EPA and DHA Inhibit Myogenesis and Downregulate the Expression of Muscle-related Genes in C2C12 Myoblasts
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Fatty Acids Preparation
2.2. Cell Proliferation Assay
2.3. Edu Assays
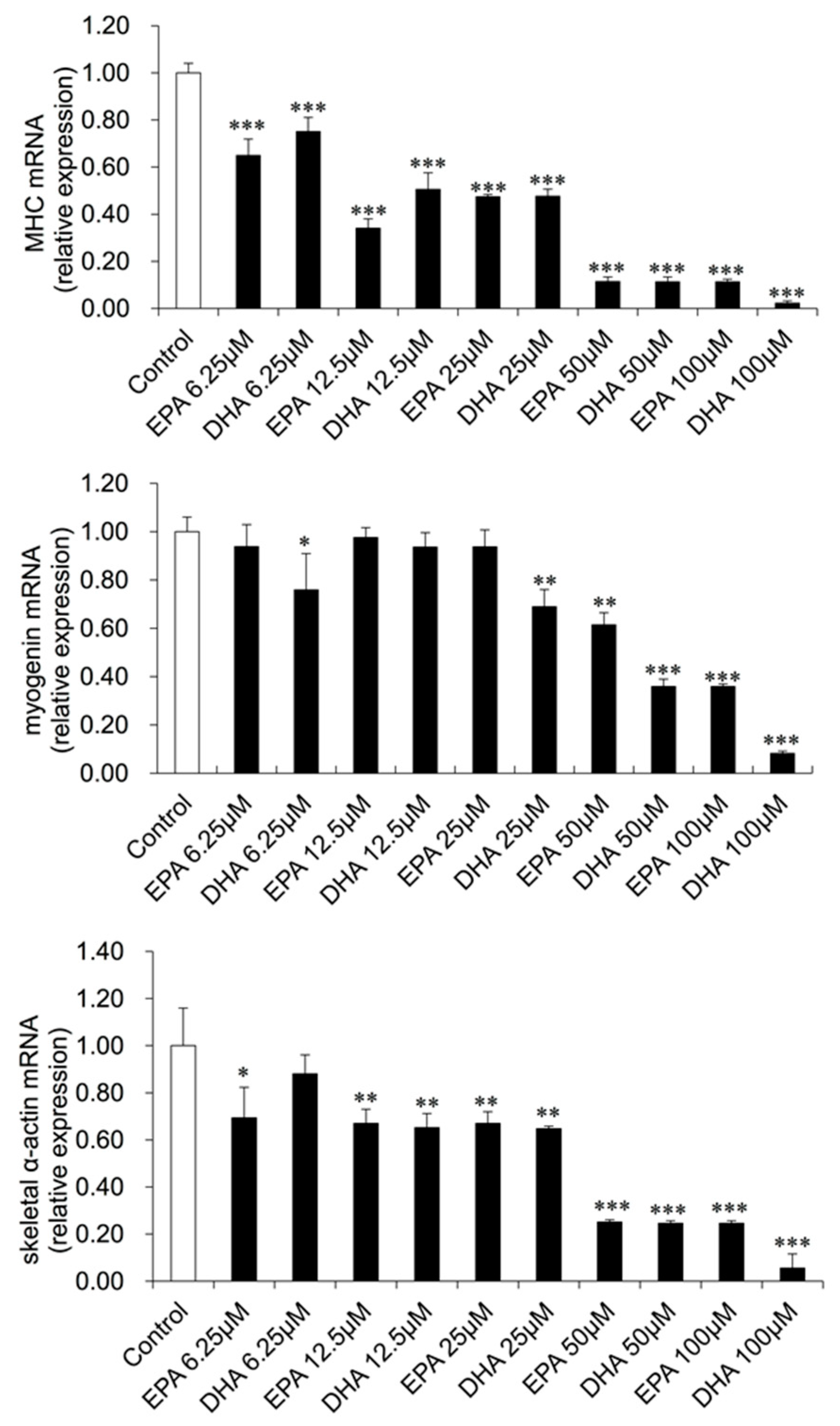
2.4. qRT-PCR Measurement
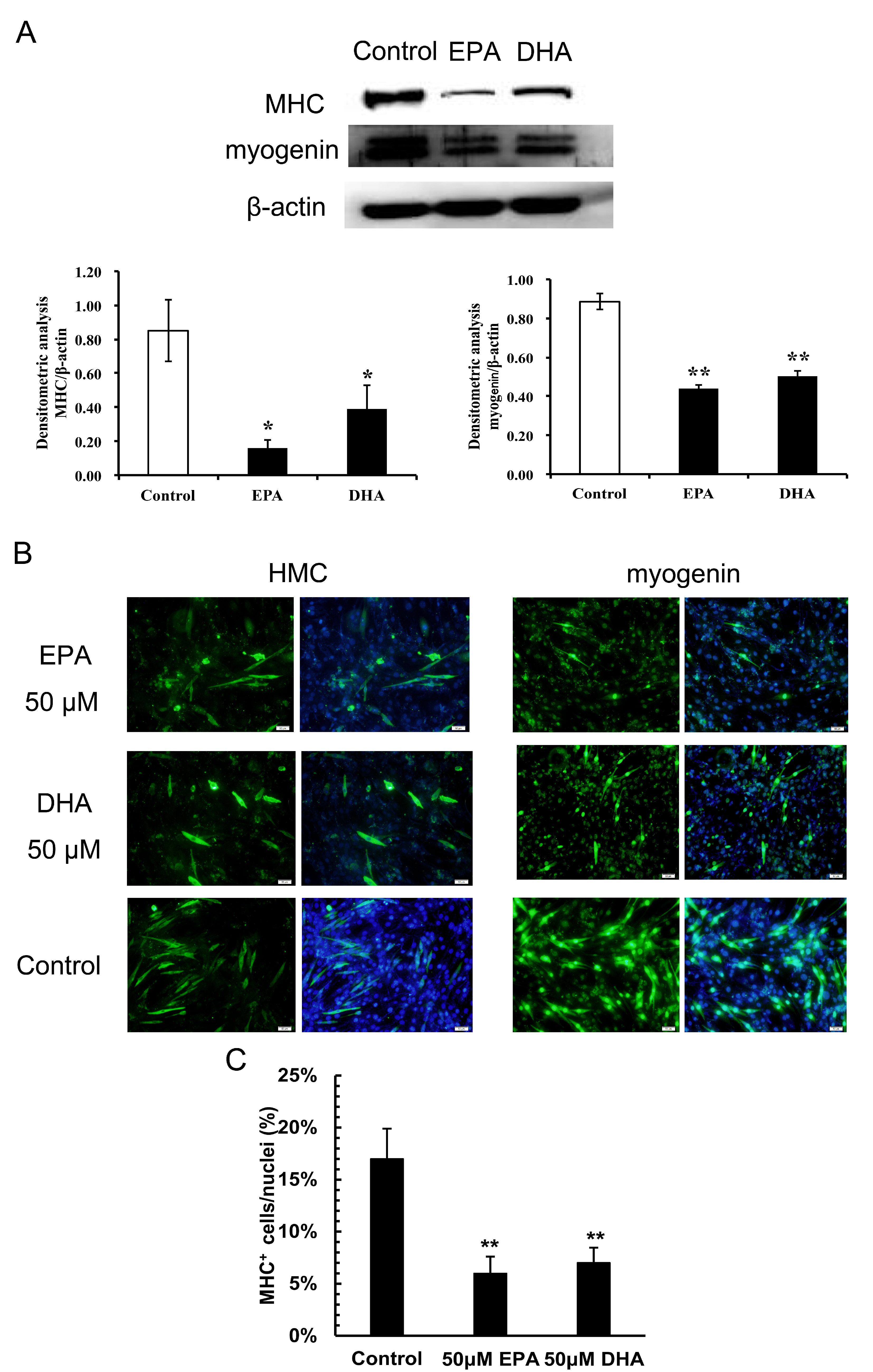
2.5. Immunoblotting
2.6. Immunofluorescence
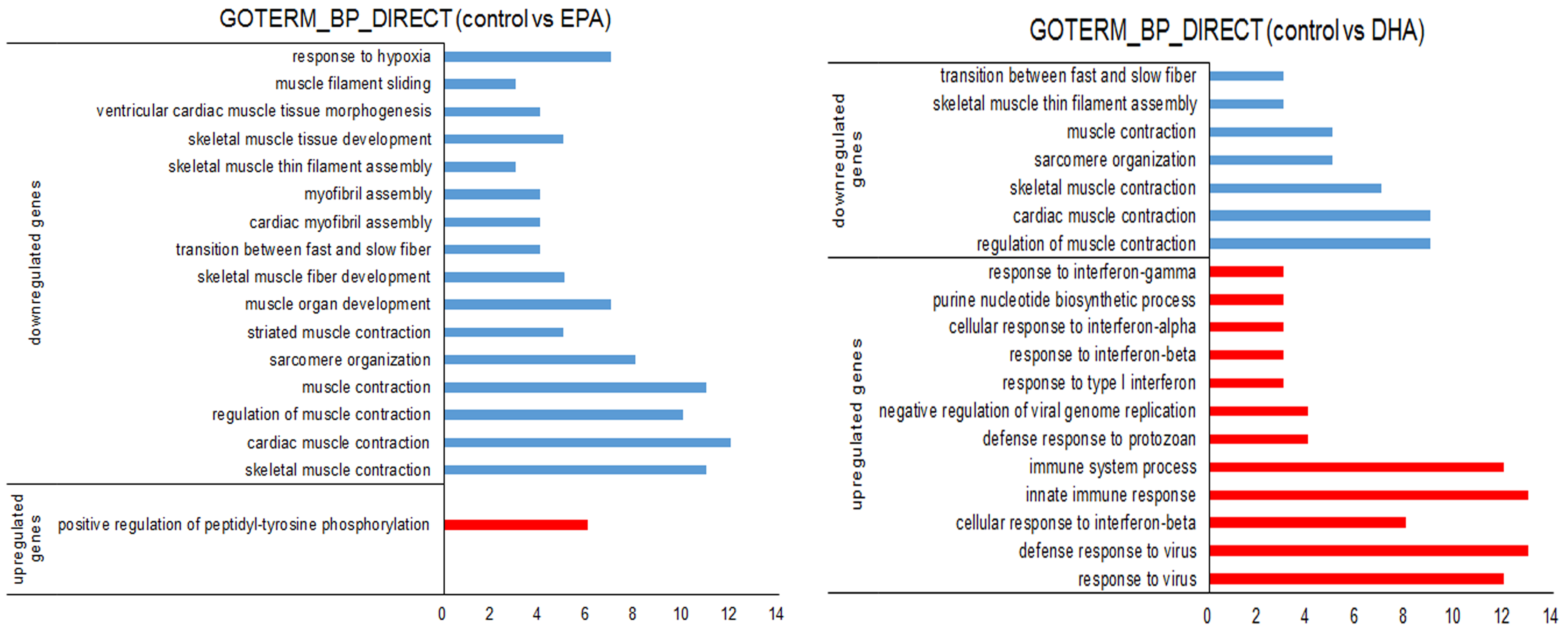
2.7. RNA-sequencing and Data Analysis
2.8. Statistical Analysis
3. Results
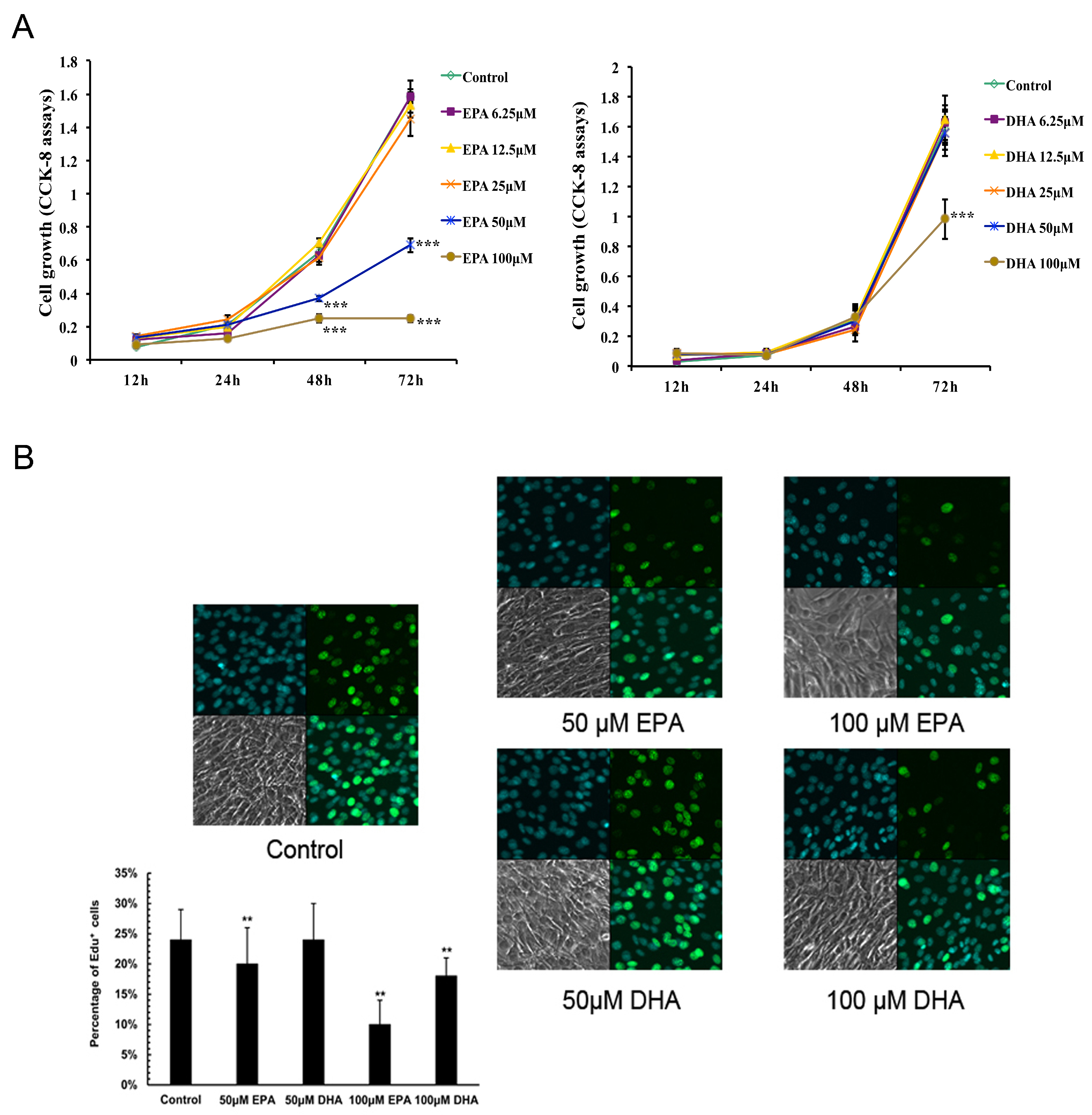
3.1. Inhibitory Effects of EPA and DHA on C2C12 Myoblast Proliferation
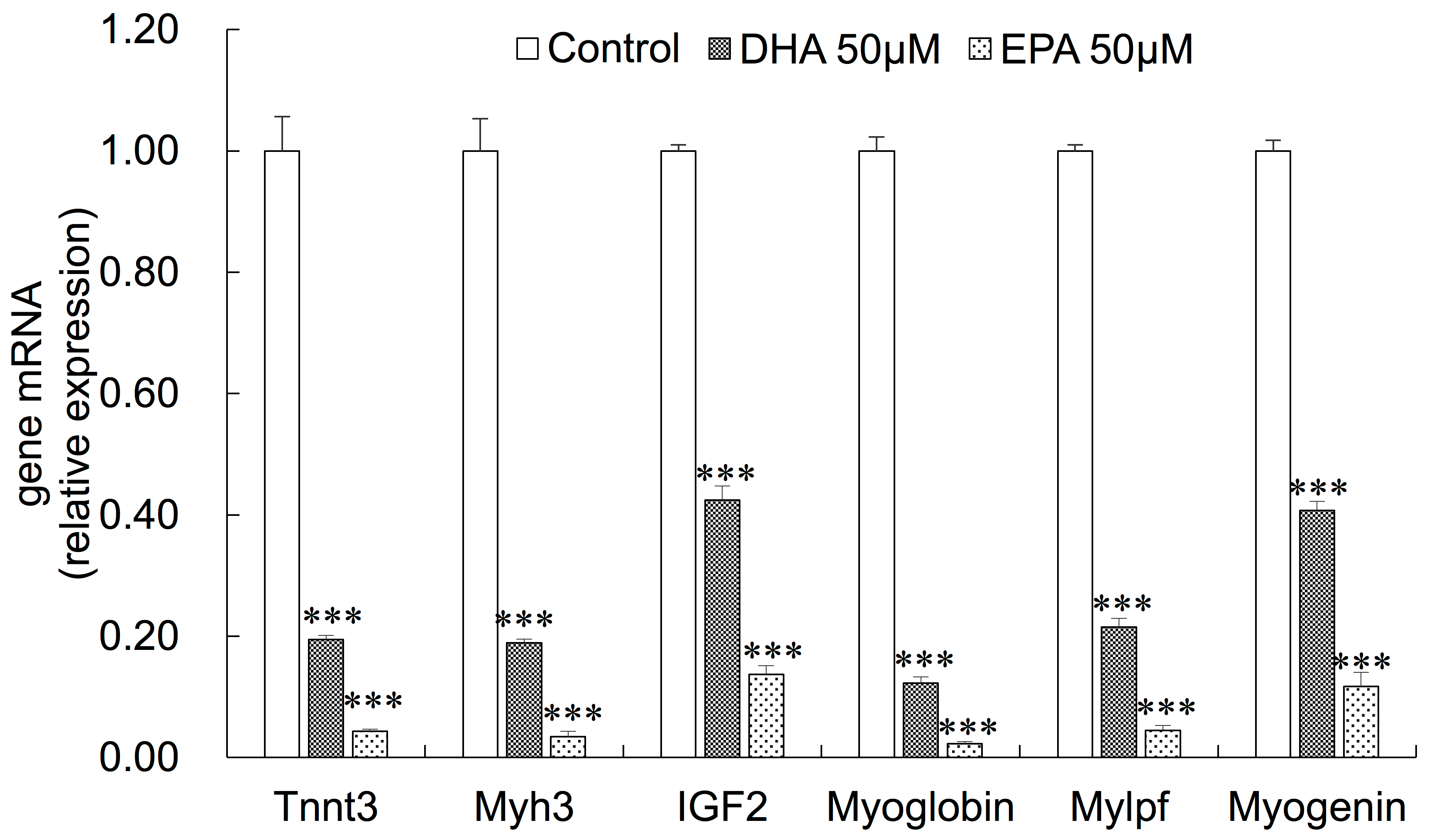
3.2. Inhibitory Effects of EPA and DHA on C2C12 Myoblast Differentiation
3.3. Gene Expression Changes in EPA- or DHA-treated C2C12 Cells
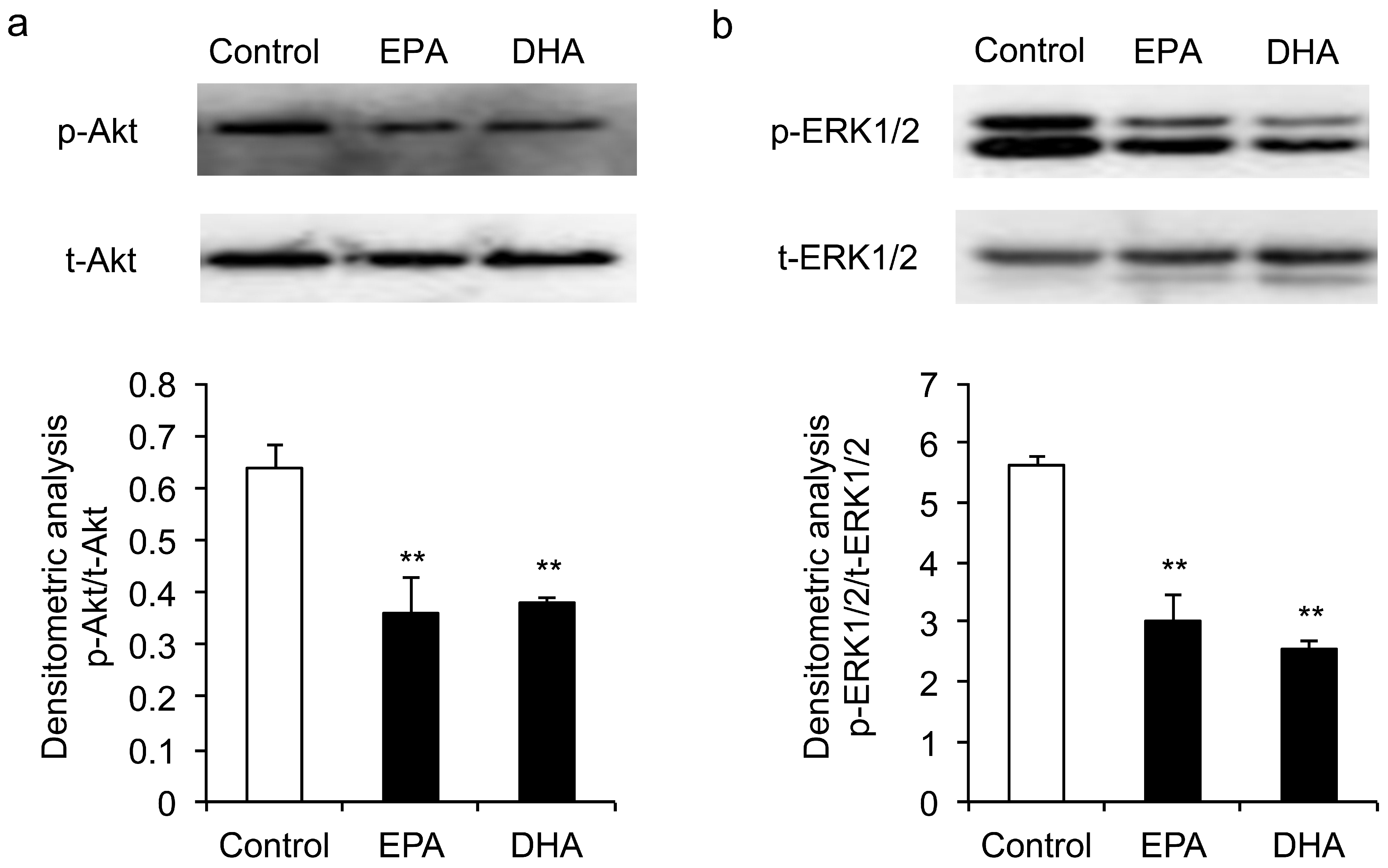
3.4. Inhibitory Effects of EPA and DHA on the Phosphorylation of ERK1/2 and Akt
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Buckingham, M. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 2001, 11, 440–448. [Google Scholar] [CrossRef]
- Sabourin, L.A.; Rudnicki, M.A. The molecular regulation of myogenesis. Clin. Genet. 2000, 57, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, D.; Saxel, O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 1977, 270, 725–727. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Tachibana, H.; Morinaga, Y.; Fujimura, Y.; Yamada, K. Modulation of proliferation and differentiation of C2C12 skeletal muscle cells by fatty acids. Life Sci. 2009, 84, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Hurley, M.S.; Flux, C.; Salter, A.M.; Brameld, J.M. Effects