PGAP3 Associated with Hyperphosphatasia with Mental Retardation Plays a Novel Role in Brain Morphogenesis and Neuronal Wiring at Early Development
Abstract
1. Introduction
2. Methods
2.1. Study Subjects
2.2. Whole-Genome Sequencing
2.3. PGAP3 Protein Homology
2.4. Zebrafish Care and Husbandry
2.5. RNA Extraction and Reverse Transcription
2.6. Whole-Mount In Situ Hybridization of the Zebrafish Embryos
2.7. Design of Morpholino Antisense Oligos and Zebrafish Microinjection
2.8. Microscopic Observation of Zebrafish Embryos
2.9. Western Blot
2.10. Phalloidin Staining of Zebrafish Larvae
2.11. Light-Sheet Microscopy
2.12. Cresyl Violet Stain
2.13. Histopathological Examination
2.14. Oligodendrocytes Analysis Using Tg (olig2: dsRed)
2.15. Zebrafish Locomotor Behavior Measurements
2.15.1. Burst Movement Analysis at 24 hpf
2.15.2. Touch-Evoked Response Assay at 72 hpf
2.15.3. Swimming Behavior Analysis at 120 hpf
2.16. Statistical Analysis
3. Results
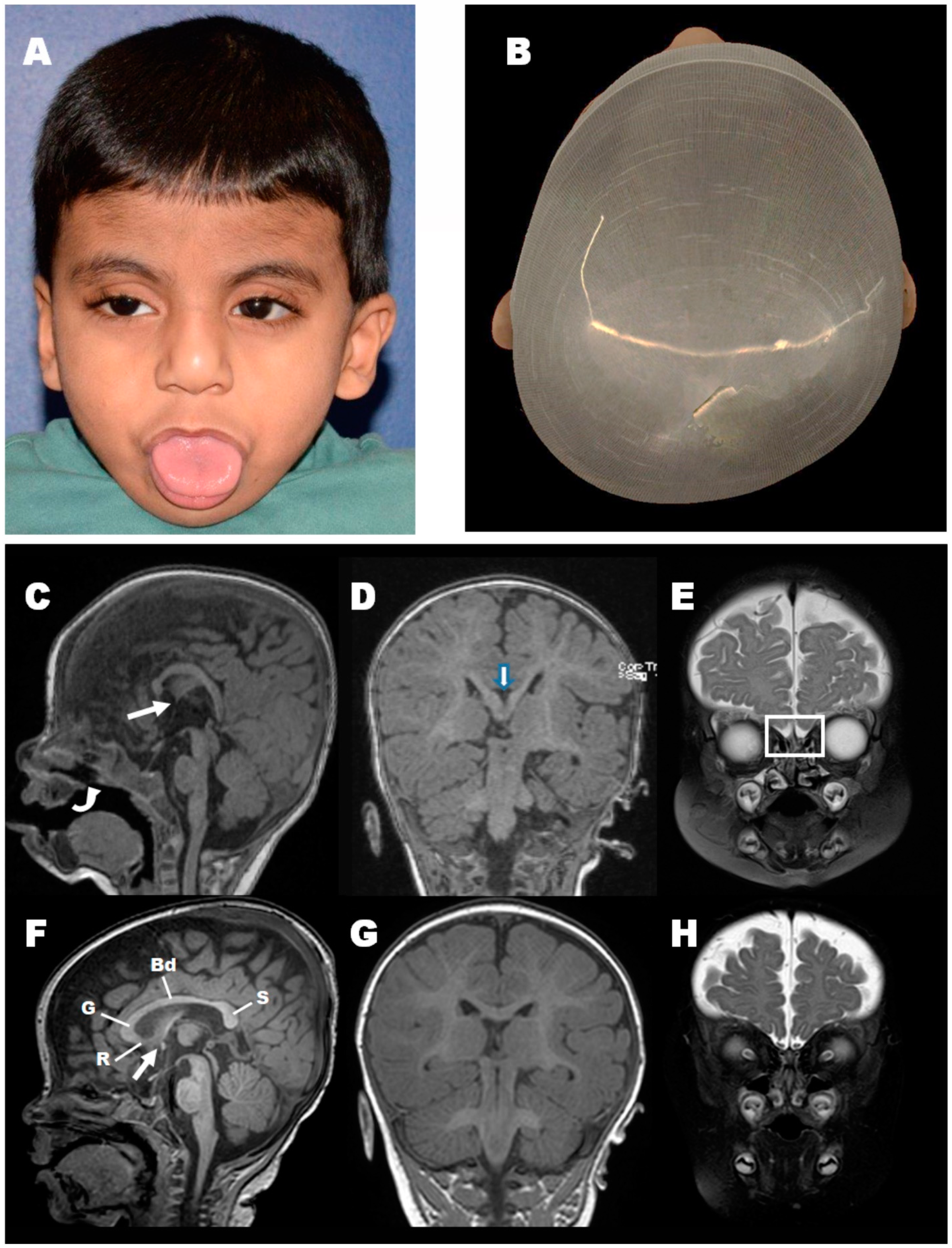
3.1. Clinical Findings
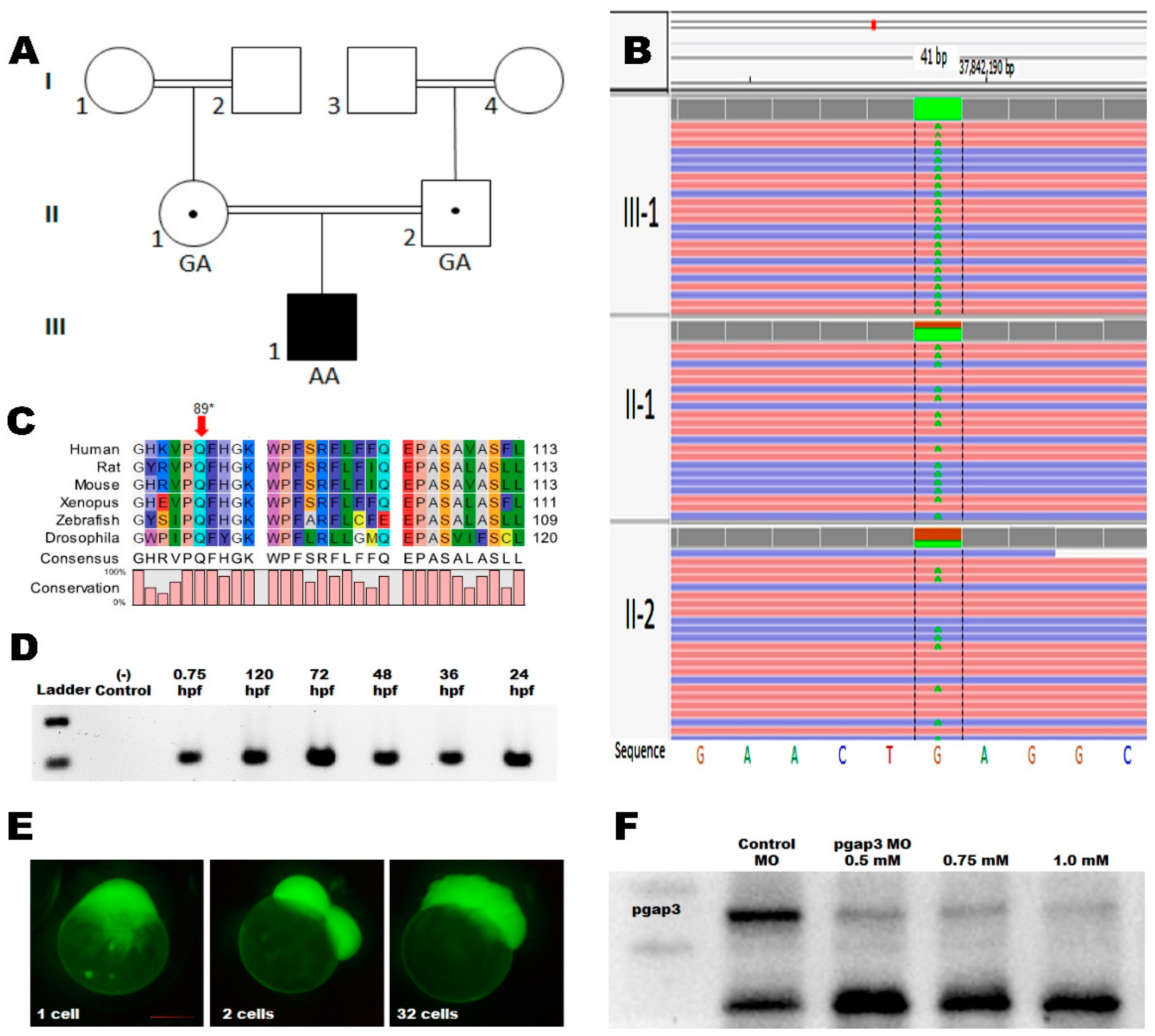
