Myosin XVI in the Nervous System
Abstract
1. Introduction
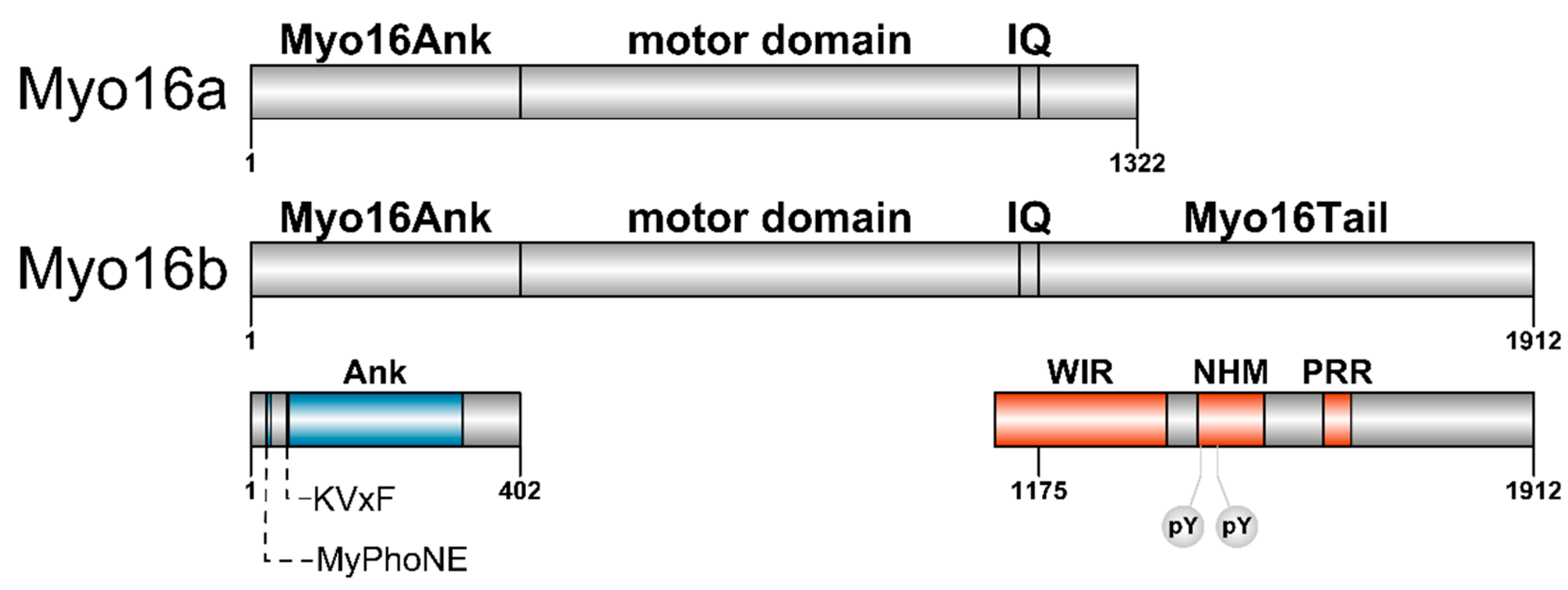
Myosin XVI
2. Interactions and Functions of Myosin XVI in the Nervous System
2.1. The N-terminal Myo16Ank Interacts with PP1c and Regulates Its Phosphatase Activity
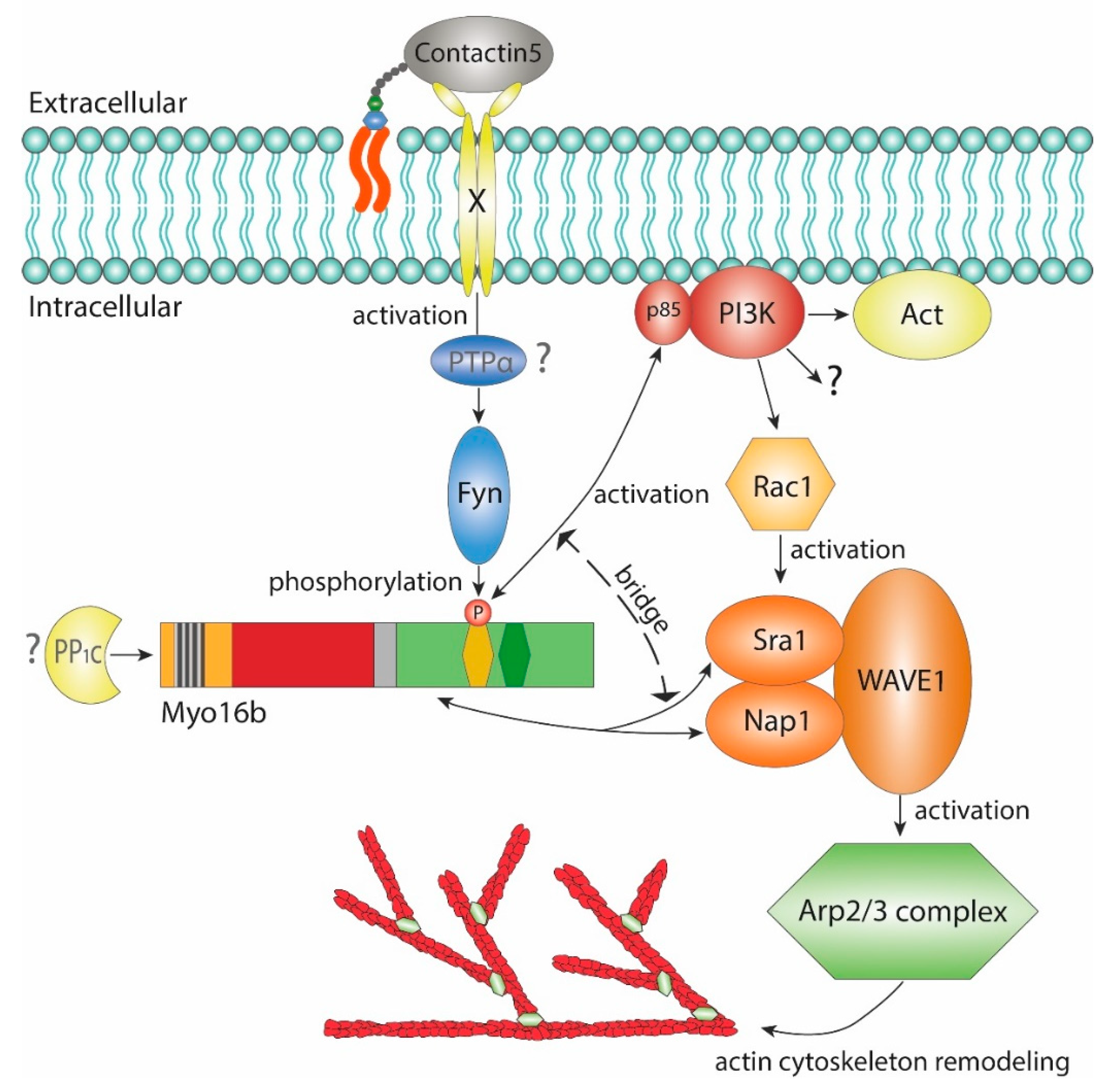
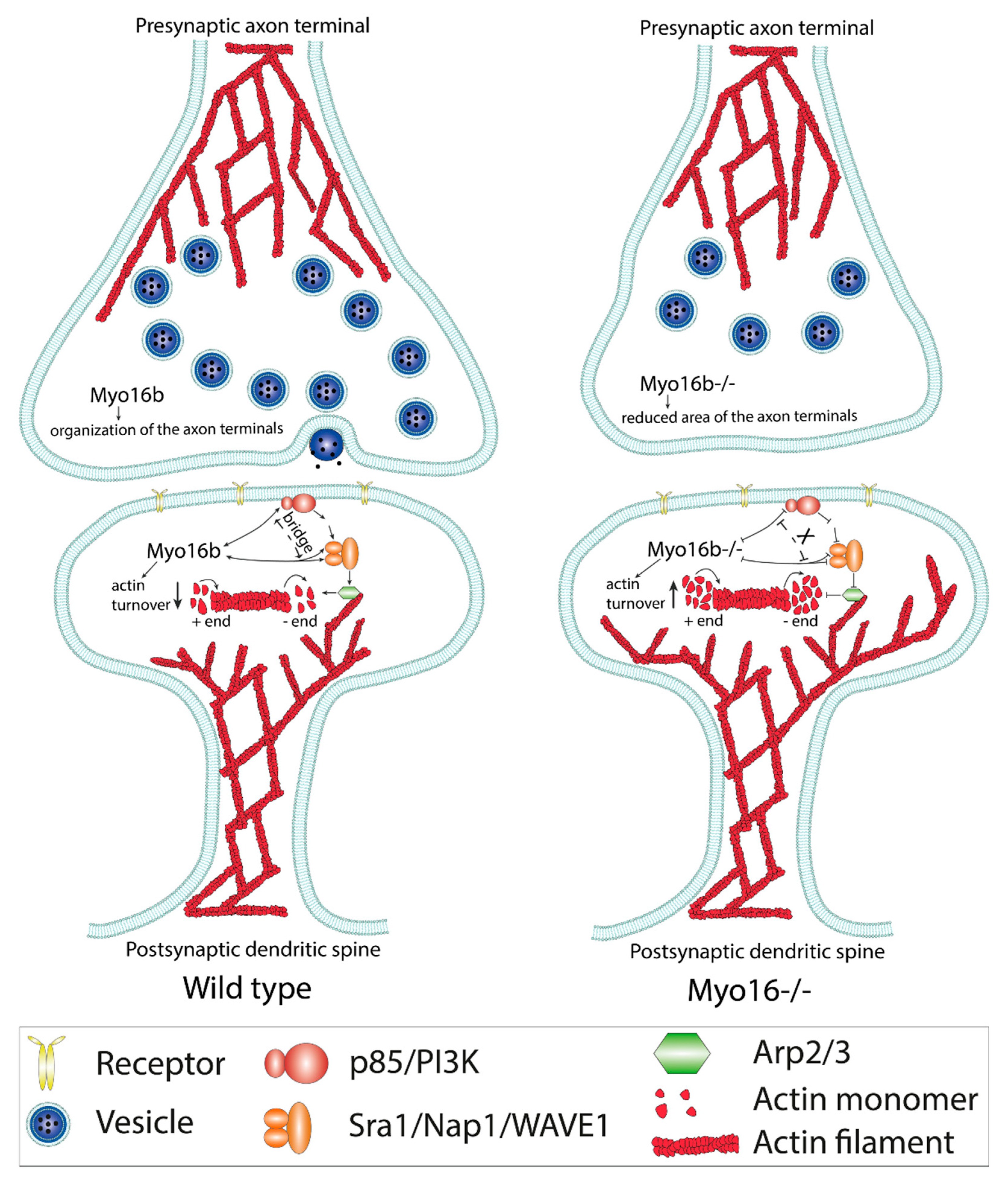
2.2. The-C terminal Myo16Tail Controls the Remodeling of the Actin Cytoskeleton by Linking PI3K and WAVE1
3. Regulatory Mechanisms Controlling the Motor Function
4. The Importance of Polyproline Sequences and Phosphorylation Sites
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Frénal, K.; Foth, B.J.; Soldati, D. Myosin Class XIV and Other Myosins in Protists. In Myosins, A Superfamily of Molecular Motors; Series: Proteins and Cell Regulation; Coluccio, L.M., Ed.; Springer Netherlands: Heidelberg, Germany, 2008; pp. 421–440. [Google Scholar]
- Coluccio, L.M.; Myosins, A. Superfamily of Molecular Motors, 2nd ed.; Series: Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar]
- Brown, M.E.; Bridgman, P.C. Myosin Function in Nervous and Sensory Systems. J. Neurobiol. 2004, 58, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Suter, D.M. Functions of Myosin Motor Proteins in the Nervous System. In Neurobiology of Actin, Advances in Neurobiology 5; Gallo, G., Lanier, L., Eds.; Springer Science and Business Media: Berlin, Germany, 2010; pp. 45–72. [Google Scholar]
- Wu, X.; Jung, G.; Hammer, J.A., 3rd. Functions of unconventional myosins. Curr. Opin. Cell Biol. 2000, 12, 42–51. [Google Scholar] [CrossRef]
- Newell-Litwa, K.A.; Horwitz, R.; Lamers, M.L. Non-Muscle myosin II in disease: Mechanisms and therapeutic opportunities. Dis. Model. Mech. 2015, 8, 1495–1515. [Google Scholar] [CrossRef] [PubMed]
- Lalwani, A.K.; Goldstein, J.A.; Kelley, M.J.; Luxford, W.; Castelein, C.M.; Mhatre, A.N. Human Nonsyndromic Hereditary Deafness DFNA17 Is Due to a Mutation in Nonmuscle Myosin MYH9. Am. J. Hum. Genet. 2000, 67, 1121–1128. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.W.; Bremner, K.H.; Vallee, R.B. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 2007, 10, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Walsh, T.; Walsh, V.; Vreugde, S.; Hertzano, R.; Shahin, H.; Haika, S.; Lee, M.K.; Kanaan, M.; King, M.-C.; Avraham, K.B. From flies’ eyes to our ears: Mutations in a human class III myosin cause progressive nonsyndromic hearing loss DFNB30. Proc. Nalt. Acad. Sci. USA 2002, 99, 7518–7523. [Google Scholar] [CrossRef]
- Chen, Y.; Tian, L.; Zhang, F.; Liu, C.; Lu, T.; Ruan, Y.; Wang, L.; Yan, H.; Yan, J.; Liu, Q.; et al. Myosin Vb gene is associated with schizophrenia in Chinese Han population. Psychiatry Res. 2013, 207, 13–18. [Google Scholar] [CrossRef]