of fatty acids on skeletal muscle cell differentiation in vitro. Br. J. Nutr. 2006, 95, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Mechanisms of action of (n-3) fatty acids. J. Nutr. 2012, 142, S592–S599. [Google Scholar] [CrossRef] [PubMed]
- Mozaffarian, D.; Wu, J.H. (n-3) fatty acids and cardiovascular health: Are effects of EPA and DHA shared or complementary? J. Nutr. 2012, 142, 614S–625S. [Google Scholar] [CrossRef]
- Calder, P.C. Omega-3 polyunsaturated fatty acids and inflammatory processes: Nutrition or pharmacology? Br. J. Clin. Pharmacol. 2013, 75, 645–662. [Google Scholar] [CrossRef]
- Liu, Y.L.; Chen, F.; Odle, J.; Lin, X.; Zhu, H.L.; Shi, H.F.; Hou, Y.Q.; Yin, J.D. Fish oil increases muscle protein mass and modulates Akt/FOXO, TLR4, and NOD signaling in weanling piglets after lipopolysaccharide challenge. J. Nutr. 2013, 143, 1331–1339. [Google Scholar] [CrossRef]
- Kamolrat, T.; Gray, S.R. The effect of eicosapentaenoic and docosahexaenoic acid on protein synthesis and breakdown in murine C2C12 myotubes. Biochem. Biophys. Res. Commun. 2013, 432, 593–598. [Google Scholar] [CrossRef]
- Capel, F.; Acquaviva, C.; Pitois, E.; Laillet, B.; Rigaudière, J.P.; Jouve, C.; Pouyet, C.; Gladine, C.; Comte, B.; Vianey, S.C.; Morio, B. DHA at nutritional doses restores insulin sensitivity in skeletal muscle by preventing lipotoxicity and inflammation. J. Nutr. Biochem. 2015, 26, 949–959. [Google Scholar] [CrossRef]
