Interplay between Cellular Autophagy and Hepatitis B Virus Replication: A Systematic Review
Abstract
1. Introduction
2. HBV-Mediated Autophagic Responses
2.1. HBV Induces the Initiation of Autophagy
2.2. HBV Promotes Phagophore Formation
2.3. HBV Activates Phagophore Expansion and Forms Autophagosomes
2.4. HBV Interferes with Autophagic Degradation
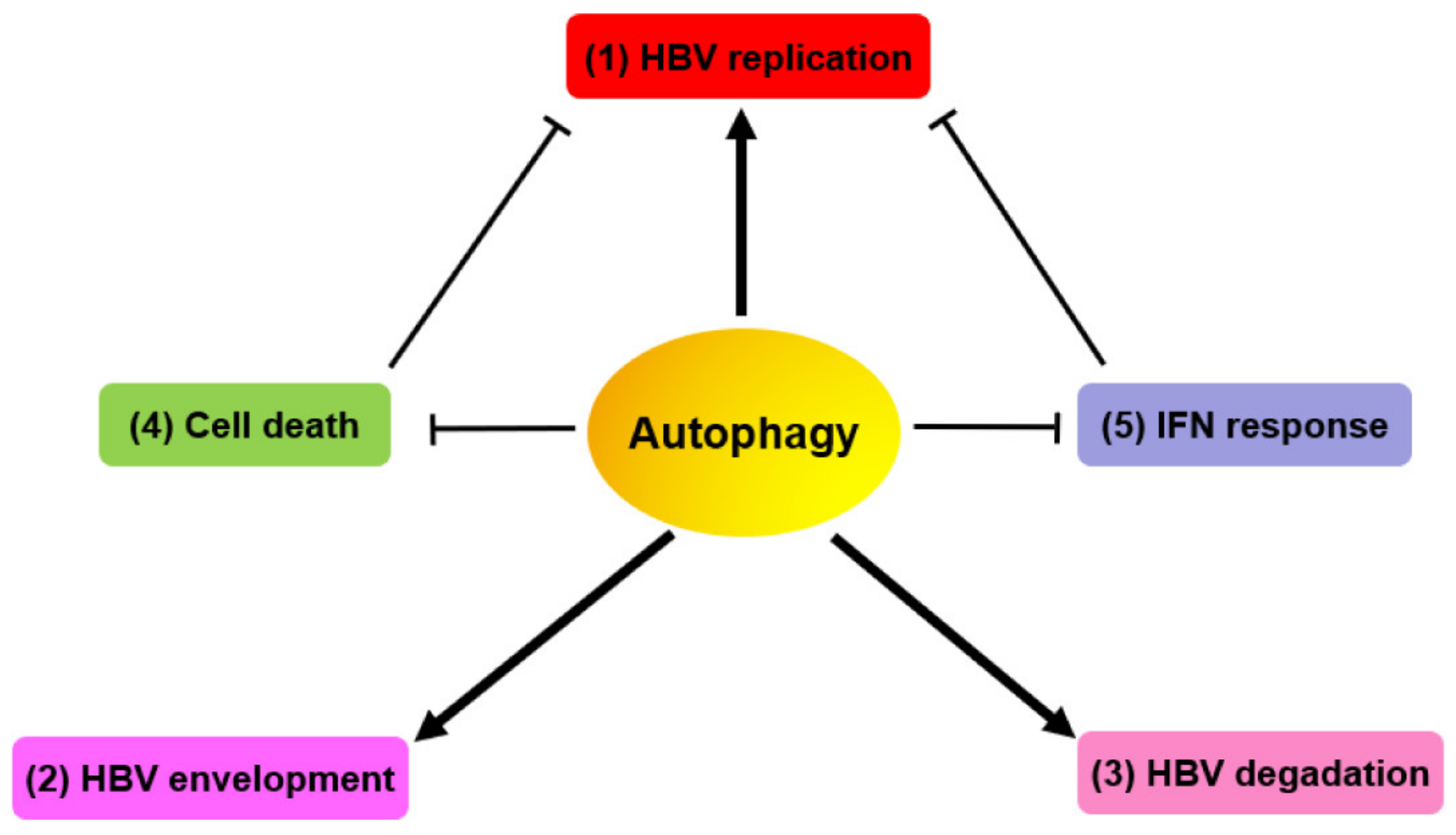
3. Autophagy-Mediated HBV Replication and Assembly
3.1. HBV Manipulates Autophagic Components for Its Replication
3.2. HBV Utilizes Autophagic Elements for its Envelopment
3.3. Late Autophagy Degrades HBV NCs/Virions and SVPs
3.4. HBV Evades Cell Death via HBX-Induced Autophagy for Persistent Infection
3.5. Autophagy Regulates HBV-Related Immune Responses
4. Autophagy-Based Anti-HBV Strategies
5. Conclusions and Perspective
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AMPK | AMP-protein activated kinase |
| ATG | Autophagy-related gene |
| BECN1 | Beclin 1 |
| HBV | Hepatitis B virus |
| HBeAg | Hepatitis B e antigen |
| MAP1LC3/LC3 | Microtubule-associated protein 1 light chain 3 |
| mTOR | Mammalian target of rapamycin |
| NC | Nucleocapsid |
| ORF | Open reading frame |
| PtdIns3K | Class III phosphatidylinositol 3-kinase |
| siRNA | Small interfering RNA |
| SVP | Subviral particle |
| ULK1 | Unc-51 like autophagy activating kinase 1 |
| UPR | Unfolded protein response |
References
- Schweitzer, A.; Horn, J.; Mikolajczyk, R.T.; Krause, G.; Ott, J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: A systematic review of data published between 1965 and 2013. Lancet 2015, 386, 1546–1555. [Google Scholar] [CrossRef]
- The Polaris Observatory Collaborators. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: A modelling study. Lancet Gastroenterol. Hepatol. 2018, 3, 383–403. [Google Scholar] [CrossRef]
- Revill, P.A.; Chisari, F.V.; Block, J.M.; Dandri, M.; Gehring, A.J.; Guo, H.; Hu, J.; Kramvis, A.; Lampertico, P.; Janssen, H.L.A.; et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol. Hepatol. 2019, 4, 545–558. [Google Scholar] [CrossRef]
- Tang, L.S.Y.; Covert, E.; Wilson, E.; Kottilil, S. Chronic Hepatitis B Infection: A Review. JAMA 2018, 319, 1802–1813. [Google Scholar] [CrossRef] [PubMed]
- Fanning, G.C.; Zoulim, F.; Hou, J.; Bertoletti, A. Therapeutic strategies for hepatitis B virus infection: Towards a cure. Nat. Rev. Drug Discov. 2019, 18, 827–844. [Google Scholar] [CrossRef]
- Bremer, C.M.; Glebe, D. The Molecular Virology of Hepatitis B Virus. Semin. Liver Dis. 2013, 33, 103–112. [Google Scholar] [CrossRef]
- Selzer, L.; Zlotnick, A. Assembly and Release of Hepatitis B Virus. Cold Spring Harb. Perspect. Med. 2015, 5, a021394. [Google Scholar] [CrossRef]
- Bruss, V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J. Virol. 1997, 71, 9350–9357. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2017, 14, 207–215. [Google Scholar] [CrossRef]
