Identification of Alternatively-Activated Pathways between Primary Breast Cancer and Liver Metastatic Cancer Using Microarray Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Gene Expression and Pathways
2.2. Gene Expression Data Pretreatment
2.3. Breast Cancer Liver Metastasis Uniquely-Enriched Pathway Analysis
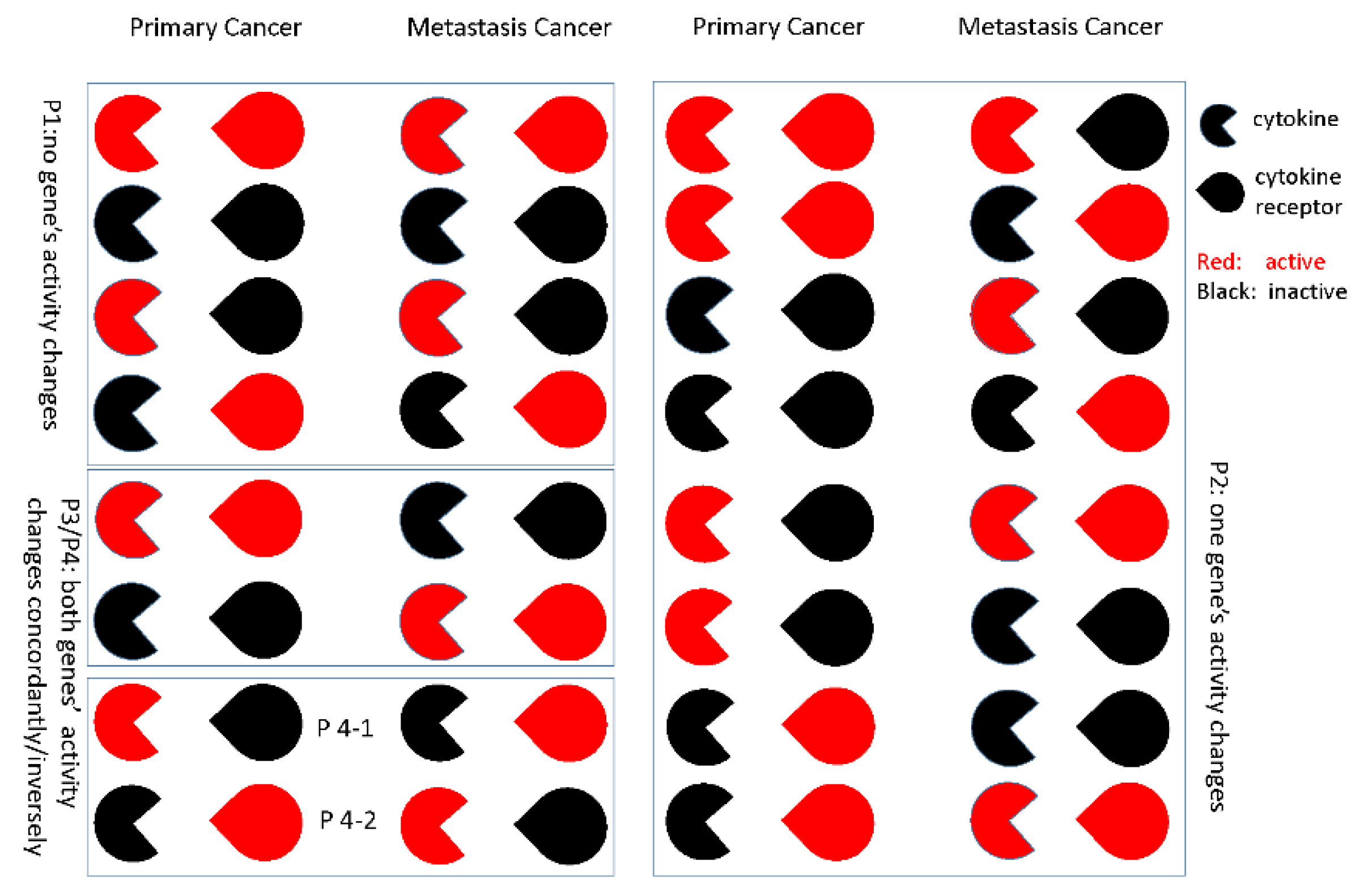
2.4. Gene-Gene Activity Alteration Patterns’ Recognition
2.5. Bayesian Network Construction
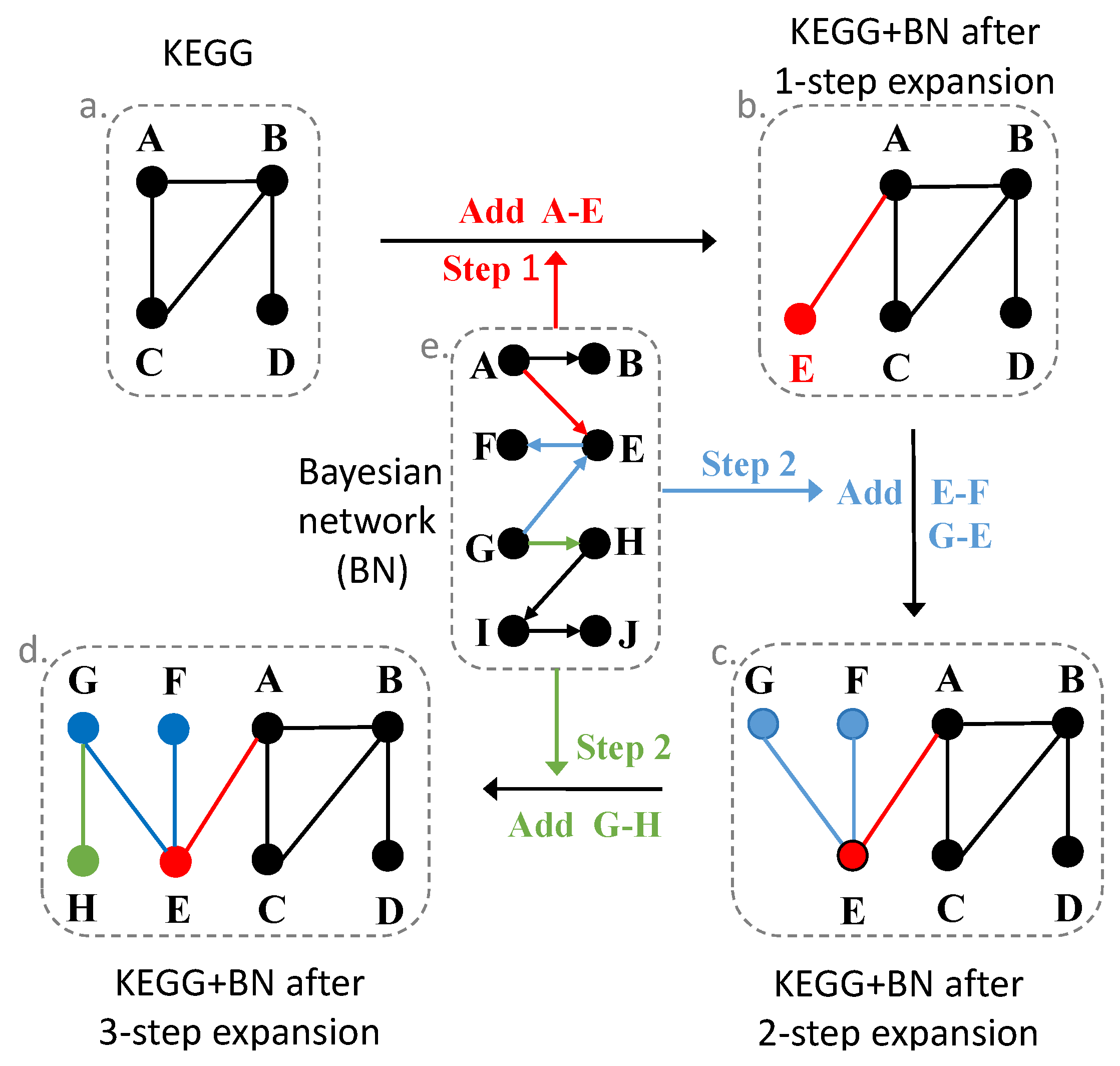
2.6. Expand Pathways with the Bayesian Network
2.7. Connectivity among Pathways: Pathway Networks
3. Results
3.1. Raw Pathway Analysis
3.2. Robustness Analysis
3.3. Expanded Network
3.4. Pathway Network
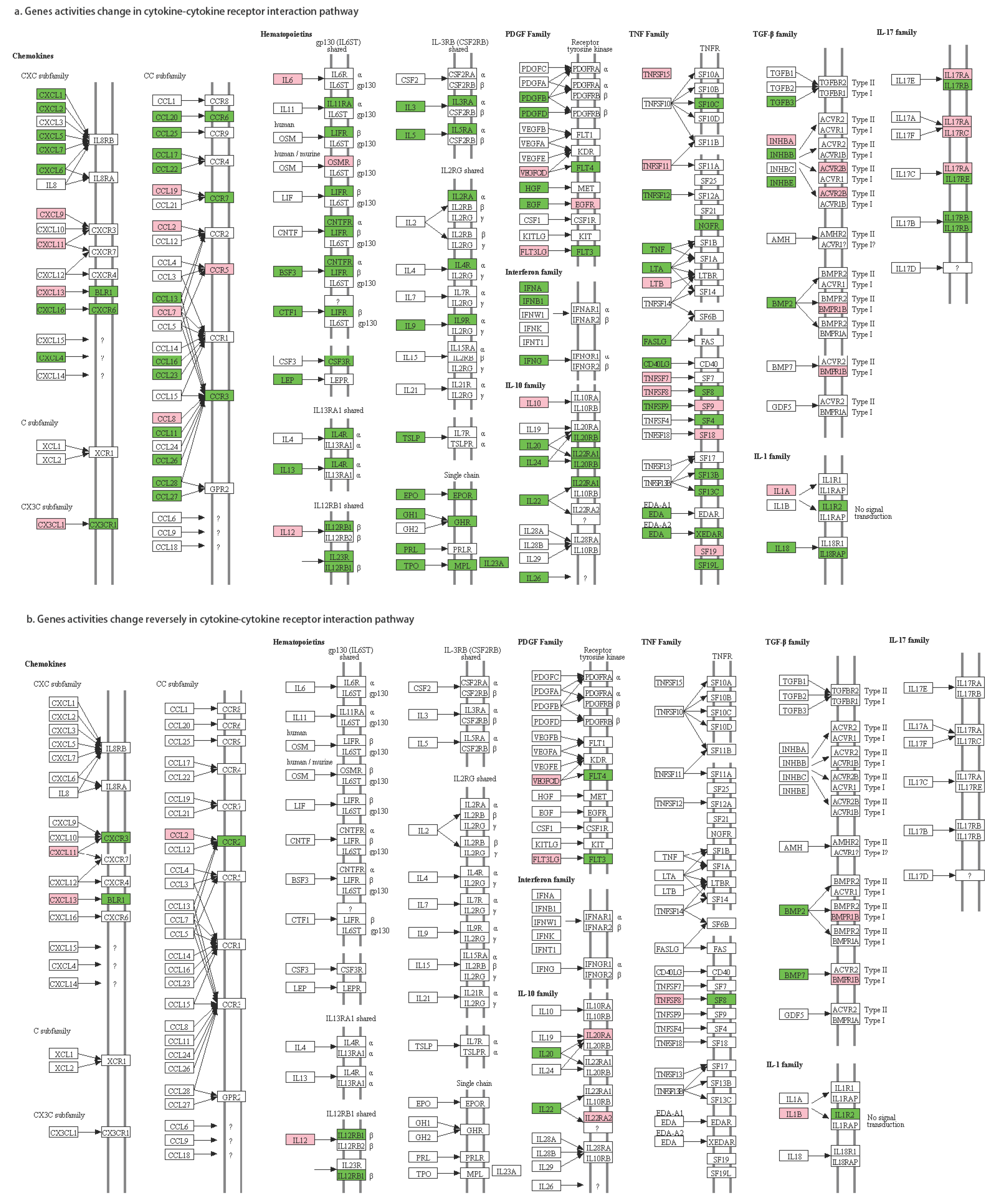
3.5. Alternatively-Activated Pathways
4. Discussions
5. Conclusions
Availability of Data and Materials
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, G.S.; Goswami, S.; Jones, J.G.; Oktay, M.H.; Condeelis, J.S. Signatures of breast cancer metastasis at a glance. J. Cell Sci. 2016, 129, 1751–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van’t Veer, L.J.; Dai, H.; Van, d.V.M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; van der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van De Vijver, M.J.; He, Y.D.; Van‘t Veer, L.J.; Dai, H.; Hart, A.A.; Voskuil, D.W.; Schreiber, G.J.; Peterse, J.L.; Roberts, C.; Marton, M.J.; et al. A Gene-Expression Signature as a Predictor of Survival in Breast Cancer. New Engl. J. Med. 2002, 347, 1999–2009. [Google Scholar] [CrossRef]
- Bueno-de-Mesquita, J.M.; Linn, S.C.; Keijzer, R.; Wesseling, J.; Nuyten, D.S.; van Krimpen, C.; Meijers, C.; de Graaf, P.W.; Bos, M.M.; Hart, A.A.; et al. Validation of 70-gene prognosis signature in node-negative breast cancer. Breast Cancer Res. Treat. 2009, 117, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Buyse, M.; Loi, S.; Veer, L.V.; Viale, G.; Delorenzi, M.; Glas, A.M.; D’Assignies, M.S.; Bergh, J.; Lidereau, R.; Ellis, P.; et al. Validation and Clinical Utility of a 70-Gene Prognostic Signature for Women with Node-Negative Breast Cancer. J. Natl. Cancer Inst. 2006, 98, 1183–1192. [Google Scholar] [CrossRef]
- Mook, S.; Schmidt, M.K.; Viale, G.; Pruneri, G.; Eekhout, I.; Floore, A.; Glas, A.M.; Bogaerts, J.; Cardoso, F.; Piccart-Gebhart, M.J.; et al. The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1-3 positive lymph nodes in an independent validation study. Breast Cancer Res Treat 2009, 116, 295–302. [Google Scholar] [CrossRef]
- Mook, S.; Schmidt, M.K.; Weigelt, B.; Kreike, B.; Eekhout, I.; van de Vijver, M.J.; Glas, A.M.; Floore, A.; Rutgers, E.J.; van’t Veer, L.J. The 70-gene prognosis signature predicts early metastasis in breast cancer patients between 55 and 70 years of age. Ann. Oncol. 2010, 21, 717–722. [Google Scholar] [CrossRef]
- Wittner, B.S.; Sgroi, D.C.; Ryan, P.D.; Bruinsma, T.J.; Glas, A.M.; Male, A.; Dahiya, S.; Habin, K.; Bernards, R.; Haber, D.A.; et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin. Cancer Res. 2008, 14, 2988–2993. [Google Scholar] [CrossRef]
