Genome-Wide Identification, Characterization and Expression Patterns of the Pectin Methylesterase Inhibitor Genes in Sorghum bicolor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and PME Activity Analysis
2.2. Identification of SbPMEI Members and Sequence Characterization
2.3. Phylogenetic Analysis, Chromosome Location and Tandem Gene Duplication
2.4. Gene Structure, GO Analysis and Protein–Protein Interaction Analysis
2.5. Cis-Acting Elements Analysis
2.6. Expression Patterns of Sorghum PMEI Genes
2.7. Abiotic Stress Treatments, RNA Isolation and qRT-PCR Analysis
3. Results
3.1. PME Activity Analysis of Six Tissues From Sorghum Bicolor
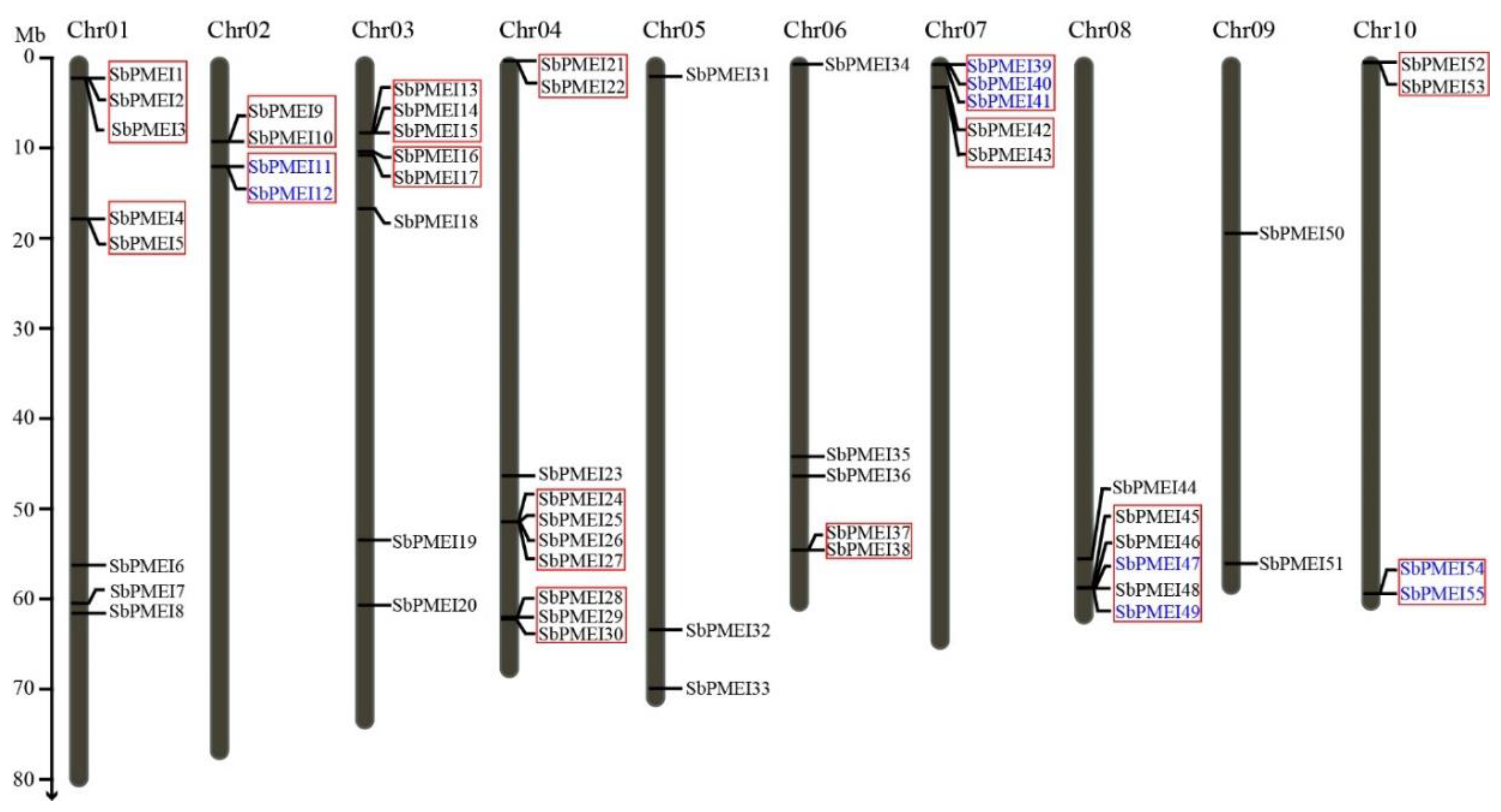
3.2. Identification, Chromosome Location and Tandem Gene Duplicates of PMEI Genes in Sorghum
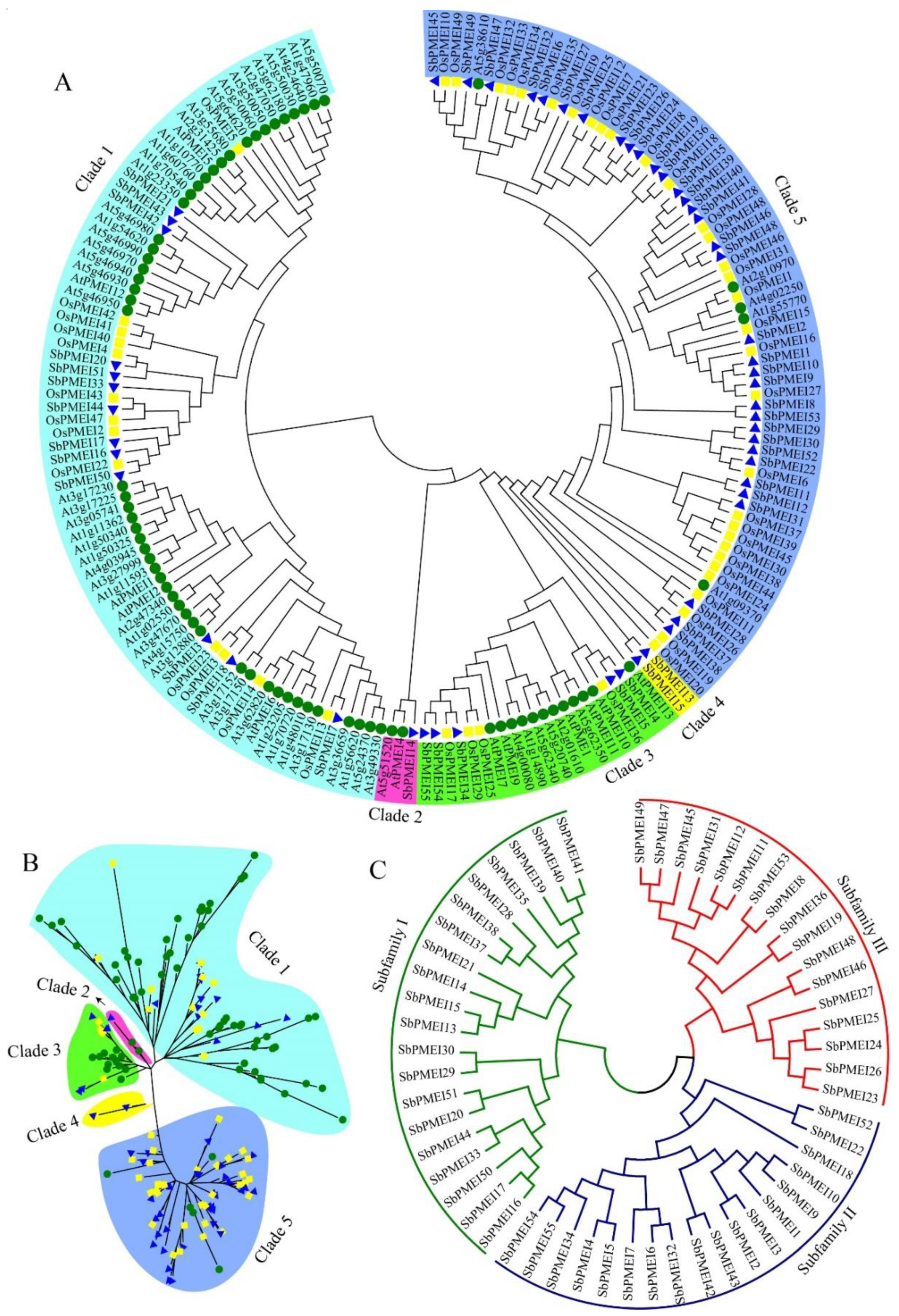
3.3. Phylogenetic Analysis and Sequence Characterizations
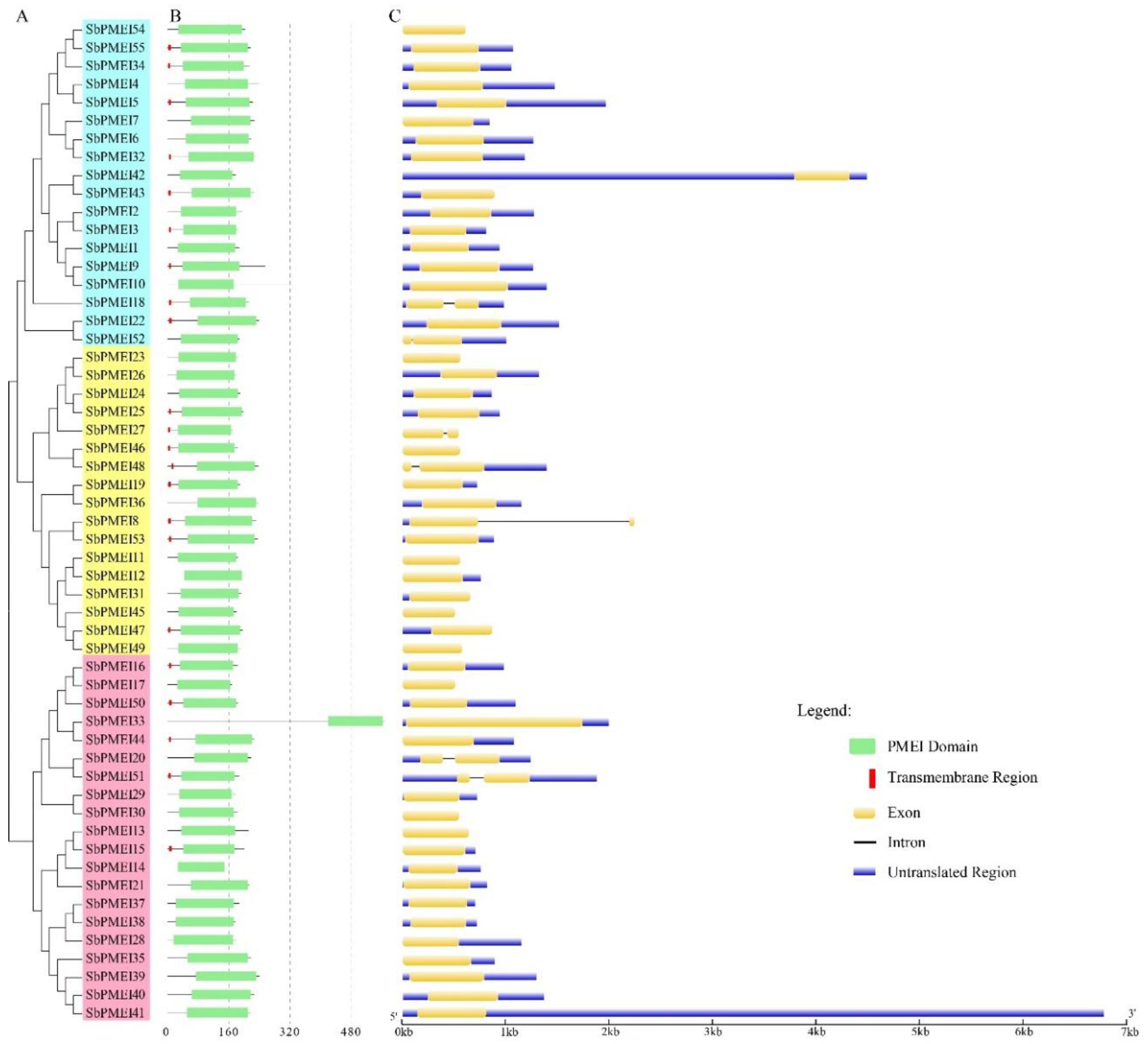
3.4. Gene Structure Analysis of SbPMEIs
3.5. GO Analysis and Protein–Protein Interaction Prediction
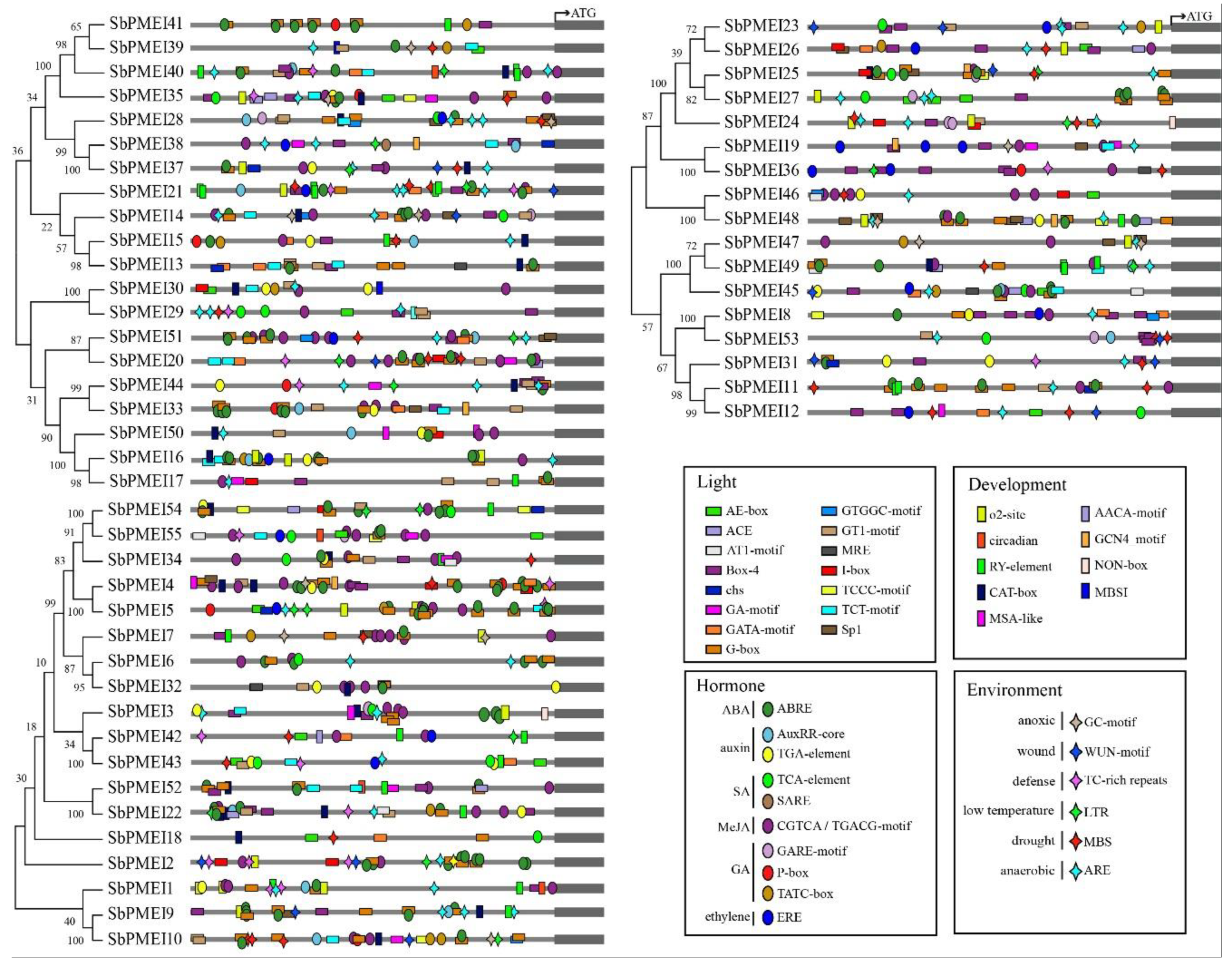
3.6. Cis-Acting Elements Detection in Promoter Regions of SbPMEI Genes
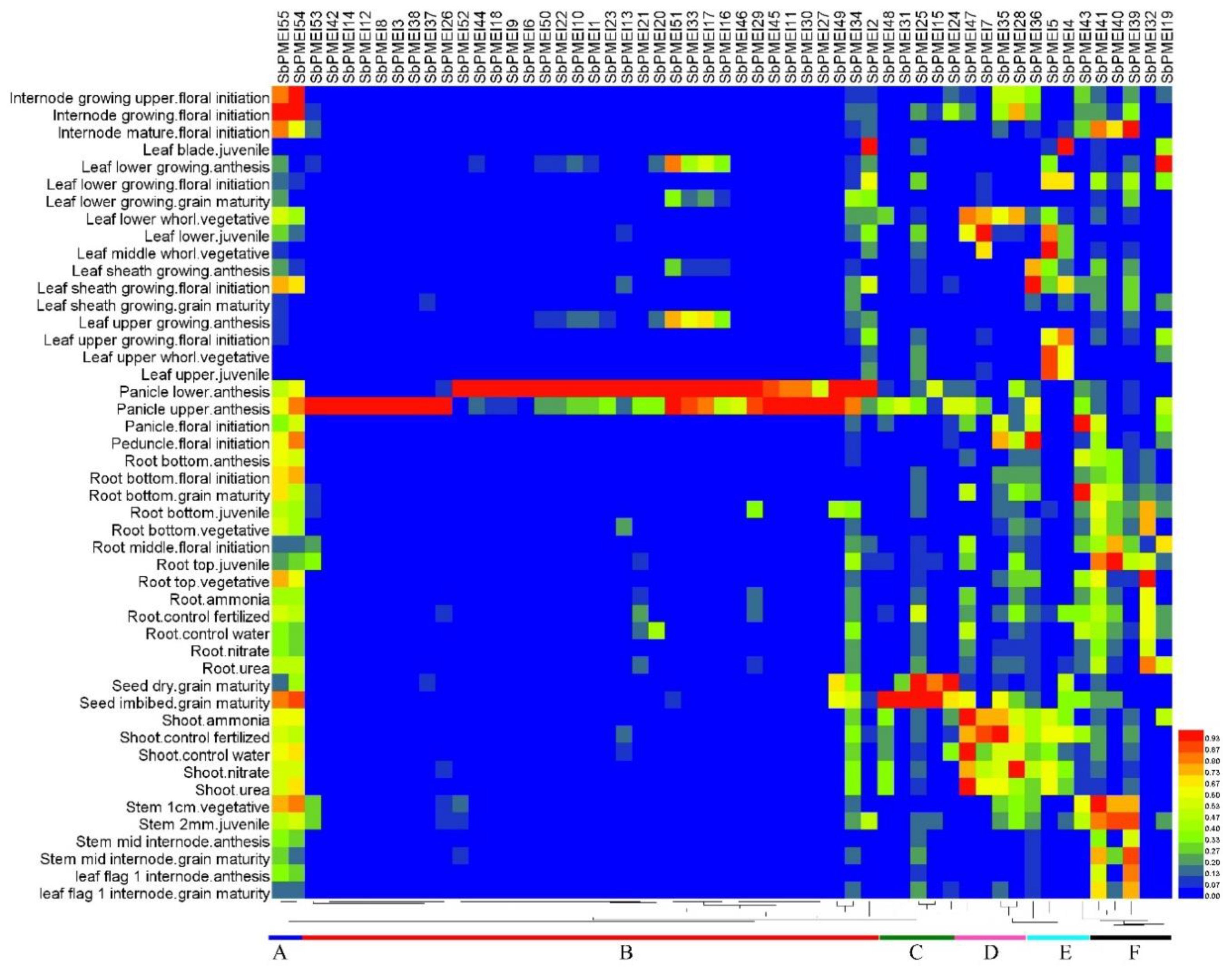
3.7. Expression Patterns of PMEI Genes in Growth Lifecycle
3.8. Expression Patters of SbPMEIs under Exogenous Hormones and Abiotic Stresses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhao, Y.; Man, Y.; Wen, J.; Guo, Y.; Lin, J. Advances in Imaging Plant Cell Walls. Trends Plant Sci. 2019, 24, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C.; Bauer, S.; Brininstool, G.M.; Facette, M.R.; Hamann, T.; Milne, J.S.; Osborne, E.; Paredez, A.R.; Persson, S.; Raab, T.K.; et al. Toward a systems approach to understanding plant-cell walls. Science 2004, 306, 2206–2211. [Google Scholar] [CrossRef] [PubMed]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef] [PubMed]
- An, S.H.; Sohn, K.H.; Choi, H.W.; Hwang, I.S.; Lee, S.C.; Hwang, B.K. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 2008, 228, 61–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Feng, J.; Wu, J.; Wang, X. BoPMEI1, a pollen-specific pectin methylesterase inhibitor, has an essential role in pollen tube growth. Planta 2010, 231, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.; Williams, J.H. Evolutionary origins of pectin methylesterase genes associated with novel aspects of angiosperm pollen tube walls. Biochem. Biophys. Res. Commun. 2017, 487, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Verma, C.; Singh, R.K.; Mishra, S. Biochemical Characterization of Pectin Methylesterase from Musa acuminata Referring to Delayed Ripening. IOSR J. Pharm. Biol. Sci. 2014, 9, 42–47. [Google Scholar] [CrossRef]