3.2. Whole-Genome Sequencing
3.3. Zebrafish Model Has Conserved pgap3 Expression through Development
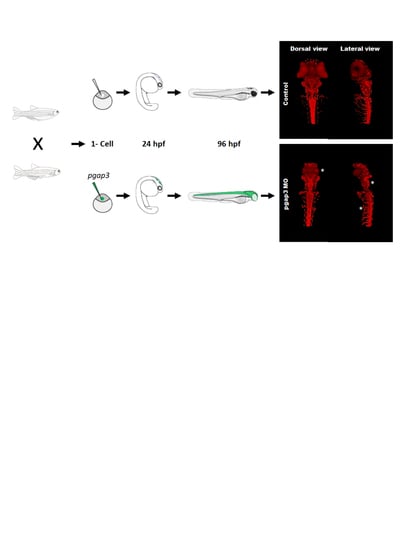
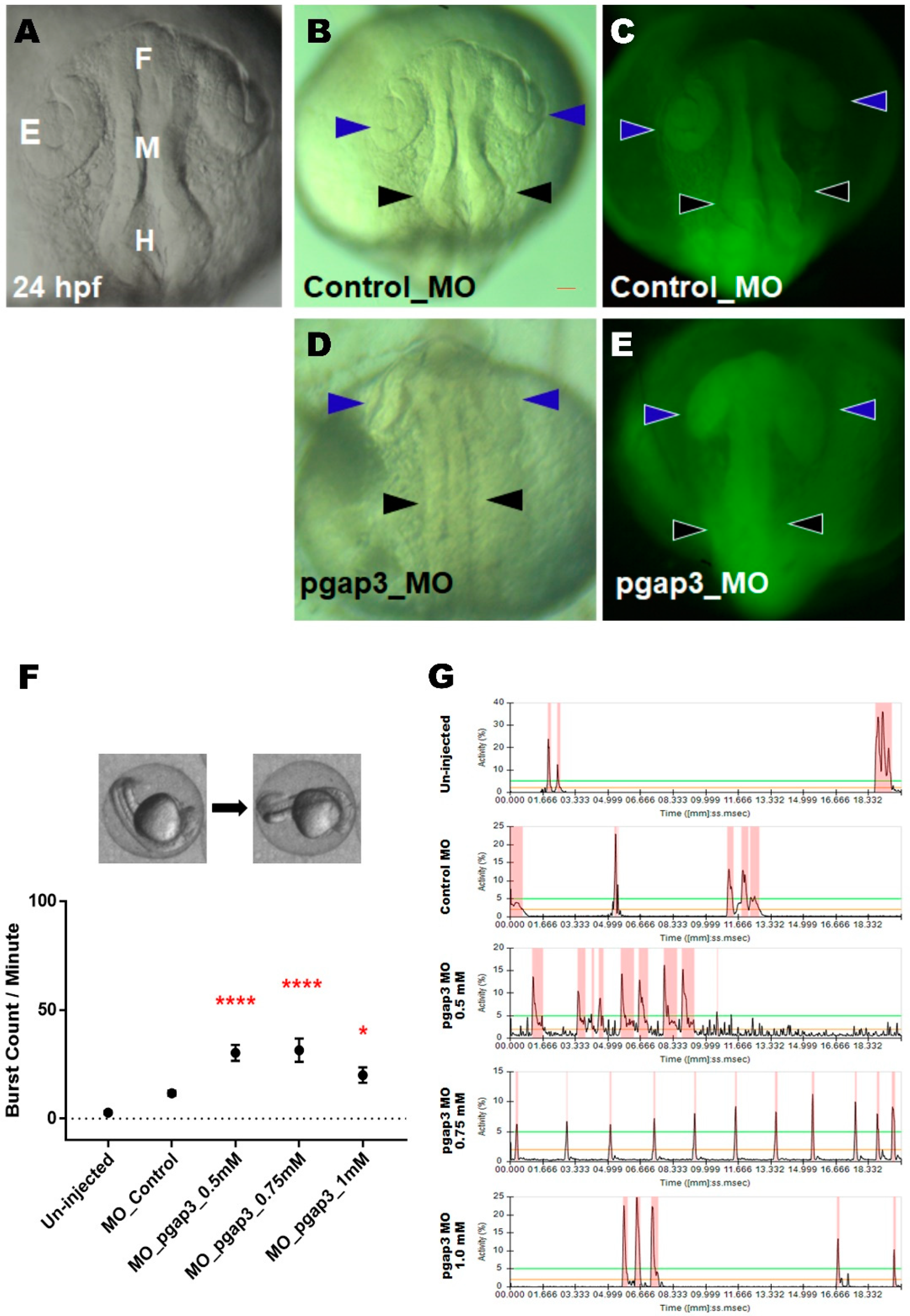
3.4. Knockdown of Pgap3 Results in Neural Tube Defects in Early Zebrafish Development
3.5. Zebrafish Pgap3 Knockdown Altered Neuronal Wiring at Early Stages of Development
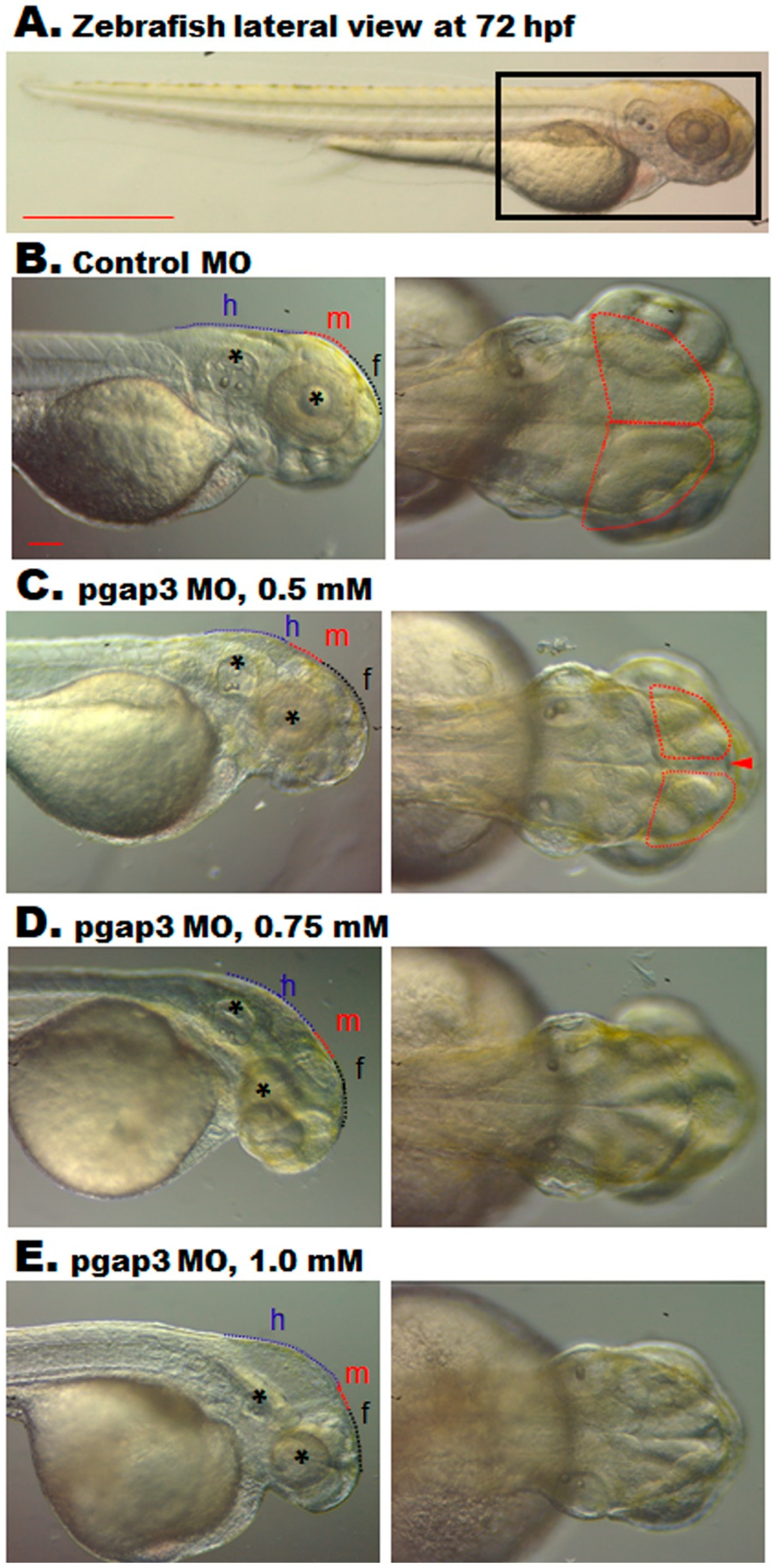
3.6. Zebrafish pgap3 Morphants Display Dysmorphic Features Resembling Human HPMRS4
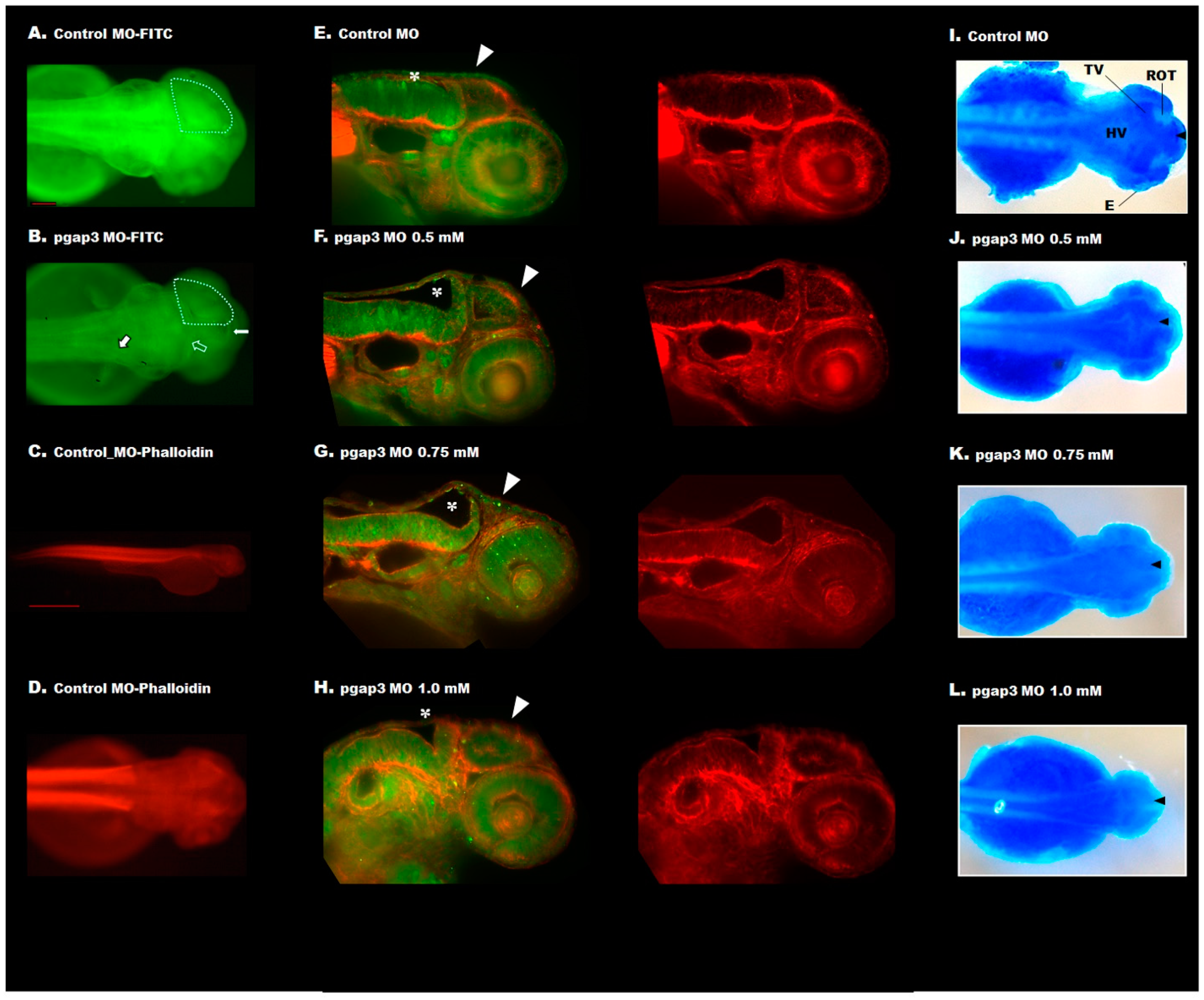
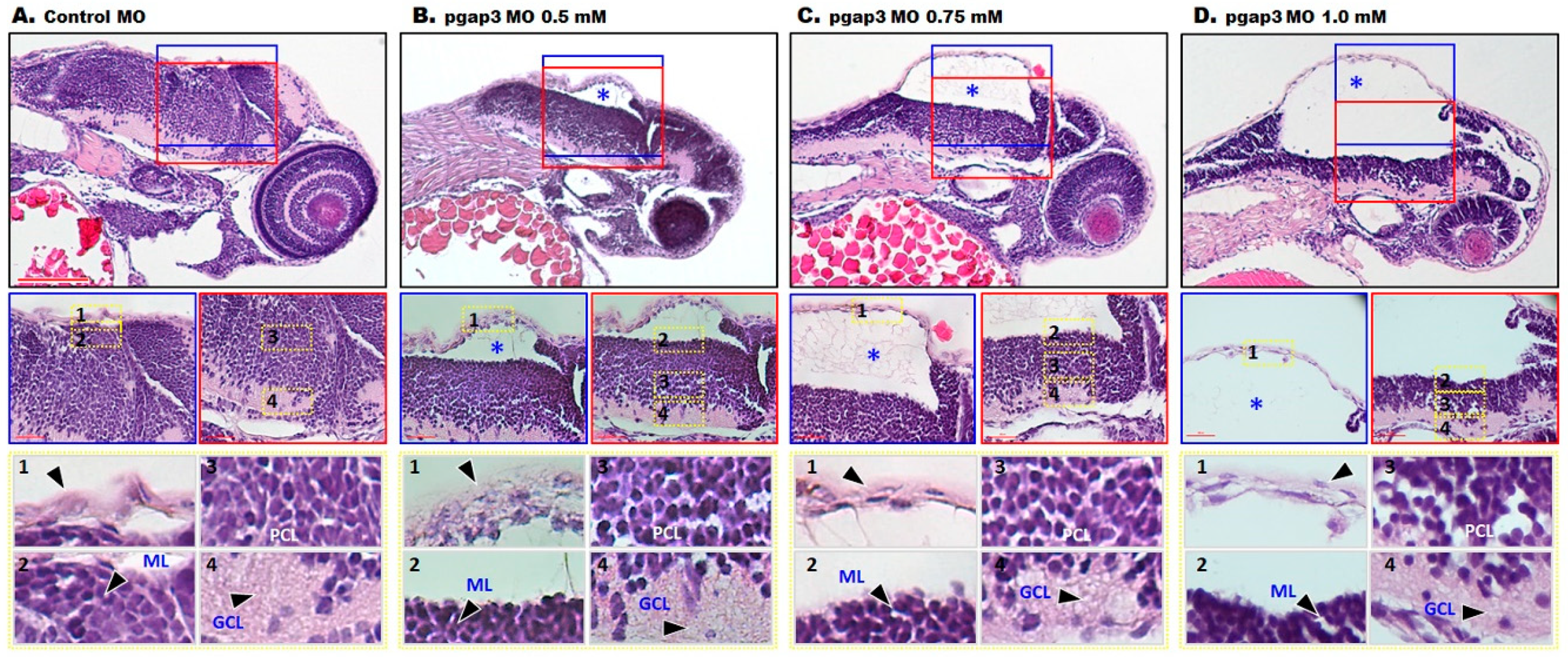
3.7. Loss of Pgap3 Resulted in Zebrafish Midbrain Tectum and Cerebellum Defects
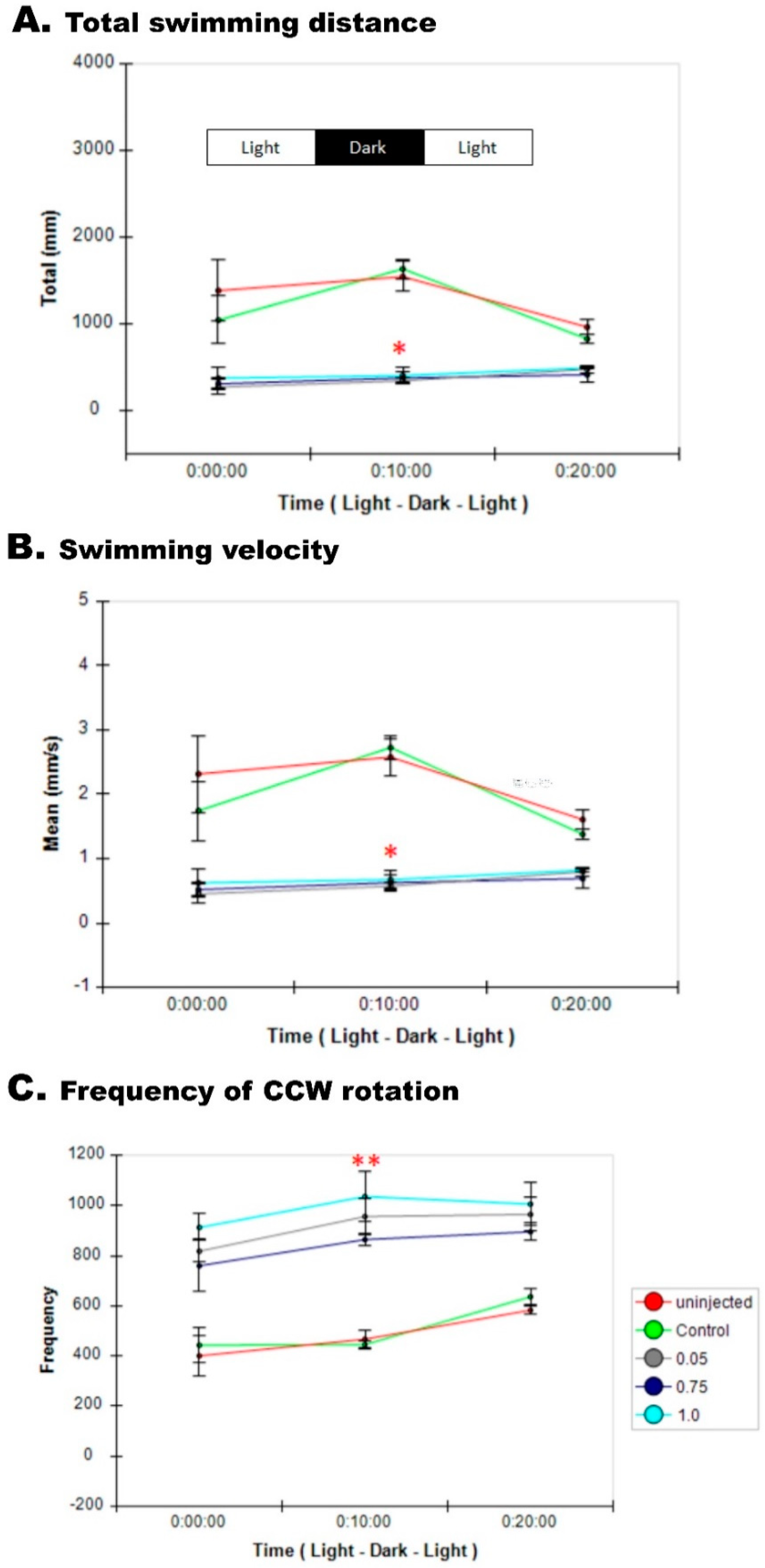
3.8. Impaired Sensory Response in Zebrafish pgap3 Morphants
3.9. Zebrafish pgap3 Morphants Display Impaired Touch Sensitivity and Seizure-Related Phenotype
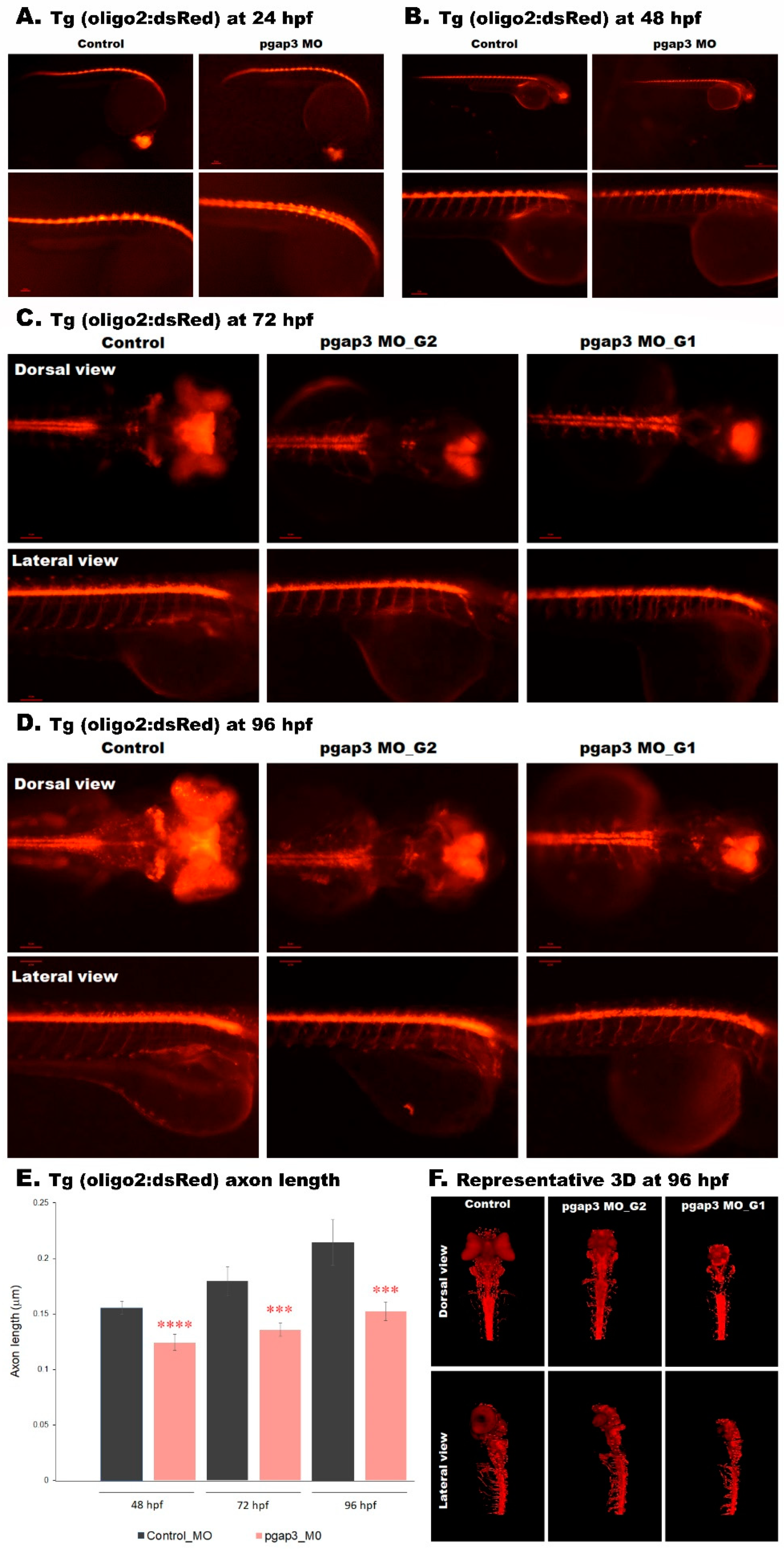
3.10. Loss of PGAP3 Function Impaired Oligodendrocytes Expression Resulting in Shorter Motor Neuron Axons
4. Discussion
5. Conclusions
Highlights
