google Scholar] [CrossRef]
- Kaiser, S.; Park, Y.K.; Franklin, J.L.; Halberg, R.B.; Yu, M.; Jessen, W.J.; Freudenberg, J.; Chen, X.; Haigis, K.; Jegga, A.G.; et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007, 8, R131. [Google Scholar] [CrossRef]
- Zeng, Q.; Phukan, S.; Xu, Y.; Sadim, M.; Rosman, D.S.; Pennison, M.; Liao, J.; Yang, G.Y.; Huang, C.C.; Valle, L.; et al. Tgfbr1 haploinsufficiency is a potent modifier of colorectal cancer development. Cancer Res. 2009, 69, 678–686. [Google Scholar] [CrossRef]
- Alberici, P.; Jagmohan-Changur, S.; De Pater, E.; Van Der Valk, M.; Smits, R.; Hohenstein, P.; Fodde, R. Smad4 haploinsufficiency in mouse models for intestinal cancer. Oncogene 2006, 25, 1841–1851. [Google Scholar] [CrossRef]
- Sodir, N.M.; Chen, X.; Park, R.; Nickel, A.E.; Conti, P.S.; Moats, R.; Bading, J.R.; Shibata, D.; Laird, P.W. Smad3 deficiency promotes tumorigenesis in the distal colon of ApcMin/+ mice. Cancer Res. 2006, 66, 8430–8438. [Google Scholar] [CrossRef]
- Aguilera, O.; Fraga, M.F.; Ballestar, E.; Paz, M.F.; Herranz, M.; Espada, J.; Garcia, J.M.; Munoz, A.; Esteller, M.; Gonzalez-Sancho, J.M. Epigenetic inactivation of the Wnt antagonist DICKKOPF-1 (DKK-1) gene in human colorectal cancer. Oncogene 2006, 25, 4116–4121. [Google Scholar] [CrossRef]
- Takaku, K.; Oshima, M.; Miyoshi, H.; Matsui, M.; Seldin, M.F.; Taketo, M.M. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell 1998, 92, 645–656. [Google Scholar] [CrossRef]
- Hamamoto, T.; Beppu, H.; Okada, H.; Kawabata, M.; Kitamura, T.; Miyazono, K.; Kato, M. Compound disruption of Smad2 accelerates malignant progression of intestinal tumors in Apc knockout mice. Cancer Res. 2002, 62, 5955–5961. [Google Scholar]
- Kunkel, T.A. Evolving views of DNA replication (in)fidelity. Cold Spring Harb. Symp. Quant. Biol. 2009, 74, 91–101. [Google Scholar] [CrossRef]
- O’Sullivan, J.N.; Bronner, M.P.; Brentnall, T.A.; Finley, J.C.; Shen, W.T.; Emerson, S.; Emond, M.J.; Gollahon, K.A.; Moskovitz, A.H.; Crispin, D.A.; et al. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat. Genet. 2002, 32, 280–284. [Google Scholar] [CrossRef]
- Poulogiannis, G.; Frayling, I.M.; Arends, M.J. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology 2010, 56, 167–179. [Google Scholar] [CrossRef]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087. [Google Scholar] [CrossRef]
- Edelmann, W.; Yang, K.; Kuraguchi, M.; Heyer, J.; Lia, M.; Kneitz, B.; Fan, K.; Brown, A.M.; Lipkin, M.; Kucherlapati, R. Tumorigenesis in Mlh1 and Mlh1/Apc1638N mutant mice. Cancer Res. 1999, 59, 1301–1307. [Google Scholar]
- de Wind, N.; Dekker, M.; Claij, N.; Jansen, L.; van Klink, Y.; Radman, M.; Riggins, G.; van der Valk, M.; van’t Wout, K.; te Riele, H. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat. Genet. 1999, 23, 359–362. [Google Scholar] [CrossRef]
- Prolla, T.A.; Baker, S.M.; Harris, A.C.; Tsao, J.L.; Yao, X.; Bronner, C.E.; Zheng, B.; Gordon, M.; Reneker, J.; Arnheim, N.; et al. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat. Genet. 1998, 18, 276–279. [Google Scholar] [CrossRef]
- Chen, P.C.; Dudley, S.; Hagen, W.; Dizon, D.; Paxton, L.; Reichow, D.; Yoon, S.R.; Yang, K.; Arnheim, N.; Liskay, R.M.; et al. Contributions by MutL homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005, 65, 8662–8670. [Google Scholar] [CrossRef]
- Lakso, M.; Pichel, J.G.; Gorman, J.R.; Sauer, B.; Okamoto, Y.; Lee, E.; Alt, F.W.; Westphal, H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc. Natl. Acad. Sci. USA 1996, 93, 5860–5865. [Google Scholar] [CrossRef]
- Kucherlapati, M.H.; Lee, K.; Nguyen, A.A.; Clark, A.B.; Hou, H., Jr.; Rosulek, A.; Li, H.; Yang, K.; Fan, K.; Lipkin, M.; et al. An Msh2 conditional knockout mouse for studying intestinal cancer and testing anticancer agents. Gastroenterology 2010, 138, 993–1002. [Google Scholar] [CrossRef]
- Reitmair, A.H.; Cai, J.C.; Bjerknes, M.; Redston, M.; Cheng, H.; Pind, M.T.; Hay, K.; Mitri, A.; Bapat, B.V.; Mak, T.W.; et al. MSH2 deficiency contributes to accelerated APC-mediated intestinal tumorigenesis. Cancer Res. 1996, 56, 2922–2926. [Google Scholar]
- Luo, F.; Brooks, D.G.; Ye, H.; Hamoudi, R.; Poulogiannis, G.; Patek, C.E.; Winton, D.J.; Arends, M.J. Conditional expression of mutated K-ras accelerates intestinal tumorigenesis in Msh2-deficient mice. Oncogene 2007, 26, 4415–4427. [Google Scholar] [CrossRef][Green Version]
