Genome-Wide Analysis of Cotton Auxin Early Response Gene Families and Their Roles in Somatic Embryogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of ARF, Aux/IAA, GH3, and SAUR Genes in Upland Cotton
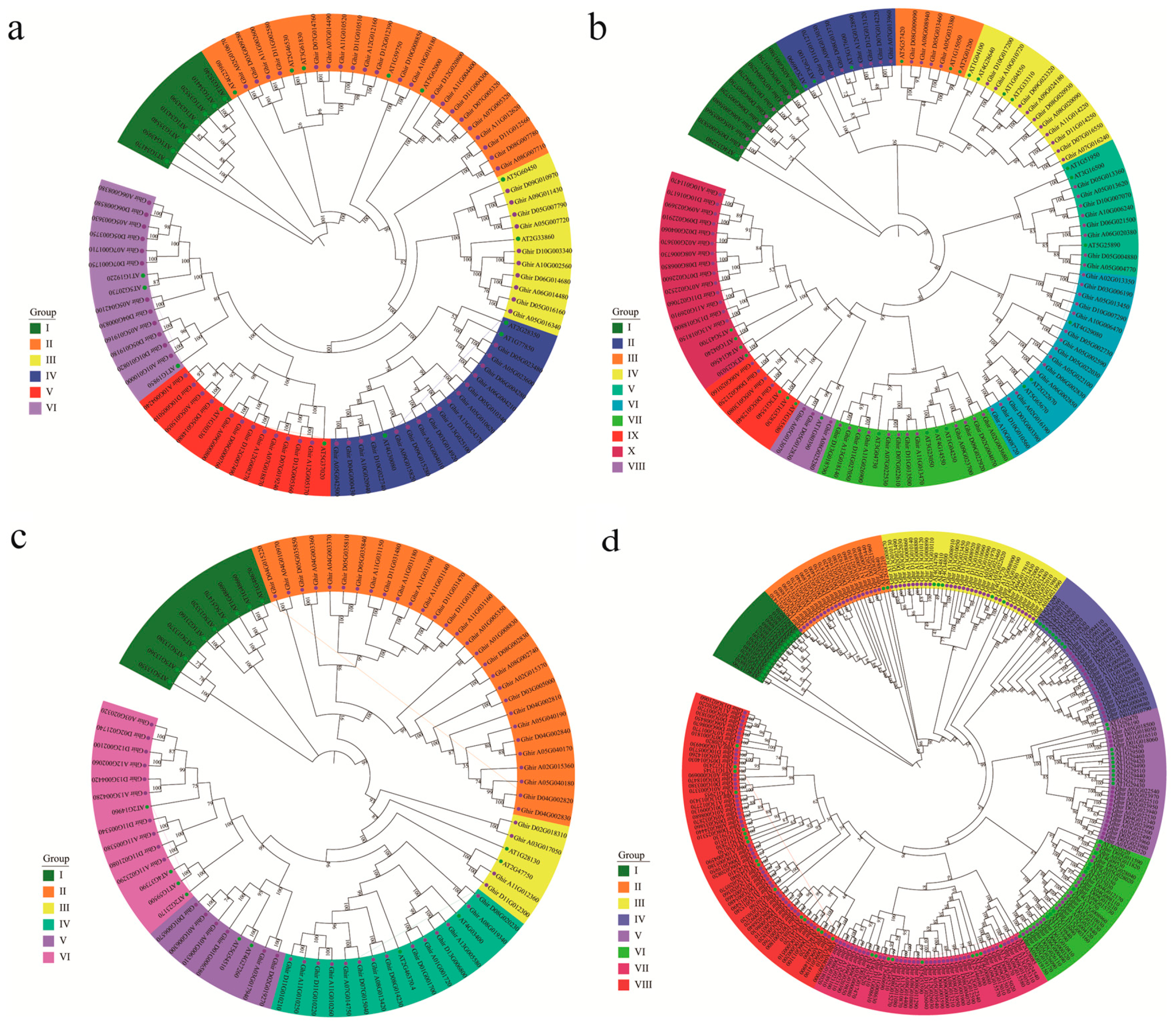
2.2. Phylogenetic Analysis and Gene Structure of Upland Cotton ARF, Aux/IAA, GH3, and SAUR Genes
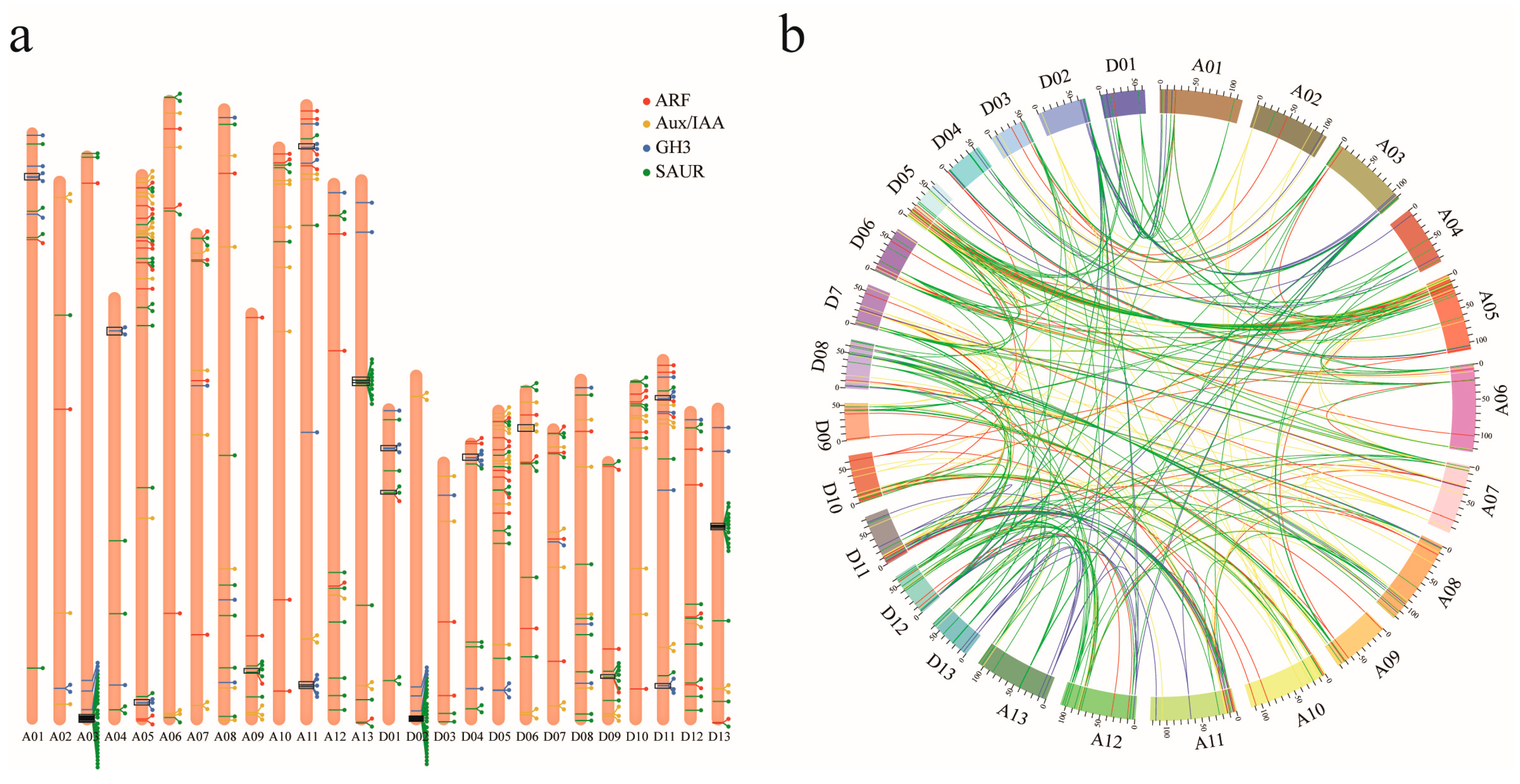
2.3. Genomic Distribution, Collinearity and Duplication Analysis of Upland Cotton ARF, Aux/IAA, GH3, and SAUR Genes
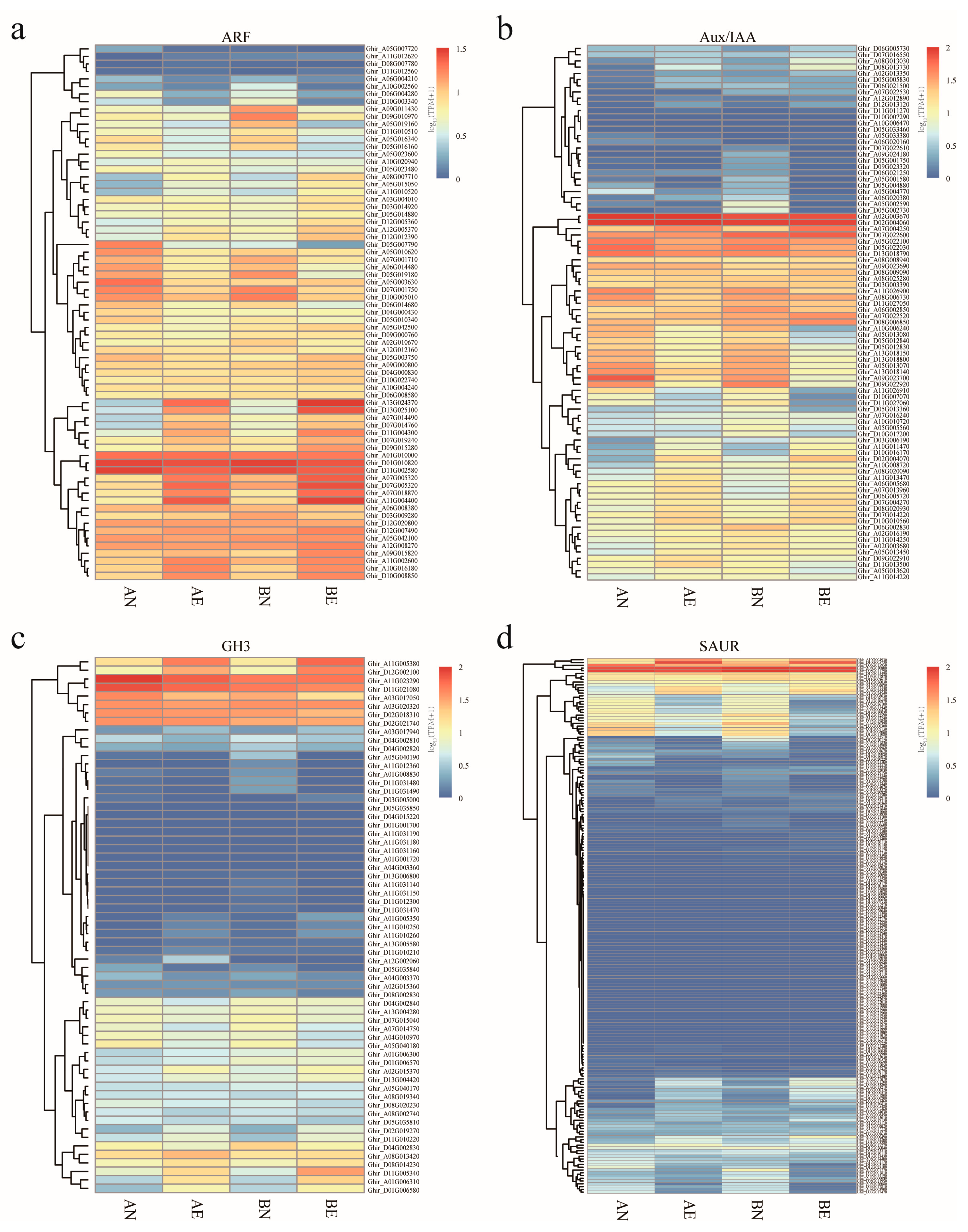
2.4. Expression Profile Analysis of ARF, Aux/IAA, GH3, and SAUR Genes during Embryogenic Ability Acquisition of Upland Cotton Callus
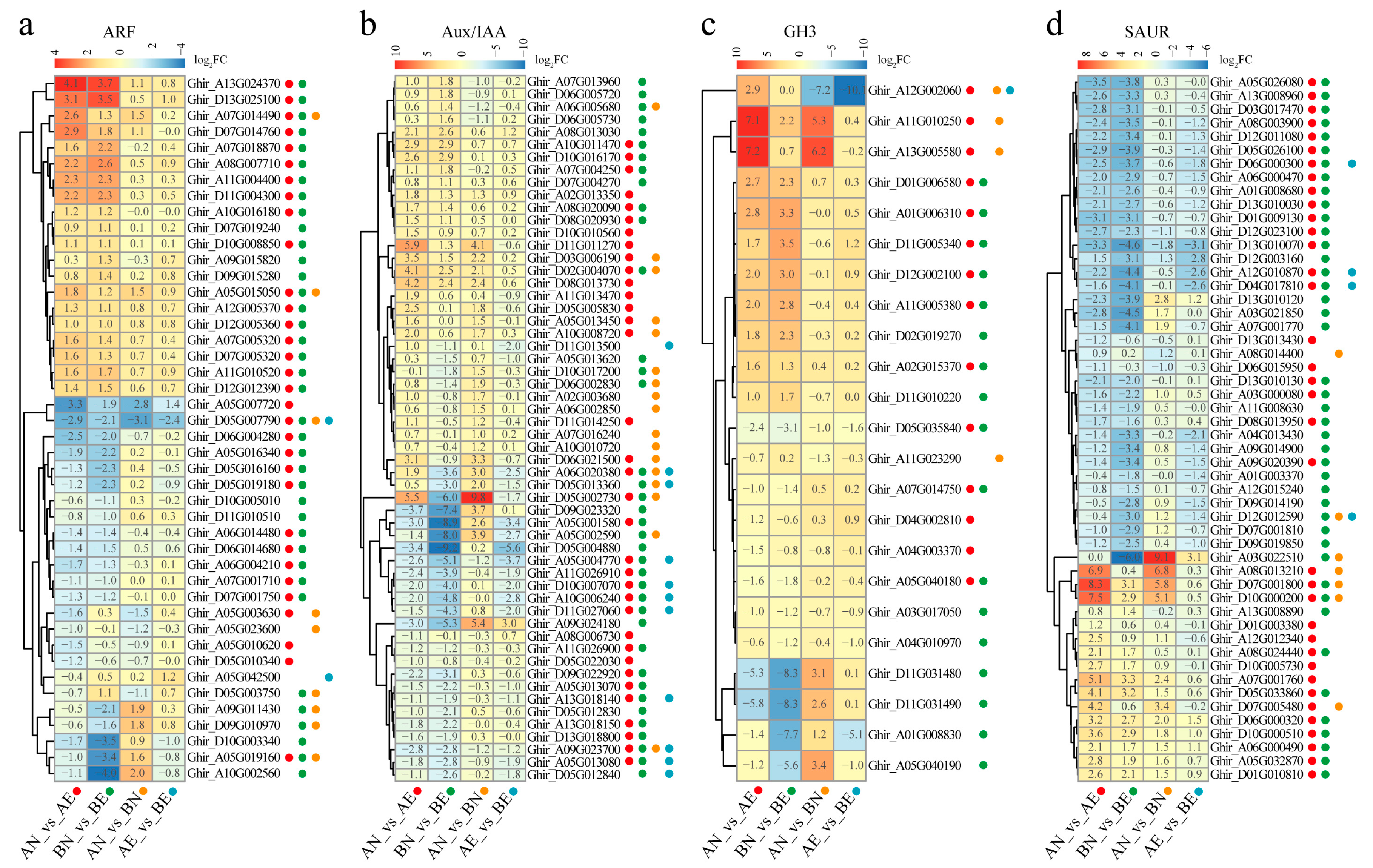
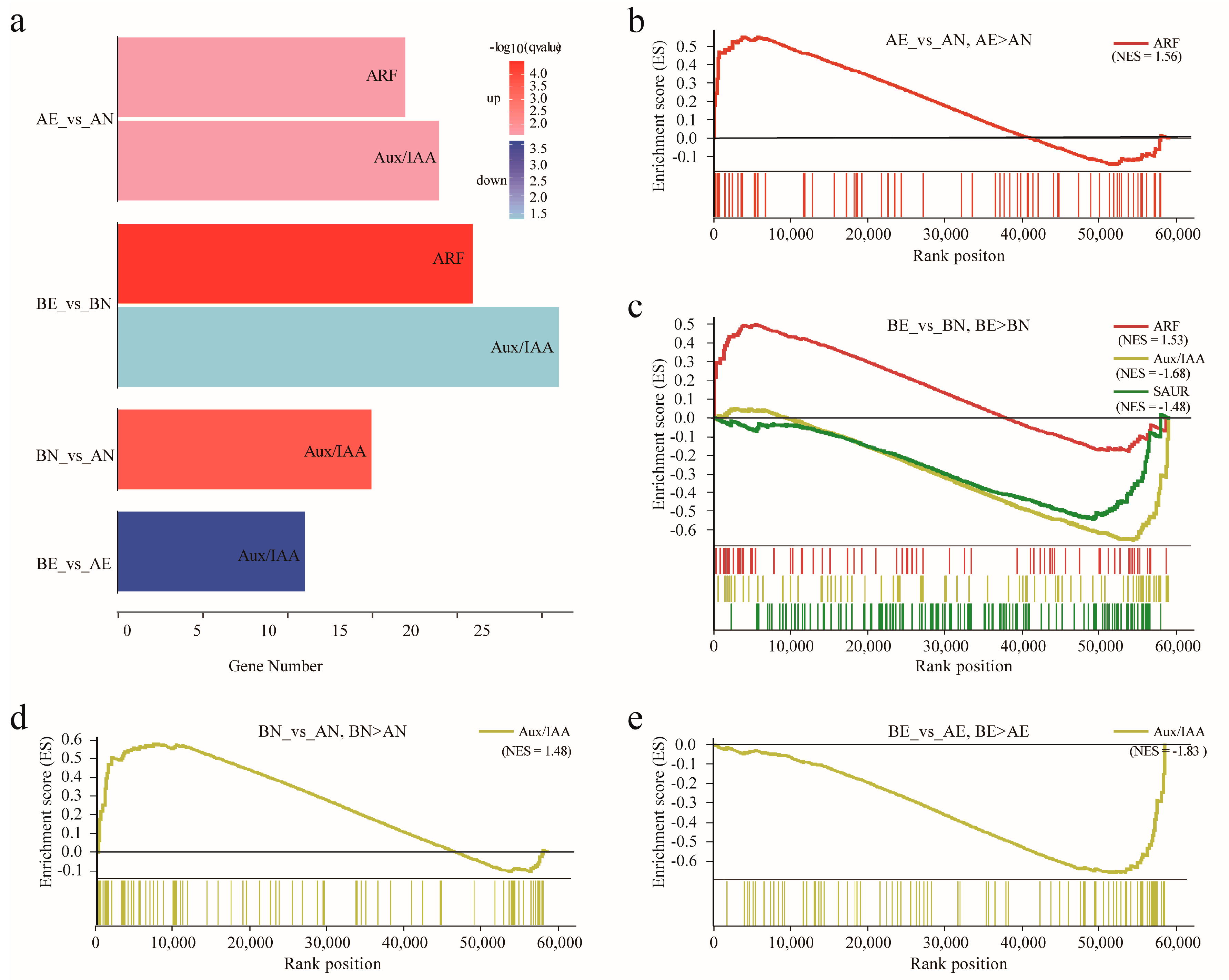
2.5. Differentially Expression Analysis
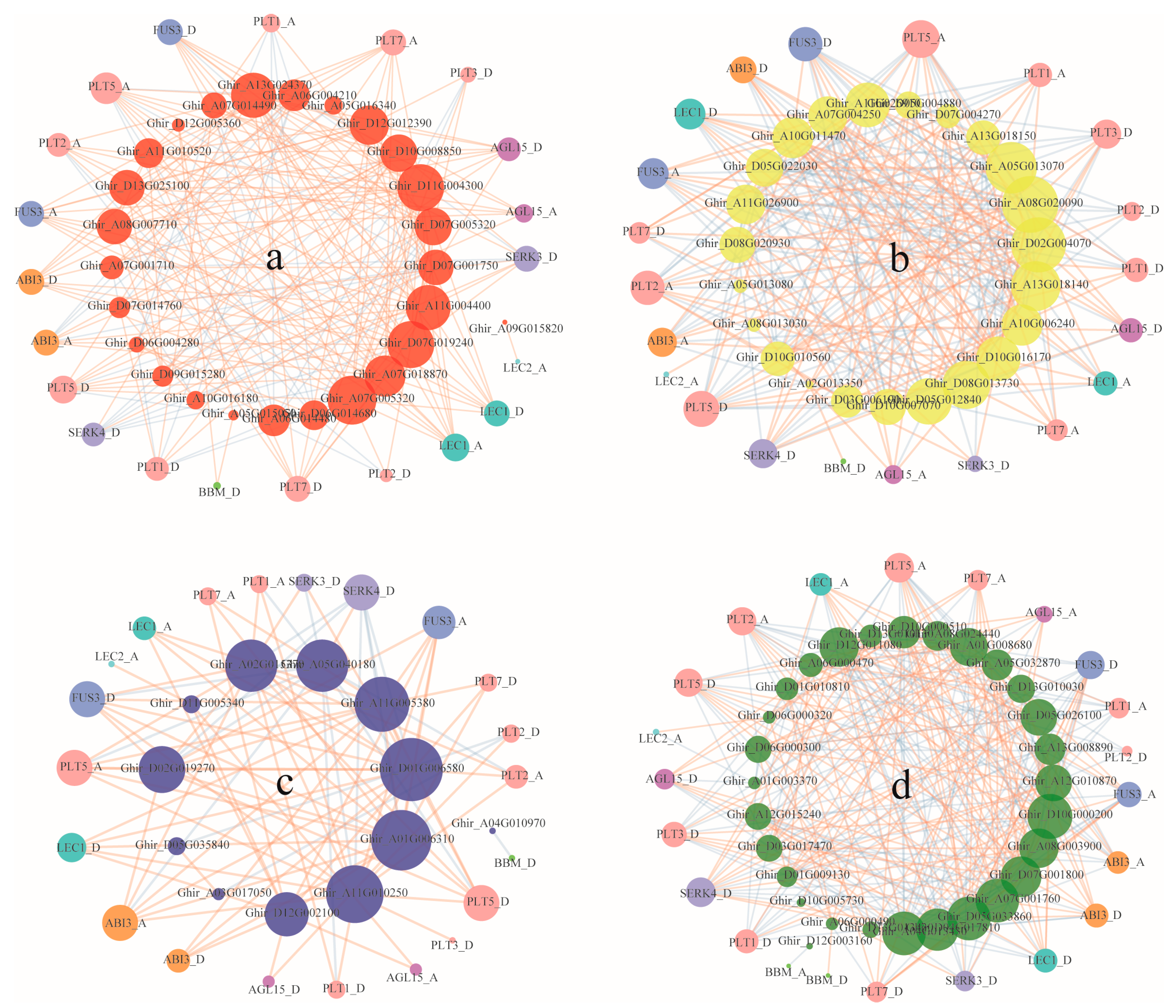
2.6. Co-Expression Network Analysis
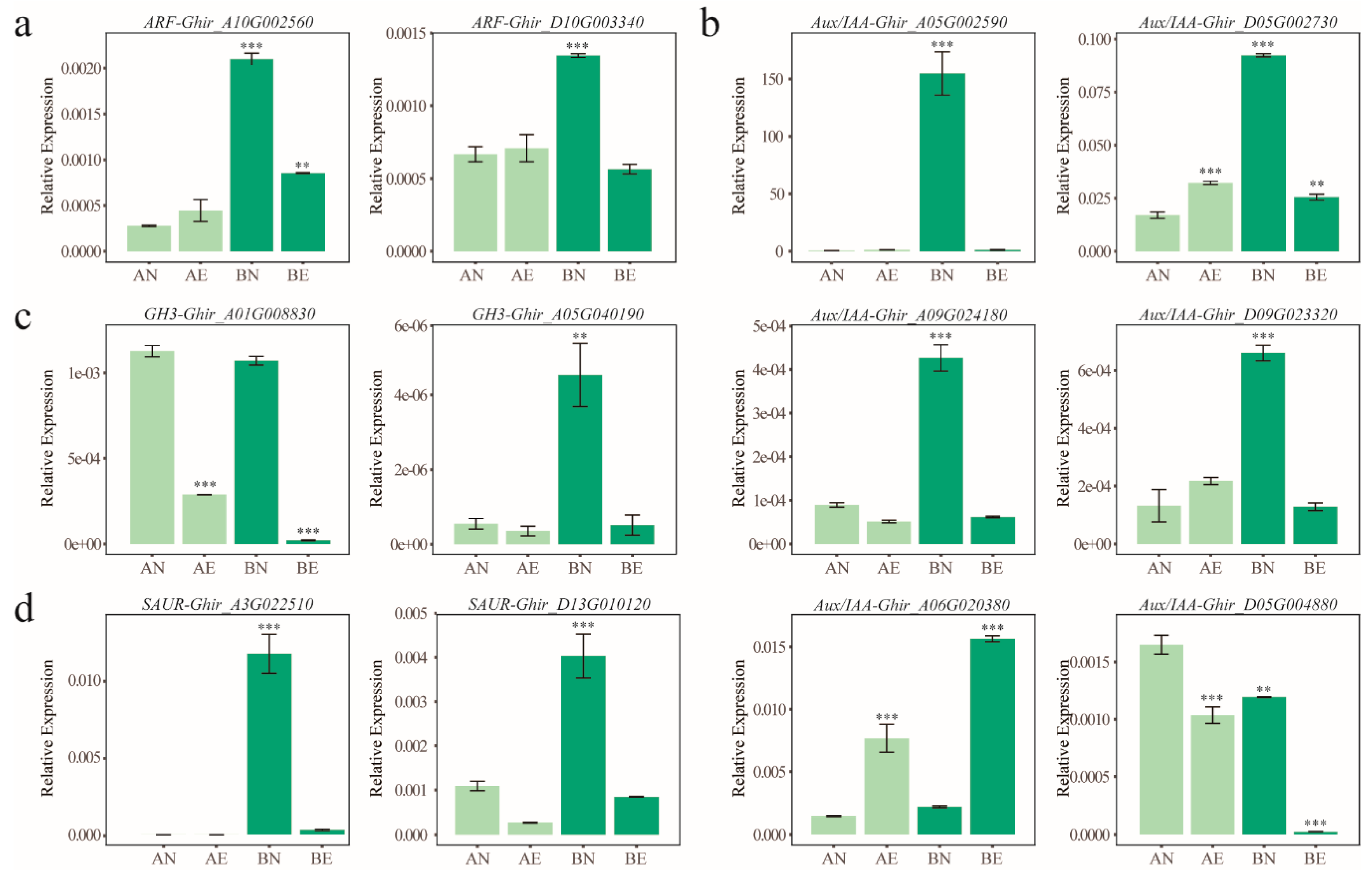
2.7. qRT-PCR Validation and Analysis
3. Results
3.1. Identification and Characterization of ARF, Aux/IAA, GH3, and SAUR Genes in Upland Cotton
3.2. Phylogenetic Analysis and Gene Structure of ARF, Aux/IAA, GH3, and SAUR Genes in Upland Cotton
3.3. Genomic Distribution and Gene Duplication of ARF, Aux/IAA, GH3, and SAUR Genes in Upland Cotton
3.4. Expression Profiling and Differentially Expression of ARF, Aux/IAA, GH3, and SAUR Genes in Upland Cotton Somatic Embryogenesis
3.5. Co-Expression of ARF, Aux/IAA, GH3, and SAUR Genes and Somatic Embryogenesis Related Genes during Cotton Somatic Embryogenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Shi, Y.-H.; Zhu, S.-W.; Mao, X.-Z.; Feng, J.-X.; Qin, Y.-M.; Zhang, L.; Cheng, J.; Wei, L.-P.; Wang, Z.-Y.; Zhu, Y.-X. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 2006, 18, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Sunilkumar, G.; Campbell, L.M.; Puckhaber, L.; Stipanovic, R.D.; Rathore, K.S. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. PNAS 2006, 103, 18054–18059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Wang, Y.; Sun, G.; Jin, S.; Zhou, T.; Meng, Z.; Zhang, R. Twenty years of research and application of transgenic cotton in China. Scientia Agricultura Sinica 2015, 48, 3372–3387. [Google Scholar]
