Ribosomal Protein uL11 as a Regulator of Metabolic Circuits Related to Aging and Cell Cycle
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Kinetics of the Growth Assay
2.3. Determination of the Mean Doubling Time
2.4. Cell Cycle Analysis
2.5. Phenotypic Analysis—a Spot Test for Sensitivity to Congo Red, Calcofluor White, MMS, and Sodium Chloride
2.6. Determination of Translational Fitness by Incorporation of 35S-Radiolabeled Methionine
2.7. Yeast Polysome Profile Analysis
2.8. Determination of Budding Lifespan
2.9. Determination of the Total Lifespan
2.10. Estimation of the Cell Volume
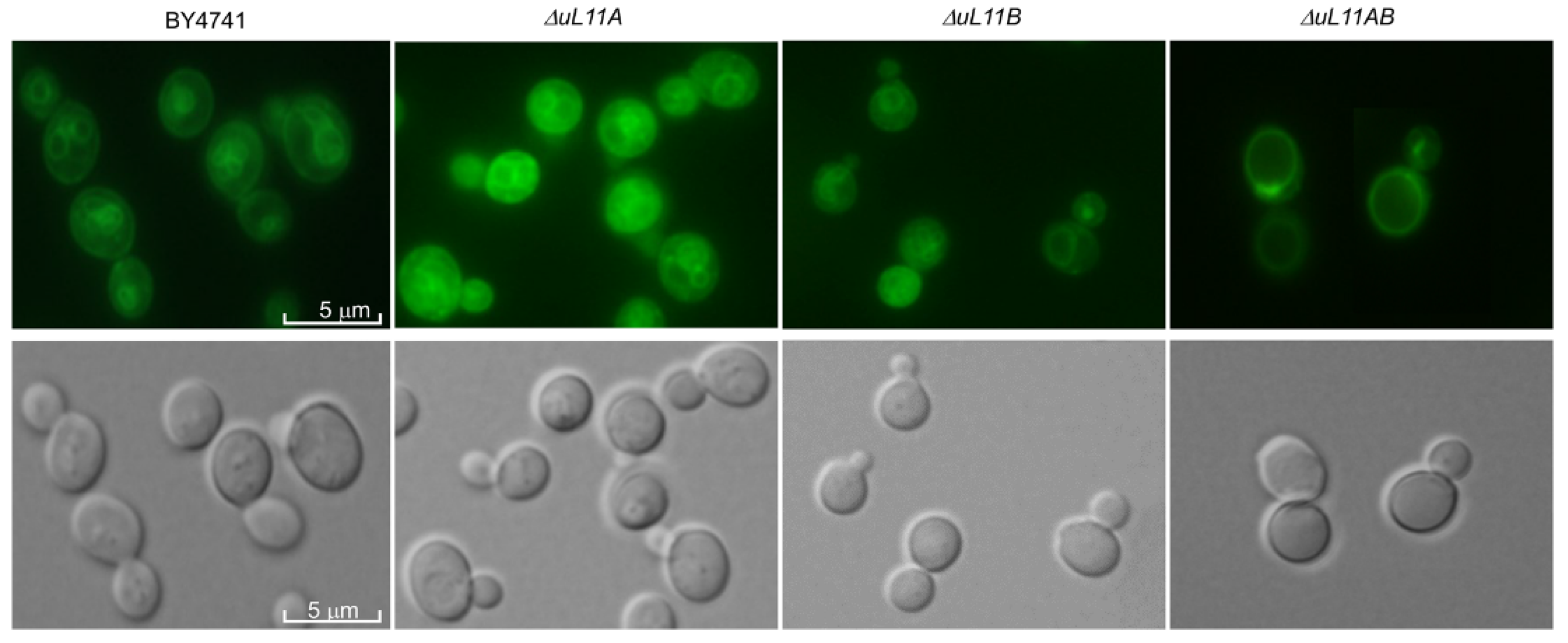
2.11. Yeast Vacuole Staining
2.12. RNA Sequencing and Analysis of RNA-Seq Data
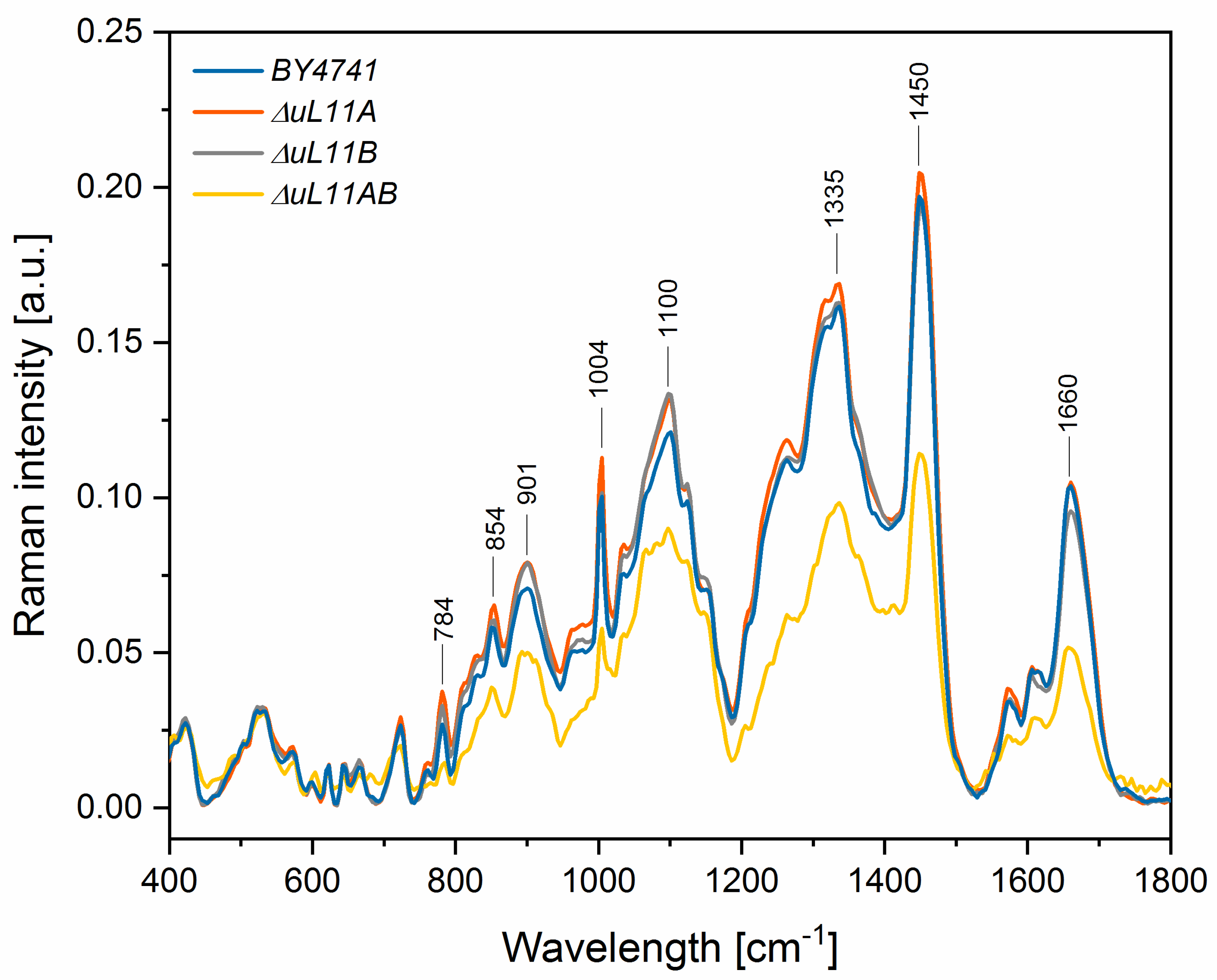
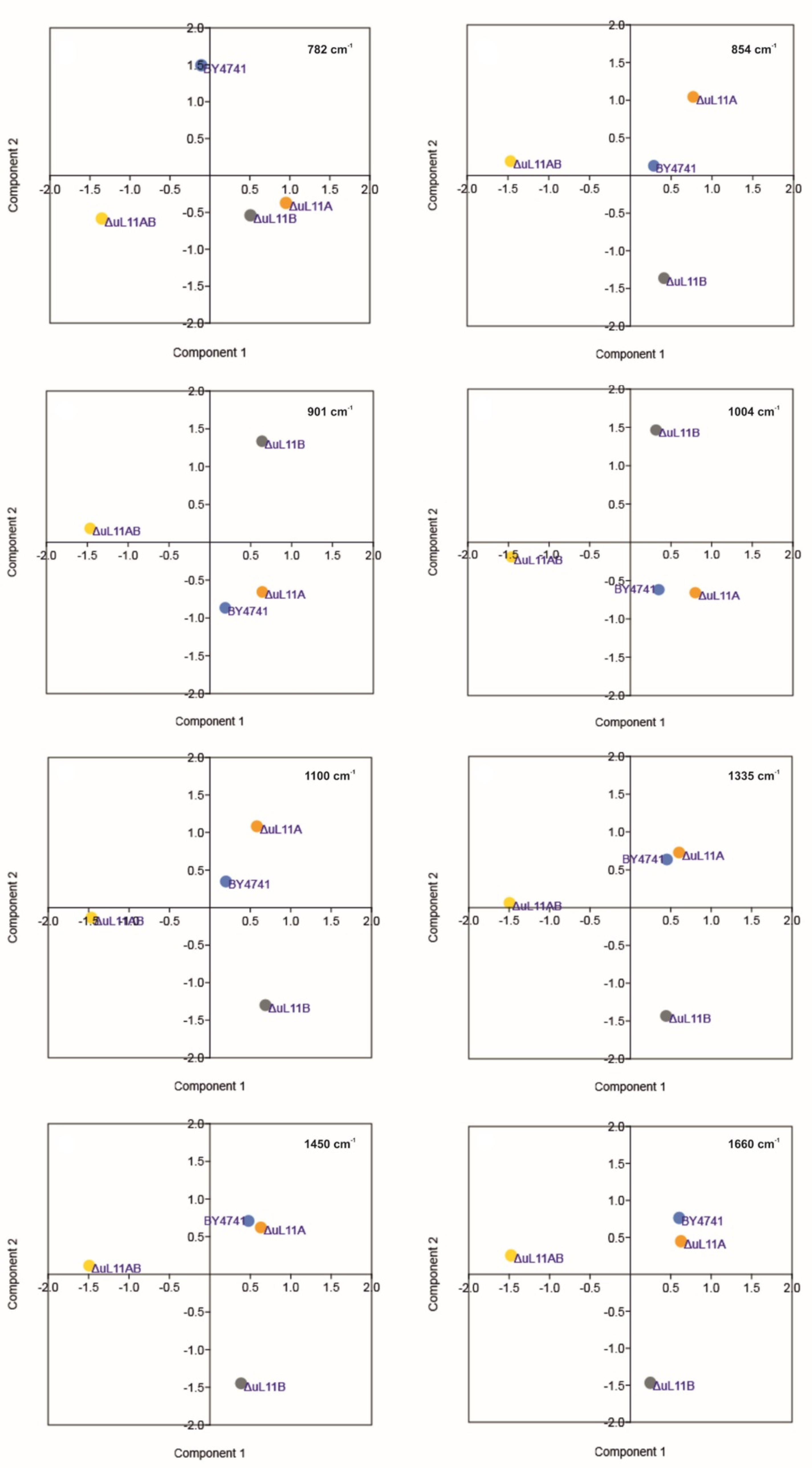
2.13. Raman Spectroscopy
2.14. Statistical Analysis
3. Results
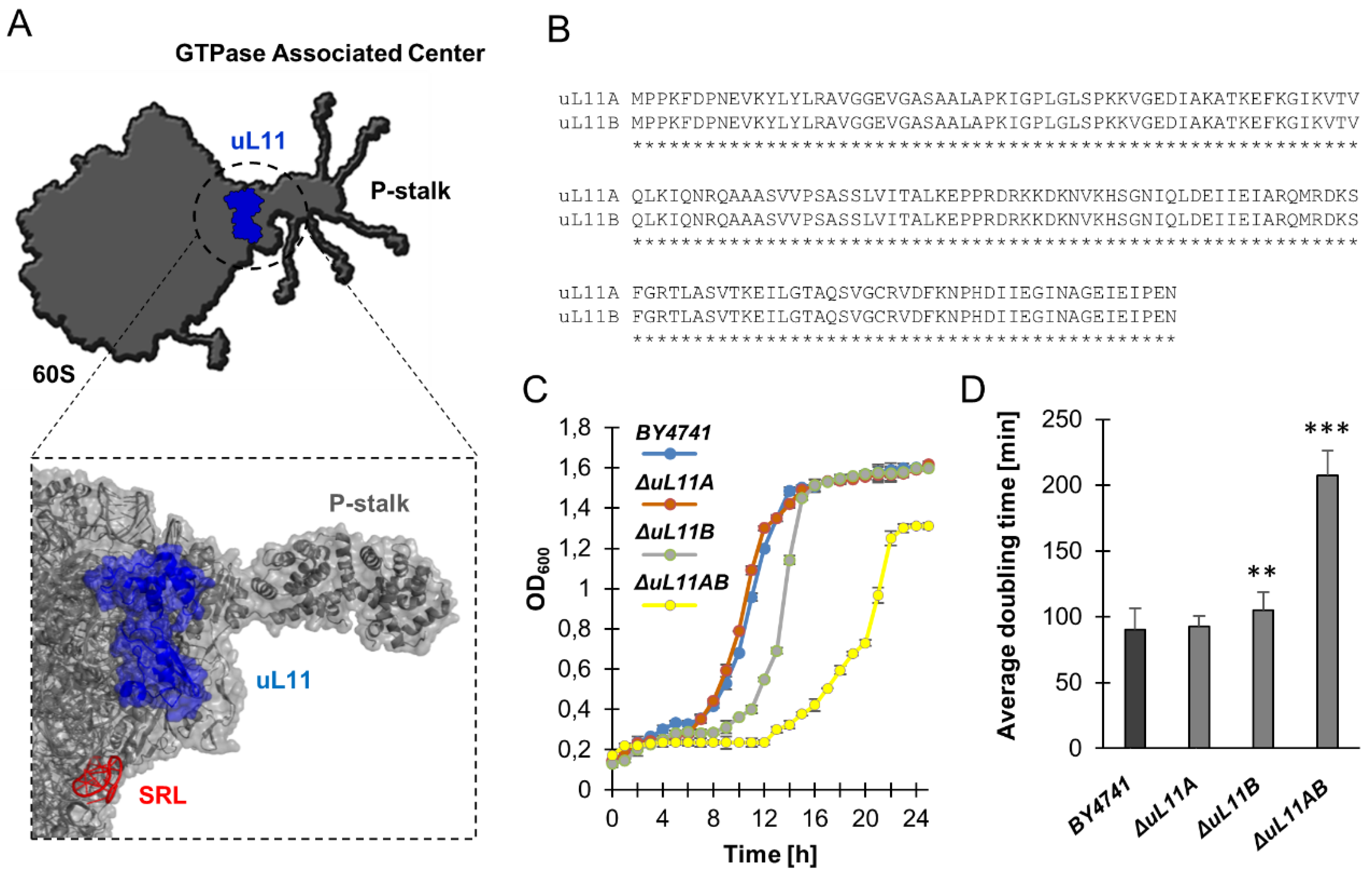
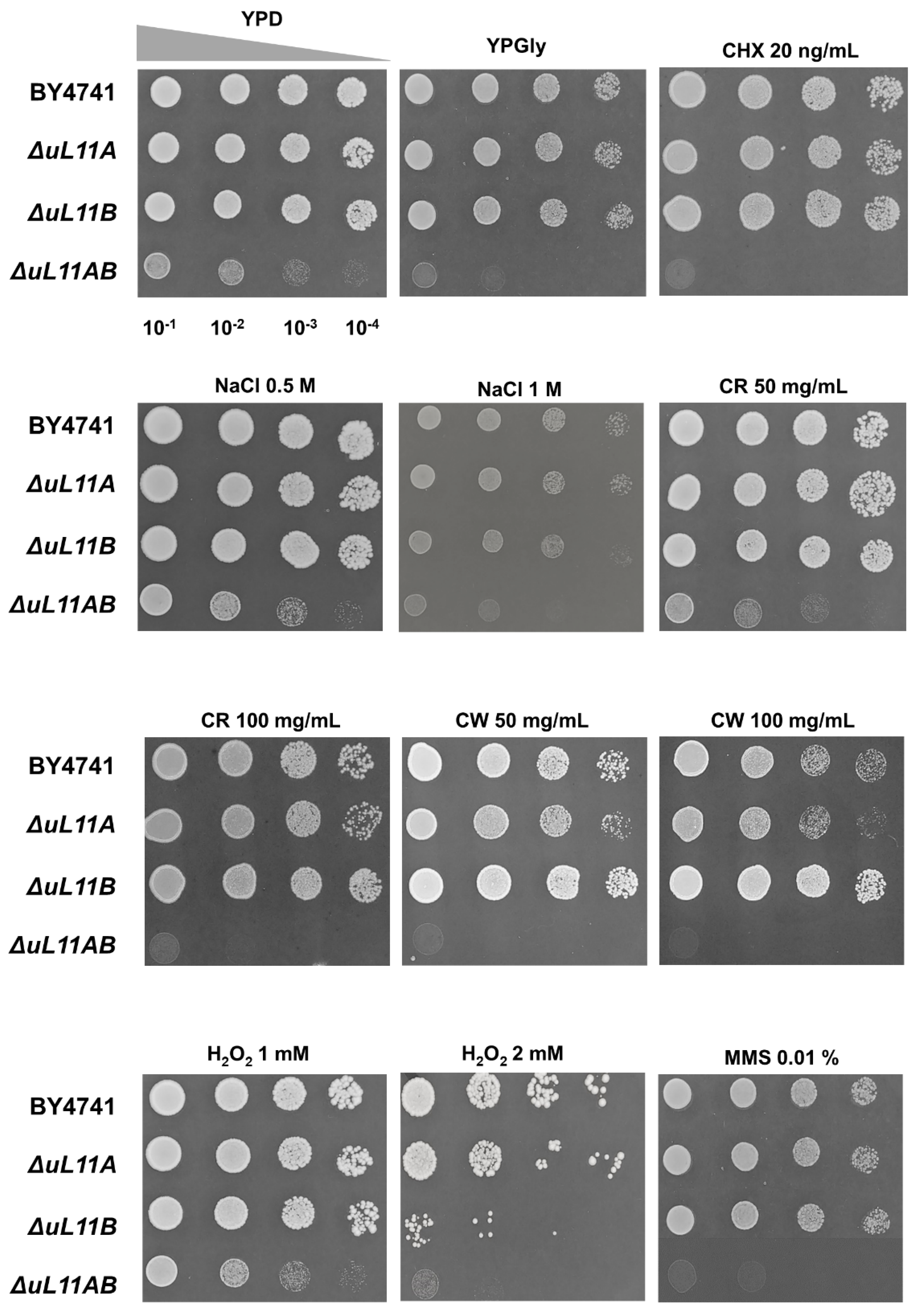
3.1. Lack of A and B isoforms of the uL11 Protein Increases the Doubling Time and Sensitivity to Factors Causing Cell Wall Disorder and Oxidative Stress
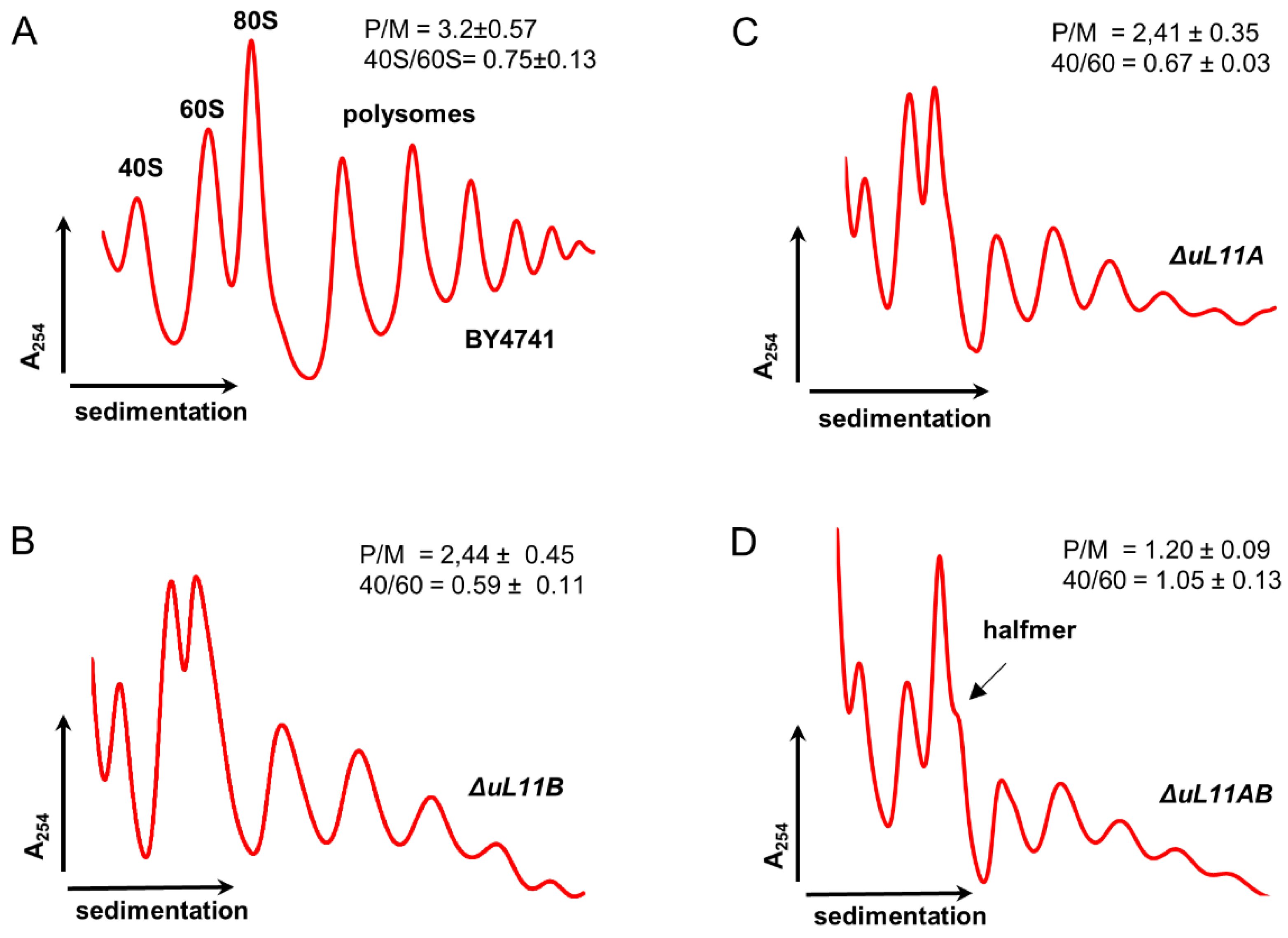
3.2. Depletion of uL11 Isoforms Negatively Affects Translation
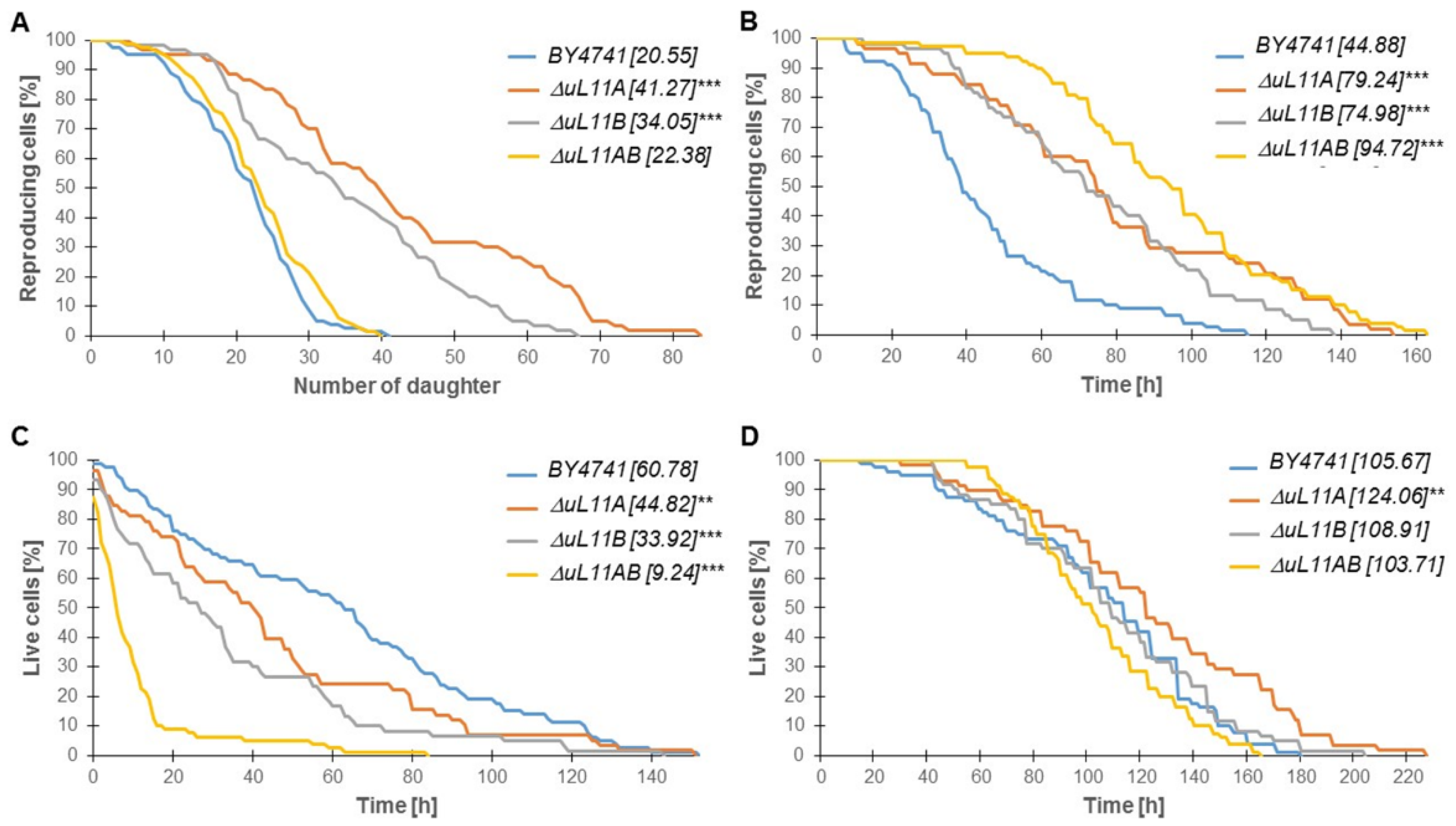
3.3. The Lifespan is Affected by the Lack of the uL11 Protein
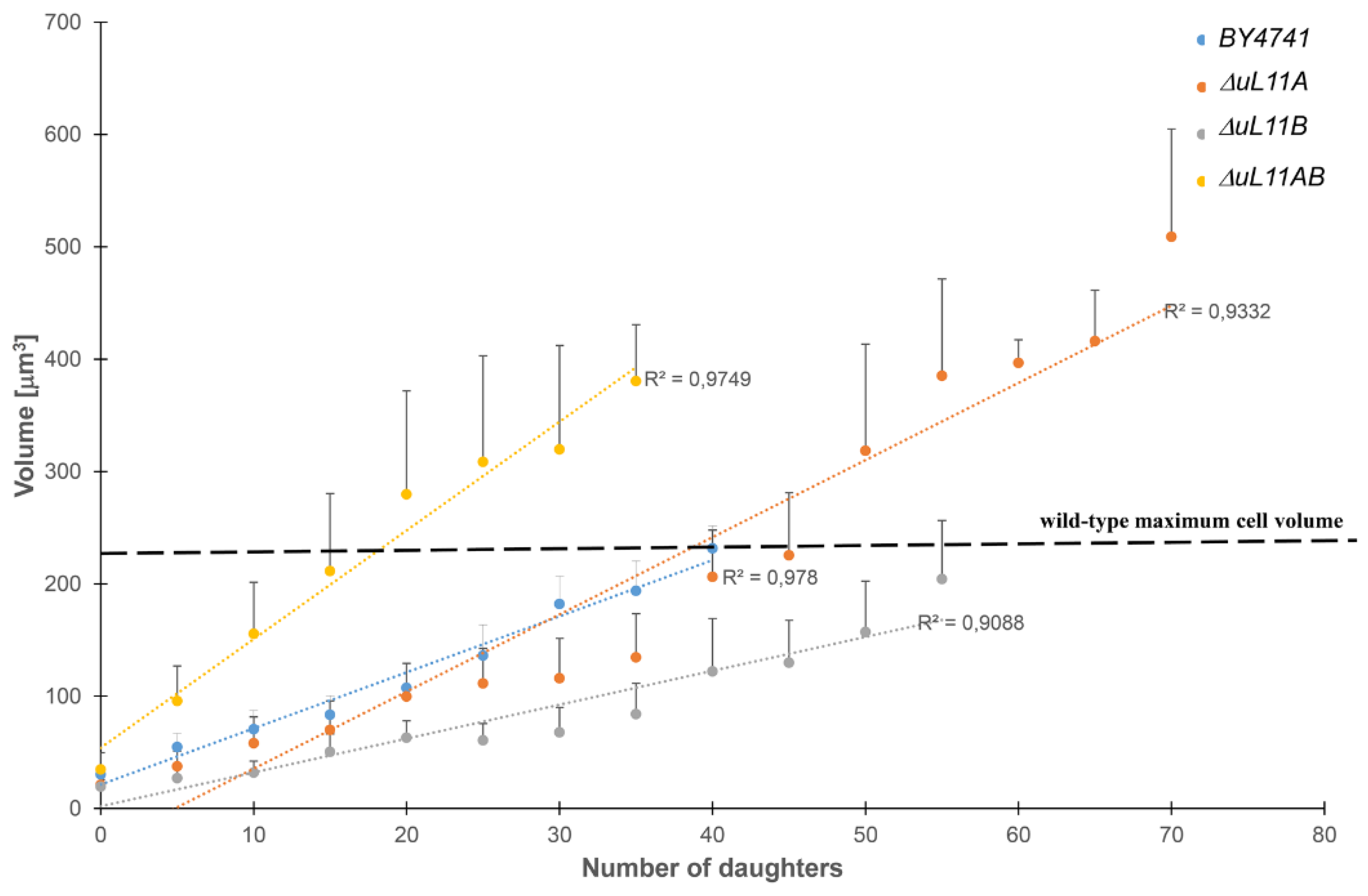
3.4. Changes in the Cell Volume During the Budding Lifespan
3.5. Deletion of uL11 Genes Affects Vacuole Biogenesis
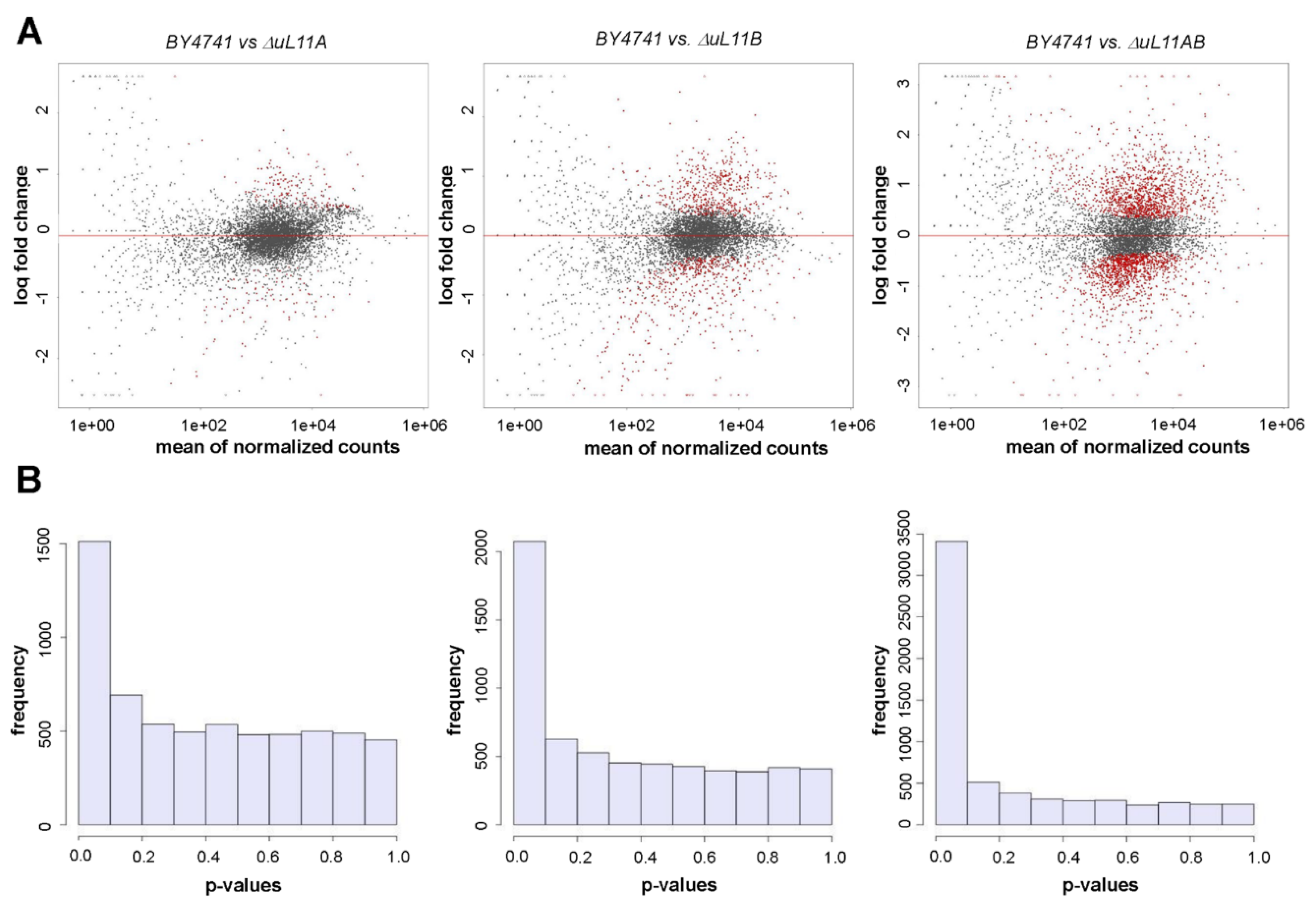
3.6. Transcriptional Changes are Stimulated by uL11 Deletion
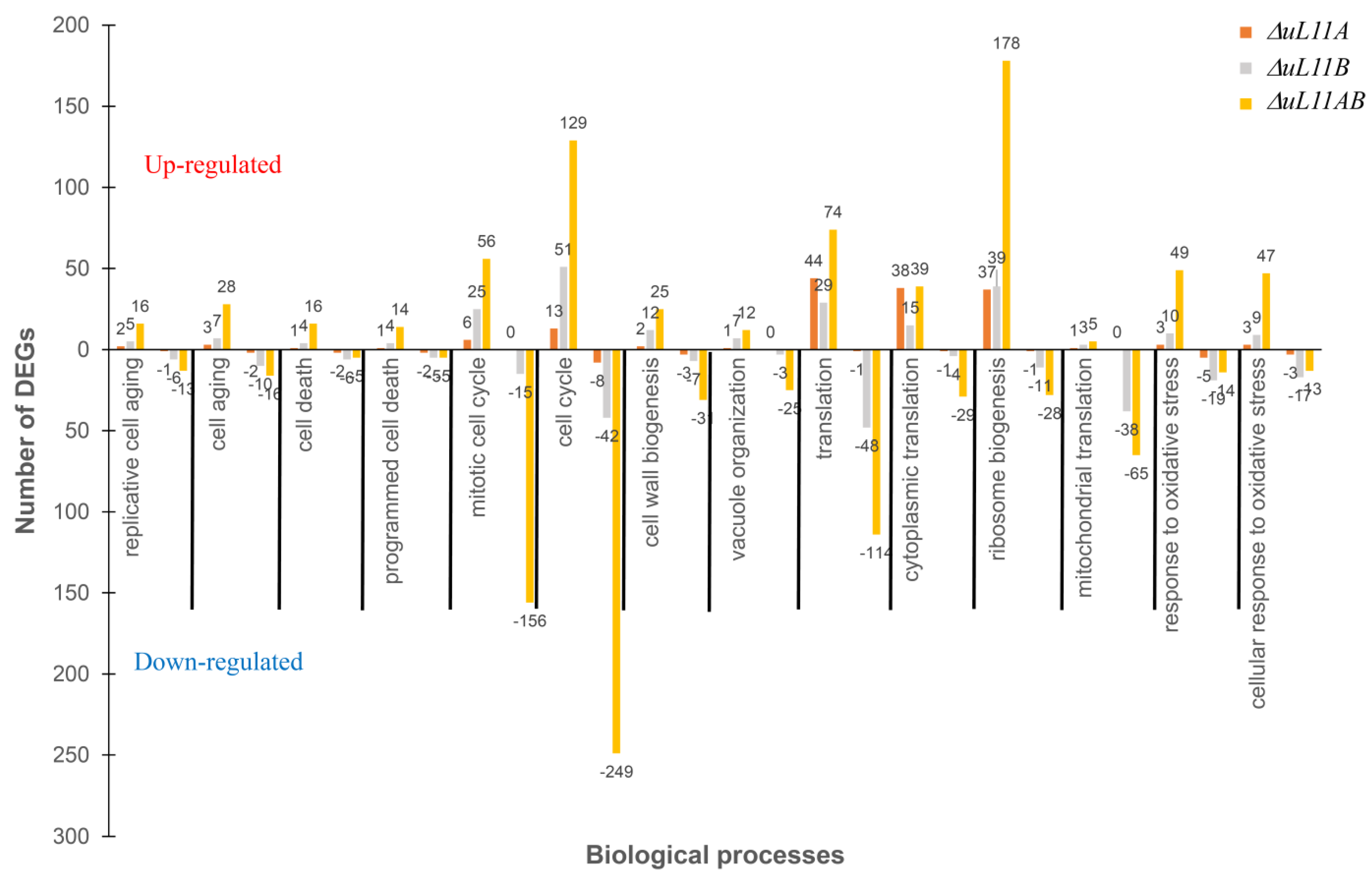
3.7. Cross-Differentially Expressed Genes (DEGs) Reveal Core Biological Processes and Pathways in Response to uL11 Deletion
3.8. Deletion of uL11 Genes Significantly Changes the Chemical Composition in Yeast
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hayflick, L. Biological Aging Is No Longer an Unsolved Problem. Ann. N. Y. Acad. Sci. 2007, 1100, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M.; Powers, R.W.; Steffen, K.K.; Westman, E.A.; Hu, D.; Dang, N.; Kerr, E.O.; Kirkland, K.T.; Fields, S.; Kennedy, B.K. Regulation of Yeast Replicative Life Span by TOR and Sch9 in Response to Nutrients. Science 2005, 310, 1193–1196. [Google Scholar] [CrossRef] [PubMed]