- Bosch, M.; Cheung, A.Y.; Hepler, P.K. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 2005, 138, 1334–1346. [Google Scholar] [CrossRef]
- Al-Qsous, S.; Carpentier, E.; Klein-Eude, D.; Burel, C.; Mareck, A.; Dauchel, H.; Gomord, V.; Balange, A.P. Identification and isolation of a pectin methylesterase isoform that could be involved in flax cell wall stiffening. Planta 2004, 219, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Wormit, A.; Usadel, B. The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs). Int. J. Mol. Sci. 2018, 19, 2878. [Google Scholar] [CrossRef]
- Ciro, B.; Domenico, C.; Alfonso, G.; Lucio, Q.; Luigi, S. A glycoprotein inhibitor of pectin methylesterase in kiwi fruit (Actinidia chinensis). Eur. J. Biochem. 1990, 193, 183–187. [Google Scholar] [CrossRef]
- Shi, D.; Ren, A.; Tang, X.; Qi, G.; Xu, Z.; Chai, G.; Hu, R.; Zhou, G.; Kong, Y. MYB52 Negatively Regulates Pectin Demethylesterification in Seed Coat Mucilage. Plant Physiol. 2018, 176, 2737–2749. [Google Scholar] [CrossRef] [Green Version]
- Wolf, S.; Grsic-Rausch, S.; Rausch, T.; Greiner, S. Identification of pollen-expressed pectin methylesterase inhibitors in Arabidopsis. FEBS Lett. 2003, 555, 551–555. [Google Scholar] [CrossRef]
- Saez-Aguayo, S.; Ralet, M.C.; Berger, A.; Botran, L.; Ropartz, D.; Marion-Poll, A.; North, H.M. PECTIN METHYLESTERASE INHIBITOR 6 promotes Arabidopsis mucilage release by limiting methylesterification of homogalacturonan in seed coat epidermal cells. Plant Cell. 2013, 25, 308–323. [Google Scholar] [CrossRef] [PubMed]
- Sénéchal, F.; L’Enfant, M.; Domon, J.M.; Rosiau, E.; Crepeau, M.J.; Surcouf, O.; Esquivel-Rodriguez, J.; Marcelo, P.; Mareck, A.; Guerineau, F.; et al. Tuning of Pectin Methylesterification: PECTIN METHYLESTERASE INHIBITOR 7 modulates the processive activity of co-expressed PECTIN METHYLESTERASE 3 in a pH-dependent manner. J. Biol. Chem. 2015, 290, 23320–23335. [Google Scholar] [CrossRef] [PubMed]