- Curtis, C. Genomic profiling of breast cancers. Curr. Opin. Obstet. Gynecol. 2015, 27, 34–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Klijn, J.; Zhang, Y.; Sieuwerts, A.; Look, M.; Yang, F.; Talantov, D.; Timmermans, M.; Meijervangelder, M.; Yu, J. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 2005, 365, 671–679. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Glick, J.H.; Gelber, R.D.; Coates, A.S.; Senn, H.-J. Meeting Highlights: International Consensus Panel on the Treatment of Primary Breast Cancer. J. Clin. Oncol. 2001, 19, 3817–3827. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health Consensus Development Panel. National Institutes of Health Consensus Development Conference statement: Adjuvant therapy for breast cancer, 1–3 November 2000. J. Natl. Cancer Inst. Monogr. 2001, 2001, 5–15. [Google Scholar] [CrossRef]
- Sgroi, D.C.; Haber, D.A.; Ryan, P.D.; Ma, X.-J.; Erlander, M.G. RE: A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell 2004, 6, 445. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef] [Green Version]
- Landemaine, T.; Jackson, A.; Rucci, N.; Sin, S.; Abad, B.M.; Sierra, A.; Boudinet, A.; Ricevuto, E.; Briffod, M.; Cherel, P.; et al. A Six-Gene Signature Predicting Breast Cancer Lung Metastasis. Cancer Res. 2008, 68, 6092–6099. [Google Scholar] [CrossRef] [PubMed]
- Patsialou, A.; Wang, Y.; Lin, J.; Whitney, K.; Goswami, S.; A Kenny, P.; Condeelis, J.S. Selective gene-expression profiling of migratory tumor cells in vivo predicts clinical outcome in breast cancer patients. Breast Cancer Res. 2012, 14, R139. [Google Scholar] [CrossRef]
- Cejalvo, J.M.; De Dueñas, E.M.; Galvan, P.; García-Recio, S.; Gasión, O.B.; Paré, L.; Antolin, S.; Martinello, R.; Blancas, I.; Adamo, B.; et al. Intrinsic Subtypes and Gene Expression Profiles in Primary and Metastatic Breast Cancer. Cancer Res. 2017, 77, 2213–2221. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, N.; Iwamoto, T.; Qi, Y.; Niikura, N.; Santarpia, L.; Yamauchi, H.; Nakamura, S.; Hortobagyi, G.N.; Pusztai, L.; Symmans, W.F.; et al. Bone metastasis-related signaling pathways in breast cancers stratified by estrogen receptor status. J. Cancer 2017, 8, 1045–1052. [Google Scholar] [CrossRef] [Green Version]
- Bidwell, B.N.; Slaney, C.Y.; Withana, N.P.; Forster, S.; Cao, Y.; Loi, S.; Andrews, D.; Mikeska, T.; E Mangan, N.; A Samarajiwa, S.; et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat. Med. 2012, 18, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Song, C.-X.; Huang, H.; Frankenberger, C.A.; Sankarasharma, D.; Gomes, S.; Chen, P.; Chen, J.; Chada, K.K.; He, C.; et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc. Natl. Acad. Sci. USA 2013, 110, 9920–9925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pai, P.; Rachagani, S.; Lakshmanan, I.; Macha, M.A.; Sheinin, Y.; Smith, L.M.; Ponnusamy, M.P.; Batra, S.K. The canonical Wnt pathway regulates the metastasis-promoting mucin MUC4 in pancreatic ductal adenocarcinoma. Mol. Oncol. 2016, 10, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Zhu, J.; Dong, L.; Chen, Y. The role of c2orf68 and PI3K/Akt/mTOR pathway in human colorectal cancer. Med. Oncol. 2014, 31, 92. [Google Scholar] [CrossRef] [PubMed]
