Abstract
Lysosomal storage diseases (LSDs) are a heterogeneous group of rare multisystem genetic disorders occurring mostly in infancy and childhood, characterized by a gradual accumulation of non-degraded substrates inside the lysosome. Although the cellular pathogenesis of LSDs is complex and still not fully understood, the approval of disease-specific therapies and the rapid emergence of novel diagnostic methods led to the implementation of extensive national newborn screening (NBS) programs in several countries. In the near future, this will help the development of standardized workflows aimed to more timely diagnose these conditions. Hereby, we report an overview of LSD diagnostic process and treatment strategies, provide an update on the worldwide NBS programs, and discuss the opportunities and challenges arising from genomics applications in screening, diagnosis, and research.
1. Introduction
Lysosomal storage diseases (LSDs) are a heterogeneous group of heritable (inborn) metabolism defects that affect the function of lysosomes. This group comprises about 70 monogenic disorders of lysosomal catabolism and is characterized by a gradual accumulation of non-degraded substrates inside the lysosome, which in turn leads to cellular dysfunction, tissue damage, and death [1]. The majority of LSDs are inherited as autosomal recessive traits (except for Fabry, Hunter, and Danon diseases that are X- linked) and are caused by mutations in genes encoding lysosomal proteins (i.e., acidic hydrolases, integral membrane proteins, and activator or carrier proteins) whose functional deficiencies trigger the pathogenetic cascade [2]. Although each disorder is rare, per se, with estimated incidences ranging from 1 in 50,000 to 1 in 250,000 live births, LSDs as a group are relatively common disorders (1:5000 live births) [1].
LSDs can be categorized by (a) the biochemical type of stored material (e.g., sphingolipidoses, mucopolysaccharidoses, glycoproteinoses); (b) the post-translational modification of lipofuscin degradation or metabolism defects; (c) the dysfunctions in membrane proteins; or (d) the altered lysosome-related organelle (LRO) defects (Figure 1).
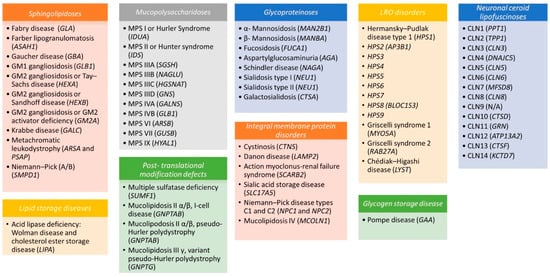
Figure 1.
Disorders of lysosomes and lysosome-related organelles (LROs). Lysosomal storage diseases (LSDs) and LROs have been subclassified according to the biochemical type of stored material (sphingolipidoses, mucopolysaccharidoses, glycoproteinoses, lipid storage diseases) or to the integral membrane proteins, post-translational modification, and lipofuscin metabolism defects. The causative gene is specified in parentheses.
Clinically, LSDs may present with a broad range of phenotypes reflecting the age of onset (e.g., more severe infantile forms vs. later/adult milder forms) [3], the extent and severity of nervous system and/or systemic involvements, and the related variability of signs and/or symptoms. In general, the disease progresses and evolves relentlessly over time. Signs and/or symptoms mainly include facial dimorphisms, psychomotor developmental delay and cognitive decline, seizures, impairment of vision, recurrent infections, muscle deficits, organomegaly, immune defects, and skeletal changes, all having a severe impact on prognosis and influencing the quality of life for patients and families [4,5].
2. The Biology of Lysosomes
Lysosomes are the key cellular organelles in macromolecule catabolism, responsible for the breakdown and recycling of a wide range of complex metabolites including glycosides, lipids, phospholipids, proteins, and nucleic acids. This catabolic function is orchestrated by approximately 60 unique acidic hydrolase enzymes (glycosidases, sulfatases, peptidases, phosphatases, lipases, and nucleases), which are located within the lysosomal lumen where the enzyme and the substrate interact with each other. While the enzymes are mainly synthesized within the endoplasmic reticulum (ER), tagged with a mannose-6-phosphate (M6P) residue in the Golgi apparatus and then trafficked into the lysosome [6,7], the substrates are transported through different routes according to the nature of the cargo. Materials from outside the cell are delivered through the endocytosis pathway via clathrin-mediated or caveolin-mediated endocytosis vesicles. The cell’s own macromolecules and metabolites are processed and degraded through autophagy, in one of its functional forms including macroautophagy, microautophagy, and chaperone-mediated autophagy [8]. Recent evidence shows that lysosomes are not only catabolic organelles, but they also function as metabolic hubs controlling nutrient sensing, amino-acid and ion homeostasis, vesicle trafficking, and cellular growth, and they establish contact sites with other organelles (e.g., mitochondria, ER, or peroxisomes) [7,9].
3. Diagnosis and Therapeutic Strategies
Diagnosis of LSDs is often a challenge for clinicians, owing to the rarity of single disorders and to the non-specificity of signs and/or symptoms, often attributed to other neurological and/or systemic diseases. Before the spread of expanded newborn screening, individuals were diagnosed years after sign and/or symptom onset, when the disease was already advanced and interventions proved less efficacious [10].
The typical diagnostic work-up for LSDs includes taking a detailed history on the clinical presentation(s) and course of disease; measuring the activity of single enzymes or protein abundance in leukocytes, fibroblasts, urine, or rehydrated dried blood spots (DBS); conducting various laboratory, ophthalmological, otolaryngology, ultrasonographic, neurophysiological, and imaging investigations [11]. When enzyme levels fall below the average, second-tier confirmatory biomarker tests and gene sequencing are performed to identify DNA-specific mutations affecting gene function (Figure 2) [2]. Moreover, prenatal diagnosis for some LSDs is emerging, using uncultured chorionic villi [12,13].

Figure 2.
Diagnostic flowchart for Gaucher and Fabry diseases. NBS, newborn screening; LysoGb1, glucosylphingosine; LysoGb3, globotriaosylphingosine; ERT, enzyme replacement therapy; SRT, substrate reduction therapy.
Major advances have been made in recent years in our understanding of the pathophysiology of LSDs [1]. This has allowed researchers to identify multiple potential clinical interventions, targeting different events in the pathogenetic cascade (Figure 3), and enabled clinicians to expand, at least for some LSDs, the opportunities for therapeutic strategies besides the supportive medical and physical therapies (e.g., management of neurological complications, ventilatory or nutritional support, orthopedic interventions in order to alleviate deformities, and several others) [14].
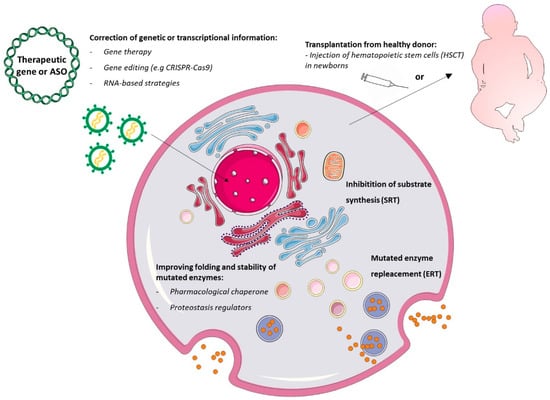
Figure 3.