- Artal-Sanz, M.; Tavernarakis, N. Mechanisms of aging and energy metabolism inCaenorhabditis elegans. Iubmb Life 2008, 60, 315–322. [Google Scholar] [CrossRef]
- Partridge, L.; Piper, M.; Mair, W.B. Dietary restriction in Drosophila. Mech. Ageing Dev. 2005, 126, 938–950. [Google Scholar] [CrossRef]
- Speakman, J.R. Body size, energy metabolism and lifespan. J. Exp. Boil. 2005, 208, 1717–1730. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Mattson, M.P. Fasting: Molecular mechanisms and clinical applications. Cell Metab. 2014, 19, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Mirisola, M.G.; Taormina, G.; Fabrizio, P.; Wei, M.; Hu, J.; Longo, V.D. Serine- and Threonine/Valine-Dependent Activation of PDK and Tor Orthologs Converge on Sch9 to Promote Aging. PLoS Genet. 2014, 10, e1004113. [Google Scholar] [CrossRef] [PubMed]
- Ruckenstuhl, C.; Netzberger, C.; Entfellner, I.; Carmona-Gutierrez, D.; Kickenweiz, T.; Stekovic, S.; Gleixner, C.; Schmid, C.; Klug, L.; Sorgo, A.G.; et al. Lifespan Extension by Methionine Restriction Requires Autophagy-Dependent Vacuolar Acidification. PLoS Genet. 2014, 10, e1004347. [Google Scholar] [CrossRef]
- Tavernarakis, N. Ageing and the regulation of protein synthesis: A balancing act? Trends Cell Boil. 2008, 18, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Blagosklonny, M.V. Rapamycin extends life- and health span because it slows aging. Aging 2013, 5, 592–598. [Google Scholar] [CrossRef]
- Zid, B.M.; Rogers, A.N.; Katewa, S.D.; Vargas, M.A.; Kolipinski, M.C.; Lu, T.A.; Benzer, S.; Kapahi, P.; Vargas, M.A. 4E-BP Extends Lifespan upon Dietary Restriction by Enhancing Mitochondrial Activity in Drosophila. Cell 2009, 139, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.N.; Chen, D.; McColl, G.; Czerwieniec, G.; Felkey, K.; Gibson, B.W.; Hubbard, A.; Melov, S.; Lithgow, G.J.; Kapahi, P. Life Span Extension via eIF4G Inhibition Is Mediated by Posttranscriptional Remodeling of Stress Response Gene Expression in C. elegans. Cell Metab. 2011, 14, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Taubert, S.; Crawford, D.; Libina, N.; Kenyon, C.; Lee, S.V. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 2007, 6, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Steffen, K.K.; Mackay, V.L.; Kerr, E.O.; Tsuchiya, M.; Hu, D.; Fox, L.A.; Dang, N.; Johnston, E.D.; Oakes, J.A.; Tchao, B.N.; et al. Yeast Life Span Extension by Depletion of 60S Ribosomal Subunits Is Mediated by Gcn4. Cell 2008, 133, 292–302. [Google Scholar] [CrossRef]