- Papadatos-Pastos, D.; Rabbie, R.; Ross, P.; Sarker, D. The role of the PI3K pathway in colorectal cancer. Crit. Rev. Oncol. 2015, 94, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Halaoui, R.; McCaffrey, L. Rewiring cell polarity signaling in cancer. Oncogene 2015, 34, 939–950. [Google Scholar] [CrossRef]
- Lee, M.J.; Ye, A.S.; Gardino, A.K.; Heijink, A.M.; Sorger, P.K.; MacBeath, G.; Yaffe, M.B. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 2012, 149, 780–794. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Bergami, P.; Huang, C.; Goydos, J.S.; Yip, D.; Bar-Eli, M.; Herlyn, M.; Smalley, K.S.M.; Mahale, A.; Eroshkin, A.; Aaronson, S.; et al. Re-wired ERK-JNK signaling pathways in melanoma. Cancer Cell 2007, 11, 447–460. [Google Scholar] [CrossRef]
- Cotton, T.B.; Nguyen, H.H.; Said, J.I.; Ouyang, Z.; Zhang, J.; Song, M. Discerning mechanistically rewired biological pathways by cumulative interaction heterogeneity statistics. Sci. Rep. 2015, 5, 9634. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Xu, L.; Zhang, R.; Huang, M.; Wang, K.; Xu, J.; Lv, H.; Shang, Z.; Zhang, M.; et al. DBGSA: A novel method of distance-based gene set analysis. J. Hum. Genet. 2012, 57, 642–653. [Google Scholar] [CrossRef]
- Minn, A.J.; Bevilacqua, E.; Yun, J.; Rosner, M.R. Identification of novel metastasis suppressor signaling pathways for breast cancer. Cell Cycle 2012, 11, 2452–2457. [Google Scholar] [CrossRef] [Green Version]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Khatri, P.; Sirota, M.; Butte, A.J. Ten Years of Pathway Analysis: Current Approaches and Outstanding Challenges. PLoS Comput. Boil. 2012, 8, e1002375. [Google Scholar] [CrossRef] [PubMed]
- Mitrea, C.; Taghavi, Z.; Bokanizad, B.; Hanoudi, S.; Tagett, R.; Donato, M.; Voichita, C.; Draghici, S. Methods and approaches in the topology-based analysis of biological pathways. Front. Physiol. 2013, 4, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Eddy, R.; Condeelis, J. The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer 2007, 7, 429–440. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.; He, W.; Tulley, S.; Gupta, G.P.; Serganova, I.; Chen, C.-R.; Manova-Todorova, K.; Blasberg, R.; Gerald, W.L.; Massagué, J. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 13909–13914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maire, V.; Nemati, F.; Richardson, M.; Vincent-Salomon, A.; Tesson, B.; Rigaill, G.; Gravier, E.; Marty-Prouvost, B.; de Koning, L.; Lang, G.; et al. Polo-like kinase 1: A potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 2013, 73, 813–823. [Google Scholar] [CrossRef]