Current lysosomal storage disorder therapeutic strategies. Gene therapy and gene editing approaches aim to introduce a functional gene or to correct the defective gene or transcript inside cells. Hematopoietic stem cell transplantation (HSCT) repopulates specific tissues and allows local release of functional lysosomal hydrolases by healthy donor cells. Substrate reduction therapy (SRT) inhibits substrate synthase at the early Golgi compartment. Enzyme replacement therapies (ERTs) deliver functional enzymes to lysosomes via the endocytosis pathway. Chaperones can stabilize the mutant enzyme and partially restore catalytic activity in the endoplasmic reticulum (ER) and overall in lysosomes. Lastly, proteostasis modifiers stabilize transcription factors within the nucleus, improve lysosomal enzyme expression and translation in the ER, reduce lysosomal membrane permeability, and improve lysosomal function.
The cornerstone for current treatment paradigms is enzyme replacement therapy (ERT), which is considered the standard of care for Gaucher, Fabry, Pompe disease, and mucopolysaccharidosis I (MPSI) [15]. Approved around the nineties by the US Food and Drug administration (FDA) Committee for ERT, the current recombinant functional enzyme is delivered via periodic intravenous infusion whose uptake occurs via the endocytosis pathway. Timely initiation of treatment is crucial for an optimal clinical outcome [16]. Major disadvantages of ERT still include the inability of the recombinant enzymes to diffuse easily in all affected tissues (in particular into the central nervous system, due to their large size), [17], the great variability in patient immune responses, and the transient therapeutic benefit [15].
Hematopoietic stem cell transplantation (HSCT) uses hematopoietic stem cells from healthy donors. These cells can repopulate specific tissues and locally release functional lysosomal hydrolases in the extracellular space and into the blood circulation [18]. Despite the high morbidity and mortality rate due to graft rejection and infection, this therapy remains the first choice of treatment for children with MPS IH (Hurler disease), since it prevents the development of neurological symptoms and increases life expectancy, especially when performed early [19].
Other strategies are directed towards restoring the equilibrium between substrate synthesis and degradation by lysosomal enzymes (the storage equation of LSDs) [7]. In particular, substrate reduction therapy (SRT) approved by the FDA in 2003 (e.g., miglustat, employed for Gaucher disease) uses small-molecule enzyme inhibitors that slow down the build-up process of macromolecules, reducing the storage amount in lysosomes, and inhibiting the biosynthetic pathways [20]. In contrast to ERT, SRT drugs are orally administered and are stable at ambient temperature. They do not generate immune reactions and cross the blood–brain barrier, but they have a slower onset of action than ERT and produce adverse effects or complications relating to drug metabolism [7].
Pharmacological chaperone therapy (PCT) (e.g., migalastat for Fabry disease) uses inhibitory molecules, which target mutant lysosomal enzymes to favor their native conformational folding, stability, catalytic activity, and correct trafficking, also extending their half-life [8]. Some studies evidenced that migalastat provides clinical results comparable to those of ERT [21]. Chaperons were also used efficaciously in combination with ERT to treat patients with Pompe disease [22]. Unfortunately, this kind of treatment is mutation-sensitive and clinically impractical owing to the great efforts needed to find an optimal drug dosage.
New-generation pharmacological strategies are rapidly advancing into clinical trials, including proteostasis modifiers and gene therapy.
Proteostasis modifiers are able to regulate the components of the proteostasis machinery (a multiple regulatory integrated system including protein synthesis, structural folding, post-translational modification, trafficking, and degradation) and have already been suggested for treating Gaucher disease and GM2 gangliosidosis (Tay–Sachs disease) [8].
LSDs are excellent candidates for gene therapy for many reasons: not only are they well-known single-gene disorders, but also the expression of the enzyme is generally not subject to complex regulatory mechanisms. In addition, even a small increase in enzymatic activity is sufficient to revert the clinical phenotype [23]. A broad range of different strategies may be exploited depending on the tissues that need to be targeted and the characteristics of the protein that must be replaced. Expression cassettes containing the functional gene may be delivered to cells via direct gene transfer (by viral-based systems) or via indirect gene transfer (by re-implanting engineered autologous patient stem cells back into the donor) [24].
Promising perspectives come from genome editing platforms (ZFN, TALEN, and CRISPR-Cas9 systems), which have recently enabled the possibility of modifying target sites within the genome in a precise manner [25,26]. The combination of nuclease-mediated genome editing with autologous hematopoietic stem cells or induced pluripotent stem cells (iPSCs) may represent a milestone for treatment of LSDs, as it would lower the overall risk of infection during treatment and avoid rejection (graft-versus-host disease) [25,26]. Both preclinical (in vitro, in vivo, and ex vivo) and clinical studies using different editing-based strategies have already been started including trials for mucopolysaccharidoses [27] and GM2-gangliosidoses [28]. The opportunities granted by RNA-based therapies are equally interesting, as they support the feasibility of reverting the LSD phenotype by partially rescuing splicing defects [29]. Indeed, about 5–19% of LSD-causing mutations affect the pre-mRNA splicing process, and in some LSDs, single splicing changes can account for up to 40–70% of pathogenic alleles [29]. Two main splicing therapy strategies have been used for LSDs: (i) modified U1 small nuclear RNAs (U1snRNA) tested in cellular models of Sanfilippo C disease (or Mucopolysaccharidosis IIIC) [30] and Fabry disease [31], and (ii) the antisense oligonucleotide (AONs) approach used in cellular models of Niemann Pick C [32] and for the late-onset form of Pompe disease [33,34].
4. LSD Worldwide Newborn Screenings and Methodological Approaches
The rationale for mandatory (expanded) NBSs has historically included serious health conditions with an effective therapy, relatively easy and reliable disease markers, and evidence for the beneficial effect of early treatment in preventing severe disabilities or even death (Wilson and Jungner screening criteria). However, many adaptations on Wilson and Jungner criteria have occurred in the last decades, reflecting better knowledge of the natural history of many such conditions, new appraisals on logistical and ethical issues, and the advances in genetic technology [35,36].