- Tang, H.; Da, L.; Mao, Y.; Li, Y.; Li, N.; Xu, Z.; Li, F.; Wang, Y.; Tiollais, P.; Li, T.; et al. Hepatitis B virus X protein sensitizes cells to starvation-induced autophagy via up-regulation of beclin 1 expression. Hepatology 2008, 49, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-T.; Chen, G.G.; Hu, B.-G.; Zhang, Z.-Y.; Yun, J.-P.; He, M.-L.; Lai, P.B.-S. Hepatitis B virus x protein induces autophagyviaactivating death-associated protein kinase. J. Viral Hepat. 2013, 21, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, Y.; Wang, Z.; Liu, K.; Wang, Y.; Ding, H.; Yuan, Z.; Liu, J. Subversion of Cellular Autophagy Machinery by Hepatitis B Virus for Viral Envelopment. J. Virol. 2011, 85, 6319–6333. [Google Scholar] [CrossRef] [PubMed]
- Xie, N.; Yuan, K.; Zhou, L.; Wang, K.; Chen, H.-N.; Lei, Y.; Lan, J.; Pu, Q.; Gao, W.; Zhang, L.; et al. PRKAA/AMPK restricts HBV replication through promotion of autophagic degradation. Autophagy 2016, 12, 1507–1520. [Google Scholar] [CrossRef] [PubMed]
- Doring, T.; Zeyen, L.; Bartusch, C. Hepatitis B Virus Subverts the Autophagy Elongation Complex Atg5-12/16L1 and Does Not Require Atg8/LC3 Lipidation for Viral Maturation. J. Virol. 2018, 92, e01513-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Fang, M.; Hu, Y.; Huang, B.; Li, N.; Chang, C.; Huang, R.; Xu, X.; Yang, Z.; Chen, Z.; et al. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy 2013, 10, 416–430. [Google Scholar] [CrossRef]
- Wang, X.; Lin, Y.; Liu, S.; Zhu, Y.; Lu, K.; Broering, R.; Lu, M. O-GlcNAcylation modulates HBV replication through regulating cellular autophagy at multiple levels. FASEB J. 2020. [Google Scholar] [CrossRef]
- Lin, Y.; Wu, C.; Wang, X.; Kemper, T.; Squire, A.; Gunzer, M.; Zhang, J.; Chen, X.; Lu, M. Hepatitis B virus is degraded by autophagosome-lysosome fusion mediated by Rab7 and related components. Protein Cell 2019, 10, 60–66. [Google Scholar] [CrossRef]
- Xie, M.; Yang, Z.; Liu, Y.; Zheng, M. The role of HBV-induced autophagy in HBV replication and HBV related-HCC. Life Sci. 2018, 205, 107–112. [Google Scholar] [CrossRef]
- Zhang, L. Autophagy in hepatitis B or C virus infection: An incubator and a potential therapeutic target. Life Sci. 2019, 242, 117206. [Google Scholar] [CrossRef]
- Lin, Y.; Wu, C.; Wang, X.; Liu, S.; Kemper, T.; Li, F.; Squire, A.; Zhu, Y.; Zhang, J.; Chen, X.; et al. Synaptosomal-associated protein 29 is required for the autophagic degradation of hepatitis B virus. FASEB J. 2019, 33, 6023–6034. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, J.; Wu, J.; Dong, M.; Shen, Z.; Lin, Y.; Li, F.; Zhang, Y.; Mao, R.; Lu, M.; et al. Host Gene SEL1L Involved in Endoplasmic Reticulum-Associated Degradation Pathway Could Inhibit Hepatitis B Virus at RNA, DNA, and Protein Levels. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.Y.; Sabatini, D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 183–203. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Deng, W.; Pang, J.; Kemper, T.; Hu, J.; Yin, J.; Zhang, J.; Lu, M. The microRNA-99 family modulates hepatitis B virus replication by promoting IGF-1R/PI3K/Akt/mTOR/ULK1 signaling-induced autophagy. Cell. Microbiol. 2017, 19, e12709. [Google Scholar] [CrossRef]