- Kimbung, S.; Kovacs, A.; Bendahl, P.O.; Malmstrom, P.; Ferno, M.; Hatschek, T.; Hedenfalk, I. Claudin-2 is an independent negative prognostic factor in breast cancer and specifically predicts early liver recurrences. Mol. Oncol. 2014, 8, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Tobin, N.P.; Harrell, J.C.; Lovrot, J.; Brage, S.E.; Stolt, M.F.; Carlsson, L.; Einbeigi, Z.; Linderholm, B.; Loman, N.; Malmberg, M.; et al. Molecular subtype and tumor characteristics of breast cancer metastases as assessed by gene expression significantly influence patient post-relapse survival. Ann. Oncol. 2015, 26, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Jia, P.; Wang, Q.; Lin, C.-C.; Li, W.-H.; Zhao, Z. Studying Tumorigenesis through Network Evolution and Somatic Mutational Perturbations in the Cancer Interactome. Mol. Boil. Evol. 2014, 31, 2156–2169. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Liu, C.; Shen, B.; Zhao, Z. Investigating cellular network heterogeneity and modularity in cancer: A network entropy and unbalanced motif approach. BMC Syst. Boil. 2016, 10 (Suppl. 3), 65. [Google Scholar] [CrossRef] [PubMed]
- Chickering, D.M. Optimal Structure Identification with Greedy Search. J. Mach. Learn. Res. 2002, 3, 47. [Google Scholar]
- Ramsey, J.D. Scaling up Greedy Causal Search for Continuous Variables. arXiv 2015, 1507, 07749v2. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhu, G.; Yan, H.H.; Pang, Y.; Jian, J.; Achyut, B.R.; Liang, X.; Weiss, J.M.; Wiltrout, R.H.; Hollander, M.C.; Yang, L. CXCR3 as a molecular target in breast cancer metastasis: Inhibition of tumor cell migration and promotion of host anti-tumor immunity. Oncotarget 2015, 6, 43408–43419. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Norsworthy, K.; Kundu, N.; Rodgers, W.H.; Gimotty, P.A.; Goloubeva, O.; Lipsky, M.; Li, Y.; Holt, D.; Fulton, A. CXCR3 expression is associated with poor survival in breast cancer and promotes metastasis in a murine model. Mol. Cancer Ther. 2009, 8, 490–498. [Google Scholar] [CrossRef] [Green Version]
- Panse, J.; Friedrichs, K.; Marx, A.; Hildebrandt, Y.; Luetkens, T.; Bartels, K.; Horn, C.; Stahl, T.; Cao, Y.; Milde-Langosch, K.; et al. Chemokine CXCL13 is overexpressed in the tumour tissue and in the peripheral blood of breast cancer patients. Br. J. Cancer 2008, 99, 930–938. [Google Scholar] [CrossRef]
- Chen, L.; Huang, Z.; Yao, G.; Lyu, X.; Li, J.; Hu, X.; Cai, Y.; Li, W.; Li, X.; Ye, C. The expression of CXCL13 and its relation to unfavorable clinical characteristics in young breast cancer. J. Transl. Med. 2015, 13, 168. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Qian, B.-Z.; Soong, D.; Cassetta, L.; Noy, R.; Sugano, G.; Kato, Y.; Li, J.; Pollard, J.W. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J. Cell Boil. 2015, 212, 1043–1059. [Google Scholar] [CrossRef]