Until 2006, when the FDA approved alfa-glucosidase for the treatment of Pompe disease [1], none of the LSDs was included in NBS programs. The concept of using DBS extracts for lysosomal enzyme testing, as well as the existence of therapeutic options and the development of new screening tests, opened up the potential for NBS in LSDs. Although widespread internationally, the inclusion of LSDs in expanded NBS is still debated as reflected by the large differences in screening programs worldwide (Figure 4 and Supplementary Table S1). The most active country is certainly the US, where the Advisory Committee on Heritable Disorders in Newborns and Children (ACHDNC) was charged to draw up national recommendations for guiding and supporting states in the development of screening programs. The ACHDNC set out the Recommended Uniform Screening Panel (or RUSP), a list of diseases including 35 core conditions and 26 secondary conditions, the Committee recommends every baby should be screened for. Among LSDs, Pompe disease and MPS I are considered the most favored for inclusion, and, since 2016, these two conditions have been included in RUSP (Figure 4) [37,38]. Meanwhile, pilot LSD screening programs have been implemented in a number of countries worldwide, including Italy [39,40]. In Northern and Central Italy, in particular, four regions (Piemonte, Veneto, Tuscany, and Umbria) started expanded NBS programs for Pompe, Fabry, Gaucher, and MPSI promoted by regional Health Government indications (Figure 4), foreshadowing its extension in Italy to a larger newborn population [41,42,43,44,45,46,47]. As a consequence, thanks to an amendment published by the Italian government in December 2018 (Gazzetta Ufficiale n. 302), LSDs are currently included in the national screening program.
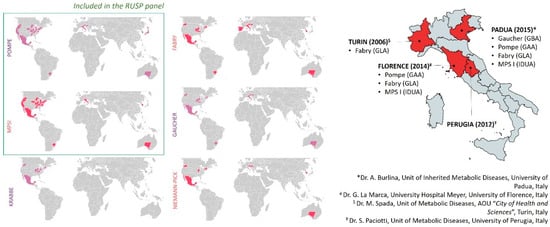
Figure 4.
Newborn screening programs (NBSs). The left side shows the worldwide distribution of NBSs including Pompe, MPSI, Krabbe, Fabry, Gaucher, and Niemann Pick diseases. Screening programs are mandated for some regions, under development, or in pilot phases for others (see Supplementary Table S1). The right side shows the pilot Italian regional screening programs for LSDs (brackets indicate the targeted genes; please refer to references reported above).
The two main platforms currently used for screening multiple LSDs are the digital microfluidic fluorometry (DMF) platform and the tandem mass spectrometry (MS/MS) platform [37,48,49,50,51]. DMF is the multiplexing advancement of the classical enzymatic fluorescent assay based on synthetic 4-methylumbelliferone (4-MU). It has been developed by Advanced Liquid Logic, Inc. (now Baebies, Inc. Durham, NC, USA) and is currently used for clinical diagnostic purposes and for NBSs. DMF is based on submicroliter droplets, which are moved on an electrode-plate chip through a process known as electrowetting. In this particular “spatial multiplexing”, each LSD enzyme reaction is performed on a single droplet under its individually optimized conditions (pH, inhibitors, buffer). The latest version simultaneously measures the activity of five enzymes to diagnose MPS-I, MPS-II, Pompe, Fabry, and Gaucher diseases. MS/MS is used to detect in a 6-plex assay the enzymatic products responsible for Pompe, MPS-I, Fabry, Gaucher, Krabbe, and Niemann-Pick-A/B diseases. The method currently commercialized as NeoLSD by PerkinElmer Corp starts with an incubation phase of the sample with a mixture of the six substrates and internal standards in a single buffer, followed by liquid–liquid extraction and flow-injection MS/MS. Detailed comparative information (space and manpower requirements, approximate costs, analytic precision) between MS/MS vs. DMF assays are listed in previous studies [37,48,49,51]. The DMF platform workflow is simpler, requires less maintenance, and generates results faster than MS/MS, providing results within the same day of specimen analysis. Conversely, MS/MS is more accurate and precise, and it can be used to assay biomarkers for which no fluorimetric methods exist [37,48,49,51].
5. Opportunities and Challenges for Genomics in LSDs
Genome-scale sequencing provides a powerful diagnostic tool for patients affected by conditions, which escape the diagnosis by traditional genetic investigation, and offers a range of new opportunities for genomic medicine. With the overwhelming entry of massive parallel sequencing (NGS) into modern medicine, scientists and clinicians started to wonder whether genomic sequencing could replace conventional biochemical tests by improving the screening of newborns or if it could just be useful as an optional supplementary tool [52]. A major driving force for this alternative approach was the rapidly decreasing price of NGS (competitive with current NBS prices) coupled with the vastly improved read depths and accuracy of sequencing platforms [53]. Considerations about the context of using sequencing information are addressed elsewhere in this issue (Figure 5).
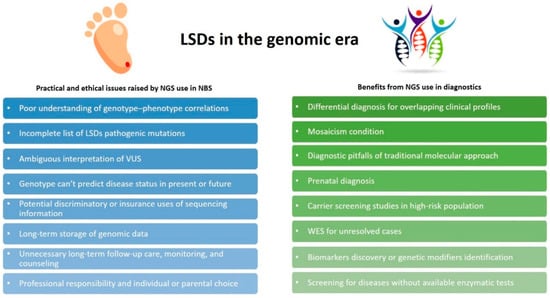
Figure 5.
Next generation sequencing (NGS) applications in LSDs. Practical and ethical issues raised by the use of NGS in NBS are shown in the blue boxes; benefits derived from NGS use in diagnostic settings are shown in the green boxes.
In a practical diagnostic setting, the goal of genome-scale sequencing is to identify genetic variants so as to provide a molecular etiology for patients’ clinical manifestations, considering all other variants as incidental findings. The high-throughput ability of NGS has been successfully used to diagnose LSDs [54,55,56,57,58,59], both in the form of exome and targeted sequencing. This is particularly useful when applied to specific diagnostic contexts, including carrier screening studies in high-risk populations (e.g., the Ashkenazi Jewish population) [60,61], prenatal diagnosis [62], unsolved cases where traditional molecular diagnostic approaches have failed [63], unclear or suspected LSD cases [64,65], as well as in defining genotype–phenotype correlations [66] or to find out genetic disease modifiers [67]. More interesting is the use of NGS to differentiate genetically heterogeneous diseases with overlapping clinical phenotypes, such as Pompe disease, limb-girdle muscular dystrophies [68,69], and Gangliosidosis [70], or to investigate mosaic conditions [71,72]. Many companies developed commercial panels and offer direct-to-consumer sequencing services for suspected LSD cases, utilizing custom panels that target few or many genes (causative genes, lysosomal pathway-related genes, or peroxisome disorder-related genes) and are based on arbitrary research (Supplementary Table S2).
In contrast, the application of genomic sequencing for NBS substantially raises a whole host of issues encompassing several disciplines, which need to be appropriately discussed. One major challenge is how to accurately interpret the clinical significance of incidental findings (variants of unknown significance, VUS), including peri-gene sequence variants, mutations localized in intronic regions and in UTRs, or synonymous variants having an impact on gene regulation. Mutational databases for LSDs are already available; however, the complete list of LSD pathogenic mutations is still expanding. Moreover, there is a lack of sufficiently large ethnicity-specific genetic datasets leading, in turn, to the risk of falling into free interpretation of VUS. A combined international effort to generate large, freely available datasets, better prediction algorithms, and standardized tests in terms of laboratory work (e.g., sequencing platforms, read depths, mean and minimum coverage) should be regarded as of primary importance to solve data interpretation [53]. More importantly, there is still a poor understanding of genotype-phenotype correlations. Most LSD patients are complex heterozygotes, the pathogenicity of many alleles is still unknown, and genotype characterization is not enough to predict disease status in the present or the future [53]. NGS might reveal patients in their early years with adult-onset disease, who might not require treatment for decades, if not at all. Further concerns regard the professional responsibility and individual or parental choices about the types of findings that should be reported, potential discriminatory or insurance uses of sequencing information, long-term storage of genomic data, unnecessary interventions, and costly long-term follow-up care, monitoring, and counseling [52,73].