- This study of a novel PGAP3 variant represents the first HPMRS4 case reported with unique clinical features of decreased fetal intrauterine movements and olfactory bulb agenesis
- Functional modeling of the PGAP3 deficit revealed the impact on neural tube development and expansion at early stages of zebrafish development
- PGAP3 plays a novel role in brain morphogenesis
- PGAP3 is essential for neuronal oligodendrocytes migration, distribution, and myelination in early stages of development
- PGAP3 Knockdown resulted in motor neuron deficits, sensory, and locomotor impairments
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Howard, M.F.; Murakami, Y.; Pagnamenta, A.T.; Daumer-Haas, C.; Fischer, B.; Hecht, J.; Keays, D.A.; Knight, S.J.; Kölsch, U.; Krüger, U. Mutations in PGAP3 impair GPI-anchor maturation, causing a subtype of hyperphosphatasia with mental retardation. Am. J. Hum. Genet. 2014, 94, 278–287. [Google Scholar] [CrossRef]
- Altassan, R.; Fox, S.; Poulin, C.; Buhas, D. Hyperphosphatasia with mental retardation syndrome, expanded phenotype of PIGL related disorders. Mol. Genet. Metab. Rep. 2018, 15, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Bellai-Dussault, K.; Nguyen, T.T.M.; Baratang, N.V.; Jimenez-Cruz, D.A.; Campeau, P.M. Clinical variability in inherited glycosylphosphatidylinositol deficiency disorders. Clin. Genet. 2019, 95, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Horn, D.; Krawitz, P.; Mannhardt, A.; Korenke, G.C.; Meinecke, P. Hyperphosphatasia-mental retardation syndrome due to PIGV mutations: Expanded clinical spectrum. Am. J. Med Genet. Part A 2011, 155, 1917–1922. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hamid, M.; Issa, M.; Otaify, G.; Abdel-Ghafar, S.; Elbendary, H.; Zaki, M. PGAP3-related hyperphosphatasia with mental retardation syndrome: Report of 10 new patients and a homozygous founder mutation. Clin. Genet. 2018, 93, 84–91. [Google Scholar] [CrossRef]
- Sakaguchi, T.; Žigman, T.; Ramadža, D.P.; Omerza, L.; Pušeljić, S.; Hrvaćanin, Z.E.; Miyake, N.; Matsumoto, N.; Barić, I. A novel PGAP3 mutation in a Croatian boy with brachytelephalangy and a thin corpus callosum. Hum. Genome