- Wilson, D.N.; Nierhaus, K.H. Ribosomal Proteins in the Spotlight. Crit. Rev. Biochem. Mol. Boil. 2005, 40, 243–267. [Google Scholar] [CrossRef]
- De La Cruz, J.; Karbstein, K.; Woolford, J.L. Functions of ribosomal proteins in assembly of eukaryotic ribosomes in vivo. Annu. Rev. Biochem. 2015, 84, 93–129. [Google Scholar] [CrossRef]
- Dalla-Venezia, N.; Vincent, A.; Marcel, V.; Catez, F.; Diaz, J.-J. Emerging Role of Eukaryote Ribosomes in Translational Control. Int. J. Mol. Sci. 2019, 20, 1226. [Google Scholar] [CrossRef]
- Planta, R.J.; Mager, W.H. The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 1998, 14, 471–477. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Shields, D.C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 1997, 387, 708–713. [Google Scholar] [CrossRef]
- Gerst, J.E. Pimp My Ribosome: Ribosomal Protein Paralogs Specify Translational Control. Trends Genet. 2018, 34, 832–845. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef]
- Ray, P.S.; Arif, A.; Fox, P.L. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem. Sci. 2007, 32, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.W.; Green, R. Ribosomopathies: There’s strength in numbers. Science 2017, 358, eaan2755. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liao, W.-J.; Liao, J.-M.; Liao, P.; Lu, H. Ribosomal proteins: Functions beyond the ribosome. J. Mol. Cell Boil. 2015, 7, 92–104. [Google Scholar] [CrossRef]
- Warner, J.R.; McIntosh, K.B. How Common Are Extraribosomal Functions of Ribosomal Proteins? Mol. Cell 2009, 34, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Ghulam, M.M.; Catala, M.; Elela, S.A. Differential expression of duplicated ribosomal protein genes modifies ribosome composition in response to stress. Nucleic Acids Res. 2019, 48, 1954–1968. [Google Scholar] [CrossRef]
- Dean, E.J.; Davis, J.C.; Davis, R.W.; Petrov, D. Pervasive and Persistent Redundancy among Duplicated Genes in Yeast. PLoS Genet. 2008, 4, e1000113. [Google Scholar] [CrossRef]