- Dong, J.; Wyss, A.; Yang, J.; Price, T.R.; Nicolas, A.; Nalls, M.; Tranah, G.; Franceschini, N.; Xu, Z.; Schulte, C.; et al. Genome-Wide Association Analysis of the Sense of Smell in U.S. Older Adults: Identification of Novel Risk Loci in African-Americans and European-Americans. Mol. Neurobiol. 2017, 54, 8021–8032. [Google Scholar] [CrossRef]
- Pastural, E.; Barrat, F.J.; Dufourcq-Lagelouse, R.; Certain, S.; Sanal, O.; Jabado, N.; Seger, R.; Griscelli, C.; Fischer, A.; Saint Basile de, G. Griscelli disease maps to chromosome 15q21 and is associated with mutations in the myosin-Va gene. Nat. Genet. 1997, 16, 289–292. [Google Scholar] [CrossRef]
- Avraham, K.B.; Hasson, T.; Steel, K.P.; Kingsley, D.M.; Russell, L.B.; Mooseker, M.S.; Copeland, N.G.; Jenkins, N.A. The mouse Snell’s waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat. Genet. 1995, 11, 369–374. [Google Scholar] [CrossRef]
- Liu, X.Z.; Walsh, J.; Tamagawa, Y.; Kitamura, K.; Nishizawa, M.; Steel, K.P.; Brown, S.D.M. Autosomal dominant non syndromic deafness caused by a mutation in the myosin 7A gene. Nat. Genet. 1997, 17, 268–269. [Google Scholar] [CrossRef] [PubMed]
- Weil, D.; Blanchard, S.; Kaplan, J.; Guilford, P.; Gibson, F.; Walsh, J.; Mbruru, P.; Valera, A.; Levilliers, J.; Weston, M.D.; et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature 1995, 374, 60–61. [Google Scholar] [CrossRef] [PubMed]
- Weil, D.; Kussel, P.; Blanchard, S.; Levy, G.; Levi-Acobas, F.; Drira, M.; Ayadi, H.; Petit, C. The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat. Genet. 1997, 16, 191–193. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Z.; Walsh, J.; Mburu, P.; Kendrick-Jones, J.; Cope, M.J.T.V.; Steel, K.P.; Brown, S.D.M. Mutations in the myosin VIIA gene caue non syndromic recessive deafness. Nat. Genet. 1997, 16, 188–190. [Google Scholar] [CrossRef]
- Gibson, F.; Walsh, J.; Mburu, P.; Varela, A.; Brown, K.A.; Antonio, M.; Beisel, K.W.; Steel, K.P.; Brown, S.D.M. A type VII myosin encoded by the mouse deafness gene shaker-1. Lett. Nat. 1995, 374, 62–64. [Google Scholar] [CrossRef]
- Probst, F.J.; Fridell, R.A.; Raphael, Y.; Saunders, T.L.; Wang, A.; Liang, Y.; Morell, R.J.; Touchman, J.W.; Lyons, R.H.; Noben-Trauth, K.; et al. Correction of Deafness in shaker-2 Mice by an Unconventional Myosin in a BAC Transgene. Science 1998, 280, 1444–1447. [Google Scholar] [CrossRef]
- Wang, A.; Liang, Y.; Fridell, R.A.; Probst, F.J.; Wilcox, E.R.; Touchman, J.W.; Morton, C.C.; Morell, R.J.; Noben-trauth, K.; Camper, A.; et al. Association of Unconventional Myosin MYO15 Mutations with Human Nonsyndromic Deafness DFNB3. Sci. New Ser. 1998, 280, 1447–1451. [Google Scholar] [CrossRef]
- Gross, J.A.; Pacis, A.; Chen, G.G.; Drupals, M.; Lutz, P.E.; Barreiro, L.B.; Turecki, G. Gene-body 5-hydroxymethylation is associated with gene expression changes in the prefrontal cortex of depressed individuals. Transl. Psychiatry 2017, 7, e1119. [Google Scholar] [CrossRef]