Nonetheless, attempts to expand the use of DNA sequencing in NBS have been carried out using customized NGS panels targeting few relevant genes or alternatively using a WES approach and bioinformatics analysis ad hoc [74,75]. On the other hand, genotyping-based NBS seems the better solution for high-risk neonates admitted to the neonatal intensive care unit (NICU), or for those disorders with no existing biochemical marker [76,77]. For example, in the case of nephropathic cystinosis, PCR- and NGS-based analyses have been used for NBS [78]. Therefore, as outlined by the Newborn Sequencing in Genomic Medicine and Public Health (NSIGHT) consortium, “sequencing technology can be beneficially used in newborns when its use is nuanced and attentive to context” [73].
6. Second-Tier Confirmatory Biomarkers: Which One?
The increasing number of NBS pilot studies worldwide has revealed an unexpectedly high number of false-positive samples and has highlighted the need to introduce second-tier testing in order to reduce the recall rate and assist in disease diagnosis. Several primary or secondary accumulating metabolites (i.e., molecules directly or indirectly enhanced as a result of defective lysosomal function) have been proposed as candidate biomarkers, as they are easily detectable in plasma or urine [3,79]. Lysosphingolipids for sphingolipidoses (LysoGb1 for Gaucher disease, LysoGb3 for Fabry disease, LysoSM and LysoSM509 for Niemann-Pick disease type A/B and C, GalSph for Krabbe disease), heparan and dermatan sulphates for MPSs, and glucose tetrasaccharide for Pompe have been proposed as candidate confirmatory biomarkers for differentiating patients with pathogenic mutations, pseudodeficiency alleles, and/or benign variants at the time of screening [39,80,81,82,83,84,85]. However, uncertainties remain in the use of these metabolic biomarkers, and further studies are necessary to achieve the development of definitive pipelines.
For the past few years, the search for new biomarkers has shifted towards microRNAs [86,87,88], opening an interesting perspective for genomics applications. Indeed, specific patterns of circulating miRNAs can identify patients with Fabry or Pompe diseases, and they could be useful for predicting the evolution of the disease or for assessing responses to therapy as well. Some of these miRNAs, in particular, were associated with heart problems and endothelial dysfunctions in Fabry patients, or they significantly correlated with phenotype severity, muscle dysfunctions, and Pompe disease-related patho/pathways (autophagy, muscle regeneration, muscle atrophy) [86,87,88].
7. The Importance of a Timely Diagnosis
As anticipated in the introduction, LSDs are rare genetic diseases not frequently encountered in the medical practice, and they often receive inadequate clinical and social consideration compared to other disorders. Nevertheless, taken together, they affect a certain percentage of the overall population (i.e., 1:5000 live births) mostly in infancy or childhood, although patients with late-onset/milder phenotypes are expected and represent the most subtle and difficult cases to identify. Indeed, while positive-NBS children with a pediatric onset of LSDs are followed by expert pediatricians, late-onset patients without a previous clinical history may be misdiagnosed for many years. Unfortunately, no pathognomonic traits for LSDs exist, as they are characterized by a non-specific, likely equivocal multisystemic symptomatology that combines systemic manifestations with overlapping neurological signs, and they are often indistinguishable even biochemically (e.g., MPSIII/Sanfilippo syndromes A-B-C-D, Table 1). These clinical and practical issues aggravate the quality of life for patients and families, as they not only suffer from their disease manifestations but also undergo continuous psychological stress caused by the uncertainty of both diagnostic responses, uncertain outcomes, and non-resolutive therapeutic solutions.

Table 1.
Disease name, causative genes, age of onset, accumulating substrates, and main clinical manifestations of MPS.
Nowadays, genetic analysis represents the only valid aid to rapidly diagnose and differentiate suspected LSDs cases, allowing causative mutations to be identified. The need to introduce broad and ad hoc designed genetic tests (e.g., targeted gene panels) in diagnostic workflows is becoming increasingly clear in order to easily identify LSDs and draw up informative guidelines that can properly direct both clinicians and geneticists towards the right criteria for results interpretation and diagnostic report writing. Timely diagnosis through genetic/genomic applications is key to halt disease progression, reduce psychological burden, optimize clinical management, and provide appropriate genetic counseling. Moreover, molecular profiling and genomic sequencing information may prompt the design of novel therapeutic drugs targeting specific mutations. The development of more effective treatments will open the possibility for new clinical trials that address stratified patient subclasses and will pave the way to personalized medicine. This, in the near future, will improve the quality of life of patients and their families and reduce both direct and indirect (e.g., care-givers services) costs to national health services and families.
8. Conclusions
LSDs are multisystem disorders with heterogeneous genetic profiles and overlapping clinical manifestations. Although they are monogenic diseases, the procedures and/or protocols for early diagnosis, management, and implementation of newborn screening programs (NBSs) deserve greater attention from the scientific community. Many efforts and elaboration phases by institutional networks and researcher partnerships are ongoing, purposing the production of benefits for patients with these rare, life-threatening diseases. MetabERN (Metabolic European Reference Networkgene, available at https://metab.ern-net.eu/), for example, is a European initiative aimed at developing a real-time consultation platform for clinical decision-making processes, and it fosters translational research programmers across inherited metabolic disease. IMI (Innovative Medicine Initiative, an EU public–private partnership), instead, is a funding project for Horizon 2020 with the goal of shortening the path to diagnosis by using newborn/pediatric genetic screening via application of advanced digital technologies (Call name H2020-JTI-IMI2-2020-23-two-stage). Application of molecular NGS-based testing (either in the form of WES or targeted gene panels) in this perspective represents a real and valuable aid to provide timely and correct diagnosis, detect carriership status, and ensure genetic counseling for family planning, thus improving the overall standards of care for patients and families. Consolidation of existing fragmented efforts will result in improved clinical and patient-oriented outcomes, increase public understanding around rare diseases, and potentially lead to better rare disease policies as well as improved value-based healthcare.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/9/8/1902/s1, Table S1: LSDs newborn screening programs worldwide, Table S2: NGS panels for LSDs diagnosis.
Author Contributions
Conceptualization, A.P., M.R. and S.C.; investigation, V.L.C. and M.G.; original drafting preparation, V.L.C.; re-drafting, reviewing, and editing, M.R., A.P. and S.C.; supervision, S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the joint project between IRIB-CNR and SANOFI “Early diagnosis of some lysosomal diseases: analysis of the clinical utility and diagnostic validity of genomic techniques for their molecular diagnosis. Assessments of the implications of the inclusion of lysosomal diseases in the context of a national neonatal screening program”.