- Guo, H.; Zhou, T.; Jiang, D.; Cuconati, A.; Xiao, G.-H.; Block, T.M.; Guo, J.-T. Regulation of Hepatitis B Virus Replication by the Phosphatidylinositol 3-Kinase-Akt Signal Transduction Pathway. J. Virol. 2007, 81, 10072–10080. [Google Scholar] [CrossRef]
- Teng, C.-F.; Wu, H.-C.; Tsai, H.-W.; Shiah, H.-S.; Huang, W.; Su, I.-J. Novel feedback inhibition of surface antigen synthesis by mammalian target of rapamycin (mTOR) signal and its implication for hepatitis B virus tumorigenesis and therapy. Hepatology 2011, 54, 1199–1207. [Google Scholar] [CrossRef]
- Lee, Y.I.; Kang-Park, S.; Do, S.-I.; Lee, Y.I. The Hepatitis B Virus-X Protein Activates a Phosphatidylinositol 3-Kinase-dependent Survival Signaling Cascade. J. Biol. Chem. 2001, 276, 16969–16977. [Google Scholar] [CrossRef]
- Yen, C.J.; Lin, Y.J.; Yen, C.S.; Tsai, H.-W.; Tsai, T.-F.; Chang, K.-Y.; Lin, P.-W.; Chiang, C.-W.; Chang, T.-T. Hepatitis B virus X protein upregulates mTOR signaling through IKKbeta to increase cell proliferation and VEGF production in hepatocellular carcinoma. PLoS ONE 2012, 7, e41931. [Google Scholar] [CrossRef]
- Teng, C.-F.; Wu, H.-C.; Shyu, W.-C.; Jeng, L.-B.; Su, I.-J. Pre-S2 Mutant-Induced Mammalian Target of Rapamycin Signal Pathways as Potential Therapeutic Targets for Hepatitis B Virus-Associated Hepatocellular Carcinoma. Cell Transplant. 2017, 26, 429–438. [Google Scholar] [CrossRef]
- Wang, X.; Huo, B.; Liu, J.; Huang, X.; Zhang, S.; Feng, T. Hepatitis B virus X reduces hepatocyte apoptosis and promotes cell cycle progression through the Akt/mTOR pathway in vivo. Gene 2019, 691, 87–95. [Google Scholar] [CrossRef]
- Yang, J.-C.; Teng, C.-F.; Wu, H.-C.; Tsai, H.-W.; Chuang, H.-C.; Tsai, T.-F.; Hsu, Y.-H.; Huang, W.; Wu, L.-W.; Su, I.-J. Enhanced expression of vascular endothelial growth factor-A in ground glass hepatocytes and its implication in hepatitis B virus hepatocarcinogenesis. Hepatology 2009, 49, 1962–1971. [Google Scholar] [CrossRef] [PubMed]
- Rawat, S.; Bouchard, M.J. The Hepatitis B Virus (HBV) HBx Protein Activates AKT To Simultaneously Regulate HBV Replication and Hepatocyte Survival. J. Virol. 2014, 89, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Bagga, S.; Rawat, S.; Ajenjo, M.; Bouchard, M.J. Hepatitis B virus (HBV) X protein-mediated regulation of hepatocyte metabolic pathways affects viral replication. Virology 2016, 498, 9–22. [Google Scholar] [CrossRef]
- Li, L.; Chen, Y.; Gibson, S. Starvation-induced autophagy is regulated by mitochondrial reactive oxygen species leading to AMPK activation. Cell. Signal. 2013, 25, 50–65. [Google Scholar] [CrossRef]
- Wang, X.; Lin, Y.; Kemper, T.; Chen, J.; Yuan, Z.; Liu, S.; Zhu, Y.; Broering, R.; Lu, M. AMPK and Akt/mTOR signalling pathways participate in glucose-mediated regulation of hepatitis B virus replication and cellular autophagy. Cell. Microbiol. 2019, 22, e13131. [Google Scholar] [CrossRef]