- Tachtsis, B.; Camera, D.; Lacham-Kaplan, O. Potential roles of n-3 PUFAs during skeletal muscle growth and regeneration. Nutrients 2018, 10, 309. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, X.; Zhang, P.; Zhu, L.; Zhao, T.; Liu, S.; Wu, Y.; Chen, X.; Fan, M. Extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase pathway is involved in inhibition of myogenic differentiation of myoblasts by hypoxia. Exp. Physiol. 2012, 97, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Johnson, S.E. ERK2 is required for efficient terminal differentiation of skeletal myoblasts. Biochem. Biophys. Res. Commun. 2006, 345, 1425–1433. [Google Scholar] [CrossRef] [PubMed]
- Rommel, C.; Bodine, S.C.; Clarke, B.A.; Rossman, R.; Nunez, L.; Stitt, T.N.; Yancopoulos, G.D.; Glass, D.J. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001, 3, 1009–1013. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wu, Z. The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J. Biol. Chem. 2000, 275, 36750–36757. [Google Scholar] [CrossRef] [PubMed]
- Wan, M.; Li, Y.; Xue, H.; Li, Q.; Li, J. Eicosapentaenoic acid inhibits TNF-α-induced Lnk expression in human umbilical vein endothelial cells: Involvement of the PI3K/Akt pathway. J. Nutr. Biochem. 2007, 18, 17–22. [Google Scholar] [CrossRef]
- Lucie, C.; Goupille, C.; Charly, B.; Pinault, M.; Domingo, I.; Guimaraes, C.; Bougnoux, P.; Chevalier, S.; Mahéo, K. Long chain n-3 polyunsaturated fatty acids increase the efficacy of docetaxel in mammary cancer cells by downregulating Akt and PKCε/δ-induced ERK pathways. Biochim. Biophys. Acta 2016, 1861, 380–390. [Google Scholar]