- Kuraguchi, M.; Edelmann, W.; Yang, K.; Lipkin, M.; Kucherlapati, R.; Brown, A.M. Tumor-associated Apc mutations in Mlh1−/−Apc1638N mice reveal a mutational signature of Mlh1 deficiency. Oncogene 2000, 19, 5755–5763. [Google Scholar] [CrossRef]
- Kuraguchi, M.; Yang, K.; Wong, E.; Avdievich, E.; Fan, K.; Kolodner, R.D.; Lipkin, M.; Brown, A.M.; Kucherlapati, R.; Edelmann, W. The distinct spectra of tumor-associated Apc mutations in mismatch repair-deficient Apc1638N mice define the roles of MSH3 and MSH6 in DNA repair and intestinal tumorigenesis. Cancer Res. 2001, 61, 7934–7942. [Google Scholar]
- Takeda, H.; Rust, A.G.; Ward, J.M.; Yew, C.C.; Jenkins, N.A.; Copeland, N.G. Sleeping Beauty transposon mutagenesis identifies genes that cooperate with mutant Smad4 in gastric cancer development. Proc. Natl. Acad. Sci. USA 2016, 113, E2057–E2065. [Google Scholar] [CrossRef]
- Starr, T.K.; Allaei, R.; Silverstein, K.A.; Staggs, R.A.; Sarver, A.L.; Bergemann, T.L.; Gupta, M.; O’Sullivan, M.G.; Matise, I.; Dupuy, A.J.; et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 2009, 323, 1747–1750. [Google Scholar] [CrossRef]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef]
| CMS1 | CMS2 | CMS3 | CMS4 |
|---|---|---|---|
| MSI Immune | Canonical | Metabolic | Mesenchymal |
| 14% | 37% | 13% | 23% |
| MSI high | MSI negative | Mixed MSI status | MSI low |
| CIMP high | CIMP negative | CIMP low | CIMP negative |
| SCNA low | SCNA high | SCNA moderate | SCNA high |
| BRAF mutations | TP53 mutations | KRAS mutations | TP53 mutations |
| epithelial signature | epithelial signature | mesenchymal signature | |
| Wnt and Myc target genes upregulation | enhanced metabolism | EMT activation and matrix remodeling | |
| immune infiltration | stromal infiltrationTGFβ signaling activation | ||
| worse survival after relaps | worse relaps-free and overall survival |
| Generated Allele or Strain Name | Advantages | Disadvantages | Reference | |
|---|---|---|---|---|
| CMS1 | BrafV600E | crypt hyperproliferation, high incidence of tumors, mucinous phenotype | not all the animals develop tumors | [29] |
| Mlh1-/- | 100% tumor development within 4 months | tumors develop in many other tissues, short lifespan | [30] | |
| Msh2loxP/loxP Villin-Cre | 90 % of mice developed adenomas and adenocarcinomas, tumor formation is restricted to the intestine | mosaic recombination in the tissue | [31] | |
| CMS2 | ApcMin | multiple intestinal tumors, early tumor development, recapitulates human FAP syndrome | relatively rare tumorigenesis in the colon | [32] |
| ApccKO/cKO Lgr5-EGFP-IRES-CreERT2 | inducible tumor initiation, all tumors develop during the same (and defined) time period | tamoxifen dose-dependent variability of the phenotype | [33] | |
| Catnb+/lox(ex3) Krt1-19-Cre | early tumor development, large amount of tumors, microadenomas in the colon | short lifespan due to extensive tumorigenesis | [34] | |
| ApcMin p53-/- | increased number and invasivity of intestinal tumors | tumors develop in many other tissues, short lifespan | [35] | |
| CMS3 | Apc2lox14/+ LSL-KrasG12D Rapbp1-Cre | combination of Apc and Kras mutations, adenomas in the colon | crossbreeding | [36] |
| ApcMin K-rasAsp12 Ah-Cre | increased number of intestinal tumors with higher effect in the colon | crossbreeding | [37] | |
| CMS4 | N/A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stastna, M.; Janeckova, L.; Hrckulak, D.; Kriz, V.; Korinek, V. Human Colorectal Cancer from the Perspective of Mouse Models. Genes 2019, 10, 788. https://doi.org/10.3390/genes10100788
Stastna M, Janeckova L, Hrckulak D, Kriz V, Korinek V. Human Colorectal Cancer from the Perspective of Mouse Models. Genes. 2019; 10(10):788. https://doi.org/10.3390/genes10100788
Chicago/Turabian StyleStastna, Monika, Lucie Janeckova, Dusan Hrckulak, Vitezslav Kriz, and Vladimir Korinek. 2019. "Human Colorectal Cancer from the Perspective of Mouse Models" Genes 10, no. 10: 788. https://doi.org/10.3390/genes10100788
APA StyleStastna, M., Janeckova, L., Hrckulak, D., Kriz, V., & Korinek, V. (2019). Human Colorectal Cancer from the Perspective of Mouse Models. Genes, 10(10), 788. https://doi.org/10.3390/genes10100788