- Lazar, C.; Uta, M.; Branza-Nichita, N. Modulation of the unfolded protein response by the human hepatitis B virus. Front. Microbiol. 2014, 5, 433. [Google Scholar] [CrossRef]
- Sir, N.; Tian, Y.; Chen, W.-L.; Ann, D.K.; Yen, T.-S.B.; Ou, J.-H.J. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc. Natl. Acad. Sci. USA 2010, 107, 4383–4388. [Google Scholar] [CrossRef]
- Zhong, L.; Shu, W.; Dai, W.; Gao, B.; Xiong, S. Reactive Oxygen Species-Mediated c-Jun NH2-Terminal Kinase Activation Contributes to Hepatitis B Virus X Protein-Induced Autophagy via Regulation of the Beclin-1/Bcl-2 Interaction. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Wang, J.; Chen, J.; Liu, Y.; Zeng, X.; Wei, M.; Wu, S.; Xiong, Q.; Song, F.; Yuan, X.; Xiao, Y.; et al. Hepatitis B Virus Induces Autophagy to Promote its Replication by the Axis of miR-192-3p-XIAP Through NF kappa B Signaling. Hepatology 2019, 69, 974–992. [Google Scholar] [CrossRef]
- Zhou, T.; Jin, M.; Ding, Y.; Zhang, Y.; Sun, Y.; Huang, S.; Xie, Q.; Xu, C.; Cai, W. Hepatitis B virus dampens autophagy maturation via negative regulation of Rab7 expression. Biosci. Trends 2016, 10, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Yorimitsu, T.; Klionsky, D.J. Endoplasmic reticulum stress: A new pathway to induce autophagy. Autophagy 2007, 3, 160–162. [Google Scholar] [CrossRef] [PubMed]
- Lazar, C.; Macovei, A.; Petrescu, S.; Branza-Nichita, N. Activation of ERAD Pathway by Human Hepatitis B Virus Modulates Viral and Subviral Particle Production. PLoS ONE 2012, 7, e34169. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wu, C.; Wang, X.; Liu, S.; Zhao, K.; Kemper, T.; Yu, H.; Li, M.; Zhang, J.; Chen, M.; et al. Glucosamine promotes hepatitis B virus replication through its dual effects in suppressing autophagic degradation and inhibiting MTORC1 signaling. Autophagy 2019, 16, 548–561. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Hu, J.; Shu, W.; Gao, B.; Xiong, S. Epigallocatechin-3-gallate opposes HBV-induced incomplete autophagy by enhancing lysosomal acidification, which is unfavorable for HBV replication. Cell Death Dis. 2015, 6, e1770. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Sir, N.; Kuo, C.-F.; Ann, D.K.; Ou, J.-H.J. Autophagy Required for Hepatitis B Virus Replication in Transgenic Mice. J. Virol. 2011, 85, 13453–13456. [Google Scholar] [CrossRef]
- Huang, W.; Zhao, F.; Huang, Y.; Li, X.; Zhu, S.; Hu, Q.; Chen, W.-X. Rapamycin Enhances HBV Production by Inducing Cellular Autophagy. Zahedan J. Res. Med. Sci. 2014, 14, 20719. [Google Scholar] [CrossRef]
- He, Q.; Song, X.; Huang, Y.; Huang, W.; Ye, B.; Luo, H.; Luo, H.; Wu, L.; Wang, Z.; Chen, W.-X.; et al. Dexamethasone Stimulates Hepatitis B Virus (HBV) Replication Through Autophagy. Med. Sci. Monit. 2018, 24, 4617–4624. [Google Scholar] [CrossRef]
