Substantially Altered Expression Profile of Diabetes/Cardiovascular/Cerebrovascular Disease Associated microRNAs in Children Descending from Pregnancy Complicated by Gestational Diabetes Mellitus—One of Several Possible Reasons for an Increased Cardiovascular Risk
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. BP and Echocardiography Measurements, BMI Assessment
2.3. Processing of Samples
2.4. Reverse Transcription
2.5. Relative Quantification of microRNAs
2.6. Statistical Analysis
2.7. Bioinformatics Analysis—microRNA-Target Interactions on Disease Ontologies, Human Phenotype Ontologies, and OMIM Disorders
2.8. Workflow of the Study
3. Results
3.1. Higher Incidence of Valve Problems and Heart Defects in a group of Children Descending from GDM Complicated Pregnancies
3.2. Only Just miR-21-5p Indicates a Trend Towards Differentiation between Children with Normal and Abnormal Clinical Findings (BMI, Blood Pressure, and Echocardiogram Findings) Descending from Normal Pregnancies
3.3. Only Just miR-29a-3p and miR-92a-3p Indicate a Slight Differentiation Between Children with Normal and Abnormal Clinical Findings (BMI, Blood Pressure, and Echocardiogram Findings) Descending from GDM Complicated Pregnancies
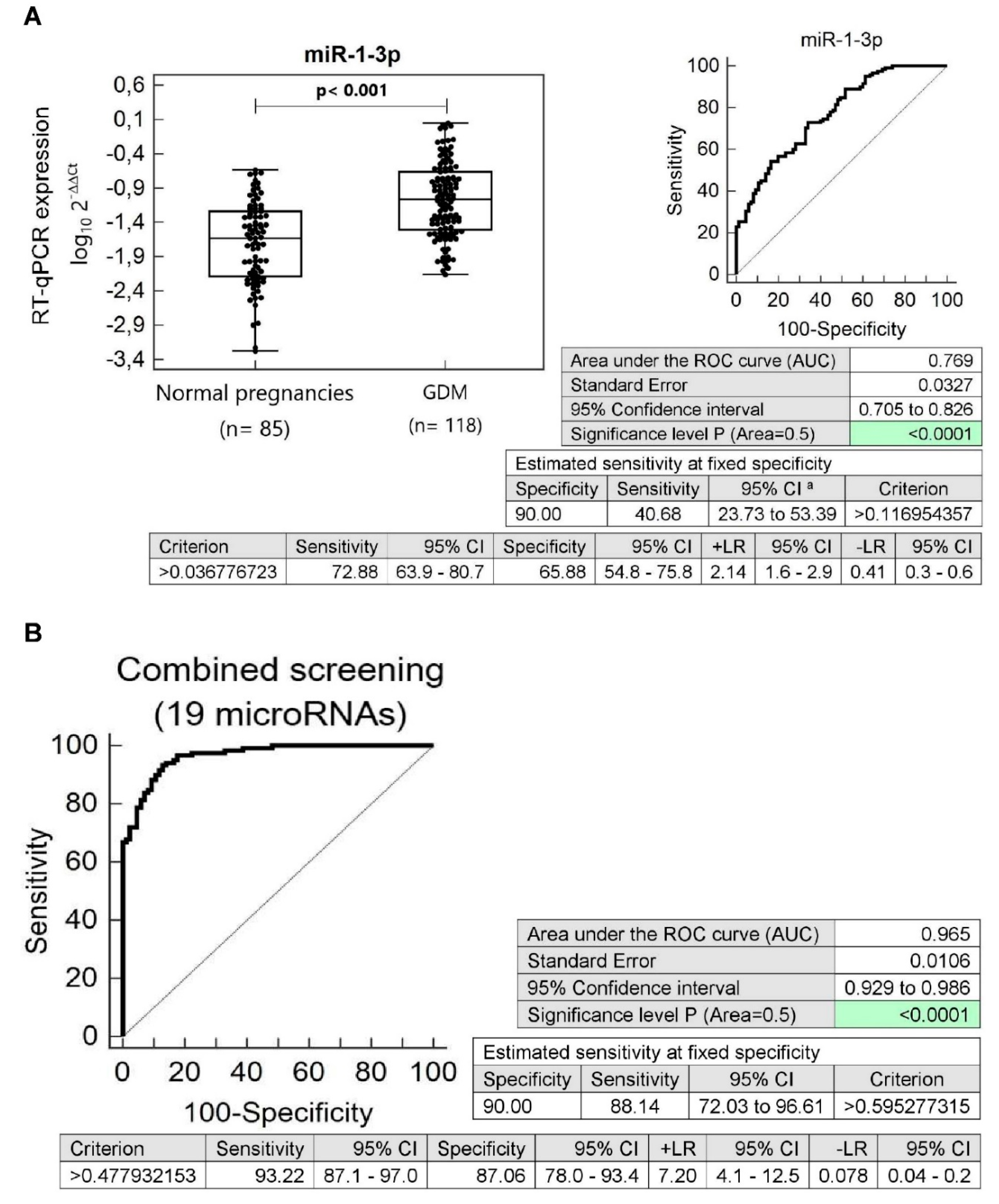
3.4. Substantially Altered Expression Profile of Diabetes/Cardiovascular/Cerebrovascular Disease Associated microRNAs in Children Descending from Pregnancy Complicated by Gestational Diabetes Mellitus
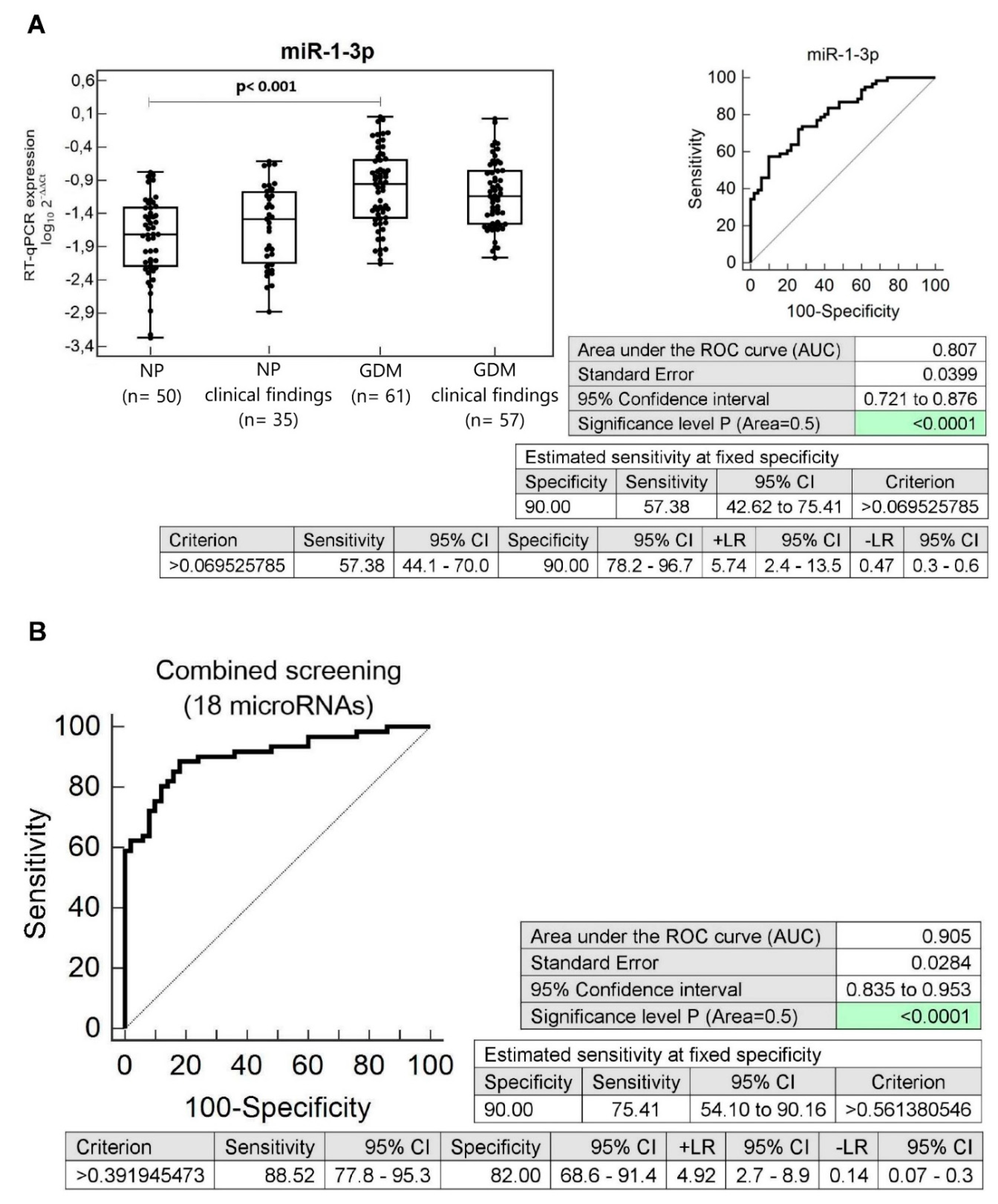
3.5. Dysregulation of Multiple microRNAs in Children Descending from GDM Complicated Pregnancies with Normal Clinical Findings (Normal BMI, Blood Pressure, and Echocardiogram Findings)
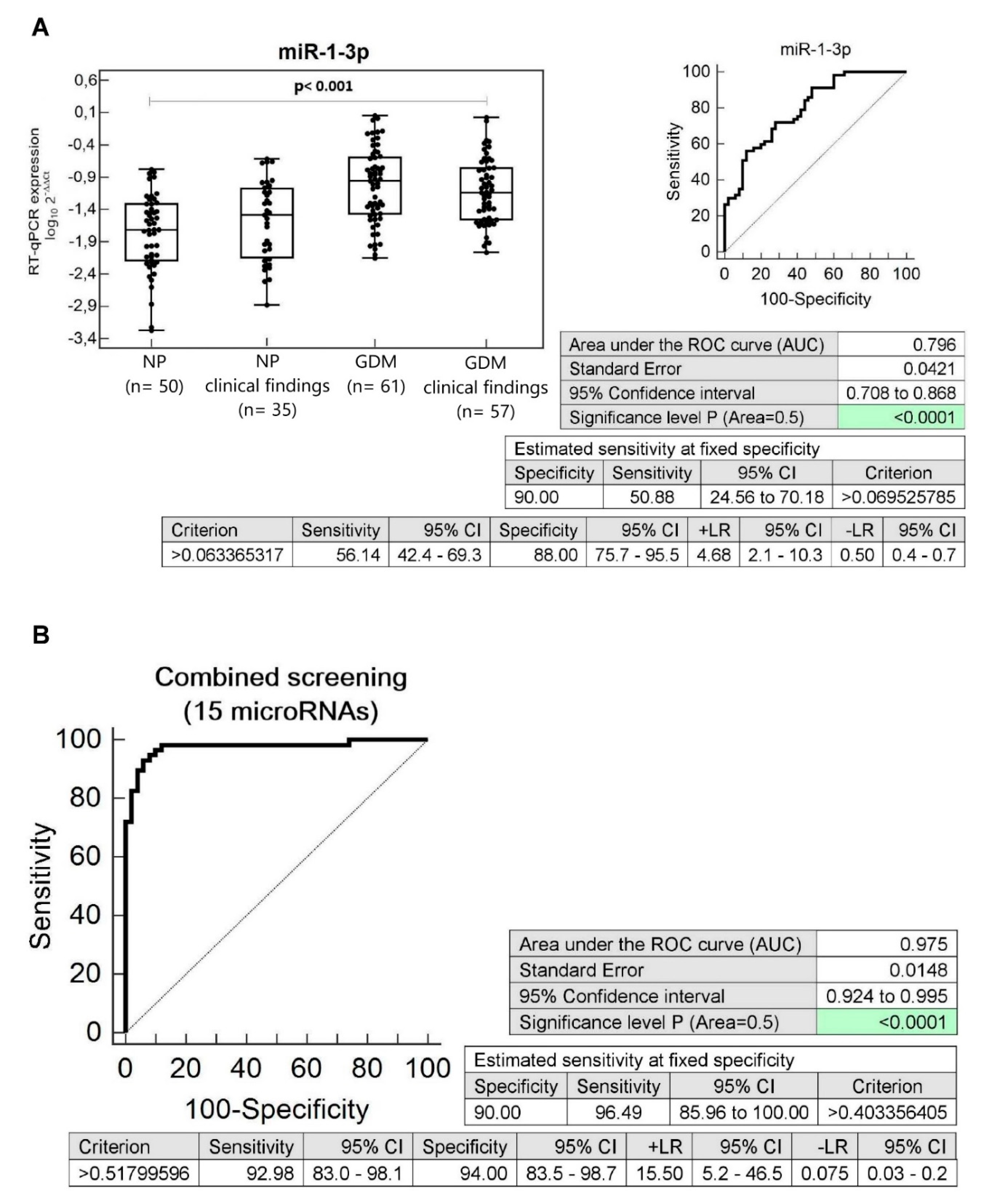
3.6. Dysregulation of Multiple microRNAs in Children Descending from GDM Complicated Pregnancies with Abnormal Clinical Findings (Abnormal BMI, Blood Pressure, and Echocardiogram Findings)
3.7. Information on microRNA-Target Interactions on Disease Ontologies, Human Phenotype Ontologies, and OMIM Disorders
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GDM | Gestational Diabetes Mellitus |
| PE | Preeclampsia |
| FGR | Fetal growth restriction |
| GH | Gestational hypertension |
| AUC | Area under the Curve |
| ROC | Receive Operating Characteristic |
| FPR | False Positive Rate |
| CI | Confidence Interval |
| LR+ | Positive Likelihood Ratio |
| LR- | Negative Likelihood Ratio |
| NP | Normal Pregnancies |
| BP | Blood Pressure |
| OGTT | Oral glucose tolerance test |
| IADPSG | International Association of Diabetes and Pregnancy Study Groups |
| BMI | Body Mass Index |
| SBP | Systolic Blood Pressure |
| DBP | Diastolic Blood Pressure |
| LDL | Low-density Lipoprotein |
| GA | Gestational age |
| CS | Caesarean Section |
| EDTA | Ethylenediaminetetraacetic Acid |
| NTC | No template control |
| NAC | No amplification control |
References
- American Diabetes Association. Gestational diabetes mellitus. Diabetes Care 2004, 27, S88–S90. [Google Scholar] [CrossRef] [PubMed]
- International Diabetes Federation. IDF Diabetes Atlas–Across the Globe. 2017. Available online: http://diabetesatlas.org/across-the-globe.html (accessed on 12 April 2020).