- Liu, C.; Tian, R.; Kong, D.; Li, F.; Shang, H.; Chen, X. Establishment and application of efficient transformation system for cotton. Scientia Agricultura Sinica 2014, 47, 4183–4197. [Google Scholar]
- Zhang, B.-H.; Liu, F.; Yao, C.-B. Plant regeneration via somatic embryogenesis in cotton. Plant Cell Tissue Organ Cult. 2000, 60, 89–94. [Google Scholar] [CrossRef]
- Leelavathi, S.; Sunnichan, V.G.; Kumria, R.; Vijaykanth, G.P.; Bhatnagar, R.K.; Reddy, V.S. A simple and rapid Agrobacterium -mediated transformation protocol for cotton (Gossypium hirsutum L.): Embryogenic calli as a source to generate large numbers of transgenic plants. Plant Cell Rep. 2004, 22, 465–470. [Google Scholar] [CrossRef]
- Li, J.; Wang, M.; Li, Y.; Zhang, Q.; Lindsey, K.; Daniell, H.; Jin, S.; Zhang, X. Multi-omics analyses reveal epigenomics basis for cotton somatic embryogenesis through successive regeneration acclimation (SRA) process. Plant Biotechnol. J. 2019, 17, 435–450. [Google Scholar] [CrossRef]
- Rajeswari, S.; Muthuramu, S.; Chandirakala, R.; Thiruvengadam, V.; Raveendran, T.S. Callus induction, somatic embryogenesis and plant regeneration in cotton (Gossypium hirsutum L.). Electron. J. Plant Breed. 2010, 1, 1186–1190. [Google Scholar]
- Jin, S.; Zhang, X.; Liang, S.; Nie, Y.; Guo, X.; Huang, C. Factors affecting transformation efficiency of embryogenic callus of Upland cotton (Gossypium hirsutum) with Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult. 2005, 81, 229–237. [Google Scholar] [CrossRef]
- Zhang, C.J.; Yu, S.X.; Fan, S.L.; Zhang, J.F.; Li, F.G. Inheritance of somatic embryogenesis using leaf petioles as explants in upland cotton. Euphytica 2011, 181, 55–63. [Google Scholar] [CrossRef]
- Deo, P.C.; Tyagi, A.P.; Taylor, M.; Harding, R.; Becker, D. Factors affecting somatic embryogenesis and transformation in modern plant breeding. S. Pac. J. Nat. App. Sci. 2010, 28, 27. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, C.; Zhang, X.; Liu, C.; Wu, Z.; Yang, Z.; Zhou, K.; Yang, X.; Li, F. Transcriptome profiling reveals auxin and cytokinin regulating somatic embryogenesis in different sister lines of cotton cultivar CCRI24. J. Integr. Plant Biol. 2013, 55, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, X.; Yuan, D.; Jin, F.; Zhang, Y.; Xu, J. Transcript profiling reveals complex auxin signalling pathway and transcription regulation involved in dedifferentiation and redifferentiation during somatic embryogenesis in cotton. BMC Plant Biol. 2012, 12, 110. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Zhang, X.; Jin, S.; Cheng, L.; Liang, S.; Hu, L.; Guo, X.; Nie, Y.; Cao, J. Chromatin reorganization and endogenous auxin/cytokinin dynamic activity during somatic embryogenesis of cultured cotton cell. Plant Cell Tissue Organ Cult. 2007, 90, 63–70. [Google Scholar] [CrossRef]
- Abel, S.; Theologis, A. Early genes and auxin action. Plant Physiol. 1996, 111, 9–17. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genetics 2009, 43, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Tiwari, S.B. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.-J.; Zhang, J.-Z. Aux/IAA gene family in plants: Molecular structure, regulation, and function. Int. J. Mole. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Tiwari, S.B.; Wang, X.-J.; Hagen, G.; Guilfoyle, T.J. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 2001, 13, 2809–2822. [Google Scholar] [CrossRef]
- Gray, W.M.; Kepinski, S.; Rouse, D.; Leyser, O.; Estelle, M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 2001, 414, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.R.; Gee, M.A.; Guilfoyle, T.J. Induction and superinduction of auxin-responsive mRNAs with auxin and protein synthesis inhibitors. J. Biol. Chem. 1990, 265, 15845–15849. [Google Scholar] [PubMed]
- McClure, B.; Guilfoyle, T. Rapid redistribution of auxin-regulated RNAs during gravitropism. Science 1989, 243, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Gray, W.M. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Cao, A.; Zheng, Y.; Yu, Y.; Wang, X.; Shao, D.; Sun, J.; Cui, B. Comparative transcriptome analysis of SE initial dedifferentiation in cotton of different SE capability. Sci. Rep. 2017, 7, 8583. [Google Scholar] [CrossRef]
- Jin, F.; Hu, L.; Yuan, D.; Xu, J.; Gao, W.; He, L.; Yang, X.; Zhang, X. Comparative transcriptome analysis between somatic embryos (SEs) and zygotic embryos in cotton: Evidence for stress response functions in SE development. Plant Biotechnol. J. 2014, 12, 161–173. [Google Scholar] [CrossRef]