- McCormick, M.A.; Delaney, J.R.; Tsuchiya, M.; Tsuchiyama, S.; Shemorry, A.; Sim, S.; Chou, A.C.-Z.; Ahmed, U.; Carr, D.; Murakami, C.J.; et al. A Comprehensive Analysis of Replicative Lifespan in 4,698 Single-Gene Deletion Strains Uncovers Conserved Mechanisms of Aging. Cell Metab. 2015, 22, 895–906. [Google Scholar] [CrossRef] [PubMed]
- Mołoń, M.; Zadrag-Tecza, R.; Bilinski, T. The longevity in the yeast Saccharomyces cerevisiae: A comparison of two approaches for assessment the lifespan. Biochem. Biophys. Res. Commun. 2015, 460, 651–656. [Google Scholar] [CrossRef]
- Borkiewicz, L.; Mołoń, M.; Molestak, E.; Grela, P.; Horbowicz-Drożdżal, P.; Wawiórka, L.; Tchórzewski, M. Functional Analysis of the Ribosomal uL6 Protein of Saccharomyces cerevisiae. Cells 2019, 8, 718. [Google Scholar] [CrossRef] [PubMed]
- Ban, N.; Beckmann, R.; Cate, J.H.D.; Dinman, J.; Dragon, F.; Ellis, S.R.; Lafontaine, D.L.J.; Lindahl, L.; Liljas, A.; Lipton, J.M.; et al. A new system for naming ribosomal proteins. Curr. Opin. Struct. Boil. 2014, 24, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Datta, P.P.; Sharma, M.R.; Qi, L.; Frank, J.; Agrawal, R.K. Interaction of the G’ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol. Cell 2005, 20, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Kavran, J.M.; Steitz, T.A. Structure of the Base of the L7/L12 Stalk of the Haloarcula marismortui Large Ribosomal Subunit: Analysis of L11 Movements. J. Mol. Boil. 2007, 371, 1047–1059. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Pevida, A.; Rodríguez-Galán, O.; Díaz-Quintana, A.J.; Kressler, D.; De La Cruz, J. Yeast Ribosomal Protein L40 Assembles Late into Precursor 60 S Ribosomes and Is Required for Their Cytoplasmic Maturation*. J. Boil. Chem. 2012, 287, 38390–38407. [Google Scholar] [CrossRef]
- Santos, C.; Ballesta, J.P. Ribosomal protein P0, contrary to phosphoproteins P1 and P2, is required for ribosome activity and Saccharomyces cerevisiae viability. J. Boil. Chem. 1994, 269, 15689–15696. [Google Scholar]
- Wawiórka, L.; Molestak, E.; Szajwaj, M.; Michalec-Wawiórka, B.; Boguszewska, A.; Borkiewicz, L.; Liudkovska, V.; Kufel, J.; Tchórzewski, M. Functional analysis of the uL11 protein impact on translational machinery. Cell Cycle 2016, 15, 1060–1072. [Google Scholar] [CrossRef]
- Imami, K.; Milek, M.; Bogdanow, B.; Yasuda, T.; Kastelic, N.; Zauber, H.; Ishihama, Y.; Landthaler, M.; Selbach, M. Phosphorylation of the Ribosomal Protein RPL12/uL11 Affects Translation during Mitosis. Mol. Cell 2018, 72, 84–98.e9. [Google Scholar] [CrossRef]