- Zgheel, F.; Alhosin, M.; Rashid, S.; Burban, M.; Auger, C.; Schini-Kerth, V.B. Redox-sensitive induction of Src/PI3-kinase/Akt and MAPKs pathways activate eNOS in response to EPA:DHA 6:1. PLoS ONE 2014, 9, e105102. [Google Scholar] [CrossRef]
- Gorjão, R.; Hirabara, S.M.; Cury-Boaventura, M.F.; de Lima, T.M.; Passos, M.E.P.; Levada-Pires, A.C.; Curi, R. Signaling Pathways Involved in the Effects of Different Fatty Acids on Interleukin-2 Induced Human Lymphocyte Proliferation. J. Clin. Cell. Immunol. 2013, 4, 171. [Google Scholar] [CrossRef]
- Luo, H.; Zhou, Y.; Hu, X.; Peng, X.; Wei, H.; Peng, J.; Jiang, S. Activation of PPARγ2 by PPARγ1 through a functional PPRE in transdifferentiation of myoblasts to adipocytes induced by EPA. Cell Cycle 2015, 14, 1830–1841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ying, Z.Z.; Tang, Z.L.; Long, L.Q.; Li, K. MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 2012, 287, 21093–21101. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmitten, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tarazona, S.; Garcíaalcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef] [PubMed]
- Picard, B.; Lefaucheur, L.; Berri, C.; Duclos, M.J. Muscle fibre ontogenesis in farm animal species. Reprod. Nutr. Dev. 2002, 42, 415–431. [Google Scholar] [CrossRef] [PubMed]
- Swanson, D.; Block, R.; Mousa, S.A. Omega-3 fatty acids EPA and DHA: Health benefits throughout life. Adv. Nutr. 2012, 3, 1–7. [Google Scholar] [CrossRef]