- Sun, R.; Tian, R.; Ma, D.; Wang, S.; Liu, C. Comparative transcriptome study provides insights into acquisition of embryogenic ability in upland cotton during somatic embryogenesis. J. Cotton Res. 2018, 1, 9. [Google Scholar] [CrossRef]
- Wang, M.; Tu, L.; Yuan, D.; Zhu, D.; Shen, C.; Li, J.; Liu, F.; Pei, L.; Wang, P.; Zhao, G.; et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 2019, 51, 224–229. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinforma. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
- Hao, Z.; Lv, D.; Ge, Y.; Shi, J.; Yu, G.; Chen, J. RIdeogram: Drawing SVG graphics to visualize and map genome-wide data in idiograms. PeerJ Preprints 2019. [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Croken, M.; Qiu, W.; White, M.W.; Kim, K. Gene Set Enrichment Analysis (GSEA) of Toxoplasma gondii expression datasets links cell cycle progression and the bradyzoite developmental program. BMC Genomics 2014, 15, 515. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Cur. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.A.; Zenser, N.; Leyser, O.; Callis, J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 2001, 13, 2349. [Google Scholar] [CrossRef] [PubMed]
- Moss, B.L.; Mao, H.; Guseman, J.M.; Hinds, T.R.; Hellmuth, A.; Kovenock, M.; Noorassa, A.; Lanctot, A.; Villalobos, L.I.A.C.; Zheng, N.; et al. Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol. 2015, 169, 803–813. [Google Scholar] [CrossRef]
- Yu, D.; Qanmber, G.; Lu, L.; Wang, L.; Li, J.; Yang, Z.; Liu, Z.; Li, Y.; Chen, Q.; Mendu, V.; et al. Genome-wide analysis of cotton GH3 subfamily II reveals functional divergence in fiber development, hormone response and plant architecture. BMC Plant Biol. 2018, 18. [Google Scholar] [CrossRef]
- Sun, R.; Wang, K.; Guo, T.; Jones, D.C.; Cobb, J.; Zhang, B.; Wang, Q. Genome-wide identification of auxin response factor (ARF) genes and its tissue-specific prominent expression in Gossypium raimondii. Funct. Integr. Genomics 2015, 15, 481–493. [Google Scholar] [CrossRef]