- Mołoń, M.; Szajwaj, M.; Tchórzewski, M.; Skoczowski, A.; Niewiadomska, E.; Zadrag-Tecza, R. The rate of metabolism as a factor determining longevity of the Saccharomyces cerevisiae yeast. AGE 2016, 38, 11. [Google Scholar] [CrossRef]
- Mołoń, M.; Zebrowski, J. Phylogenetic relationship and FTIR spectroscopy-derived lipid determinants of lifespan parameters in the Saccharomyces cerevisiae yeast. Fems Yeast Res. 2017, 17. [Google Scholar] [CrossRef]
- Minois, N.; Frajnt, M.; Wilson, C.; Vaupel, J.W. Advances in measuring lifespan in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2004, 102, 402–406. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Biliński, T.; Bartosz, G. Hypothesis: Cell volume limits cell divisions. Acta Biochim. Pol. 2006, 53, 833–835. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.-H.M.; Reyes, L.; Sohail, S.M.; Tran, N.K.; Marshall, W.F. Organelle Size Scaling of the Budding Yeast Vacuole by Relative Growth and Inheritance. Curr. Boil. 2016, 26, 1221–1228. [Google Scholar] [CrossRef]
- Rösch, P.; Harz, M.; Schmitt, M.; Popp, J. Raman spectroscopic identification of single yeast cells. J. Raman Spectrosc. 2005, 36, 377–379. [Google Scholar] [CrossRef]
- Lemma, T.; Wang, J.; Arstila, K.; Hytönen, V.P.; Toppari, J.J. Identifying yeasts using surface enhanced Raman spectroscopy. Spectrochim. Acta Part. A: Mol. Biomol. Spectrosc. 2019, 218, 299–307. [Google Scholar] [CrossRef]
- Kerker, M. Electromagnetic model for surface-enhanced Raman scattering (SERS) on metal colloids. Acc. Chem. Res. 1984, 17, 271–277. [Google Scholar] [CrossRef]
- Kneipp, K.; Wang, Y.; Kneipp, H.; Perelman, L.T.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS). Phys. Rev. Lett. 1997, 78, 1667–1670. [Google Scholar] [CrossRef]
- Kneipp, K.; Kneipp, H.; Deinum, G.; Itzkan, I.; Dasari, R.R.; Feld, M.S. Single-Molecule Detection of a Cyanine Dye in Silver Colloidal Solution Using Near-Infrared Surface-Enhanced Raman Scattering. Appl. Spectrosc. 1998, 52, 175–178. [Google Scholar] [CrossRef]
- Schmeing, T.M.; Ramakrishnan, V. What recent ribosome structures have revealed about the mechanism of translation. Nature 2009, 461, 1234–1242. [Google Scholar] [CrossRef]