- Chen, X.; Hu, Y.; Zhang, W.; Chen, K.; Hu, J.; Li, X.; Liang, L.; Cai, X.; Hu, J.; Wang, K.; et al. Cisplatin induces autophagy to enhance hepatitis B virus replication via activation of ROS/JNK and inhibition of the Akt/mTOR pathway. Free Radic. Biol. Med. 2019, 131, 225–236. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Y.; Xue, J.; Yang, Z.; Shi, Y.; Shi, Y.; Lou, G.; Wu, S.; Qi, J.; Liu, W.; et al. MicroRNA-141 Targets Sirt1 and Inhibits Autophagy to Reduce HBV Replication. Cell. Physiol. Biochem. 2017, 41, 310–322. [Google Scholar] [CrossRef]
- Doring, T.; Prange, R. Rab33B and its autophagic Atg5/12/16L1 effector assist in hepatitis B virus naked capsid formation and release. Cell. Microbiol. 2015, 17, 747–764. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Revill, P. Overview of hepatitis B viral replication and genetic variability. J. Hepatol. 2016, 64, S4–S16. [Google Scholar] [CrossRef]
- Zahoor, M.; Farhan, H. Crosstalk of Autophagy and the Secretory Pathway and Its Role in Diseases. Int. Rev. Cell Mol. Biol. 2018, 337, 153–184. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tan, D.; Cai, Y.; Reinisch, K.M.; Walz, T.; Ferro-Novick, S. A requirement for ER-derived COPII vesicles in phagophore initiation. Autophagy 2014, 10, 708–709. [Google Scholar] [CrossRef]
- Ge, L.; Zhang, M.; Kenny, S.J.; Liu, D.; Maeda, M.; Saito, K.; Mathur, A.; Xu, K.; Schekman, R. Remodeling of ER -exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 2017, 18, 1586–1603. [Google Scholar] [CrossRef]
- Rabouille, C. COPII vesicles and the expansion of the phagophore. eLife 2019, 8, 8. [Google Scholar] [CrossRef]
- Zeyen, L.; Döring, T.; Stieler, J.T.; Prange, R. Hepatitis B subviral envelope particles use the COPII machinery for intracellular transport via selective exploitation of Sec24A and Sec23B. Cell. Microbiol. 2020, 22, e13181. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, J.; Liu, L.; Zhang, J.; Wang, D.; Ma, L.; He, Y.; Liu, Y.; Liu, Z.; Wu, J. The X Protein of Hepatitis B Virus Inhibits Apoptosis in Hepatoma Cells through Enhancing the Methionine Adenosyltransferase 2A Gene Expression and ReducingS-Adenosylmethionine Production. J. Biol. Chem. 2011, 286, 17168–17180. [Google Scholar] [CrossRef]
- Shin, G.C.; Kang, H.S.; Lee, A.R.; Kim, K.H. Hepatitis B virus-triggered autophagy targets TNFRSF10B/death receptor 5 for degradation to limit TNFSF10/TRAIL response. Autophagy 2016, 12, 2451–2466. [Google Scholar] [CrossRef]
- Mao, Y.; Da, L.; Tang, H.; Yang, J.; Lei, Y.; Tiollais, P.; Li, T.; Zhao, M. Hepatitis B virus X protein reduces starvation-induced cell death through activation of autophagy and inhibition of mitochondrial apoptotic pathway. Biochem. Biophys. Res. Commun. 2011, 415, 68–74. [Google Scholar] [CrossRef]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Kunanopparat, A.; Hirankarn, N.; Kittigu, C.; Tangkijvanich, P.; Kimkong, I. Autophagy machinery impaired interferon signalling pathways to benefit hepatitis B virus replication. Asian Pac. J. Allergy Immunol. 2015, 34, 77–85. [Google Scholar] [CrossRef][Green Version]