- Bin Fang, W.; Jokar, I.; Zou, A.; Lambert, D.; Dendukuri, P.; Cheng, N. CCL2/CCR2 Chemokine Signaling Coordinates Survival and Motility of Breast Cancer Cells through Smad3 Protein- and p42/44 Mitogen-activated Protein Kinase (MAPK)-dependent Mechanisms. J. Boil. Chem. 2012, 287, 36593–36608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, J.-L.; Yang, P.-C.; Shih, J.-Y.; Yang, C.-Y.; Wei, L.-H.; Hsieh, C.-Y.; Chou, C.-H.; Jeng, Y.-M.; Wang, M.-Y.; Chang, K.-J.; et al. The VEGF-C/Flt-4 axis promotes invasion and metastasis of cancer cells. Cancer Cell 2006, 9, 209–223. [Google Scholar] [CrossRef] [Green Version]
- Varney, M.L.; Singh, R.K. VEGF-C-VEGFR3/Flt4 axis regulates mammary tumor growth and metastasis in an autocrine manner. Am. J. Cancer Res. 2015, 5, 616–628. [Google Scholar]
- Yu, H.; Zhang, S.; Zhang, R.; Zhang, L. The role of VEGF-C/D and Flt-4 in the lymphatic metastasis of early-stage invasive cervical carcinoma. J. Exp. Clin. Cancer Res. 2009, 28, 98. [Google Scholar] [CrossRef]
- Péron, J.M.; Esche, C.; Subbotin, V.M.; Maliszewski, C.; Lotze, M.T.; Shurin, M.R. FLT3-ligand administration inhibits liver metastases: Role of NK cells. J. Immunol. 1998, 161, 6164–6170. [Google Scholar]
- Hamaguchi, T.; Wakabayashi, H.; Matsumine, A.; Sudo, A.; Uchida, A. TNF inhibitor suppresses bone metastasis in a breast cancer cell line. Biochem. Biophys. Res. Commun. 2011, 407, 525–530. [Google Scholar] [CrossRef]
- Nutter, F.; Holen, I.; Brown, H.K.; Cross, S.S.; Evans, C.A.; Walker, M.; E Coleman, R.; A Westbrook, J.; Selby, P.J.; E Brown, J.; et al. Different molecular profiles are associated with breast cancer cell homing compared with colonisation of bone: Evidence using a novel bone-seeking cell line. Endocr.-Relat. Cancer 2014, 21, 327–341. [Google Scholar] [CrossRef]
- Holen, I.; Lefley, D.V.; Francis, S.E.; Rennicks, S.; Bradbury, S.; Coleman, R.E.; Ottewell, P. IL-1 drives breast cancer growth and bone metastasis in vivo. Oncotarget 2016, 7, 75571–75584. [Google Scholar] [CrossRef]
- Hsu, Y.-H.; Hsing, C.-H.; Li, C.-F.; Chan, C.-H.; Chang, M.-C.; Yan, J.-J.; Chang, M.-S. Anti–IL-20 Monoclonal Antibody Suppresses Breast Cancer Progression and Bone Osteolysis in Murine Models. J. Immunol. 2012, 188, 1981–1991. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, G.; Kim, J.-Y.; Yun, H.J.; Lim, S.-C.; Choi, H.S. Interleukin-22 promotes epithelial cell transformation and breast tumorigenesis via MAP3K8 activation. Carcinog. 2014, 35, 1352–1361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ottewell, P.D.; O’Donnell, L.; Holen, I. Molecular alterations that drive breast cancer metastasis to bone. BoneKEy Rep. 2015, 4, 643. [Google Scholar] [CrossRef] [PubMed]
- Buijs, J.T.; van der Horst, G.; van den Hoogen, C.; Cheung, H.; de Rooij, B.; Kroon, J.; Petersen, M.; van Overveld, P.G.; Pelger, R.C.; van der Pluijm, G. The BMP2/7 heterodimer inhibits the human breast cancer stem cell subpopulation and bone metastases formation. Oncogene 2012, 31, 2164–2174. [Google Scholar] [CrossRef] [PubMed]