- Di Matteo, A.; Giovane, A.; Raiola, A.; Camardella, L.; Bonivento, D.; De Lorenzo, G.; Cervone, F.; Bellincampi, D.; Tsernoglou, D. Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. Plant Cell 2005, 17, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Sénéchal, F.; Mareck, A.; Marcelo, P.; Lerouge, P.; Pelloux, J. Arabidopsis PME17 Activity can be Controlled by Pectin Methylesterase Inhibitor 4. Plant Signal Behav. 2015, 10, e983351. [Google Scholar] [CrossRef]
- Pelletier, S.; Van Orden, J.; Wolf, S.; Vissenberg, K.; Delacourt, J.; Ndong, Y.A.; Pelloux, J.; Bischoff, V.; Urbain, A.; Mouille, G.; et al. A role for pectin de-methylesterification in a developmentally regulated growth acceleration in dark-grown Arabidopsis hypocotyls. New Phytol. 2010, 188, 726–739. [Google Scholar] [CrossRef]
- Müller, K.; Levesque-Tremblay, G.; Bartels, S.; Weitbrecht, K.; Wormit, A.; Usadel, B.; Haughn, G.; Kermode, A.R. Demethylesterification of Cell Wall Pectins in Arabidopsis Plays a Role in Seed Germination. Plant Physiol. 2013, 161, 305–316. [Google Scholar] [CrossRef]
- Müller, K.; Levesque-Tremblay, G.; Fernandes, A.; Wormit, A.; Bartels, S.; Usadel, B.; Kermode, A. Overexpression of a pectin methylesterase inhibitor in Arabidopsis thaliana leads to altered growth morphology of the stem and defective organ separation. Plant Signal Behav. 2013, 8, e26464. [Google Scholar] [CrossRef]
- Pinzón-Latorre, D.; Deyholos, M.K. Pectinmethylesterases (PME) and Pectinmethylesterase Inhibitors (PMEI) Enriched during Phloem Fiber Development in Flax (Linum usitatissimum). PLoS ONE 2014, 8, e105386. [Google Scholar] [CrossRef] [PubMed]
- Reca, I.B.; Lionetti, V.; Camardella, L.; D’Avino, R.; Giardina, T.; Cervone, F.; Bellincampi, D. A functional pectin methylesterase inhibitor protein (SolyPMEI) is expressed during tomato fruit ripening and interacts with PME-1. Plant Mol. Biol. 2012, 79, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Raiola, A.; Mattei, B.; Bellincampi, D. The Grapevine VvPMEI1 Gene Encodes a Novel Functional Pectin Methylesterase Inhibitor Associated to Grape Berry Development. PLoS ONE 2015, 10, e0133810. [Google Scholar] [CrossRef] [PubMed]
- Alfonso, C.; Ciro, B.; Lucio, Q.; Domenico, C.; Luigi, S. A glycoprotein inhibitor of pectin methylesterase in kiwi fruit Purification by affinity chromatography and evidence of a ripening-related precursor. Eur. J. Biochem. 1995, 233, 926–929. [Google Scholar] [CrossRef]