- Luo, M.X.; Wong, S.H.; Chan, M.T.; Yu, L.; Yu, S.S.B.; Wu, F.; Xiao, Z.; Wang, X.; Zhang, L.; Cheng, A.S.L.; et al. Autophagy Mediates HBx-Induced Nuclear Factor-kappaB Activation and Release of IL-6, IL-8, and CXCL2 in Hepatocytes. J. Cell. Physiol. 2015, 230, 2382–2389. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, X.; Gao, Y.; Zhou, R.; Wei, M.; Dong, J.; Yan, H.; Zhao, Y. Hepatitis B Virus Inhibits Neutrophil Extracellular Trap Release by Modulating Reactive Oxygen Species Production and Autophagy. J. Immunol. 2018, 202, 805–815. [Google Scholar] [CrossRef]
- Wan, Y.; Cao, W.; Han, T.; Ren, S.; Feng, J.; Chen, T.; Wang, J.; Broering, R.; Lu, M.; Zhu, Y. Inducible Rubicon facilitates viral replication by antagonizing interferon production. Cell. Mol. Immunol. 2017, 14, 607–620. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1–phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Wang, L.; Tian, Y.; Ou, J.-H.J. HCV Induces the Expression of Rubicon and UVRAG to Temporally Regulate the Maturation of Autophagosomes and Viral Replication. PLoS Pathog. 2015, 11, e1004764. [Google Scholar] [CrossRef]
- Ma, S.; Chen, X.; Tan, Q.; Dai, S.; Li, D.; Wu, S.; Yu, Y.; Zang, G.; Tang, Z. An Engineered DC-Targeting Lentivector Induces Robust T Cell Responses and Inhibits HBV Replication in HBV Transgenic Mice via Upregulating T Cell Autophagy. Cell. Physiol. Biochem. 2018, 48, 1041–1059. [Google Scholar] [CrossRef]
- Ma, S.; Chen, X.; Tan, Q.; Li, D.; Dai, S.; Wu, S.; Yu, Y.; Zang, G.; Tang, Z. An engineered novel lentivector specifically transducing dendritic cells and eliciting robust HBV-specific CTL response by upregulating autophagy in T cells. Cell Cycle 2018, 17, 1220–1234. [Google Scholar] [CrossRef]
- Cheng, L.-S.; Li, J.; Liu, Y.; Wang, F.-P.; Wang, S.-Q.; She, W.-M.; Wu, S.-D.; Qi, X.-L.; Zhou, Y.-P.; Jiang, W. HMGB1-induced autophagy: A new pathway to maintain Treg function during chronic hepatitis B virus infection. Clin. Sci. 2017, 131, 381–394. [Google Scholar] [CrossRef]
- Zhang, J.; Fu, L.-L.; Tian, M.; Liu, H.-Q.; Li, J.-J.; Li, Y.; He, J.; Huang, J.; Ouyang, L.; Gao, H.-Y.; et al. Design and synthesis of a novel candidate compound NTI-007 targeting sodium taurocholate cotransporting polypeptide [NTCP]–APOA1–HBx–Beclin1-mediated autophagic pathway in HBV therapy. Bioorg. Med. Chem. 2015, 23, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Fan, F.; Ding, L.; Liu, J.; Su, S.; Yin, P.; Cao, M.; Zhao, W.; Hu, H.-M.; Wang, L. An autophagosome-based therapeutic vaccine for HBV infection: A preclinical evaluation. J. Transl. Med. 2014, 12, 361. [Google Scholar] [CrossRef] [PubMed]
- Abdoli, A.; Nakhaie, M.; Feizi, N.; Jeda, A.S.; Ramezani, A. Harmonized Autophagy Versus Full-Fledged Hepatitis B Virus: Victorious or Defeated. Viral Immunol. 2019, 32, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Huovila, A.P.; Eder, A.M.; Fuller, S.D. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J. Cell Biol. 1992, 118, 1305–1320. [Google Scholar] [CrossRef]