- Bowes, S.B.; Hennessy, T.R.; Umpleby, A.M.; Benn, J.J.; Jackson, N.C.; Boroujerdi, M.A.; Sönksen, P.H.; Lowy, C. Measurement of glucose metabolism and insulin secretion during normal pregnancy and pregnancy complicated by gestational diabetes. Diabetologia 1996, 39, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Di Cianni, G.; Miccoli, R.; Volpe, L.; Lencioni, C.; Del Prato, S. Intermediate metabolism in normal pregnancy and in gestational diabetes. Diabetes Metab. Res. Rev. 2003, 19, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Barbour, L.A.; McCurdy, C.E.; Hernandez, T.L.; Kirwan, J.P.; Catalano, P.M.; Friedman, J.E. Cellular mechanisms for insulin resistance in normal pregnancy and gestational diabetes. Diabetes Care 2007, 30, S112–S119, Corrected in Diabetes Care 2007, 30, 3154. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.Y.; Li, G.M.; Dong, Q.; Peng, H. MiR-577 inhibits pancreatic β-cell function and survival by targeting fibroblast growth factor 21 (FGF-21) in pediatric diabetes. Genet. Mol. Res. 2015, 14, 15462–15470. [Google Scholar] [CrossRef]
- Dias, S.; Pheiffer, C.; Abrahams, Y.; Rheeder, P.; Adam, S. Molecular Biomarkers for Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2018, 19, 2926. [Google Scholar] [CrossRef]
- Zhao, C.; Dong, J.; Jiang, T.; Shi, Z.; Yu, B.; Zhu, Y.; Chen, D.; Xu, J.; Huo, R.; Dai, J.; et al. Early Second-Trimester Serum miRNA Profiling Predicts Gestational Diabetes Mellitus. PLoS ONE 2011, 6, e23925. [Google Scholar] [CrossRef]
- Zhu, Y.; Tian, F.; Li, H.; Zhou, Y.; Lu, J.; Ge, Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int. J. Gynaecol. Obstet. 2015, 130, 49–53. [Google Scholar] [CrossRef]
- Tryggestad, J.B.; Vishwanath, A.; Jiang, S.; Mallappa, A.; Teague, A.M.; Takahashi, Y.; Thompson, D.M.; Chernausek, S.D. Influence of gestational diabetes mellitus on human umbilical vein endothelial cell miRNA. Clin. Sci. 2016, 130, 1955–1967. [Google Scholar] [CrossRef]
- Poirier, C.; Desgagné, V.; Guérin, R.; Bouchard, L. MicroRNAs in Pregnancy and Gestational Diabetes Mellitus: Emerging Role in Maternal Metabolic Regulation. Curr. Diabetes Rep. 2017, 17, 35. [Google Scholar] [CrossRef]
- Wander, P.L.; Boyko, E.J.; Hevner, K.; Parikh, V.J.; Tadesse, M.G.; Sorensen, T.K.; Williams, M.A.; Enquobahrie, D.A. Circulating early- and mid-pregnancy microRNAs and risk of gestational diabetes. Diabetes Res. Clin. Pract. 2017, 132, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.L.; Jia, Y.J.; Xing, B.H.; Shi, D.D.; Dong, X.J. Plasma microRNA-16-5p, -17-5p and -20a-5p: Novel diagnostic biomarkers for gestational diabetes mellitus. J. Obstet. Gynaecol. Res. 2017, 43, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, G.; Guarino, E.; Grieco, G.E.; Formichi, C.; Poggi, C.D.; Ceccarelli, E.; Dotta, F. Circulating microRNA (miRNA) Expression Profiling in Plasma of Patients with Gestational Diabetes Mellitus Reveals Upregulation of miRNA miR-330-3p. Front. Endocrinol. 2017, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Stirm, L.; Huypens, P.; Sass, S.; Batra, R.; Fritsche, L.; Brucker, S.; Abele, H.; Hennige, A.M.; Theis, F.; Beckers, J.; et al. Maternal Whole Blood Cell miRNA-340 Is Elevated in Gestational Diabetes and Inversely Regulated by Glucose and Insulin. Sci. Rep. 2018, 8, 1366. [Google Scholar] [CrossRef]