- Church, M.W.; Jen, K.L.; Dowhan, L.M.; Adams, B.R.; Hotra, J.W. Excess and deficient omega-3 fatty acid during pregnancy and lactation cause impaired neural transmission in rat pups. Neurotoxicol. Teratol. 2008, 30, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.A.; Raja, J.S.; Hoffman, M.L.; Zinn, S.A.; Govoni, K.E. Poor maternal nutrition inhibits muscle development in ovine offspring. J. Anim. Sci. Biotechnol. 2014, 5, 79–89. [Google Scholar] [CrossRef]
- Peng, Y.; Zheng, Y.; Zhang, Y.; Zhao, J.; Chang, F.; Lu, T.; Zhang, R.; Li, Q.; Hu, X.; Li, N. Different effects of omega-3 fatty acids on the cell cycle in C2C12 myoblast proliferation. Mol. Cell. Biochem. 2012, 367, 165–173. [Google Scholar] [CrossRef]
- Hsueh, T.Y.; Baum, J.I.; Huang, Y. Effect of eicosapentaenoic acid and docosahexaenoic acid on myogenesis and mitochondrial biosynthesis during murine skeletal muscle cell differentiation. Front. Nutr. 2018, 5, 15. [Google Scholar] [CrossRef]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 2006, 7, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, P.A. The roles of PPARs in adipocyte differentiation. Prog. Lipid Res. 2001, 40, 269–281. [Google Scholar] [CrossRef]
- Tontonoz, P.; Spiegelman, B.M. Fat and beyond: The diverse biology of PPARgamma. Annu. Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Matsusue, K.; Peters, J.M.; Gonzalez, F.J. PPARbeta/delta potentiates PPARgamma-stimulated adipocyte differentiation. FASEB J. 2004, 18, 1477–1479. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Sun, T.; Bookout, A.L.; Downes, M.; Yu, R.T.; Evans, R.M.; Mangelsdorf, D.J. A nuclear receptor atlas: 3T3-L1 adipogenesis. Mol. Endocrinol. 2005, 19, 2437–2450. [Google Scholar] [CrossRef] [PubMed]