- Liu, Y.F.; Sowell, S.M.; Luo, Y.; Chaubey, A.; Cameron, R.S.; Kim, H.G.; Srivastava, A.K. Autism and intellectual disability-associated KIRREL3 interacts with neuronal proteins MAP1B and MYO16 with potential roles in neurodevelopment. PLoS ONE 2015, 10, e0123106. [Google Scholar] [CrossRef]
- Rodriguez-Murillo, L.; Xu, B.; Roos, J.L.; Abecasis, G.R.; Gogos, J.A.; Karayiorgou, M. Fine mapping on chromosome 13q32-34 and brain expression analysis implicates MYO16 in schizophrenia. Neuropsychopharmacology 2014, 39, 934–943. [Google Scholar] [CrossRef][Green Version]
- Kao, C.; Chen, H.; Chen, H.; Yang, J.; Huang, M.; Chiu, Y.; Lin, S.; Lee, Y.; Liu, C.; Chuang, L.; et al. Identification of Susceptible Loci and Enriched Pathways for Bipolar II Disorder Using Genome-Wide Association Studies. Int. J. Neuropsychopharmacol. 2016, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.C.; Pauls, D.L.; Lange, C.; Sasanfar, R.; Santangelo, S.L. Sex-specific association of a common variant of the XG gene with autism spectrum disorders. Am. J. Med. Genet. Part. B Neuropsychiatr. Genet. 2013, 162, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.G.; Liu, C.; Cameron, P.L.; Cameron, R.S. Myr 8, a novel unconventional myosin expressed during brain development associates with the protein phosphatase catalytic subunits 1alpha and 1gamma1. J. Neurosci. 2001, 21, 7954–7968. [Google Scholar] [CrossRef]
- Thompson, R.F.; Langford, G.M. Myosin superfamily evolutionary history. Anat. Rec. 2002, 268, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Odronitz, F.; Kollmar, M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007, 8, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Sebé-Pedrós, A.; Grau-Bové, X.; Richards, T.A.; Ruiz-Trillo, I. Evolution and classification of myosins, a paneukaryotic whole-genome approach. Genome Biol. Evol. 2014, 6, 290–305. [Google Scholar] [CrossRef]
- Yokoyama, K.; Tezuka, T.; Kotani, M.; Nakazawa, T.; Hoshina, N.; Shimoda, Y.; Kakuta, S.; Sudo, K.; Watanabe, K.; Iwakura, Y.; et al. NYAP: A phosphoprotein family that links PI3K to WAVE1 signalling in neurons. EMBO J. 2011, 30, 4739–4754. [Google Scholar] [CrossRef]
- Bugyi, B.; Kengyel, A. Myosin XVI. In Myosins, A Superfamily of Molecular Motors; Series: Advances in Experimental Medicine and Biology; Lynne, M.C., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 405–419. [Google Scholar]
- Bähler, M. Are class III and class IX myosins motorized signalling molecules? Biochim. Biophys. Acta-Mol. Cell Res. 2000, 1496, 52–59. [Google Scholar] [CrossRef]
- Sellers, J.R. Myosins: A diverse superfamily. Biochim Biophys Acta-Mol. Cell Res. 2000, 1496, 3–22. [Google Scholar] [CrossRef]
- Bird, J.E.; Takagi, Y.; Billington, N.; Strub, M.-P.; Sellers, J.R.; Friedman, T.B. Chaperone-enhanced purification of unconventional myosin 15, a molecular motor specialized for stereocilia protein trafficking. Proc. Nalt. Acad. Sci. USA 2014, 111, 1–6. [Google Scholar] [CrossRef]
