A Portal to Visualize Transcriptome Profiles in Mouse Models of Neurological Disorders
Abstract
1. Introduction
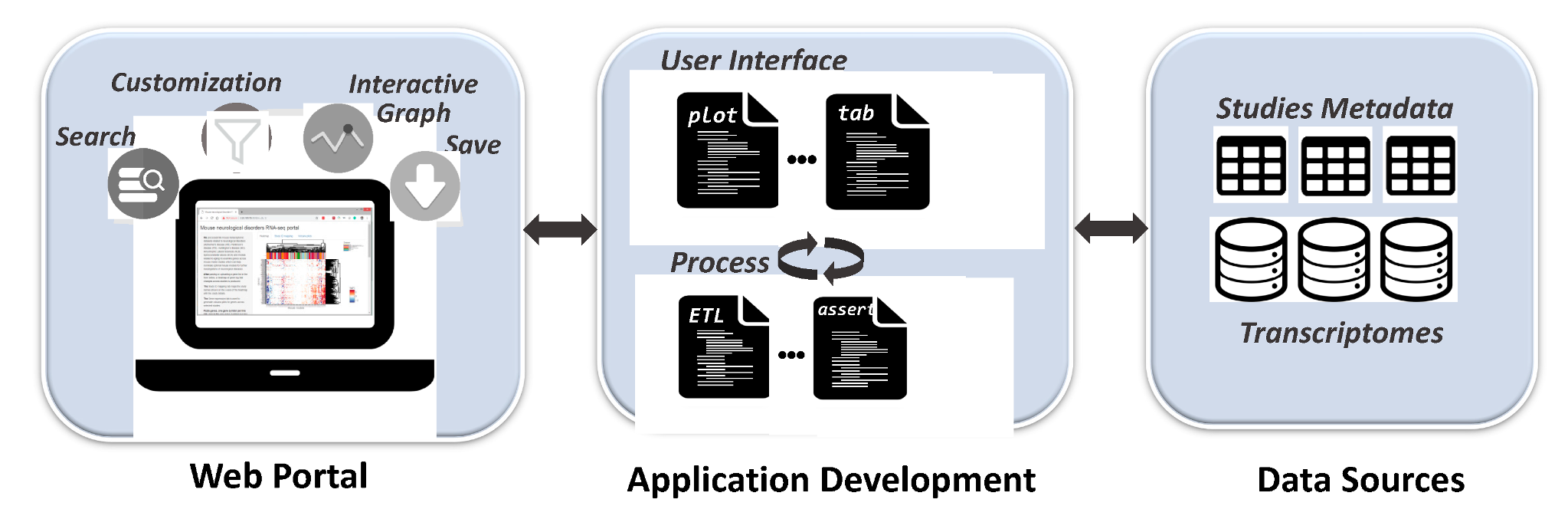
2. Materials and Methods
2.1. Infrastructure
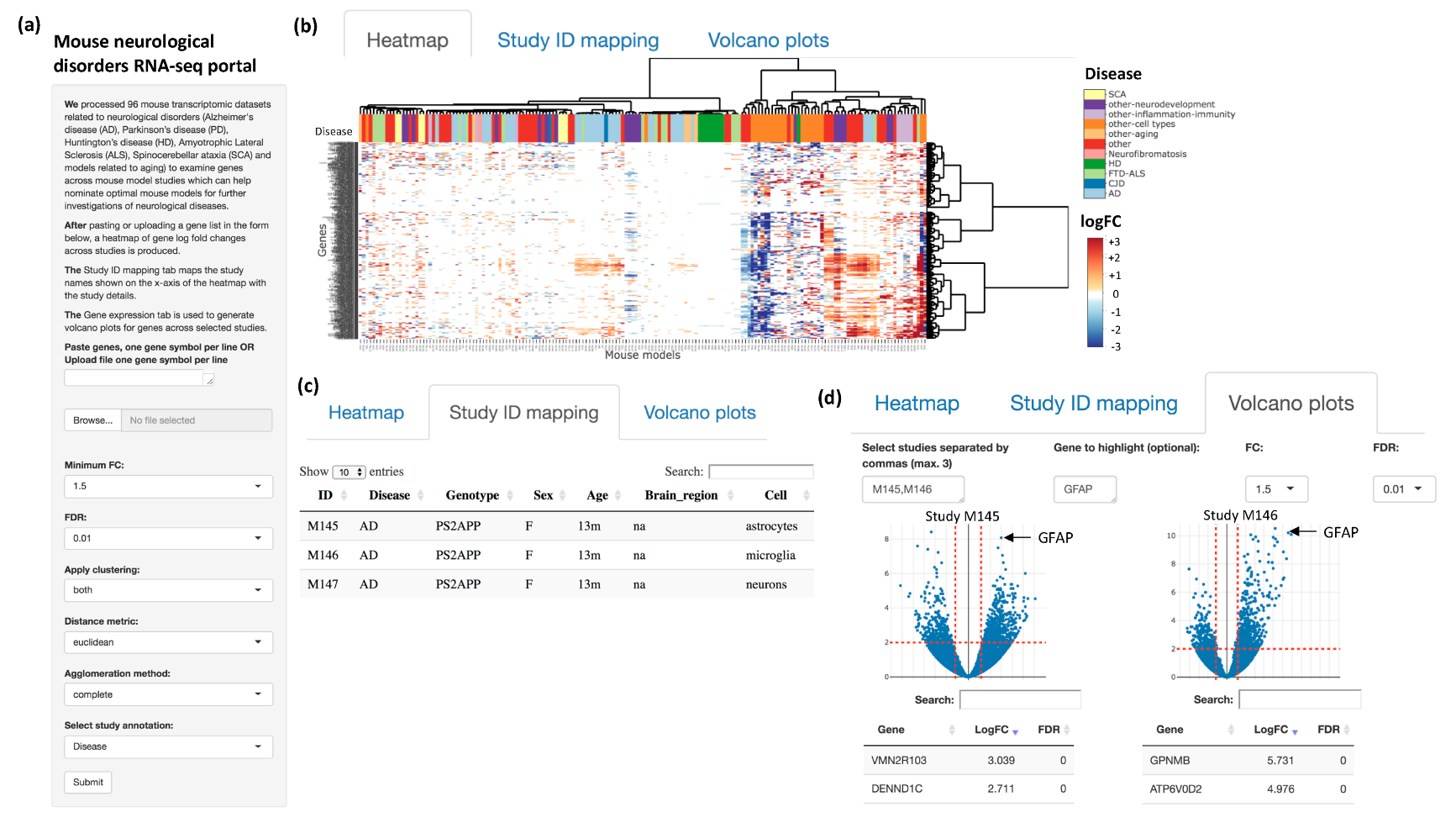
2.2. Web Interface
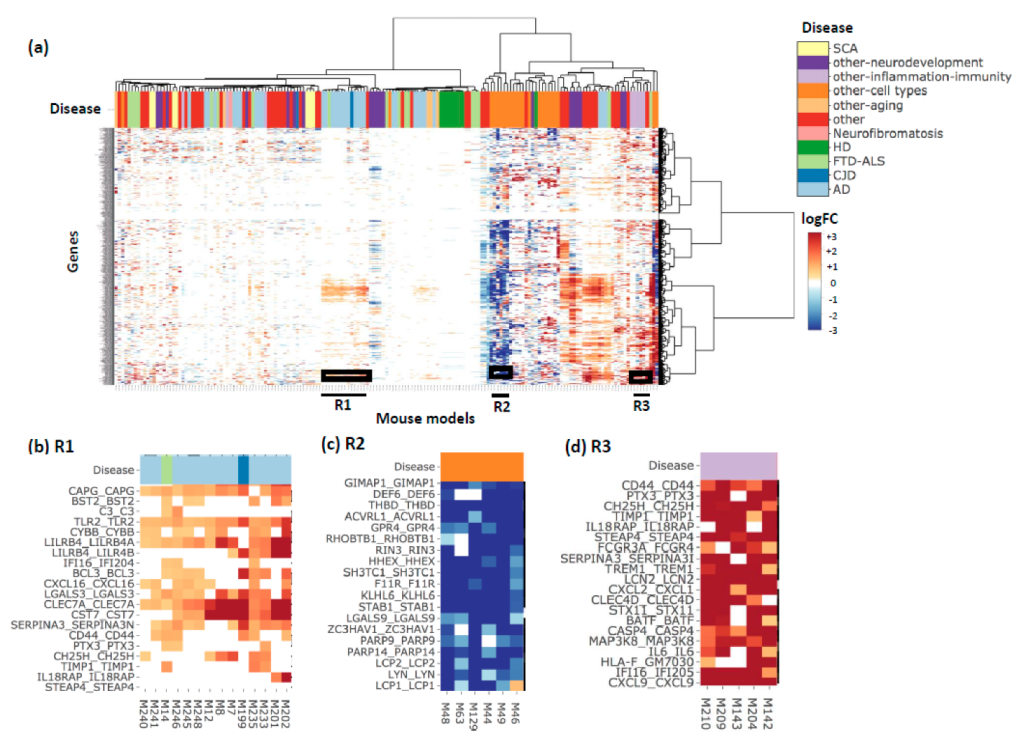
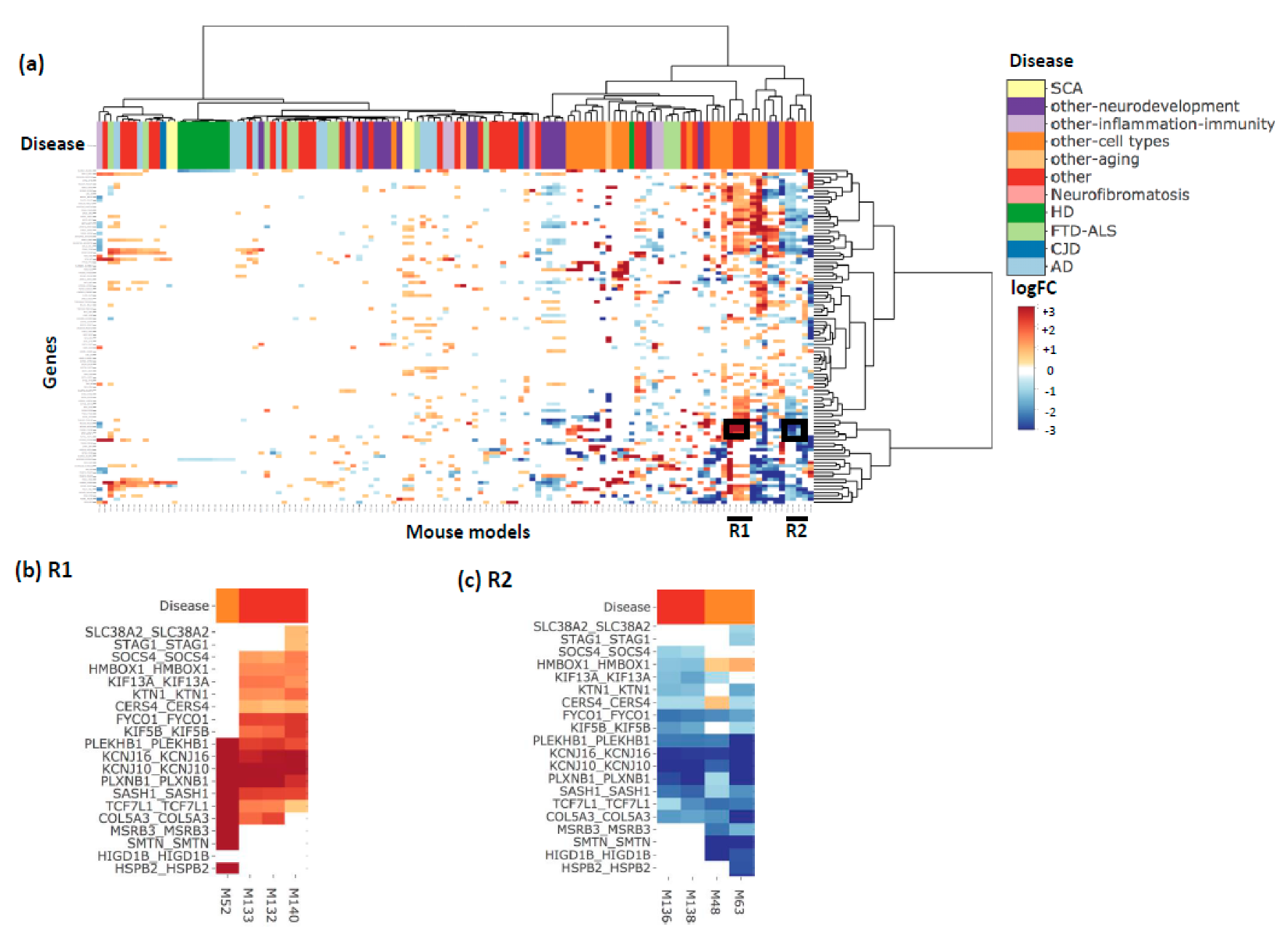
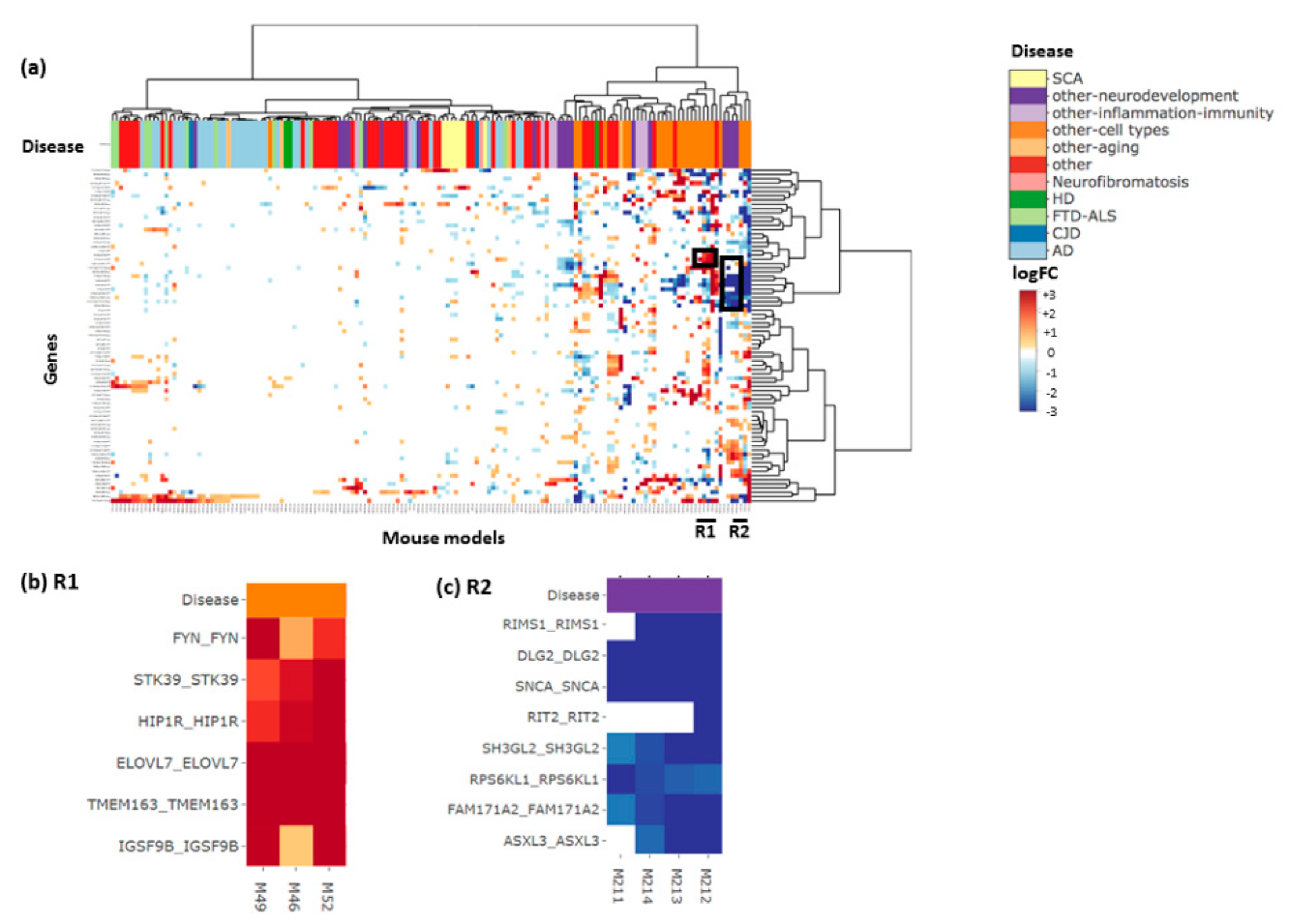
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, K.; Sloan, S.A.; Bennett, M.