- Tontonoz, P.; Graves, R.A.; Budavari, A.I.; Erdjument-Bromage, H.; Lui, M.; Hu, E.; Tempst, P.; Spiegelman, B.M. Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPAR7 and RXRa. Nucleic Acids Res. 1994, 22, 5628–5634. [Google Scholar] [CrossRef] [PubMed]
- Hu, E.; Liang, P.; Spiegelman, B.M. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J. Biol. Chem. 1996, 271, 10697–10703. [Google Scholar] [CrossRef] [PubMed]
- Erbay, E.; Park, I.H.; Nuzzi, P.D.; Schoenherr, C.J.; Chen, J. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J. Cell Biol. 2003, 163, 931–936. [Google Scholar] [CrossRef]
- Jiao, S.; Ren, H.; Li, Y.; Zhou, J.; Duan, C.; Lu, L. Differential regulation of IGF-I and IGF-II gene expression in skeletal muscle cells. Mol. Cell. Biochem. 2013, 373, 107–113. [Google Scholar] [CrossRef]
- Carter, E.J.; Cosgrove, R.A.; Gonzalez, I.; Eisemann, J.H.; Lovett, F.A.; Cobb, L.J.; Pell, J.M. MEK5 and ERK5 are mediators of the pro-myogenic actions of IGF-2. J. Cell Sci. 2009, 122, 3104–3112. [Google Scholar] [CrossRef]
- Callan, A.C.; Milne, E. Involvement of the IGF system in fetal growth and childhood cancer: An overview of potential mechanisms. Cancer Causes Control 2009, 20, 1783–1798. [Google Scholar] [CrossRef] [PubMed]
- Denys, A.; Hichami, A.; Khan, N.A. Eicosapentaenoic acid and docosahexaenoic acid modulate MAP kinase (ERK1/ERK2) signaling in human T cells. J. Lipid Res. 2001, 42, 2015–2020. [Google Scholar]
- Ohashi, K.; Nagata, Y.; Wada, E.; Zammit, K.; Ohashi, P.S.; Shiozuka, M.; Matsuda, R. Zinc promotes proliferation and activation of myogenic cells via the PI3K/Akt and ERK signaling cascade. Exp. Cell Res. 2015, 333, 228–237. [Google Scholar] [CrossRef]