- Buijs, J.T.; Henriquez, N.V.; Van Overveld, P.G.; Que, I.; Schwaninger, R.; Rentsch, C.; Dijke, P.T.; Driouch, K.; Lidereau, R.; Vukicevic, S.; et al. Bone Morphogenetic Protein 7 in the Development and Treatment of Bone Metastases from Breast Cancer. Cancer Res. 2007, 67, 8742–8751. [Google Scholar] [CrossRef] [Green Version]
- Buijs, J.T.; Petersen, M.; Van Der Horst, G.; Van Der Pluijm, G. Bone morphogenetic proteins and its receptors; therapeutic targets in cancer progression and bone metastasis? Curr. Pharm. Des. 2010, 16, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- McClintick, J.N.; Edenberg, H.J. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinform. 2006, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Strauß, L.; Ruffing, U.; Abdulla, S.; Alabi, A.; Akulenko, R.; Garrine, M.; Germann, A.; Grobusch, M.P.; Helms, V.; Herrmann, M.; et al. Detecting Staphylococcus aureus Virulence and Resistance Genes: A Comparison of Whole-Genome Sequencing and DNA Microarray Technology. J. Clin. Microbiol. 2016, 54, 1008–1016. [Google Scholar] [CrossRef]
- Gendelman, R.; Xing, H.; Mirzoeva, O.K.; Sarde, P.; Curtis, C.; Feiler, H.S.; McDonagh, P.; Gray, J.W.; Khalil, I.; Korn, W.M. Bayesian Network Inference Modeling Identifies TRIB1 as a Novel Regulator of Cell-Cycle Progression and Survival in Cancer Cells. Cancer Res. 2017, 77, 1575–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Z.-Q.; Guo, P.; Si, S.-B.; Geng, Z.-M.; Chen, C.; Cong, L.-L. Analysis of prognostic factors for survival after surgery for gallbladder cancer based on a Bayesian network. Sci. Rep. 2017, 7, 293. [Google Scholar] [CrossRef] [PubMed]
- Cantor, J.R.; Sabatini, D.M. Cancer cell metabolism: One hallmark, many faces. Cancer Discov. 2012, 2, 881–898. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Qin, Y.; Liu, S. Cytokines, breast cancer stem cells (BCSCs) and chemoresistance. Clin. Transl. Med. 2018, 7, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.J.; Adachi, I. Serum interleukin-6 levels correlate to tumor progression and prognosis in metastatic breast carcinoma. Anticancer. Res. 1999, 19, 1427–1432. [Google Scholar] [PubMed]
- Bachelot, T.; Ray-Coquard, I.; Menetrier-Caux, C.; Rastkha, M.; Duc, A.; Blay, J.-Y. Prognostic value of serum levels of interleukin 6 and of serum and plasma levels of vascular endothelial growth factor in hormone-refractory metastatic breast cancer patients. Br. J. Cancer 2003, 88, 1721–1726. [Google Scholar] [CrossRef] [Green Version]
- Bozcuk, H.; Uslu, G.; Samur, M.; Yıldız, M.; Ozben, T.; Ozdogan, M.; Artaç, M.; Altunbas, H.; Akan, I.; Savas, B. Tumour necrosis factor-alpha, interleukin-6, and fasting serum insulin correlate with clinical outcome in metastatic breast cancer patients treated with chemotherapy. Cytokine 2004, 27, 58–65. [Google Scholar] [CrossRef]