L.; Scholze, A.R.; O’Keeffe, S.; Phatnani, H.P.; Guarnieri, P.; Caneda, C.; Ruderisch, N.; et al. An RNA-Sequencing Transcriptome and Splicing Database of Glia, Neurons, and Vascular Cells of the Cerebral Cortex. J. Neurosci. 2014, 34, 11929–11947. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sloan, S.A.; Clarke, L.E.; Caneda, C.; Plaza, C.A.; Blumenthal, P.D.; Vogel, H.; Steinberg, G.K.; Edwards, M.S.B.; Li, G.; et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 2016, 89, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Hayamizu, T.F.; Finger, J.H.; Bello, S.M.; McCright, I.J.; Xu, J.; Baldarelli, R.M.; Beal, J.S.; Campbell, J.; Corbani, L.E.; et al. The mouse Gene Expression Database (GXD): 2019 update. Nucleic Acids Res. 2019, 47, D774–D779. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366. [Google Scholar] [CrossRef] [PubMed]
- Wan, Y.W.; Al-Ouran, R.; Mangleburg, C.G.; Lee, T.V.; Allison, K.; Neuner, S.; Kaczorowski, C.; Phillip, V.; Howell, G.; Martini-Stoica, H.; et al. Functional Dissection of Alzheimer’s Disease Brain Gene Expression Signatures in Humans and Mouse Models. bioRxiv 2019. [Google Scholar] [CrossRef]
- Chang, W.; Cheng, J.; Allaire, J.; Xie, Y.; McPherson, J. Shiny: Web Application Framework for R. R Package Version 1.2.0. Available online: https://cran.r-project.org/web/packages/shiny/index.html (accessed on 1 March 2019).