- Meyuhas, O. Ribosomal Protein S6 Phosphorylation. Int. Rev. Cell Mol. Biol. 2015, 320, 41–73. [Google Scholar] [CrossRef]
- Uechi, T.; Tanaka, T.; Kenmochi, N. A Complete Map of the Human Ribosomal Protein Genes: Assignment of 80 Genes to the Cytogenetic Map and Implications for Human Disorders. Genomics 2001, 72, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Barakat, A.; Szick-Miranda, K.; Chang, I.F.; Guyot, R.; Blanc, G.; Cooke, R.; Delseny, M.; Bailey-Serres, J. The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 2001, 127, 398–415. [Google Scholar] [CrossRef] [PubMed]
- Parenteau, J.; Lavoie, M.; Catala, M.; Malik-Ghulam, M.; Gagnon, J.; Elela, S.A. Preservation of Gene Duplication Increases the Regulatory Spectrum of Ribosomal Protein Genes and Enhances Growth under Stress. Cell Rep. 2015, 13, 2516–2526. [Google Scholar] [CrossRef] [PubMed]
- Komili, S.; Farny, N.G.; Roth, F.P.; Silver, P.A. Functional Specificity among Ribosomal Proteins Regulates Gene Expression. Cell 2007, 131, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Diaconu, M.; Kothe, U.; Schlunzen, F.; Fischer, N.; Harms, J.M.; Tonevitsky, A.G.; Stark, H.; Rodnina, M.V.; Wahl, M.C. Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation. Cell 2005, 121, 991–1004. [Google Scholar] [CrossRef]
- Wimberly, B.T.; Guymon, R.; McCutcheon, J.P.; White, S.W.; Ramakrishnan, V. A Detailed View of a Ribosomal Active Site. Cell 1999, 97, 491–502. [Google Scholar] [CrossRef]
- Krokowski, D.; Boguszewska, A.; Abramczyk, D.; Liljas, A.; Tchórzewski, M.; Grankowski, N. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol. Microbiol. 2006, 60, 386–400. [Google Scholar] [CrossRef] [PubMed]
- Steffen, K.K.; McCormick, M.A.; Pham, K.M.; Mackay, V.L.; Delaney, J.R.; Murakami, C.J.; Kaeberlein, M.; Kennedy, B.K. Ribosome Deficiency Protects Against ER Stress in Saccharomyces cerevisiae. Genetics 2012, 191, 107–118. [Google Scholar] [CrossRef]
- Mołoń, M.; Zadrag-Tecza, R. Effect of temperature on replicative aging of the budding yeast Saccharomyces cerevisiae. Biogerontology 2015, 17, 347–357. [Google Scholar] [CrossRef]
- Maitra, N.; He, C.; Blank, H.M.; Tsuchiya, M.; Schilling, B.; Kaeberlein, M.; Aramayo, R.; Kennedy, B.K.; Polymenis, M. Translational control of one-carbon metabolism underpins ribosomal protein phenotypes in cell division and longevity. eLife 2020, 9, e53127. [Google Scholar] [CrossRef]