- Lionetti, V.; Raiola, A.; Camardella, L.; Giovane, A.; Obel, N.; Pauly, M.; Favaron, F.; Cervone, F.; Bellincampi, D. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 2007, 143, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Fabri, E.; DeCaroli, M.; Hansen, A.R.; Willats, W.G.; Piro, G.; Bellincampi, D. Three Pectin Methylesterase Inhibitors Protect Cell Wall Integrity for Arabidopsis Immunity to Botrytis. Plant Physiol. 2017, 173, 1844–1863. [Google Scholar] [CrossRef]
- Volpi, C.; Janni, M.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. The Ectopic Expression of a Pectin Methyl Esterase Inhibitor Increases Pectin Methyl Esterification and Limits Fungal Diseases in Wheat. Mol. Plant Microbe Interact. 2011, 24, 1012–1019. [Google Scholar] [CrossRef]
- Tundo, S.; Kalunke, R.; Janni, M.; Volpi, C.; Lionetti, V.; Bellincampi, D.; Favaron, F.; D’Ovidio, R. PvPGIP2 Accumulation in Specific Floral Tissues but Not in the Endosperm Limits Fusarium graminearum Infection in Wheat. Mol. Plant Microbe Interact. 2016, 29, 629–639. [Google Scholar] [CrossRef]
- Wang, M.; Yuan, D.; Gao, W.; Li, Y.; Tan, J.; Zhang, X. A comparative genome analysis of PME and PMEI families reveals the evolution of pectin metabolism in plant cell walls. PLoS ONE 2013, 8, e72082. [Google Scholar] [CrossRef]
- Li, X.; Hamyat, M.; Liu, C.; Salman, A.; Gao, X.; Guo, C.; Wang, Y.; Guo, Y. Identification and Characterization of the WOX Family Genes in Five Solanaceae Species Reveal Their Conserved Roles in Peptide Signaling. Genes 2018, 9, 260. [Google Scholar] [CrossRef]
- Liu, T.; Yu, H.; Xiong, X.; Yu, Y.; Yue, X.; Liu, J.; Cao, J. Genome-Wide Identification and Characterization of Pectin Methylesterase Inhibitor Genes in Brassica oleracea. Int. J. Mol. Sci. 2018, 19, 3388. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xu, Z.; Ding, A.; Kong, Y. Genome-Wide Identification and Expression Profiling Analysis of the Xyloglucan Endotransglucosylase/Hydrolase Gene Family in Tobacco (Nicotiana tabacum L.). Genes 2018, 9, 273. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.M.; Bateman, A.; Binns, D.; Bork, P.; Das, U.; Daugherty, L.; Duquenne, L.; et al. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Ding, A.; Marowa, P.; Kong, Y. Genome-wide identification of the expansin gene family in tobacco (Nicotiana tabacum). Mol. Genet. Genom. 2016, 291, 1891–1907. [Google Scholar] [CrossRef] [PubMed]