- Xiao, G.; He, P.; Zhao, P.; Liu, H.; Zhang, L.; Pang, C.; Yu, J. Genome-wide identification of the GhARF gene family reveals that GhARF2 and GhARF18 are involved in cotton fibre cell initiation. J. Exp. Bot. 2018, 69, 4323–4337. [Google Scholar] [CrossRef]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nature Biotechnol. 2015, 33, 531. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, G.; Geng, Y.; Wu, M.; Pei, W.; Zhai, H.; Zang, X.; Li, X.; Zhang, J.; Yu, S.; et al. A genome-wide analysis of the small auxin-up RNA (SAUR) gene family in cotton. BMC Genomics 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi, B.; Masoomi-Aladizgeh, F.; Shariatpanahi, M.E.; Azadi, P.; Keshavarz-Alizadeh, M. Molecular characterization and expression analysis of SERK1 and SERK2 in Brassica napus L.: Implication for microspore embryogenesis and plant regeneration. Plant Cell Rep. 2016, 35, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Elhiti, M.; Stasolla, C.; Wang, A. Molecular regulation of plant somatic embryogenesis. In Vitro Cell. Dev. Biol., Plant 2013, 49, 631–642. [Google Scholar] [CrossRef]
- Horstman, A.; Bemer, M.; Boutilier, K. A transcriptional view on somatic embryogenesis. Regeneration 2017, 4, 201–216. [Google Scholar] [CrossRef] [PubMed]
- Horstman, A.; Li, M.; Heidmann, I.; Weemen, M.; Chen, B.; Muiño, J.M.; Angenent, G.C.; Boutilier, K. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 2017, 175, 848–857. [Google Scholar] [CrossRef]
- Bouchabke-Coussa, O.; Obellianne, M.; Linderme, D.; Montes, E.; Maia-Grondard, A.; Vilaine, F.; Pannetier, C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013, 32, 675–686. [Google Scholar] [CrossRef]
- Su, Y.H.; Zhao, X.Y.; Liu, Y.B.; Zhang, C.L.; O’Neill, S.D.; Zhang, X.S. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 2009, 59, 448–460. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, X.; Lin, Y.; Xie, G.; Fu, F.; Liu, H.; Wang, J.; Gao, S.; Lan, H.; Rong, T. Characterization of a ZmSERK gene and its relationship to somatic embryogenesis in a maize culture. Plant Cell Tissue Organ Cult. 2011, 105, 29–37. [Google Scholar] [CrossRef]
- Zheng, W.; Zhang, X.; Yang, Z.; Wu, J.; Li, F.; Duan, L.; Liu, C.; Lu, L.; Zhang, C.; Li, F. AtWuschel promotes formation of the embryogenic callus in Gossypium hirsutum. PLoS ONE 2014, 9, e87502. [Google Scholar] [CrossRef]
- Gil, P.; Liu, Y.; Orbovic, V.; Verkamp, E.; Poff, K.L.; Green, P.J. Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol. 1994, 104, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Okushima, Y. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [PubMed]