- Prange, R. Host factors involved in hepatitis B virus maturation, assembly, and egress. Med. Microbiol. Immunol. 2012, 201, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Zeyen, L.; Döring, T.; Prange, R. Hepatitis B Virus Exploits ERGIC-53 in Conjunction with COPII to Exit Cells. Cells 2020, 9, 1889. [Google Scholar] [CrossRef]
- Chua, C.E.L.; Lim, Y.S.; Lee, M.G.; Tang, B.L. Non-classical membrane trafficking processes galore. J. Cell. Physiol. 2012, 227, 3722–3730. [Google Scholar] [CrossRef]
- Kimura, T.; Jia, J.; Kumar, S.; Choi, S.W.; Gu, Y.; Mudd, M.; Dupont, N.; Jiang, S.; Peters, R.; Farzam, F.; et al. Dedicated SNARE s and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 2016, 36, 42–60. [Google Scholar] [CrossRef]
- Novellino, L.; Rossi, R.L.; Bonino, F.; Cavallone, D.; Abrignani, S.; Pagani, M.; Brunetto, M.R. Circulating Hepatitis B Surface Antigen Particles Carry Hepatocellular microRNAs. PLoS ONE 2012, 7, e31952. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Mao, R.-C.; Zhang, Y.-M.; Zhang, Y.-J.; Liu, H.-Y.; Qin, Y.-L.; Lu, M.-J.; Zhang, J.-M. Serum microRNA-124 is a novel biomarker for liver necroinflammation in patients with chronic hepatitis B virus infection. J. Viral Hepat. 2014, 22, 128–136. [Google Scholar] [CrossRef]
- Li, F.; Zhou, P.; Deng, W.; Wang, J.; Mao, R.; Zhang, Y.; Li, J.; Yu, J.; Yang, F.; Huang, Y.; et al. Serum microRNA-125b correlates with hepatitis B viral replication and liver necroinflammation. Clin. Microbiol. Infect. 2016, 22, 384.e1–384.e10. [Google Scholar] [CrossRef] [PubMed]
- Winther, T.N.; Heiberg, I.L.; Bang-Berthelsen, C.H.; Pociot, F.; Hogh, B. Hepatitis B Surface Antigen Quantity Positively Correlates with Plasma Levels of microRNAs Differentially Expressed in Immunological Phases of Chronic Hepatitis B in Children. PLoS ONE 2013, 8, e80384. [Google Scholar] [CrossRef] [PubMed]
- Pollicino, T.; Cacciola, I.; Saffioti, F.; Raimondo, G. Hepatitis B virus PreS/S gene variants: Pathobiology and clinical implications. J. Hepatol. 2014, 61, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Pollicino, T.; Amaddeo, G.; Restuccia, A.; Raffa, G.; Alibrandi, A.; Cutroneo, G.; Favaloro, A.; Maimone, S.; Squadrito, G.; Raimondo, G. Impact of hepatitis B virus (HBV) preS/S genomic variability on HBV surface antigen and HBV DNA serum levels. Hepatology 2012, 56, 434–443. [Google Scholar] [CrossRef]
- Chen, B.-F. Hepatitis B virus pre-S/S variants in liver diseases. World J. Gastroenterol. 2018, 24, 1507–1520. [Google Scholar] [CrossRef]
- Cao, L.; Wu, C.; Shi, H.; Gong, Z.; Zhang, E.; Wang, H.; Zhao, K.; Liu, S.; Li, S.; Gao, X.; et al. Coexistence of Hepatitis B Virus Quasispecies Enhances Viral Replication and the Ability To Induce Host Antibody and Cellular Immune Responses. J. Virol. 2014, 88, 8656–8666. [Google Scholar] [CrossRef]