- Galili, T.; O’Callaghan, A.; Sidi, J.; Sievert, C. heatmaply: An R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 2018, 34, 1600–1602. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Gaiteri, C.; Bodea, L.G.; Wang, Z.; McElwee, J.; Podtelezhnikov, A.A.; Zhang, C.; Xie, T.; Tran, L.; Dobrin, R.; et al. Integrated Systems Approach Identifies Genetic Nodes and Networks in Late-Onset Alzheimer’s Disease. Cell 2013, 153, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Mostafavi, S.; Gaiteri, C.; Sullivan, S.E.; White, C.C.; Tasaki, S.; Xu, J.; Taga, M.; Klein, H.U.; Patrick, E.; Komashko, V.; et al. A molecular network of the aging human brain provides insights into the pathology and cognitive decline of Alzheimer’s disease. Nat. Neurosci. 2018, 21, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Solga, A.C.; Pong, W.W.; Walker, J.; Wylie, T.; Magrini, V.; Apicelli, A.J.; Griffith, M.; Griffith, O.L.; Kohsaka, S.; Wu, G.F.; et al. RNA-sequencing reveals oligodendrocyte and neuronal transcripts in microglia relevant to central nervous system disease: Comparative Microglia Transcriptomal Analysis. Glia 2015, 63, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Brichta, L.; Shin, W.; Jackson-Lewis, V.; Blesa, J.; Yap, E.L.; Walker, Z.; Zhang, J.; Roussarie, J.P.; Alvarez, M.J.; Califano, A.; et al. Identification of neurodegenerative factors using translatome–regulatory network analysis. Nat. Neurosci. 2015, 18, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Expanding Parkinson’s disease genetics: Novel risk loci, genomic context, causal insights and heritable risk. bioRxiv 2018. [Google Scholar] [CrossRef]
- Hanamsagar, R.; Alter, M.D.; Block, C.S.; Sullivan, H.; Bolton, J.L.; Bilbo, S.D. Generation of a microglial developmental index in mice and in humans reveals a sex difference in maturation and immune reactivity: HANAMSAGAR. Glia 2017, 65, 1504–1520. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Ouran, R.; Wan, Y.-W.; Mangleburg, C.G.; Lee, T.V.; Allison, K.; Shulman, J.M.; Liu, Z. A Portal to Visualize Transcriptome Profiles in Mouse Models of Neurological Disorders. Genes 2019, 10, 759. https://doi.org/10.3390/genes10100759
Al-Ouran R, Wan Y-W, Mangleburg CG, Lee TV, Allison K, Shulman JM, Liu Z. A Portal to Visualize Transcriptome Profiles in Mouse Models of Neurological Disorders. Genes. 2019; 10(10):759. https://doi.org/10.3390/genes10100759
Chicago/Turabian StyleAl-Ouran, Rami, Ying-Wooi Wan, Carl Grant Mangleburg, Tom V. Lee, Katherine Allison, Joshua M. Shulman, and Zhandong Liu. 2019. "A Portal to Visualize Transcriptome Profiles in Mouse Models of Neurological Disorders" Genes 10, no. 10: 759. https://doi.org/10.3390/genes10100759
APA StyleAl-Ouran, R., Wan, Y.-W., Mangleburg, C. G., Lee, T. V., Allison, K., Shulman, J. M., & Liu, Z. (2019). A Portal to Visualize Transcriptome Profiles in Mouse Models of Neurological Disorders. Genes, 10(10), 759. https://doi.org/10.3390/genes10100759