- Kengyel, A.; Bécsi, B.; Kónya, Z.; Sellers, J.R.; Erdődi, F.; Nyitrai, M. Ankyrin domain of myosin 16 influences motor function and decreases protein phosphatase catalytic activity. Eur. Biophys. J. 2015, 44, 207–218. [Google Scholar] [CrossRef]
- Taft, M.; Latham, S. Myosin XVIII. In Myosins, A Superfamily of Molecular Motors; Series: Advances in Experimental Medicine and Biology; Lynne, M.C., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 421–438. [Google Scholar]
- Hartshorne, D.J.; Ito, M.; Erdödi, F. Myosin light chain phosphatase: Subunit composition, interactions and regulation. J. Muscle Res. Cell Motil. 1998, 19, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.T.W. Protein phosphatase 1 – targeted in many directions. J. Cell Sci. 2020, 115, 241–256. [Google Scholar]
- Bollen, M. Combinatorial control of protein phosphatase-1. TRENDS Biochem. Sci. 2001, 26, 426–430. [Google Scholar] [CrossRef]
- Hartman, M.A.; Finan, D.; Sivaramakrishnan, S.; Spudich, J.A. Principles of Unconventional Myosin Function and Targeting. Annu. Rev. Cell Dev. Biol. 2016, 27, 133–155. [Google Scholar] [CrossRef] [PubMed]
- Krendel, M.; Mooseker, M.S. Myosins: Tails (and Heads) of Functional Diversity. Physiology 2005, 20, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Foth, B.J.; Goedecke, M.C.; Soldati, D. New insights into myosin evolution and classification. Proc. Nalt. Acad. Sci. USA 2006, 103, 3681–3686. [Google Scholar] [CrossRef]
- Kengyel, A.; Telek, E.; Kónya, Z.; Bécsi, B.; Erdődi, F.; Nyitrai, M. Interactions and Functions of Myosin 16 Domains. Biophys. J. 2017, 112, 267a. [Google Scholar] [CrossRef]
- Salter, M.W.; Kalia, L.V. SRC kinases: A hub for NMDA receptor regulation. Nat. Rev. Neurosci. 2004, 5, 317–328. [Google Scholar] [CrossRef]
- Mercati, O.; Danckaert, A.; André-Leroux, G.; Bellinzoni, M.; Gouder, L.; Watanabe, K.; Shimoda, Y.; Grailhe, R.; De Chaumont, F.; Bourgeron, T.; et al. Contactin 4, -5 and -6 differentially regulate neuritogenesis while they display identical PTPRG binding sites. Biol. Open 2013, 2, 324–334. [Google Scholar] [CrossRef]
- Tettamanti, G.; Kasahara, K.; Watanabe, K.; Kozutsumi, Y.; Oohira, A.; Yamamoto, T.; Sanai, Y. Association of GPI-Anchored Protein TAG-1 with Src-Family Kinase Lyn in Lipid Rafts of Cerebellar Granule Cells. Neurochem. Res. 2002, 27, 823–829. [Google Scholar]
- Zeng, L.; D’alessandri, L.; Kalousek, M.B.; Vaughan, L.; Pallen, C.J. Protein Tyrosine Phosphatase (PTP ) and Contactin Form a Novel Neuronal Receptor Complex Linked to the Intracellular Tyrosine Kinase fyn. J. Cell Biol. 1999, 147, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Castellano, E.; Downward, J. Ras interaction with PI3K: More than just another effector pathway. Genes Cancer 2011, 2, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Vivanco, I.; Sawyers, C.L. The phosphatidylinositol 3-kinase-AKT pathway in humancancer. Nat. Rev. Cancer 2002, 2, 489–501. [Google Scholar] [CrossRef]