- Remington, D.L. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 2004, 135, 1738–1752. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Tyagi, A.K.; Sharma, A.K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genetics Genomics 2011, 285, 245–260. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Agarwal, P.; Tyagi, A.K.; Sharma, A.K. Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Mol. Genetics Genomics 2012, 287, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, S.; He, Y.; Guan, X.; Zhu, X.; Cheng, L.; Wang, J.; Lu, G. Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 2012, 509, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Peng, Z.; Liu, S.; He, Y.; Cheng, L.; Kong, F.; Wang, J.; Lu, G. Genome-wide analysis of Aux/IAA gene family in Solanaceae species using tomato as a model. Mol. Genet Genomics 2012, 287, 295–311. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Dong, C.; Ma, Y.; Deng, L.; He, S.; Yi, S.; Lv, Q.; Zheng, Y. Comprehensive analysis of SAUR gene family in citrus and its transcriptional correlation with fruitlet drop from abscission zone A. Funct. Integr. Genomics 2015, 15, 729–740. [Google Scholar] [CrossRef]
- Wu, J.; Liu, S.; Guan, X.; Chen, L.; He, Y.; Wang, J.; Lu, G. Genome-wide identification and transcriptional profiling analysis of auxin response-related gene families in cucumber. BMC Res. Notes 2014, 7, 218. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 2006, 6, 47–59. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Tyagi, A.K.; Khurana, J.P. The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genomics 2006, 6, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hao, X.; Cao, J. Small auxin upregulated RNA (SAUR) gene family in maize: Identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum: SAUR gene family in maize. J. Integr. Plant Biol. 2014, 56, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Yue, R.; Tao, S.; Yang, Y.; Zhang, L.; Xu, M.; Wang, H.; Shen, C. Genome-wide identification, expression analysis of auxin-responsive GH3 family genes in maize (Zea mays L.) under abiotic stresses: Genome-wide analysis of ZmGH3 gene family. J. Integr. Plant Biol. 2015, 57, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Jiang, H.; Chen, W.; Qian, Y.; Ma, Q.; Cheng, B.; Zhu, S. Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul. 2011, 63, 225–234. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, D.; Bian, Y.; Lv, Y.; Xie, Q. Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays. L.). Mol. Biol. Rep. 2010, 37, 3991–4001. [Google Scholar] [CrossRef]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef]
- Yan, Y.; Stolz, S.; Chetelat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 2007, 19, 2470–2483. [Google Scholar] [CrossRef]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.-S.; Amasino, R.; Scheres, B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef]
- Horstman, A.; Willemsen, V.; Boutilier, K.; Heidstra, R. AINTEGUMENTA-LIKE proteins: Hubs in a plethora of networks. Trends Plant Sci. 2014, 19, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Jha, P.; Kumar, V. BABY BOOM (BBM): A candidate transcription factor gene in plant biotechnology. Biotechnol. Lett. 2018, 40, 1467–1475. [Google Scholar] [CrossRef] [PubMed]
| Gene Family | Domains/Motifs | Number of Genes |
|---|---|---|
| ARF | 71 | |
| DBD, ARF-AD a, Aux/IAA | 24 | |
| DBD, ARF-RD a, Aux/IAA | 47 | |
| Aux/IAA | 86 | |
| Domains I, II, III, IV | 67 | |
| Domains I, II, III | 1 | |
| Domains II, III, IV | 13 | |
| Domains I, II | 1 | |
| Domains I, IV | 1 | |
| Domains II, III | 1 | |
| Domains III, IV | 2 | |
| GH3 | 63 | |
| GH3 | 47 | |
| 2 × GH3 | 8 | |
| GH3, JAR1 | 8 | |
| SAUR | 194 | |
| SAUR, Motifs 1 b, 2, 3 | 167 | |
| SAUR, Motifs 1, 2 | 9 | |
| SAUR, Motifs 1, 3 | 2 | |
| SAUR, Motifs 2, 3 | 12 | |
| SAUR, Motif 2 | 3 | |
| SAUR, Motif 3 | 1 |
| DEGs | AN_vs_AE | BN_vs_BE | AN_vs_BN | AE_vs_BE | Total | |
|---|---|---|---|---|---|---|
| ARF | 32 | 38 | 9 | 2 | 44 | |
| up | 17 | 21 | 5 | 1 | - | |
| down | 15 | 17 | 4 | 1 | - | |
| Aux/IAA | 35 | 38 | 17 | 11 | 56 | |
| up | 19 | 12 | 15 | 0 | - | |
| down | 16 | 26 | 2 | 11 | - | |
| GH3 | 14 | 17 | 4 | 1 | 23 | |
| up | 9 | 8 | 2 | 0 | - | |
| down | 5 | 9 | 2 | 1 | - | |
| SAUR | 36 | 43 | 7 | 4 | 52 | |
| up | 15 | 10 | 6 | 0 | - | |
| down | 21 | 33 | 1 | 4 | - |
| Auxin Early Response Family | DEGs | Number (Percentage) of DEGs Co-Expressed with | |||