- McCormick, R.F.; Truong, S.K.; Sreedasyam, A.; Jenkins, J.; Shu, S.; Sims, D.; Kennedy, M.; Amirebrahimi, M.; Weers, B.D.; McKinley, B.; et al. The Sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2018, 93, 338–354. [Google Scholar] [CrossRef]
- Hong, M.; Kim, D.Y.; Lee, T.G.; Jeon, W.B.; Seo, Y.W. Functional characterization of pectin methylesterase inhibitor (PMEI) in wheat. Genes Genet. Syst. 2010, 85, 97–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudhakar, R.P.; Srinivas, R.D.; Sivasakthi, K.; Bhatnagar-Mathur, P.; Vadez, V.; Sharma, K.K. Evaluation of Sorghum [Sorghum bicolor (L.)] Reference Genes in Various Tissues and under Abiotic Stress Conditions for Quantitative Real-Time PCR Data Normalization. Front. Plant Sci. 2016, 7, 529. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. PNAS 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.Y.; Nguyen, H.P.; Eom, S.H.; Lee, C. Integrative analysis of pectin methylesterase (PME) and PME inhibitors in tomato (Solanum lycopersicum): Identification, tissue-specific expression, and biochemical characterization. Plant Physiol. Biochem. 2018, 132, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.Y.; Nguyen, H.P.; Lee, C. Genome-wide identification and expression analysis of rice pectin methylesterases: Implication of functional roles of pectin modification in rice physiology. J. Plant Physiol. 2015, 183, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.P.; Jeong, H.Y.; Kim, H.; Kim, Y.C.; Lee, C. Molecular and biochemical characterization of rice pectin methylesterase inhibitors (OsPMEIs). Plant Physiol. Biochem. 2016, 101, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.; Thu, S.; Balasubramanian, V.; Cobos, C.; Disasa, T.; Mendu, V. Identification, Characterization, and Expression Analysis of Cell Wall Related Genes in Sorghum bicolor (L.) Moench, a Food, Fodder, and Biofuel Crop. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Yu, H.; Xiong, X.; Yue, X.; Yu, Y.; Huang, L.; Cao, J. Genome-Wide Identification, Molecular Evolution, and Expression Profiling Analysis of Pectin Methylesterase Inhibitor Genes in Brassica campestris ssp. chinensis. Int. J. Mol. Sci. 2018, 19, 1338. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Sequence ID | Subfamily | Signal Position | ORF (bp) | Subcellular Localization | Predicted Features | No. of Intron | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Plant-mPLoc | WOLF PSORT | Length (aa) | NM (kDa) | pI | ||||||