- Bugyi, B.; Carlier, M.-F. Control of Actin Filament Treadmilling in Cell Motility. Annu. Rev. Biophys. 2010, 39, 449–470. [Google Scholar] [CrossRef]
- Carver, R.J.; Chen, B.; Chou, H.-T.; Brautigam, C.A.; Xing, W.; Yang, S.; Henry, L.; Doolittle, L.K.; Walz, T.; Rosen, M.K. Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. eLife 2017, 6, e29795. [Google Scholar]
- Roesler, M.K.; Lombino, F.L.; Freitag, S.; Schweizer, M.; Hermans-Borgmeyer, I.; Schwarz, J.R.; Kneussel, M.; Wagner, W. Myosin XVI Regulates Actin Cytoskeleton Dynamics in Dendritic Spines of Purkinje Cells and Affects Presynaptic Organization. Front. Cell Neurosci. 2019, 13, 330. [Google Scholar] [CrossRef]
- Koberstein, J.N.; Poplawski, S.G.; Wimmer, M.E.; Porcari, G.; Kao, C.; Gomes, B.; Risso, D.; Hakonarson, H.; Zhang, N.R.; Schultz, R.T.; et al. Learning-dependent chromatin remodeling highlights noncoding regulatory regions linked to autism. Sci. Signal. 2018, 11, eaan6500. [Google Scholar] [CrossRef]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an Online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef]
- Batters, C.; Veigel, C. Mechanics and Activation of Unconventional Myosins. Traffic 2016, 17, 860–871. [Google Scholar] [CrossRef]
- Heissler, S.M.; Sellers, J.R. Various Themes of Myosin Regulation. J. Mol. Biol. 2016, 428, 1927–1946. [Google Scholar] [CrossRef] [PubMed]
- Fili, N.; Toseland, C.P. Unconventional myosins: How regulation meets function. Int. J. Mol. Sci 2020, 21, 67. [Google Scholar] [CrossRef] [PubMed]
- Bement, W.M.; Mooseker, M.S. TEDS Rule: A Molecular Rationale for Differential Regulation of Myosins by Phosphorylation of the Heavy Chain Head. Cell Motil. Cytoskeleton 1995, 31, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Brzeska, H.; Knaus, U.G.; Wang, Z.-Y.; Bokoch, G.M.; Korn, E.D. p21-activated kinase has substrate specificity similar to Acanthamoeba myosin I heavy chain kinase and activates Acanthamoeba myosin I. Proc. Nalt. Acad. Sci. USA 1997, 94, 1092–1095. [Google Scholar] [CrossRef]
- Brzeska, H.; Lynch, T.J.; Martin, B.; Korn, E.D. The Localization and Sequence of the Phosphorylation Sites of Acantharnoeba Myosins I. J. Biol. Chem. 1989, 264, 19340–19348. [Google Scholar]
- Dürrwang, U.; Fujita-Becker, S.; Erent, M.; Kull, F.J.; Tsiavaliaris, G.; Geeves, M.A.; Manstein, D.J. Dictyostelium myosin-IE is a fast molecular motor involved in phagocytosis. J. Cell Sci. 2006, 119, 550–558. [Google Scholar] [CrossRef]
- Fujita-Becker, S.; Dürrwang, U.; Erent, M.; Clark, R.J.; Geeves, M.A.; Manstein, D.J. Changes in Mg2+ ion concentration and heavy chain phosphorylation regulate the motor activity of a class I myosin. J. Biol. Chem. 2005, 280, 6064–6071. [Google Scholar] [CrossRef]