- Yu, M.; Wang, H.; Xu, Y.; Yu, D.; Li, D.; Liu, X.; Du, W. Insulin-like growth factor-1 (IGF-1) promotes myoblast proliferation and skeletal muscle growth of embryonic chickens via the PI3K/Akt signalling pathway. Cell Biol. Int. 2015, 39, 910–922. [Google Scholar] [CrossRef]
- Lee, S.J.; Hwang, J.; Jeong, H.J.; Yoo, M.; Go, G.Y.; Lee, J.R.; Leem, Y.E.; Park, J.W.; Seo, D.W.; Kim, Y.K.; et al. PKN2 and Cdo interact to activate AKT and promote myoblast differentiation. Cell Death Dis. 2016, 7, e2431. [Google Scholar] [CrossRef]
- White, M.F. Insulin signaling in health and disease. Science 2003, 302, 1710–1711. [Google Scholar] [CrossRef] [PubMed]
- Elshewy, H.M.; Lee, M.H.; Obeid, L.M.; Jaffa, A.A.; Luttrell, L.M. The insulin-like growth factor type 1 and insulin-like growth factor type 2/mannose-6-phosphate receptors independently regulate ERK1/2 activity in HEK293 cells. J. Biol. Chem. 2007, 282, 26150–26157. [Google Scholar] [CrossRef] [PubMed]
- Florini, J.R.; Magri, K.A.; Ewton, D.Z.; James, P.L.; Grindstaff, K.; Rotwein, P.S. “Spontaneous” differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J. Biol. Chem. 1991, 266, 15917–15923. [Google Scholar]
| Gene | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| MHC | CGCCCACCTGGAGCGGATGA | CTTGCGGTCCTCCTCGGTCTGGT |
| Myogenin | CGGTGGAGGATATGTCTGTTG | GGTGTTAGCCTTATGTGAATGG |
| Skeletal α-actin | CAGAGCAAGCGAGGTATCC | GTCCCCAGAATCCAACACG |
| Myh3 | ATGAGCGGCGTGTTAAGGA | ATTGACTTGCGATTCTGCGATA |
| Mylpf | TTTCCATCTGGAGCTACTGC | ATAATGCCATCCCTGTTCTG |