Acknowledgments
The authors gratefully acknowledge Cristina Calì, Alfia Corsino, Maria Patrizia D’Angelo, and Francesco Marino for administrative and technical support; prof. Rosemary Ready (University of Catania, Italy) is gratefully acknowledged for editing the second draft of the present study.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the writing of the manuscript, or in the decision to publish the results.
References
- Platt, F.M.; d’Azzo, A.; Davidson, B.L.; Neufeld, E.F.; Tifft, C.J. Lysosomal storage diseases. Nat. Rev. Dis. Primers 2018, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.Y.; Bodamer, O.A.; Watson, M.S.; Wilcox, W.R. Lysosomal storage diseases: Diagnostic confirmation and management of presymptomatic individuals. Genet. Med. 2011, 13, 457–484. [Google Scholar] [CrossRef] [PubMed]
- Aerts, J.M.F.G.; Kallemeijn, W.W.; Wegdam, W.; Joao Ferraz, M.; van Breemen, M.J.; Dekker, N.; Kramer, G.; Poorthuis, B.J.; Groener, J.E.M.; Cox-Brinkman, J.; et al. Biomarkers in the diagnosis of lysosomal storage disorders: Proteins, lipids, and inhibodies. J. Inherit. Metab. Dis. 2011, 34, 605–619. [Google Scholar] [CrossRef]
- Alcalay, R.N.; Wolf, P.; Levy, O.A.; Kang, U.J.; Waters, C.; Fahn, S.; Ford, B.; Kuo, S.H.; Vanegas, N.; Shah, H.; et al. Alpha galactosidase A activity in Parkinson’s disease. Neurobiol. Dis. 2018, 112, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Eisenstein, M. Lysosomal storage disorders: 4 big questions. Nature 2016, 537, S165. [Google Scholar] [CrossRef]
- Ferreira, C.R.; Gahl, W.A. Lysosomal storage diseases. Transl. Sci. Rare Dis. 2017, 2, 1–71. [Google Scholar] [CrossRef]
- Parenti, G.; Andria, G.; Ballabio, A. Lysosomal storage diseases: From pathophysiology to therapy. Annu. Rev. Med. 2015, 66, 471–486. [Google Scholar] [CrossRef]
- Mohamed, F.E.; Al-Gazali, L.; Al-Jasmi, F.; Ali, B.R. Pharmaceutical Chaperones and Proteostasis Regulators in the Therapy of Lysosomal Storage Disorders: Current Perspective and Future Promises. Front Pharm. 2017, 8, 448. [Google Scholar] [CrossRef]
- Marques, A.R.A.; Saftig, P. Lysosomal storage disorders—Challenges, concepts and avenues for therapy: Beyond rare diseases. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef]
- Kingma, S.D.K.; Bodamer, O.A.; Wijburg, F.A. Epidemiology and diagnosis of lysosomal storage disorders; challenges of screening. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 145–157. [Google Scholar] [CrossRef]
- Mokhtariye, A.; Hagh-Nazari, L.; Varasteh, A.R.; Keyfi, F. Diagnostic methods for Lysosomal Storage Disease. Rep. Biochem. Mol. Biol. 2019, 7, 119–128. [Google Scholar] [PubMed]
- Verma, J.; Bijarnia-Mahay, S.; Verma, I.C. Prenatal Diagnosis of Lysosomal Storage Disorders Using Chorionic Villi. In Lysosomes; Humana Press: New York, NY, USA, 2017; Volume 1594, pp. 265–291. [Google Scholar]
- Li, D.; Lin, Y.; Huang, Y.; Zhang, W.; Jiang, M.; Li, X.; Zhao, X.; Sheng, H.; Yin, X.; Su, X.; et al. Early prenatal diagnosis of lysosomal storage disorders by enzymatic and molecular analysis. Prenat. Diagn. 2018, 38, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Parenti, G.; Pignata, C.; Vajro, P.; Salerno, M. New strategies for the treatment of lysosomal storage diseases (review). Int. J. Mol. Med. 2013, 31, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Li, M. Enzyme Replacement Therapy: A Review and Its Role in Treating Lysosomal Storage Diseases. Pediatr. Ann. 2018, 47, e191–e197. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, R.H. Enzyme replacement therapy for lysosomal storage diseases. Curr. Opin. Pediatr. 2011, 23, 588–593. [Google Scholar] [CrossRef]
- Poswar, F.O.; Vairo, F.; Burin, M.; Michelin-Tirelli, K.; Brusius-Facchin, A.C.; Kubaski, F.; Souza, C.F.M.; Baldo, G.; Giugliani, R. Lysosomal diseases: Overview on current diagnosis and treatment. Genet. Mol. Biol. 2019, 42, 165–177. [Google Scholar] [CrossRef]
- Beck, M. Treatment strategies for lysosomal storage disorders. Dev. Med. Child Neurol. 2018, 60, 13–18. [Google Scholar] [CrossRef]
- Lund, T.C. Hematopoietic stem cell transplant for lysosomal storage diseases. Pediatr. Endocrinol. Rev. 2013, 11, 91–98. [Google Scholar]
- Coutinho, M.F.; Santos, J.I.; Alves, S. Less Is More: Substrate Reduction Therapy for Lysosomal Storage Disorders. Int. J. Mol. Sci. 2016, 17, 1065. [Google Scholar] [CrossRef]
- Hughes, D.A.; Nicholls, K.; Shankar, S.P.; Sunder-Plassmann, G.; Koeller, D.; Nedd, K.; Vockley, G.; Hamazaki, T.; Lachmann, R.; Ohashi, T.; et al. Oral pharmacological chaperone migalastat compared with enzyme replacement therapy in Fabry disease: 18-month results from the randomised phase III ATTRACT study. J. Med. Genet. 2017, 54, 288–296. [Google Scholar] [CrossRef]
- Kishnani, P.; Tarnopolsky, M.; Roberts, M.; Sivakumar, K.; Dasouki, M.; Dimachkie, M.M.; Finanger, E.; Goker-Alpan, O.; Guter, K.A.; Mozaffar, T.; et al. Duvoglustat HCl Increases Systemic and Tissue Exposure of Active Acid α-Glucosidase in Pompe Patients Co-administered with Alglucosidase α. Mol. Ther. 2017, 25, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Nagree, M.S.; Scalia, S.; McKillop, W.M.; Medin, J.A. An update on gene therapy for lysosomal storage disorders. Expert Opin. Biol. Ther. 2019, 19, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Sands, M.S.; Davidson, B.L. Gene therapy for lysosomal storage diseases. Mol. Ther. 2006, 13, 839–849. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, T.G.; da Silveira Matte, U.; Giugliani, R.; Baldo, G. Genome Editing: Potential Treatment for Lysosomal Storage Diseases. Curr. Stem Cell Rep. 2015, 1, 9–15. [Google Scholar] [CrossRef]