- Attanapola, S.L.; Alexander, C.J.; Mulvihill, D.P. Ste20-kinase-dependent TEDS-site phosphorylation modulates the dynamic localisation and endocytic function of the fission yeast class I myosin, Myo1. J. Cell Sci. 2009, 122, 3856–3861. [Google Scholar] [CrossRef]
- Liu, X.; Lee, D.Y.; Cai, S.; Yu, S.; Shu, S.; Levine, R.L.; Korn, E.D. Regulation of the actin-activated MgATPase activity of Acanthamoeba myosin II by phosphorylation of serine 639 in motor domain loop 2. Proc. Nalt. Acad. Sci. USA 2012, 110, E23–E32. [Google Scholar] [CrossRef]
- Komaba, S.; Inoue, A.; Maruta, S.; Hosoya, H.; Ikebe, M. Determination of human myosin III as a motor protein having a protein kinase activity. J. Biol. Chem. 2003, 278, 21352–21360. [Google Scholar] [CrossRef]
- Quintero, O.A.; Unrath, W.C.; Stevens, S.M.; Manor, U.; Kachar, B.; Yengo, C.M. Myosin 3A kinase activity is regulated by phosphorylation of the kinase domain activation loop. J. Biol. Chem. 2013, 288, 37126–37137. [Google Scholar] [CrossRef] [PubMed]
- Kambara, T.; Komaba, S.; Ikebe, M. Human myosin III is a motor having an extremely high affinity for actin. J. Biol. Chem. 2006, 281, 37291–37301. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, M.; Homma, K.; Saito, J.; Inoue, A.; Ikebe, R.; Ikebe, M. Dual Regulation of Mammalian Myosin VI Motor Function. J. Biol. Chem. 2001, 276, 39600–39607. [Google Scholar] [CrossRef] [PubMed]
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef]
- Cameron, R.S.; Liu, C.; Pihkala, J.P.S. Myosin 16 levels fluctuate during the cell cycle and are downregulated in response to DNA replication stress. Cytoskeleton 2013, 70, 328–348. [Google Scholar] [CrossRef]
- Hornbeck, P.; Zhang, B.; Murray, B.; M Kornhauser, J.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic. Acids Res. 2014, 43, 512–520. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Heissler, S.M.; Sellers, J.R. Myosin light chains: Teaching old dogs new tricks. Bioarchitecture 2014, 4, 169–188. [Google Scholar] [CrossRef]
- Milton, D.L.; Schneck, A.N.; Ziech, D.A.; Ba, M.; Facemyer, K.C.; Halayko, A.J.; Baker, J.E.; Gerthoffer, W.T.; Cremo, C.R. Direct evidence for functional smooth muscle myosin II in the 10S self-inhibited monomeric conformation in airway smooth muscle cells. Proc. Nalt. Acad. Sci. USA 2011, 108, 1421–1426. [Google Scholar] [CrossRef]
- Wang, F.; Thirumurugan, K.; Stafford, W.F.; Hammer, J.A.; Knight, P.J.; Sellers, J.R. Regulated Conformation of Myosin V. J. Biol. Chem. 2004, 279, 2333–2336. [Google Scholar] [CrossRef]
- Batters, C.; Brack, D.; Ellrich, H.; Averbeck, B.; Veigel, C. Calcium can mobilize and activate myosin-VI. Proc. Nalt. Acad. Sci. USA 2016, E1162–E1169. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Baboolal, T.G.; Siththanandan, V.; Chen, M.; Walker, M.L.; Knight, P.J.; Peckham, M.; Sellers, J.R. A FERM domain autoregulates Drosophila myosin 7a activity. Proc. Nalt. Acad. Sci. USA 2009, 106, 4189–4194. [Google Scholar] [CrossRef]