- Banta, L.M.; Robinson, J.S.; Klionsky, D.J.; Emr, S.D. Organelle assembly in yeast: Characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Boil. 1988, 107, 1369–1383. [Google Scholar] [CrossRef] [PubMed]
- Baba, M.; Takeshige, K.; Baba, N.; Ohsumi, Y. Ultrastructural analysis of the autophagic process in yeast: Detection of autophagosomes and their characterization. J. Cell Boil. 1994, 124, 903–913. [Google Scholar] [CrossRef] [PubMed]
- Bonangelino, C.J.; Nau, J.J.; Duex, J.E.; Brinkman, M.; Wurmser, A.E.; Gary, J.D.; Emr, S.D.; Weisman, L.S. Osmotic stress–induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J. Cell Boil. 2002, 156, 1015–1028. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Weisman, L.S. The vacuole/lysosome is required for cell-cycle progression. eLife 2015, 4, e08160. [Google Scholar] [CrossRef]
- Deryło, K.; Michalec-Wawiórka, B.; Krokowski, D.; Wawiórka, L.; Hatzoglou, M.; Tchórzewski, M. The uL10 protein, a component of the ribosomal P-stalk, is released from the ribosome in nucleolar stress. Biochim. Et Biophys. Acta (Bba) - Bioenerg. 2018, 1865, 34–47. [Google Scholar] [CrossRef]
- Tchórzewski, M.; Boldyreff, B.; Grankowski, N. Extraribosomal function of the acidic ribosomal P1-protein YP1alpha from Saccharomyces cerevisiae. Acta Biochim. Pol. 1999, 46, 901–910. [Google Scholar] [CrossRef] [PubMed]
- Martín-Villanueva, S.; Fernández-Pevida, A.; Kressler, D.; De La Cruz, J.; Villanueva, M.-; Pevida, F.-; La Cruz, D. The Ubiquitin Moiety of Ubi1 Is Required for Productive Expression of Ribosomal Protein eL40 in Saccharomyces cerevisiae. Cells 2019, 8, 850. [Google Scholar] [CrossRef] [PubMed]
- Hury, J.; Nagaswamy, U.; Larios-Sanz, M.; Fox, G.E. Ribosome origins: The relative age of 23S rRNA Domains. Orig. Life Evol. Biosph. 2006, 36, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.; Gao, H.; Sengupta, J.; Gao, N.; Taylor, D.J. The process of mRNA-tRNA translocation. Proc. Natl. Acad. Sci. USA 2007, 104, 19671–19678. [Google Scholar] [CrossRef] [PubMed]
- Grela, P.; Bernadó, P.; Svergun, D.I.; Kwiatowski, J.; Abramczyk, D.; Grankowski, N.; Tchórzewski, M. Structural Relationships Among the Ribosomal Stalk Proteins from the Three Domains of Life. J. Mol. Evol. 2008, 67, 154–167. [Google Scholar] [CrossRef]