- Christensen, C.; Choy, F. A Prospective Treatment Option for Lysosomal Storage Diseases: CRISPR/Cas9 Gene Editing Technology for Mutation Correction in Induced Pluripotent Stem Cells. Diseases 2017, 5, 6. [Google Scholar] [CrossRef]
- Poletto, E.; Baldo, G.; Gomez-Ospina, N. Genome Editing for Mucopolysaccharidoses. Int. J. Mol. Sci. 2020, 21, 500. [Google Scholar] [CrossRef]
- Ou, L.; Przybilla, M.J.; Tăbăran, A.-F.; Overn, P.; O’Sullivan, M.G.; Jiang, X.; Sidhu, R.; Kell, P.J.; Ory, D.S.; Whitley, C.B. A novel gene editing system to treat both Tay–Sachs and Sandhoff diseases. Gene Ther. 2020, 27, 226–236. [Google Scholar] [CrossRef]
- Dardis, A.; Buratti, E. Impact, Characterization, and Rescue of Pre-mRNA Splicing Mutations in Lysosomal Storage Disorders. Genes 2018, 9, 73. [Google Scholar] [CrossRef]
- Matos, L.; Canals, I.; Dridi, L.; Choi, Y.; Prata, M.J.; Jordan, P.; Desviat, L.R.; Pérez, B.; Pshezhetsky, A.V.; Grinberg, D.; et al. Therapeutic strategies based on modified U1 snRNAs and chaperones for Sanfilippo C splicing mutations. Orphanet J. Rare Dis. 2014, 9. [Google Scholar] [CrossRef]
- Ferri, L.; Covello, G.; Caciotti, A.; Guerrini, R.; Denti, M.A.; Morrone, A. Double-target Antisense U1snRNAs Correct Mis-splicing Due to c.639+861C>T and c.639+919G>A GLA Deep Intronic Mutations. Mol. Ther. Nucleic Acids 2016, 5, e380. [Google Scholar] [CrossRef]
- Rodríguez-Pascau, L.; Coll, M.J.; Vilageliu, L.; Grinberg, D. Antisense oligonucleotide treatment for a pseudoexon-generating mutation in theNPC1gene causing Niemann-Pick type C disease. Hum. Mutat. 2009, 30, E993–E1001. [Google Scholar] [CrossRef] [PubMed]
- Aung-Htut, M.T.; Ham, K.A.; Tchan, M.; Johnsen, R.; Schnell, F.J.; Fletcher, S.; Wilton, S.D. Splice modulating antisense oligonucleotides restore some acid-alpha-glucosidase activity in cells derived from patients with late-onset Pompe disease. Sci. Rep. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- van der Wal, E.; Bergsma, A.J.; Pijnenburg, J.M.; van der Ploeg, A.T.; Pijnappel, W.W.M.P. Antisense Oligonucleotides Promote Exon Inclusion and Correct the Common c.-32-13T>G GAA Splicing Variant in Pompe Disease. Mol. Ther. Nucleic Acids 2017, 7, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.M.G.; Jungner, G. Principles and Practice of Screening for Disease; World Health Organization: Geneva, Switzerland, 1968; p. 164. [Google Scholar]
- Andermann, A. Revisting wilson and Jungner in the genomic age: A review of screening criteria over the past 40 years. Bull. World Health Organ. 2008, 86, 317–319. [Google Scholar] [CrossRef] [PubMed]
- Gelb, M. Newborn Screening for Lysosomal Storage Diseases: Methodologies, Screen Positive Rates, Normalization of Datasets, Second-Tier Tests, and Post-Analysis Tools. Int. J. Neonatal Screen. 2018, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Anderson, S. Newborn Screening for Lysosomal Storage Disorders. J. Pediatric Health Care 2018, 32, 285–294. [Google Scholar] [CrossRef]
- Schielen, P.; Kemper, E.; Gelb, M. Newborn Screening for Lysosomal Storage Diseases: A Concise Review of the Literature on Screening Methods, Therapeutic Possibilities and Regional Programs. Int. J. Neonatal Screen. 2017, 3, 6. [Google Scholar] [CrossRef]
- Taruscio, D.; Carbone, P.; Polizzi, A. Expanded Newborn Screening: A Chess Board Motif in Public Health. J. Pediatr. Biochem. 2016, 6, 66–70. [Google Scholar]
- Burlina, A.B.; Polo, G.; Salviati, L.; Duro, G.; Zizzo, C.; Dardis, A.; Bembi, B.; Cazzorla, C.; Rubert, L.; Zordan, R.; et al. Newborn screening for lysosomal storage disorders by tandem mass spectrometry in North East Italy. J. Inherit. Metab. Dis. 2018, 41, 209–219. [Google Scholar] [CrossRef]
- Burlina, A.; Gueraldi, D.; Polo, G.; Rubert, L.; Cazzorla, C.; Giuliani, A.; Burlina, A. Newborn screening for MPS I: The clinical benefit. Mol. Genet. Metab. 2020, 129, S35–S36. [Google Scholar] [CrossRef]
- Burlina, A.; Polo, G.; Gueraldi, D.; Rubert, L.; Cazzorla, C.; Giuliani, A.; Burlina, A. High incidence of Gaucher disease in northeast Italy: Results from lysosomal newborn screening. Mol. Genet. Metab. 2020, 129, S36. [Google Scholar] [CrossRef]
- Paciotti, S.; Persichetti, E.; Pagliardini, S.; Deganuto, M.; Rosano, C.; Balducci, C.; Codini, M.; Filocamo, M.; Menghini, A.R.; Pagliardini, V.; et al. First pilot newborn screening for four lysosomal storage diseases in an Italian region: Identification and analysis of a putative causative mutation in the GBA gene. Clin. Chim. Acta. 2012, 413, 1827–1831. [Google Scholar] [CrossRef] [PubMed]
- Spada, M.; Pagliardini, S.; Yasuda, M.; Tukel, T.; Thiagarajan, G.; Sakuraba, H.; Ponzone, A.; Desnick, R.J. High incidence of later-onset fabry disease revealed by newborn screening. Am. J. Hum. Genet. 2006, 79, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Scolamiero, E.; Casetta, B.; Malvagia, S.; Tanigawa, T.; Forni, G.; Funghini, S.; Mura, M.; Raspini, F.; Poggiali, S.; la Marca, G. Development of a fast LC-MS/MS protocol for combined measurement of six LSDs on dried blood spot in a newborn screening program. J. Pharm. Biomed. Anal. 2019, 165, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Forni, G.; Malvagia, S.; Funghini, S.; Scolamiero, E.; Mura, M.; Della Bona, M.; Villanelli, F.; Damiano, R.; la Marca, G. LC-MS/MS method for simultaneous quantification of heparan sulfate and dermatan sulfate in urine by butanolysis derivatization. Clin. Chim. Acta 2019, 488, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Gelb, M.H.; Lukacs, Z.; Ranieri, E.; Schielen, P. Newborn Screening for Lysosomal Storage Disorders: Methodologies for Measurement of Enzymatic Activities in Dried Blood Spots. Int. J. Neonatal Screen. 2019, 5, 1. [Google Scholar] [CrossRef]