| Myoglobin | CTCCTAAGTCCCAGTCCATTT | CACTCCCTCTAAGCAACCCT |
| Tnnt3 | GCCCTCATTGACAGCCACTT | CTCCTCCGCCAATCTGTTCT |
| IGF2 | CGCTTCAGTTTGTCTGTTCG | AGGTAGACACGTCCCTCTCG |
| Pparg | ATGGAGCCTAACTTTGAGTT | CAGCAGGTTGTCTTGGATG |
| Ppard | TTCAGAGGACCAGCCACAG | GGAGACAGCAAGAACAGGAG |
| C/EBPα | CGTCTAAGATGAGGGAGTCAGG | CAGATGGAGGAGCACAGAG |
| aP2 | CGACAGGAAGGTGAAGAGCA | CGACAGGAAGGTGAAGAGCA |
| Adipoq | ATCATTATGACGGCAGCAC | CAGATGGAGGAGCACAGAG |
| β-actin | CAGCCTTCCTTCTTGGGTAT | TGGCATAGAGGTCTTTACGG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Xu, X.; Liu, Y.; Zhang, L.; Odle, J.; Lin, X.; Zhu, H.; Wang, X.; Liu, Y. EPA and DHA Inhibit Myogenesis and Downregulate the Expression of Muscle-related Genes in C2C12 Myoblasts. Genes 2019, 10, 64. https://doi.org/10.3390/genes10010064
Zhang J, Xu X, Liu Y, Zhang L, Odle J, Lin X, Zhu H, Wang X, Liu Y. EPA and DHA Inhibit Myogenesis and Downregulate the Expression of Muscle-related Genes in C2C12 Myoblasts. Genes. 2019; 10(1):64. https://doi.org/10.3390/genes10010064
Chicago/Turabian StyleZhang, Jing, Xin Xu, Yan Liu, Lin Zhang, Jack Odle, Xi Lin, Huiling Zhu, Xiuying Wang, and Yulan Liu. 2019. "EPA and DHA Inhibit Myogenesis and Downregulate the Expression of Muscle-related Genes in C2C12 Myoblasts" Genes 10, no. 1: 64. https://doi.org/10.3390/genes10010064
APA StyleZhang, J., Xu, X., Liu, Y., Zhang, L., Odle, J., Lin, X., Zhu, H., Wang, X., & Liu, Y. (2019). EPA and DHA Inhibit Myogenesis and Downregulate the Expression of Muscle-related Genes in C2C12 Myoblasts. Genes, 10(1), 64. https://doi.org/10.3390/genes10010064