| SbPMEI1 | Sobic.001G024300 | II | 1-24 | 564 | C.M | chlo: 9, extr: 4 | 187 | 19.30 | 5.48 | 0 |
| SbPMEI2 | Sobic.001G024400 | II | 1-31 | 588 | C.M | chlo: 9, extr: 4 | 195 | 20.85 | 4.68 | 0 |
| SbPMEI3 | Sobic.001G024900 | II | 1-34 | 723 | C.M, Nuc | chlo: 14 | 240 | 24.76 | 8.86 | 0 |
| SbPMEI4 | Sobic.001G198700 | II | 1-26 | 720 | C.M, Nuc | chlo: 9, vacu: 3, mito: 1 | 239 | 24.29 | 5.09 | 0 |
| SbPMEI5 | Sobic.001G198800 | II | 1-24 | 672 | C.M, Nuc | chlo: 12, mito: 1 | 223 | 22.49 | 5.87 | 0 |
| SbPMEI6 | Sobic.001G288400 | II | 1-23 | 660 | C.M | chlo: 11, extr: 2 | 219 | 21.93 | 8.83 | 0 |
| SbPMEI7 | Sobic.001G323801 | II | 1-20 | 681 | C.M | chlo: 11, nucl: 1, mito: 1 | 227 | 22.88 | 8.72 | 0 |
| SbPMEI8 | Sobic.001G328900 | III | 1-19 | 714 | Nuc | extr: 12, vacu: 1 | 231 | 24.31 | 8.18 | 1 |
| SbPMEI9 | Sobic.002G091200 | I | 1-25 | 768 | C.M | chlo: 13 | 255 | 25.91 | 4.69 | 0 |
| SbPMEI10 | Sobic.002G091300 | I | 1-22 | 948 | Nuc | chlo: 7, extr: 6 | 315 | 30.34 | 4.15 | 0 |
| SbPMEI11 | Sobic.002G102800 | III | 1-23 | 555 | C.M | extr: 5, vacu: 5, chlo: 1, cyto: 1, plas: 1 | 184 | 18.64 | 6.24 | 0 |
| SbPMEI12 | Sobic.002G103000 | III | 1-38 | 579 | C.M | extr: 5, chlo: 4, mito: 2, E.R._plas: 2, plas: 1.5 | 198 | 20.15 | 4.86 | 0 |
| SbPMEI13 | Sobic.003G099500 | I | 1-32 | 639 | Nuc | chlo: 9, mito: 2, vacu: 2 | 212 | 23.01 | 10.39 | 0 |
| SbPMEI14 | Sobic.003G099600 | I | 1-23 | 477 | C.M | chlo: 9, mito: 2, extr: 2 | 158 | 17.11 | 5.72 | 0 |
| SbPMEI15 | Sobic.003G100100 | I | 1-30 | 603 | C.M, Nuc | chlo: 11, vacu: 2 | 200 | 21.41 | 9.32 | 0 |
| SbPMEI16 | Sobic.003G114800 | I | 1-25 | 552 | C.M | extr: 5, chlo: 3, cyto: 3, vacu: 2 | 183 | 19.92 | 9.10 | 0 |
| SbPMEI17 | Sobic.003G114900 | I | 1-22 | 507 | C.M | extr: 9, vacu: 3, chlo: 1 | 168 | 18.06 | 4.75 | 0 |
| SbPMEI18 | Sobic.003G147700 | II | 1-42 | 594 | C.M | E.R.: 2.5, E.R._plas: 2.5, chlo: 2, mito: 2, extr: 2, vacu: 2, plas: 1.5, nucl: 1 | 213 | 22.19 | 4.57 | 1 |
| SbPMEI19 | Sobic.003G203900 | III | 1-27 | 570 | C.M | chlo: 4, extr: 4, vacu: 4, mito: 1 | 189 | 19.61 | 9.50 | 0 |
| SbPMEI20 | Sobic.003G270700 | I | 1-25 | 657 | C.M | chlo: 9, extr: 4 | 218 | 22.85 | 4.62 | 1 |
| SbPMEI21 | Sobic.004G002700 | I | 1-37 | 642 | C.M | extr: 8, plas: 2, golg: 2, chlo: 1 | 213 | 23.48 | 9.00 | 0 |
| SbPMEI22 | Sobic.004G002800 | II | 1-40 | 720 | C.M | chlo: 3, mito: 2, vacu: 2, E.R.: 2, nucl: 1, cyto: 1, plas: 1, extr: 1 | 239 | 25.16 | 4.79 | 0 |