|---|---|---|---|---|---|
| ARF | Aux/IAA | GH3 | SAUR | ||
| ARF | 44 | 38 (86.4%) | 39 (88.6%) | 37 (84.1%) | 40 (90.9%) |
| Aux/IAA | 56 | 39 (69.6%) | 41 (73.2%) | 39 (69.6%) | 41 (73.2%) |
| GH3 | 23 | 18 (78.3%) | 19 (82.6%) | 17 (73.9%) | 20 (87.0%) |
| SAUR | 52 | 48 (92.3%) | 49 (94.2%) | 45 (86.5%) | 49 (94.2%) |
| SE Related Genes | Number (Percentage) of Co-Expressed Auxin Early Response Genes | |||
|---|---|---|---|---|
| ARF | AuxIAA | GH3 | SAUR | |
| ABI3 | 14 (53.8%) | 15 (65.2%) | 8 (66.7%) | 18 (62.1%) |
| AGL15 | 8 (30.8%) | 8 (34.8%) | 3 (25.0%) | 9 (31.0%) |
| FUS3 | 12 (46.2%) | 13 (56.5%) | 8 (66.7%) | 16 (55.2%) |
| LEC1 | 10 (38.5%) | 11 (47.8%) | 5 (41.7%) | 12 (41.4%) |
| LEC2 | 1 (3.8%) | 1 (4.3%) | 1 (8.3%) | 2 (6.9%) |
| PLT1 | 8 (30.8%) | 8 (34.85) | 3 (25.0%) | 11 (37.9%) |
| PLT2 | 8 (30.8%) | 14 (60.9%) | 6 (50.0%) | 14 (48.3%) |
| PLT3 | 5 (19.2%) | 10 (43.5%) | 1 (8.3%) | 12 (41.4%) |
| PLT7 | 10 (38.5%) | 7 (30.4%) | 3 (25.0%) | 9 (31.0%) |
| BBM | 2 (7.7%) | 1 (4.3%) | 1 (8.3%) | 2 (6.9%) |
| PLT5 | 12 (46.2%) | 17 (73.9%) | 7 (58.3%) | 16 (55.2%) |
| WUS | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| SERK3 | 9 (34.6%) | 5 (21.7%) | 3 (25.0%) | 9 (31.0%) |
| SERK4 | 8 (30.8%) | 10 (43.5%) | 7 (58.3%) | 15 (51.7) |
| Total | 26 | 23 | 12 | 29 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, R.; Wang, S.; Ma, D.; Li, Y.; Liu, C. Genome-Wide Analysis of Cotton Auxin Early Response Gene Families and Their Roles in Somatic Embryogenesis. Genes 2019, 10, 730. https://doi.org/10.3390/genes10100730
Sun R, Wang S, Ma D, Li Y, Liu C. Genome-Wide Analysis of Cotton Auxin Early Response Gene Families and Their Roles in Somatic Embryogenesis. Genes. 2019; 10(10):730. https://doi.org/10.3390/genes10100730
Chicago/Turabian StyleSun, Ruibin, Shaohui Wang, Dan Ma, Yilin Li, and Chuanliang Liu. 2019. "Genome-Wide Analysis of Cotton Auxin Early Response Gene Families and Their Roles in Somatic Embryogenesis" Genes 10, no. 10: 730. https://doi.org/10.3390/genes10100730
APA StyleSun, R., Wang, S., Ma, D., Li, Y., & Liu, C. (2019). Genome-Wide Analysis of Cotton Auxin Early Response Gene Families and Their Roles in Somatic Embryogenesis. Genes, 10(10), 730. https://doi.org/10.3390/genes10100730