- Gelb, M.H.; Scott, C.R.; Turecek, F. Newborn screening for lysosomal storage diseases. Clin. Chem. 2015, 61, 335–346. [Google Scholar] [CrossRef]
- Yu, C.; Sun, Q.; Zhou, H. Enzymatic Screening and Diagnosis of Lysosomal Storage Diseases. N. Am. J. Med. Sci. 2013, 6, 186–193. [Google Scholar] [CrossRef]
- Millington, D.; Bali, D. Current State of the Art of Newborn Screening for Lysosomal Storage Disorders. Int. J. Neonatal Screen. 2018, 4, 24. [Google Scholar] [CrossRef]
- Berg, J.S.; Agrawal, P.B.; Bailey, D.B., Jr.; Beggs, A.H.; Brenner, S.E.; Brower, A.M.; Cakici, J.A.; Ceyhan-Birsoy, O.; Chan, K.; Chen, F.; et al. Newborn Sequencing in Genomic Medicine and Public Health. Pediatrics 2017, 139, e20162252. [Google Scholar] [CrossRef]
- Friedman, E. Next generation sequencing for newborn screening: Are we there yet? Genet. Res. 2015, 97, e17. [Google Scholar] [CrossRef] [PubMed]
- Schiffmann, R.; Yubero, D.; Brandi, N.; Ormazabal, A.; Garcia-Cazorla, À.; Pérez-Dueñas, B.; Campistol, J.; Ribes, A.; Palau, F.; Artuch, R.; et al. Targeted Next Generation Sequencing in Patients with Inborn Errors of Metabolism. PLoS ONE 2016, 11, e0156359. [Google Scholar]
- Málaga, D.R.; Brusius-Facchin, A.C.; Siebert, M.; Pasqualim, G.; Saraiva-Pereira, M.L.; de Souza, C.F.M.; Schwartz, I.V.D.; Matte, U.; Giugliani, R. Sensitivity, advantages, limitations, and clinical utility of targeted next-generation sequencing panels for the diagnosis of selected lysosomal storage disorders. Genet. Mol. Biol. 2019, 42, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Gheldof, A.; Seneca, S.; Stouffs, K.; Lissens, W.; Jansen, A.; Laeremans, H.; Verloo, P.; Schoonjans, A.S.; Meuwissen, M.; Barca, D.; et al. Clinical implementation of gene panel testing for lysosomal storage diseases. Mol. Genet. Genom. Med. 2018, e00527. [Google Scholar] [CrossRef]
- Komlosi, K.; Sólyom, A.; Beck, M. The Role of Next-Generation Sequencing in the Diagnosis of Lysosomal Storage Disorders. J. Inborn Errors Metab. Screen. 2016, 4, 232640981666937. [Google Scholar] [CrossRef]
- Mori, M.; Haskell, G.; Kazi, Z.; Zhu, X.; DeArmey, S.M.; Goldstein, J.L.; Bali, D.; Rehder, C.; Cirulli, E.T.; Kishnani, P.S. Sensitivity of whole exome sequencing in detecting infantile- and late-onset Pompe disease. Mol. Genet. Metab. 2017, 122, 189–197. [Google Scholar] [CrossRef]
- Di Fruscio, G.; Schulz, A.; De Cegli, R.; Savarese, M.; Mutarelli, M.; Parenti, G.; Banfi, S.; Braulke, T.; Nigro, V.; Ballabio, A. Lysoplex: An efficient toolkit to detect DNA sequence variations in the autophagy-lysosomal pathway. Autophagy 2015, 11, 928–938. [Google Scholar] [CrossRef]
- Arjunan, A.; Litwack, K.; Collins, N.; Charrow, J. Carrier screening in the era of expanding genetic technology. Genet. Med. 2016, 18, 1214–1217. [Google Scholar] [CrossRef]
- Hoffman, J.D.; Greger, V.; Strovel, E.T.; Blitzer, M.G.; Umbarger, M.A.; Kennedy, C.; Bishop, B.; Saunders, P.; Porreca, G.J.; Schienda, J.; et al. Next-generation DNA sequencing of HEXA: A step in the right direction for carrier screening. Mol. Genet. Genom. Med. 2013, 1, 260–268. [Google Scholar] [CrossRef]
- Yoshida, S.; Kido, J.; Matsumoto, S.; Momosaki, K.; Mitsubuchi, H.; Shimazu, T.; Sugawara, K.; Endo, F.; Nakamura, K. Prenatal diagnosis of Gaucher disease using next-generation sequencing. Pediatr. Int. 2016, 58, 946–949. [Google Scholar] [CrossRef]
- Tsai, A.C.-H.; Hung, Y.-W.; Harding, C.; Koeller, D.M.; Wang, J.; Wong, L.-J.C. Next generation deep sequencing corrects diagnostic pitfalls of traditional molecular approach in a patient with prenatal onset of Pompe disease. Am. J. Med Genet. Part A 2017, 173, 2500–2504. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, A.; D’Avanzo, F.; Bertoldi, L.; Zampieri, G.; Feltrin, E.; De Pascale, F.; Rampazzo, A.; Forzan, M.; Valle, G.; Tomanin, R. Setup and Validation of a Targeted Next-Generation Sequencing Approach for the Diagnosis of Lysosomal Storage Disorders. J. Mol. Diagn. 2020, 22, 488–502. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhang, Y.; Gedvilaite, E.; Loh, J.W.; Lin, T.; Liu, X.; Liu, C.-G.; Kumar, D.; Donnelly, R.; Raymond, K.; et al. Using whole-exome sequencing to investigate the genetic bases of lysosomal storage diseases of unknown etiology. Hum. Mutat. 2017, 38, 1491–1499. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Pan, J.; Linpeng, S.; Li, Z.; Tan, H.; Wu, L. Identification of Five Novel Mutations Causing Rare Lysosomal Storage Diseases. Med. Sci. Monit. 2019, 25, 7634–7644. [Google Scholar] [CrossRef]
- Davidson, B.A.; Hassan, S.; Garcia, E.J.; Tayebi, N.; Sidransky, E. Exploring genetic modifiers of Gaucher disease: The next horizon. Hum. Mutat. 2018, 39, 1739–1751. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, J.A.; Guecaimburu Ehuletche, M.d.R.; Perna, A.; Dubrovsky, A.; Franca, M.C.; Vargas, S.; Hegde, M.; Claeys, K.G.; Straub, V.; Daba, N.; et al. The Latin American experience with a next generation sequencing genetic panel for recessive limb-girdle muscular weakness and Pompe disease. Orphanet J. Rare Dis. 2020, 15, 11. [Google Scholar] [CrossRef]