| SbPMEI23 | Sobic.004G147700 | III | 1-25 | 558 | C.M | vacu: 7, E.R.: 3, plas: 2, extr: 2 | 185 | 19.22 | 5.09 | 0 |
| SbPMEI24 | Sobic.004G167700 | III | 1-27 | 570 | C.M | extr: 9, chlo: 2, vacu: 2 | 189 | 19.62 | 5.07 | 0 |
| SbPMEI25 | Sobic.004G167800 | III | 1-35 | 600 | C.M | chlo: 13 | 199 | 20.66 | 5.89 | 0 |
| SbPMEI26 | Sobic.004G167900 | III | 1-22 | 543 | C.M | extr: 13 | 180 | 18.85 | 5.43 | 0 |
| SbPMEI27 | Sobic.004G168000 | III | 1-25 | 513 | C.M | extr: 10, chlo: 1, mito: 1, vacu: 1 | 170 | 18.17 | 5.90 | 1 |
| SbPMEI28 | Sobic.004G277100 | I | 1-19 | 543 | C.M | extr: 6, chlo: 3, vacu: 3, nucl: 1 | 180 | 18.79 | 4.46 | 0 |
| SbPMEI29 | Sobic.004G277600 | I | 1-27 | 537 | C.M | chlo: 8, extr: 5 | 178 | 18.87 | 4.98 | 0 |
| SbPMEI30 | Sobic.004G277700 | I | 1-26 | 546 | Nuc | extr: 12, chlo: 1 | 183 | 18.91 | 5.45 | 0 |
| SbPMEI31 | Sobic.005G022600 | III | 1-23 | 588 | C.M | extr: 11, chlo: 1, cyto: 1 | 193 | 19.47 | 6.09 | 0 |
| SbPMEI32 | Sobic.005G160700 | II | 1-30 | 696 | C.M | chlo: 13 | 231 | 23.52 | 8.42 | 0 |
| SbPMEI33 | Sobic.005G212900 | I | 1-24 | 1707 | Nuc | vacu: 7, nucl: 2, extr: 2, golg: 2 | 568 | 59.99 | 5.26 | 0 |
| SbPMEI34 | Sobic.006G004300 | II | 1-27 | 645 | C.M | chlo: 7, extr: 3, vacu: 3 | 214 | 22.83 | 10.66 | 0 |
| SbPMEI35 | Sobic.006G079100 | I | 1-21 | 660 | C.M | extr: 9, vacu: 3, chlo: 2 | 219 | 22.88 | 8.24 | 0 |
| SbPMEI36 | Sobic.006G094000 | III | 1-57 | 714 | C.M | chlo: 10, mito: 3 | 237 | 25.01 | 9.47 | 0 |
| SbPMEI37 | Sobic.006G194800 | I | 1-21 | 567 | C.M | chlo: 6, extr: 6, cyto: 1 | 188 | 19.84 | 6.42 | 0 |
| SbPMEI38 | Sobic.006G194900 | I | 1-21 | 534 | C.M | extr: 8, chlo: 4, nucl: 1 | 177 | 18.27 | 6.72 | 0 |
| SbPMEI39 | Sobic.007G008000 | I | 1-25 | 723 | C.M, Nuc | chlo: 13 | 240 | 23.74 | 7.69 | 0 |
| SbPMEI40 | Sobic.007G008100 | I | 1-25 | 681 | C.M, Nuc | chlo: 12, cyto: 1 | 226 | 22.94 | 8.32 | 0 |
| SbPMEI41 | Sobic.007G008200 | I | 1-18 | 660 | C.M | chlo: 12, mito: 1 | 219 | 21.84 | 6.99 | 0 |
| SbPMEI42 | Sobic.007G036000 | I | 1-24 | 537 | C.M | extr: 10, chlo: 1, plas: 1, vacu: 1 | 178 | 18.77 | 4.51 | 0 |
| SbPMEI43 | Sobic.007G036200 | I | 1-24 | 552 | Nuc | extr: 11, chlo: 1, vacu: 1 | 183 | 20.05 | 5.19 | 0 |
| SbPMEI44 | Sobic.008G129800 | I | 1-31 | 684 | C.M | extr: 10, chlo: 1, cyto: 1, mito: 1 | 227 | 23.45 | 9.17 | 0 |