- Britschgi, A.; Bill, A.; Brinkhaus, H.; Rothwell, C.; Clay, I.; Duss, S.; Rebhan, M.; Raman, P.; Guy, C.T.; Wetzel, K.; et al. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc. Natl. Acad. Sci. USA 2013, 110, E1026–E1034. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.J.; Monteith, G.R.; Roberts-Thomson, S.J. Calcium transport and signaling in the mammary gland: Targets for breast cancer. Biochim. Biophys. Acta 2006, 1765, 235–255. [Google Scholar] [CrossRef]
- Wakefield, L.M.; Roberts, A.B. TGF-beta signaling: Positive and negative effects on tumorigenesis. Curr. Opin. Genet. Dev. 2002, 12, 22–29. [Google Scholar] [CrossRef]
- Buck, M.B.; Knabbe, C. TGF-Beta Signaling in Breast Cancer. Ann. N. Y. Acad. Sci. 2006, 1089, 119–126. [Google Scholar] [CrossRef]
- Polanowska, J.; Le Cam, L.; Orsetti, B.; Vallès, H.; Fabbrizio, E.; Fajas, L.; Taviaux, S.; Theillet, C.; Sardet, C. Human E2F5 gene is oncogenic in primary rodent cells and is amplified in human breast tumors. Genes Chromosom. Cancer 2000, 28, 126–130. [Google Scholar] [CrossRef]
- Kothandaraman, N.; Bajic, V.B.; Brendan, P.N.; Huak, C.Y.; Keow, P.B.; Razvi, K.; Salto-Tellez, M.; Choolani, M. E2F5 status significantly improves malignancy diagnosis of epithelial ovarian cancer. BMC Cancer 2010, 10, 64. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yim, S.-H.; Xu, H.-D.; Jung, S.-H.; Yang, S.Y.; Hu, H.-J.; Jung, C.-K.; Chung, Y.-J. A potential oncogenic role of the commonly observed E2F5 overexpression in hepatocellular carcinoma. World J. Gastroenterol. 2011, 17, 470–477. [Google Scholar] [CrossRef] [PubMed]
- Khaleel, S.S.; Andrews, E.H.; Ung, M.; DiRenzo, J.; Cheng, C. E2F4 regulatory program predicts patient survival prognosis in breast cancer. Breast Cancer Res. 2014, 16, 486. [Google Scholar] [CrossRef] [PubMed]




| Pathway Resource | Number of Significant Primary Cancer-Related Pathways | Number of Significant Metastatic Cancer-Related Pathways |
|---|---|---|
| Raw pathways | 17 | 24 |
| Expanded pathways by primary cancer Bayesian network 1 time | 22 | 13 |
| Expanded pathways by primary cancer Bayesian network 2 times | 41 | 0 |
| Expanded pathways by primary cancer Bayesian network 3 time | 55 | 0 |
| Expanded pathways by metastatic cancer Bayesian network 1 time | 11 | 34 |
| Expanded pathways by metastatic cancer Bayesian network 2 times | 5 | 49 |
| Expanded pathways by metastatic cancer Bayesian network 3 time | 1 | 48 |
| Type | Cytokine | Cytokine Receptor | Pattern | Function |
|---|---|---|---|---|
| Chemokines | CXCL11 | CXCR3 | 10→01 | CXCR3 is a molecular target in breast cancer metastasis [48,49] |
| CXCL13 | BLR1 | 10→01 | CXCL13 is overexpressed in breast cancer patients [50,51] | |
| CCL2 | CCR2 | 10→01 | the CCL2-triggered chemokine cascade in macrophages promotes metastatic seeding of breast cancer cells, thereby amplifying the pathology already extant in the system [52] CCL2/CCR2 chemokine signaling coordinates the survival and motility of breast cancer cells with implications on the metastatic process [53] | |