- Ueno, T.; Komatsu, M. Autophagy in the liver: Functions in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 170–184. [Google Scholar] [CrossRef]
- Allaire, M.; Rautou, P.-E.; Codogno, P.; Lotersztajn, S. Autophagy in liver diseases: Time for translation? J. Hepatol. 2019, 70, 985–998. [Google Scholar] [CrossRef]
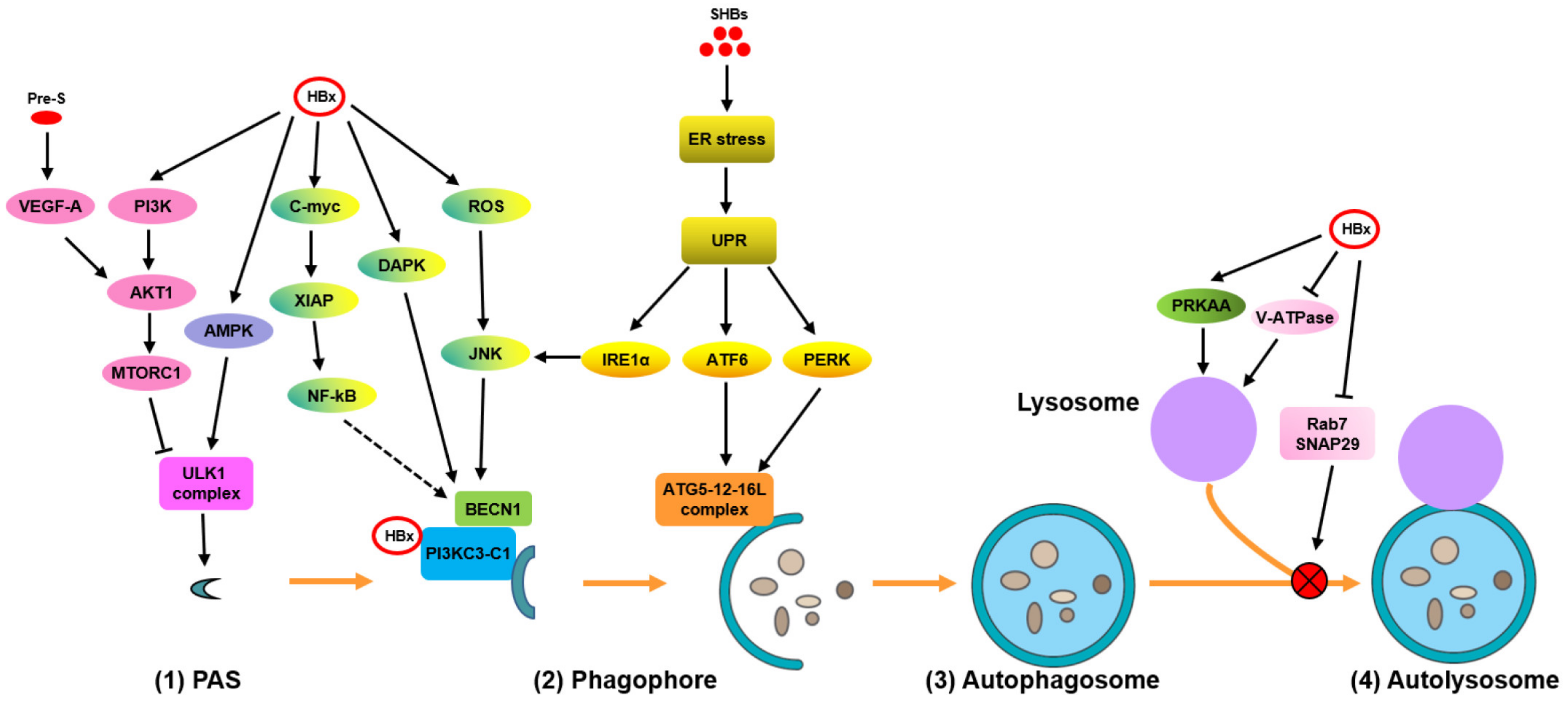
| HBV Proteins. | Models | Mechanisms | Effects | References |
|---|---|---|---|---|
| HBx | HepG2.2.15 and HepAD38 cells | Activate AMPK by ROS accumulation | Induce autophagy initiation (?) | [14] |
| Promote autophagic degradation | ||||
| HBx | Primary rat hepatocytes; HepG2.2.15 cells and primary hepatocytes | Activate AMPK signaling | Induce autophagy initiation | [34,36] |
| HBx | L02, Chang, HepG2, and BEL-7404 cells | Directly transactivates BECN1 promoter activity and upregulates its expression during starvation | Promote phagophore formation | [11] |
| HBx | Chang cells | Increase the activity of DAPK in a BECN1-associated pathway | Promote phagophore formation | [12] |
| SHBs | Huh7 cells | Activate IRE1α/XBP1/BECN1 axis | Promote phagophore formation | [13,37] |
| HBx | Huh7.5 cells | Directly bind to PtdIns3K and enhance its enzymatic activity | Promote phagophore formation | [38] |
| HBx | HepG2 cells | Dissociate BECN1 and Bcl-2 via the ROS/JNK signaling pathway | Promote phagophore formation | [39] |
| HBx | HepG2.2.15 and Huh7 cells; primary human hepatocytes; hydrodynamic-based HBV mouse model | Activate BECN1-mediated autophagy through C-myc/miR-192-3p/XIAP/NF-κB axis | Promote phagophore formation | [40] |
| SHBs | Huh7 cells | Activate PERK/eIF2α and ATF6/GRP78/94 signaling to enhance their interaction with the autophagy-associated proteins ATG5, ATG12, and/or ATG16L | Activate phagophore expansion and form autophagosomes | [13,37] |
| HBx | Huh7 cells | Impair lysosome maturation by inhibiting its acidification | Interfere with autophagic degradation | [16] |
| HBV | HepG2.2.15 and Huh7 cells | Block the fusion of autophagosomes with lysosomes by decreasing the expression of Rab7 and SNAP29 | Interfere with autophagic degradation | [18,21,41] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.; Zhao, Z.; Huang, A.; Lu, M. Interplay between Cellular Autophagy and Hepatitis B Virus Replication: A Systematic Review. Cells 2020, 9, 2101. https://doi.org/10.3390/cells9092101
Lin Y, Zhao Z, Huang A, Lu M. Interplay between Cellular Autophagy and Hepatitis B Virus Replication: A Systematic Review. Cells. 2020; 9(9):2101. https://doi.org/10.3390/cells9092101
Chicago/Turabian StyleLin, Yong, Zhenyu Zhao, Ailong Huang, and Mengji Lu. 2020. "Interplay between Cellular Autophagy and Hepatitis B Virus Replication: A Systematic Review" Cells 9, no. 9: 2101. https://doi.org/10.3390/cells9092101
APA StyleLin, Y., Zhao, Z., Huang, A., & Lu, M. (2020). Interplay between Cellular Autophagy and Hepatitis B Virus Replication: A Systematic Review. Cells, 9(9), 2101. https://doi.org/10.3390/cells9092101