| SbPMEI45 | Sobic.008G154500 | III | 1-22 | 507 | C.M | chlo: 10, cyto: 3 | 180 | 17.97 | 5.28 | 0 |
| SbPMEI46 | Sobic.008G154600 | III | 1-19 | 555 | C.M | extr: 11, vacu: 3 | 184 | 18.65 | 4.59 | 0 |
| SbPMEI47 | Sobic.008G154700 | III | 1-26 | 591 | C.M | extr: 8, chlo: 3, cyto: 2 | 196 | 19.03 | 5.71 | 0 |
| SbPMEI48 | Sobic.008G154801 | III | 1-49 | 711 | Nuc | chlo: 5, cyto: 2, mito: 2, nucl: 1, plas: 1, extr: 1, vacu: 1 | 237 | 23.96 | 6.58 | 1 |
| SbPMEI49 | Sobic.008G154900 | III | 1-23 | 576 | C.M | chlo: 5, extr: 5, cyto: 2, mito: 1 | 191 | 18.71 | 4.90 | 0 |
| SbPMEI50 | Sobic.009G091600 | I | 1-30 | 555 | C.M | extr: 7, chlo: 3, vacu: 3 | 184 | 19.99 | 9.24 | 0 |
| SbPMEI51 | Sobic.009G216400 | III | 1-24 | 567 | C.M, Nuc | extr: 8, vacu: 4, chlo: 1 | 188 | 20.66 | 5.83 | 1 |
| SbPMEI52 | Sobic.010G006900 | II | 1-31 | 567 | C.M | extr: 13 | 188 | 19.46 | 5.36 | 1 |
| SbPMEI53 | Sobic.010G007600 | I | 1-26 | 708 | Nuc | extr: 5, chlo: 4, vacu: 4 | 235 | 25.08 | 4.93 | 0 |
| SbPMEI54 | Sobic.010G261000 | II | 1-21 | 609 | C.M | chlo: 6, extr: 3, cyto: 2, vacu: 2 | 202 | 20.55 | 9.35 | 0 |
| SbPMEI55 | Sobic.010G261100 | II | 1-29 | 654 | C.M | chlo: 14 | 217 | 21.62 | 9.60 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, A.; Ahmed, R.I.; Chen, H.; Han, L.; Sun, J.; Ding, A.; Guo, Y.; Kong, Y. Genome-Wide Identification, Characterization and Expression Patterns of the Pectin Methylesterase Inhibitor Genes in Sorghum bicolor. Genes 2019, 10, 755. https://doi.org/10.3390/genes10100755
Ren A, Ahmed RI, Chen H, Han L, Sun J, Ding A, Guo Y, Kong Y. Genome-Wide Identification, Characterization and Expression Patterns of the Pectin Methylesterase Inhibitor Genes in Sorghum bicolor. Genes. 2019; 10(10):755. https://doi.org/10.3390/genes10100755
Chicago/Turabian StyleRen, Angyan, Rana Imtiaz Ahmed, Huanyu Chen, Linhe Han, Jinhao Sun, Anming Ding, Yongfeng Guo, and Yingzhen Kong. 2019. "Genome-Wide Identification, Characterization and Expression Patterns of the Pectin Methylesterase Inhibitor Genes in Sorghum bicolor" Genes 10, no. 10: 755. https://doi.org/10.3390/genes10100755
APA StyleRen, A., Ahmed, R. I., Chen, H., Han, L., Sun, J., Ding, A., Guo, Y., & Kong, Y. (2019). Genome-Wide Identification, Characterization and Expression Patterns of the Pectin Methylesterase Inhibitor Genes in Sorghum bicolor. Genes, 10(10), 755. https://doi.org/10.3390/genes10100755