| PDG family | VEGFC | FLT4 | 10→01 | activation of the VEGF-C/Flt-4 axis enhances mobility of cancer cells and contributes to the promotion of metastasis in animals [54] VEGF-C-VEGFR3/Flt4 axis regulates mammary tumor growth and metastasis in an autocrine manner [55] VEGF-C/D and Flt-4 may play an important role in the process of lymphatic metastasis of early-stage invasive cervical carcinoma through paracrine and autocrine mechanisms [56] |
| FLT3LG | FLT3 | 10→01 | FLT3-ligand administration inhibits liver metastases [57] | |
| TNF family | TNFSF8 | TNFRSF8 | 10→01 | TNF inhibitor suppresses bone metastasis in a breast cancer cell line [58] |
| IL-1 family | IL1B | IL1R2 | 10→01 | IL-1B is a potential biomarker for predicting breast cancer patients at increased risk for developing bone metastasis [59] IL-1 drives breast cancer growth and bone metastasis in vivo [60] |
| IL-10 family | IL20 | IL20RA | 01→10 | IL-20 plays pivotal roles in the tumor progression of breast cancer; IL-20 may be a novel target in treating breast tumor-induced osteolysis [61] |
| IL22 | IL22RA2 | 01→10 | IL-22 promotes epithelial cell transformation and breast tumorigenesis [62] | |
| TGF-b family | BMP2 | BMPR1B | 01→10 | Bone morphogenic proteins are related to driving breast cancer metastasis to bone [63] The BMP2/7 heterodimer inhibits the human breast cancer stem cell subpopulation and bone metastases’ formation [64] TGF-β activity is controlled by the expression of BMP7; BMP7 expression has been shown to be inversely proportional to the tumorigenicity and invasive behavior of MDA-MB-231 breast cancer cells [65] The presence of high levels of BMP7 expression in primary tumors has been strongly associated with accelerated bone metastasis, especially from ductal carcinomas [66] |
| BMP7 | ||||
| Hematopoietic | IL12A | IL12RB1 | 10→01 | NA |
| IL12B | 10→01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Li, J.; Liu, E.; Kinnebrew, G.; Zhang, X.; Stover, D.; Huo, Y.; Zeng, Z.; Jiang, W.; Cheng, L.; et al. Identification of Alternatively-Activated Pathways between Primary Breast Cancer and Liver Metastatic Cancer Using Microarray Data. Genes 2019, 10, 753. https://doi.org/10.3390/genes10100753
Wang L, Li J, Liu E, Kinnebrew G, Zhang X, Stover D, Huo Y, Zeng Z, Jiang W, Cheng L, et al. Identification of Alternatively-Activated Pathways between Primary Breast Cancer and Liver Metastatic Cancer Using Microarray Data. Genes. 2019; 10(10):753. https://doi.org/10.3390/genes10100753
Chicago/Turabian StyleWang, Limei, Jin Li, Enze Liu, Garrett Kinnebrew, Xiaoli Zhang, Daniel Stover, Yang Huo, Zhi Zeng, Wanli Jiang, Lijun Cheng, and et al. 2019. "Identification of Alternatively-Activated Pathways between Primary Breast Cancer and Liver Metastatic Cancer Using Microarray Data" Genes 10, no. 10: 753. https://doi.org/10.3390/genes10100753