Var. 2018, 5, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Scott, E.K.; Baier, H. The cellular architecture of the larval zebrafish tectum, as revealed by gal4 enhancer trap lines. Front. Neural Circuits 2009, 3, 13. [Google Scholar] [CrossRef]
- Nozaki, M.; Ohishi, K.; Yamada, N.; Kinoshita, T.; Nagy, A.; Takeda, J. Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab. Investig. A J. Tech. Methods Pathol. 1999, 79, 293–299. [Google Scholar]
- Kozol, R.A.; Abrams, A.J.; James, D.M.; Buglo, E.; Yan, Q.; Dallman, J.E. Function over form: Modeling groups of inherited neurological conditions in zebrafish. Front. Mol. Neurosci. 2016, 9, 55. [Google Scholar] [CrossRef]
- Sakai, C.; Ijaz, S.; Hoffman, E.J. Zebrafish models of neurodevelopmental disorders: Past, present, and future. Front. Mol. Neurosci. 2018, 11, 294. [Google Scholar] [CrossRef]
- Vaz, R.; Hofmeister, W.; Lindstrand, A. Zebrafish Models of Neurodevelopmental Disorders: Limitations and Benefits of Current Tools and Techniques. Int. J. Mol. Sci. 2019, 20, 1296. [Google Scholar] [CrossRef] [PubMed]
- Brand, M.; Heisenberg, C.-P.; Jiang, Y.-J.; Beuchle, D.; Lun, K.; Furutani-Seiki, M.; Granato, M.; Haffter, P.; Hammerschmidt, M.; Kane, D.A. Mutations in zebrafish genes affecting the formation of the boundary between midbrain and hindbrain. Development 1996, 123, 179–190. [Google Scholar] [PubMed]
- Kalueff, A.V.; Stewart, A.M.; Gerlai, R. Zebrafish as an emerging model for studying complex brain disorders. Trends Pharmacol. Sci. 2014, 35, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Stiles, J.; Jernigan, T.L. The basics of brain development. Neuropsychol. Rev. 2010, 20, 327–348. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Consortium, G.P. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P. The mutational constraint spectrum quantified from variation in 141,456 humans. bioRxiv 2020, 531210. [Google Scholar] [CrossRef]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef]
- Exome, E.V.S.N.G. Sequencing Project (ESP). Seattle, WA, USA. Available online: http://evs.gs.washington.edu/EVS/ (accessed on 22 January 2019).