| Strain | Cell Cycle Phase | |
|---|---|---|
| BY4741 | G1 [%] | 17.04 ± 0.39 |
| G2 [%] | 71.94 ± 0.45 | |
| S [%] | 11.02 ± 0.65 | |
| ∆uL11A | G1 [%] | 28.85 ± 0.76 |
| G2 [%] | 63.05 ± 1.34 | |
| S [%] | 8.10 ± 0.76 | |
| ∆uL11B | G1 [%] | 19.82 ± 1.88 |
| G2 [%] | 72.09 ± 0.42 | |
| S [%] | 8.09 ± 0.98 | |
| ∆uL11AB | G1 [%] | 41.88 ± 0.51 |
| G2 [%] | 33.43 ± 0.59 | |
| S [%] | 24.67 ± 1.77 |
| Peak Position [Raman Shift, cm−1] | Raman Spectroscopy–Peaks Intensity [a.u.] | Vibrations | |||
|---|---|---|---|---|---|
| BY4741 | ∆uL11A | ∆uL11B | ∆uL11AB | ||
| 784 | 0.027 | 0.038 *** | 0.033 ** | 0.014 *** | stretching (O–P–O) RNA |
| 854 | 0.058 | 0.065 * | 0.060 | 0.038 *** | Polysaccharides, phospholipids |
| 901 | 0.071 | 0.079 * | 0.079 * | 0.050 *** | symmetric benzene/pyrrole in-phase and out-of-phase breathing mode of tryptophan and phenylalanine |
| 1004 | 0.100 | 0.113 ** | 0.102 | 0.056 *** | |
| 1100 | 0.121 | 0.132 | 0.133 * | 0.089 *** | PO2 in nucleic acid |
| 1335 | 0.161 | 0.169 | 0.163 | 0.098 *** | C-H deformation vibrations from proteins |
| 1450 | 0.196 | 0.205 | 0.194 | 0.114 *** | C-H deformation vibrations from lipids |
| 1660 | 0.104 | 0.105 | 0.096 * | 0.051 *** | Amide I: mainly C=O stretching vibrations and contributions of N–H bending vibrations |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mołoń, M.; Molestak, E.; Kula-Maximenko, M.; Grela, P.; Tchórzewski, M. Ribosomal Protein uL11 as a Regulator of Metabolic Circuits Related to Aging and Cell Cycle. Cells 2020, 9, 1745. https://doi.org/10.3390/cells9071745
Mołoń M, Molestak E, Kula-Maximenko M, Grela P, Tchórzewski M. Ribosomal Protein uL11 as a Regulator of Metabolic Circuits Related to Aging and Cell Cycle. Cells. 2020; 9(7):1745. https://doi.org/10.3390/cells9071745
Chicago/Turabian StyleMołoń, Mateusz, Eliza Molestak, Monika Kula-Maximenko, Przemysław Grela, and Marek Tchórzewski. 2020. "Ribosomal Protein uL11 as a Regulator of Metabolic Circuits Related to Aging and Cell Cycle" Cells 9, no. 7: 1745. https://doi.org/10.3390/cells9071745
APA StyleMołoń, M., Molestak, E., Kula-Maximenko, M., Grela, P., & Tchórzewski, M. (2020). Ribosomal Protein uL11 as a Regulator of Metabolic Circuits Related to Aging and Cell Cycle. Cells, 9(7), 1745. https://doi.org/10.3390/cells9071745