- Lévesque, S.; Auray-Blais, C.; Gravel, E.; Boutin, M.; Dempsey-Nunez, L.; Jacques, P.-E.; Chenier, S.; Larue, S.; Rioux, M.-F.; Al-Hertani, W.; et al. Diagnosis of late-onset Pompe disease and other muscle disorders by next-generation sequencing. Orphanet J. Rare Dis. 2016, 11, 8. [Google Scholar] [CrossRef]
- Mahdieh, N.; Mikaeeli, S.; Tavasoli, A.R.; Rezaei, Z.; Maleki, M.; Rabbani, B. Genotype, phenotype and in silico pathogenicity analysis of HEXB mutations: Panel based sequencing for differential diagnosis of gangliosidosis. Clin. Neurol. Neurosurg. 2018, 167, 43–53. [Google Scholar] [CrossRef]
- in’t Groen, S.L.M.; de Faria, D.O.S.; Iuliano, A.; van den Hout, J.M.P.; Douben, H.; Dijkhuizen, T.; Cassiman, D.; Witters, P.; Barba Romero, M.-Á.; de Klein, A.; et al. Novel GAA Variants and Mosaicism in Pompe Disease Identified by Extended Analyses of Patients with an Incomplete DNA Diagnosis. Mol. Ther. Methods Clin. Dev. 2020, 17, 337–348. [Google Scholar] [CrossRef]
- Pianese, L.; Fortunato, A.; Silvestri, S.; Solano, F.G.; Burlina, A.; Burlina, A.P.; Ragno, M. Maternal germline mosaicism in Fabry disease. Neurol. Sci. 2019, 40, 1279–1281. [Google Scholar] [CrossRef]
- Johnston, J.; Lantos, J.D.; Goldenberg, A.; Chen, F.; Parens, E.; Koenig, B.A.; members of the, N.E.; Policy Advisory, B. Sequencing Newborns: A Call for Nuanced Use of Genomic Technologies. Hastings Cent. Rep. 2018, 48, S2–S6. [Google Scholar] [CrossRef] [PubMed]
- van Campen, J.C.; Sollars, E.S.A.; Thomas, R.C.; Bartlett, C.M.; Milano, A.; Parker, M.D.; Dawe, J.; Winship, P.R.; Peck, G.; Grafham, D.; et al. Next Generation Sequencing in Newborn Screening in the United Kingdom National Health Service. Int. J. Neonatal Screen. 2019, 5, 40. [Google Scholar] [CrossRef] [PubMed]
- Stark, Z.; Tan, T.Y.; Chong, B.; Brett, G.R.; Yap, P.; Walsh, M.; Yeung, A.; Peters, H.; Mordaunt, D.; Cowie, S.; et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet. Med. 2016, 18, 1090–1096. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lim, J.; Shin, J.E.; Eun, H.S.; Park, M.S.; Park, K.I.; Namgung, R.; Lee, J.S. Implementation of a Targeted Next-Generation Sequencing Panel for Constitutional Newborn Screening in High-Risk Neonates. Yonsei Med. J. 2019, 60, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Kingsmore, S.F. Newborn testing and screening by whole-genome sequencing. Genet. Med. 2016, 18, 214–216. [Google Scholar] [CrossRef][Green Version]
- Fleige, T.; Burggraf, S.; Czibere, L.; Häring, J.; Glück, B.; Keitel, L.M.; Landt, O.; Harms, E.; Hohenfellner, K.; Durner, J.; et al. Next generation sequencing as second-tier test in high-throughput newborn screening for nephropathic cystinosis. Eur. J. Hum. Genet. 2019, 28, 193–201. [Google Scholar] [CrossRef]
- Bobillo Lobato, J.; Jiménez Hidalgo, M.; Jiménez Jiménez, L. Biomarkers in Lysosomal Storage Diseases. Diseases 2016, 4, 40. [Google Scholar] [CrossRef]
- Burlina, A.B.; Polo, G.; Rubert, L.; Gueraldi, D.; Cazzorla, C.; Duro, G.; Salviati, L.; Burlina, A.P. Implementation of Second-Tier Tests in Newborn Screening for Lysosomal Disorders in North Eastern Italy. Int. J. Neonatal Screen. 2019, 5, 24. [Google Scholar] [CrossRef]
- Polo, G.; Burlina, A.P.; Ranieri, E.; Colucci, F.; Rubert, L.; Pascarella, A.; Duro, G.; Tummolo, A.; Padoan, A.; Plebani, M.; et al. Plasma and dried blood spot lysosphingolipids for the diagnosis of different sphingolipidoses: A comparative study. Clin. Chem. Lab. Med. 2019, 57, 1863–1874. [Google Scholar] [CrossRef]
- Johnson, B.; Mascher, H.; Mascher, D.; Legnini, E.; Hung, C.Y.; Dajnoki, A.; Chien, Y.-H.; Maródi, L.; Hwu, W.-L.; Bodamer, O.A. Analysis of Lyso-Globotriaosylsphingosine in Dried Blood Spots. Ann. Lab. Med. 2013, 33, 274. [Google Scholar] [CrossRef]
- Spada, M.; Kasper, D.; Pagliardini, V.; Biamino, E.; Giachero, S.; Porta, F. Metabolic progression to clinical phenotype in classic Fabry disease. Ital. J. Pediatr. 2017, 43, 1. [Google Scholar] [CrossRef]
- Chien, Y.-H.; Bodamer, O.A.; Chiang, S.-C.; Mascher, H.; Hung, C.; Hwu, W.-L. Lyso-globotriaosylsphingosine (lyso-Gb3) levels in neonates and adults with the Fabry disease later-onset GLA IVS4+919G>A mutation. J. Inherit. Metab. Dis. 2012, 36, 881–885. [Google Scholar] [CrossRef]
- Kuchar, L.; Sikora, J.; Gulinello, M.E.; Poupetova, H.; Lugowska, A.; Malinova, V.; Jahnova, H.; Asfaw, B.; Ledvinova, J. Quantitation of plasmatic lysosphingomyelin and lysosphingomyelin-509 for differential screening of Niemann-Pick A/B and C diseases. Anal. Biochem. 2017, 525, 73–77. [Google Scholar] [CrossRef]
- Tarallo, A.; Carissimo, A.; Gatto, F.; Nusco, E.; Toscano, A.; Musumeci, O.; Coletta, M.; Karali, M.; Acampora, E.; Damiano, C.; et al. microRNAs as biomarkers in Pompe disease. Genet. Med. 2018, 21, 591–600. [Google Scholar] [CrossRef]
- Carrasco-Rozas, A.; Fernández-Simón, E.; Lleixà, M.C.; Belmonte, I.; Pedrosa-Hernandez, I.; Montiel-Morillo, E.; Nuñez-Peralta, C.; Llauger Rossello, J.; Segovia, S.; De Luna, N.; et al. Identification of serum microRNAs as potential biomarkers in Pompe disease. Ann. Clin. Transl. Neurol. 2019, 6, 1214–1224. [Google Scholar] [CrossRef]
- Cammarata, G.; Scalia, S.; Colomba, P.; Zizzo, C.; Pisani, A.; Riccio, E.; Montalbano, M.; Alessandro, R.; Giordano, A.; Duro, G. A pilot study of circulating microRNAs as potential biomarkers of Fabry disease. Oncotarget 2018, 9, 27333–27345. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).