- Da’as, S.I.; Coombs, A.J.; Balci, T.B.; Grondin, C.A.; Ferrando, A.A.; Berman, J.N. The zebrafish reveals dependence of the mast cell lineage on Notch signaling in vivo. Blood the J. Am. Soc. Hematol. 2012, 119, 3585–3594. [Google Scholar] [CrossRef] [PubMed]
- Da’as, S.I.; Yalcin, H.C.; Nasrallah, G.K.; Mohamed, I.A.; Nomikos, M.; Yacoub, M.H.; Fakhro, K.A. Functional characterization of human myosin-binding protein C3 variants associated with hypertrophic cardiomyopathy reveals exon-specific cardiac phenotypes in zebrafish model. J. Cell. Physiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xu, X. Immunostaining of dissected zebrafish embryonic heart. J. Vis. Exp. 2012, 10, e3510. [Google Scholar] [CrossRef]
- Garbuzova-Davis, S.; Rodrigues, M.C.; Hernandez-Ontiveros, D.G.; Tajiri, N.; Frisina-Deyo, A.; Boffeli, S.M.; Abraham, J.V.; Pabon, M.; Wagner, A.; Ishikawa, H. Blood-brain barrier alterations provide evidence of subacute diaschisis in an ischemic stroke rat model. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Basnet, R.M.; Guarienti, M.; Memo, M. Zebrafish embryo as an in vivo model for behavioral and pharmacological characterization of methylxanthine drugs. Int. J. Mol. Sci. 2017, 18, 596. [Google Scholar] [CrossRef]
- Sztal, T.E.; Ruparelia, A.A.; Williams, C.; Bryson-Richardson, R.J. Using touch-evoked response and locomotion assays to assess muscle performance and function in zebrafish. J. Vis. Exp. 2016, e54431. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Hou, Y.-Y.; Sun, M.-Z.; Zhang, C.-Y.; Bai, G.; Zhao, X.; Feng, X.-Z. Behavioural screening of zebrafish using neuroactive traditional Chinese medicine prescriptions and biological targets. Sci. Rep. 2014, 4, 1–9. [Google Scholar] [CrossRef]
- Abi Farraj, L.; Khatoun, W.D.; Abou Chebel, N.; Wakim, V.; Dawali, K.; Ghassibe-Sabbagh, M. Clinical, genetic, and molecular characterization of hyperphosphatasia with mental retardation: A case report and literature review. Diagn. Pathol. 2019, 14, 123. [Google Scholar] [CrossRef] [PubMed]
- Doğan, Ö.A.; Demir, G.Ü.; Kosukcu, C.; Taskiran, E.Z.; Simsek-Kiper, P.Ö.; Utine, G.E.; Alikaşifoğlu, M.; Boduroğlu, K. Hyperphosphatasia with mental retardation syndrome type 4 In two siblings-expanding the phenotypic and mutational spectrum. Eur. J. Med Genet. 2019, 62, 103535. [Google Scholar] [CrossRef]
- Fakhro, K.A.; Robay, A.; Rodrigues-Flores, J.L.; Mezey, J.G.; Al-Shakaki, A.A.; Chidiac, O.; Stadler, D.; Malek, J.A.; Imam, A.B.; Sheikh, A. Point of Care Exome Sequencing Reveals Allelic and Phenotypic Heterogeneity Underlying Mendelian disease in Qatar. Hum. Mol. Genet. 2019, 28, 3970–3981. [Google Scholar] [CrossRef]
- Fakhro, K.A.; Staudt, M.R.; Ramstetter, M.D.; Robay, A.; Malek, J.A.; Badii, R.; Al-Marri, A.A.-N.; Khalil, C.A.; Al-Shakaki, A.; Chidiac, O. The Qatar genome: A population-specific tool for precision medicine in the Middle East. Hum. Genome Var. 2016, 3, 1–7. [Google Scholar] [CrossRef]
- Gutzman, J.H.; Graeden, E.G.; Lowery, L.A.; Holley, H.S.; Sive, H. Formation of the zebrafish midbrain–hindbrain boundary constriction requires laminin-dependent basal constriction. Mech. Dev. 2008, 125, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Lowery, L.A.; De Rienzo, G.; Gutzman, J.H.; Sive, H. Characterization and classification of zebrafish brain morphology mutants. Adv. Integr. Anat. Evol. Biol. 2009, 292, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Richendrfer, H.; Creton, R.; Colwill, R.M. The embryonic zebrafish as a model system to study the effects of environmental toxicants on behavior. In Zebrafish; Nova Science Publishers: New York, NY, USA, 2014; pp. 245–264. [Google Scholar]
- Kalueff, A.V.; Gebhardt, M.; Stewart, A.M.; Cachat, J.M.; Brimmer, M.; Chawla, J.S.; Craddock, C.; Kyzar, E.J.; Roth, A.; Landsman, S. Towards a comprehensive catalog of zebrafish behavior 1.0 and beyond. Zebrafish 2013, 10, 70–86. [Google Scholar] [CrossRef] [PubMed]
- Saint-Amant, L.; Drapeau, P. Time course of the development of motor behaviors in the zebrafish embryo. J. Neurobiol. 1998, 37, 622–632. [Google Scholar] [CrossRef]
- Schröter, C.; Herrgen, L.; Cardona, A.; Brouhard, G.J.; Feldman, B.; Oates, A.C. Dynamics of zebrafish somitogenesis. Develop. Dynamic. 2008, 237, 545–553. [Google Scholar] [CrossRef]
- Schoots, A.M.; Meijer, R.C.; Denucé, J.M. Dopaminergic regulation of hatching in fish embryos. Dev. Biol. 1983, 100, 59–63. [Google Scholar] [CrossRef]
- Stewart, A.M.; Desmond, D.; Kyzar, E.; Gaikwad, S.; Roth, A.; Riehl, R.; Collins, C.; Monnig, L.; Green, J.; Kalueff, A.V. Perspectives of zebrafish models of epilepsy: What, how and where next? Brain Res. Bull. 2012, 87, 135–143. [Google Scholar] [CrossRef]
- Rowitch, D.H. Glial specification in the vertebrate neural tube. Nat. Rev. Neurosci. 2004, 5, 409. [Google Scholar] [CrossRef]
- Balobaid, A.; Ben-Omran, T.; Ramzan, K.; Altassan, R.; Almureikhi, M.; Musa, S.; Al-Hashmi, N.; Al-Owain, M.; Al-Zaidan, H.; Al-Hassnan, Z. Delineating the phenotypic spectrum of hyperphosphatasia with mental retardation syndrome 4 in 14 patients of Middle-Eastern origin. Am. J. Med Genet. Part A 2018, 176, 2850–2857. [Google Scholar] [CrossRef]