- Umeki, N.; Jung, H.S.; Sakai, T.; Sato, O.; Ikebe, R.; Ikebe, M. Phospholipid-dependent regulation of the motor activity of myosin X. Nat. Struct. Mol. Biol. 2011, 18, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signaling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Kay, B.K.; Williamson, M.P.; Sudol, M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000, 14, 231–241. [Google Scholar] [CrossRef]
- Stapley, B.J.; Creamer, T.P. A survey of left-handed polyproline II helices. Protein Sci. 1999, 8, 587–595. [Google Scholar] [CrossRef]
- Alexei, A.A.; Michael, J.E.S. Left-handed polyproline II helices commonly occur in globular proteins. J. Mol. Biol. 1993, 229, 472–493. [Google Scholar]
- Adam, L.R.; Trevor, P.C. Polyproline II helical structure in protein unfolded states: Lysine peptides revisited. Protein Sci. 2002, 11, 980–985. [Google Scholar]
- Williamson, M.P. The structure and function of proline-rich regions in proteins. Biochem. J. 1994, 297, 249–260. [Google Scholar] [CrossRef]
- Steven, L.P.; Jasbinder, S.S. Mitogen-activated protein kinases: Versatile transducers for cell signaling. Trends Biochem. Sci. 1992, 17, 233–238. [Google Scholar]
- Florian, P.; James, A.B. Serine and Threonine Phosphorlyation. In Basic Neurochemistry, American Society for Neurochemistry; Elsevier Inc.: Amsterdam, The Netherlands, 2012; pp. 467–492. [Google Scholar]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic. Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.S.; Miller, W.T. Cooperative activation of Src family kinases by SH3 and SH2 ligands. Cancer Lett. 2007, 257, 116–123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gerbec, Z.J.; Thakar, M.S.; Malarkannan, S. The Fyn-ADAP axis: Cytotoxicity versus cytokine production in killer cells. Front. Immunol. 2015, 6, 472. [Google Scholar] [CrossRef] [PubMed]
- Saksela, K.; Permi, P. SH3 domain ligand binding: What’s the consensus and where’s the specificity? FEBS Lett. 2012, 586, 2609–2614. [Google Scholar] [CrossRef]
- Vig, A.T.; Földi, I.; Szikora, S.; Migh, E.; Gombos, R.; Tóth, M.Á.; Huber, T.; Pintér, R.; Talián, G.C.; Mihály, J.; et al. The activities of the C-terminal regions of the formin protein Disheveled-associated activator of morphogenesis (DAAM) in actin dynamics. J. Biol. Chem. 2017, 292, 13566–13583. [Google Scholar] [CrossRef]
- Holt, M.R.; Koffer, A. Cell motility: Proline-rich proteins promote protrusions. Trends Cell Biol. 2001, 11, 38–46. [Google Scholar] [CrossRef]
- Michaelsen, K.; Murk, K.; Zagrebelsky, M.; Dreznjak, A.; Jockusch, B.M.; Rothkegel, M.; Korte, M. Fine-tuning of neuronal architecture requires two profilin isoforms. Proc. Nalt. Acad. Sci. USA 2010, 107, 15780–15785. [Google Scholar] [CrossRef]