- Knaus, A.; Awaya, T.; Helbig, I.; Afawi, Z.; Pendziwiat, M.; Abu-Rachma, J.; Thompson, M.D.; Cole, D.E.; Skinner, S.; Annese, F. Rare noncoding mutations extend the mutational spectrum in the PGAP3 subtype of hyperphosphatasia with mental retardation syndrome. Hum. Mutat. 2016, 37, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Pagnamenta, A.T.; Murakami, Y.; Anzilotti, C.; Titheradge, H.; Oates, A.J.; Morton, J.; Study, D.; Kinoshita, T.; Kini, U.; Taylor, J.C. A homozygous variant disrupting the PIGH start-codon is associated with developmental delay, epilepsy, and microcephaly. Hum. Mutat. 2018, 39, 822–826. [Google Scholar] [CrossRef]
- Trenouth, M. Shape changes during human fetal craniofacial growth. J. Anat. 1984, 139, 639. [Google Scholar] [PubMed]
- Fagard, J.; Esseily, R.; Jacquey, L.; O’Regan, K.; Somogyi, E. Fetal origin of sensorimotor behavior. Front. Neurorobotics 2018, 12, 23. [Google Scholar] [CrossRef]
- Ackerman, S. Discovering the Brain; National Academies Press: Washington, DC, USA, 1992; ISBN 978-0-309-46799-5. [Google Scholar] [CrossRef]
- Burrows, D.; Samarut, É.; Liu, J.; Baraban, S.; Richardson, M.; Meyer, M.; Rosch, R. Imaging epilepsy in larval zebrafish. Eur. J. Paediatr. Neurol. 2020, 24, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Turrini, L.; Fornetto, C.; Marchetto, G.; Müllenbroich, M.; Tiso, N.; Vettori, A.; Resta, F.; Masi, A.; Mannaioni, G.; Pavone, F. Optical mapping of neuronal activity during seizures in zebrafish. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Káradóttir, R.; Attwell, D. Neurotransmitter receptors in the life and death of oligodendrocytes. Neuroscience 2007, 145, 1426–1438. [Google Scholar] [CrossRef]
| Clinical Feature | Proband II-1 | Previously Reported Cases with Loss-Off-Function Variants | ||||
|---|---|---|---|---|---|---|
| This Study | Abi Farraj et al. 2019 [29] | Abdel-Hamid et al. 2018 [5] | Howard et al. 2014 [1] | Dogan et al. 2019 [30] | Abdel-Hamid et al. 2018 [5] | |
| p.Gln89* | p.C68LfsX88 | p.M135Hfs*28 | p.Leu147Profs*16, p.Asp305Gly | p.Tyr169* | p.D273Sfs*37 | |
| DD, developmental delay; ID, intellectual disability | yes | yes | yes | yes | yes | yes |
| Neurological abnormalities (seizures, hypotonia) | yes, only hypotonia | yes | yes | yes | yes, only hypotonia | yes |
| Dysmorphic facial features | yes, cleft palate | yes, cleft palate | yes | yes | yes, cleft palate | yes |
| Cranial shape anomalies | yes, plagiocephaly | no | no | no | yes, postnatal microcephaly | no |
| Deafness | no | no | yes, hearing loss | N/A | no | no |
| Ophthalmological anomalies | yes, poor vision | no | yes, optic nerve pallor | N/A | no | no |
| Cardiac anomalies | no | yes, Congenital Heart Defect (CHD) | yes, patent ductus arteriosus (PDA) and atrial septal defect (ASD) | N/A | yes, left ventricle hypertrophy | no |
| GU, genitourinary malformation | no | yes, | yes, undescended testicles | N/A | no | yes, hypoplastic clitoris, absent labia minora |
| GI, gastrointestinal anomalies including GERD gastroesophageal reflux disease | yes, swallowing dysfunction | yes, esophagitis, hiatal hernia, petechial gastritis, nodular duodenitis | yes, inguinal hernia, hepatosplenomegaly | N/A | yes, dysphagia, umbilical hernia | no |
| Nephrocalcinosis | no | N/A | no | N/A | no | no |
| Teeth anomalies | no | N/A | yes, double row teeth | N/A | no | no |
| Nail anomalies | no | no | no | N/A | yes, brittle hypoplastic nails | no |
| Short fingers or hands | no | no | no | no | no | no |
| Hand/feet anomalies | Yes, hands clawing, feet talipes deformity | yes, clinodactyly of 5th digit | no | no | no | no |
| Skeletal findings | yes | no | yes | no | no | yes, pectus excavatum |
| Brain MRI/CT anomalies | yes, thin corpus callosum | yes, left temporal lobe bleeding | yes, thin corpus callosum | N/A | yes, thin corpus callosum | no |
| Prenatal finding | yes, decreased IUM | no | N/A | N/A | no | N/A |
| Elevated serum alkaline phosphatase | yes | yes | yes | yes | yes | yes |
| other | tongue thrust posture, mild left ear constriction | skin hyperpigmentation | squint, protruding tongue, nystagmus | - | ear pit, nystagmus, Pectusexcavatum, sparse hair | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Da’as, S.I.; Aamer, W.; Hasan, W.; Al-Maraghi, A.; Al-Kurbi, A.; Kilani, H.; AlRayahi, J.; Zamel, K.; Stotland, M.A.; Fakhro, K.A. PGAP3 Associated with Hyperphosphatasia with Mental Retardation Plays a Novel Role in Brain Morphogenesis and Neuronal Wiring at Early Development. Cells 2020, 9, 1782. https://doi.org/10.3390/cells9081782
Da’as SI, Aamer W, Hasan W, Al-Maraghi A, Al-Kurbi A, Kilani H, AlRayahi J, Zamel K, Stotland MA, Fakhro KA. PGAP3 Associated with Hyperphosphatasia with Mental Retardation Plays a Novel Role in Brain Morphogenesis and Neuronal Wiring at Early Development. Cells. 2020; 9(8):1782. https://doi.org/10.3390/cells9081782
Chicago/Turabian StyleDa’as, Sahar I., Waleed Aamer, Waseem Hasan, Aljazi Al-Maraghi, Alya Al-Kurbi, Houda Kilani, Jehan AlRayahi, Khaled Zamel, Mitchell A. Stotland, and Khalid A. Fakhro. 2020. "PGAP3 Associated with Hyperphosphatasia with Mental Retardation Plays a Novel Role in Brain Morphogenesis and Neuronal Wiring at Early Development" Cells 9, no. 8: 1782. https://doi.org/10.3390/cells9081782
APA StyleDa’as, S. I., Aamer, W., Hasan, W., Al-Maraghi, A., Al-Kurbi, A., Kilani, H., AlRayahi, J., Zamel, K., Stotland, M. A., & Fakhro, K. A. (2020). PGAP3 Associated with Hyperphosphatasia with Mental Retardation Plays a Novel Role in Brain Morphogenesis and Neuronal Wiring at Early Development. Cells, 9(8), 1782. https://doi.org/10.3390/cells9081782