- Pinto-Costa, R.; Sousa, S.C.; Leite, S.C.; Nogueira-Rodrigues, J.; Da Silva, T.F.; Machado, D.; Marques, J.; Costa, A.C.; Liz, M.A.; Bartolini, F.; et al. Profilin 1 delivery tunes cytoskeletal dynamics toward CNS axon regeneration. J. Clin. Investig. 2020, 130, 2024–2040. [Google Scholar] [CrossRef]
- Neidt, E.M.; Scott, B.J.; Kovar, D.R. Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J. Biol. Chem. 2009, 284, 673–684. [Google Scholar] [CrossRef]
- Shao, J.; Welch, W.J.; Diprospero, N.A.; Diamond, M.I. Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol. Cell Biol. 2008, 28, 5196–5208. [Google Scholar] [CrossRef]
- Sathish, K.; Padma, B.; Munugalavadla, V.; Bhargavi, V.; Radhika, K.V.N.; Wasia, R.; Sairam, M.; Singh, S.S. Phosphorylation of profilin regulates its interaction with actin and poly (L-proline). Cell Signal. 2004, 16, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Diamond, M.I. Protein phosphatase 1 dephosphorylates profilin-1 at ser-137. PLoS ONE 2012, 7, e32802. [Google Scholar] [CrossRef] [PubMed]

| Myosin 1 | |||||||
|---|---|---|---|---|---|---|---|
| IIA IIB | IIIA | VA VB | VI | VIIA | XV | XVI | |
| Myosin encoding gene | NMMHC-IIA NMMHC-IIB | MYO3A | MYO5A MYO5B | MYO6 | MYO7A | MYO15 | MYO16 |
| Alzheimer disease | IIB | ||||||
| Autism | IIB | XVI | |||||
| Schizophrenia | VB | XVI | |||||
| Bipolar disorder subtype II | XVI | ||||||
| Major depressive disorder | XVI | ||||||
| Non-syndromic deafness | IIA | IIIA | VIIA | XV | |||
| Snell’s waltzer syndrome | VI | ||||||
| Usher syndrome | VIIA | ||||||
| Griscelli disease | VA | ||||||
| Lissencephaly | IIB | ||||||
| Sense of smell | VB | ||||||
| Reference | [6,7,8] | [9] | [10,11,12] | [13] | [14,15,16,17,18] | [19,20] | [21,22,23,24,25] |
| Motor Domain | |||||||
|---|---|---|---|---|---|---|---|
| Consensus Amino Acid 1 | P-Loop | DLLAK Motif | |||||
| (497–504 aa) | (722–742 aa) | S948 | S992 | K1029 | Y1075GY1077 | Y1090 | |
| * * : | : | . | . | * | * | * | |
| Hs | S[GERGSGKS] | DMIIRRHTIQIAEFFRDLLAK | S | S | K | YGY | Y |
| Mm | S[GERGSGKT] | DVIIRRHTIQMAAFYRDLLAK | S | S | K | SGY | Y |
| Rn | S[GERGSGKT] | DVIIRRHTTQIAAFYRDLLAK | S | S | K | YGY | Y |
| Gg | S[GESGSGKT] | DMIVRRHTIEMAEFYRDLLAK | S | S | K | YGY | Y |
| Xt | S[GESGSGKT] | DMITRRHSVDTAEFYRDLLAK | S | G | K | YGY | Y |
| Dr | S[GESGSGKS] | DVITRRHTVEMSNHHRDLLTK | C | N | K | YGY | Y |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Telek, E.; Kengyel, A.; Bugyi, B. Myosin XVI in the Nervous System. Cells 2020, 9, 1903. https://doi.org/10.3390/cells9081903
Telek E, Kengyel A, Bugyi B. Myosin XVI in the Nervous System. Cells. 2020; 9(8):1903. https://doi.org/10.3390/cells9081903
Chicago/Turabian StyleTelek, Elek, András Kengyel, and Beáta Bugyi. 2020. "Myosin XVI in the Nervous System" Cells 9, no. 8: 1903. https://doi.org/10.3390/cells9081903
APA StyleTelek, E., Kengyel, A., & Bugyi, B. (2020). Myosin XVI in the Nervous System. Cells, 9(8), 1903. https://doi.org/10.3390/cells9081903