- He, Y.; Bai, J.; Liu, P.; Dong, J.; Tang, Y.; Zhou, J.; Han, P.; Xing, J.; Chen, Y.; Yu, X. miR-494 Protects Pancreatic β-cell Function by Targeting PTEN in Gestational Diabetes Mellitus. EXCLI J. 2017, 16, 1297–1307. [Google Scholar]
- Pheiffer, C.; Dias, S.; Rheeder, P.; Adam, S. Decreased Expression of Circulating miR-20a-5p in South African Women with Gestational Diabetes Mellitus. Mol. Diagn. Ther. 2018, 22, 345–352. [Google Scholar] [CrossRef]
- Tagoma, A.; Alnek, K.; Kirss, A.; Uibo, R.; Haller-Kikkatalo, K. MicroRNA profiling of second trimester maternal plasma shows upregulation of miR-195-5p in patients with gestational diabetes. Gene 2018, 672, 137–142. [Google Scholar] [CrossRef]
- Lamadrid-Romero, M.; Solís, K.H.; Cruz-Reséndiz, M.S.; Pérez, J.E.; Díaz, N.F.; Flores-Herrera, H.; García-López, G.; Perichart, O.; Reyes-Muñoz, E.; Arenas-Huertero, F.; et al. Central nervous system development-related microRNAs levels increase in the serum of gestational diabetic women during the first trimester of pregnancy. Neurosci. Res. 2018, 130, 8–22. [Google Scholar] [CrossRef]
- Hocaoglu, M.; Demirer, S.; Senturk, H.; Turgut, A.; Komurcu-Bayrak, E. Differential expression of candidate circulating microRNAs in maternal blood leukocytes of the patients with preeclampsia and gestational diabetes mellitus. Pregnancy Hypertens. 2019, 17, 5–11. [Google Scholar] [CrossRef]
- Yoffe, L.; Polsky, A.; Gilam, A.; Raff, C.; Mecacci, F.; Ognibene, A.; Crispi, F.; Gratacós, E.; Kanety, H.; Mazaki-Tovi, S.; et al. Early Diagnosis of Gestational Diabetes Mellitus Using Circulating microRNAs. Eur. J. Endocrinol. 2019, 181, 565–577. [Google Scholar] [CrossRef]
- Kawasaki, M.; Arata, N.; Miyazaki, C.; Mori, R.; Kikuchi, T.; Ogawa, Y.; Ota, E. Obesity and abnormal glucose tolerance in offspring of diabetic mothers: A systematic review and meta-analysis. PLoS ONE 2018, 13, e0190676. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Vargas, L.; Addison, S.S.; Nistala, R.; Kurukulasuriya, D.; Sowers, J.R. Gestational Diabetes and the Offspring: Implications in the Development of the Cardiorenal Metabolic Syndrome in Offspring. Cardiorenal Med. 2012, 2, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, N.M.; Visser, G.H.A.; van Rossem, L.; Biesma, D.H.; Wit, J.M.; de Valk, H.W. Long-term BMI and growth profiles in offspring of women with gestational diabetes. Diabetologia 2018, 61, 1037–1045. [Google Scholar] [CrossRef]
- Bianco, M.E.; Josefson, J.L. Hyperglycemia During Pregnancy and Long-Term Offspring Outcomes. Curr. Diabetes Rep. 2020, 19, 143. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Jang, H.C.; Park, H.K.; Cho, N.H. Early manifestation of cardiovascular disease risk factors in offspring of mothers with previous history of gestational diabetes mellitus. Diabetes Res. Clin. Pract. 2007, 78, 238–245. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, S.; Li, W.; Leng, J.; Wang, L.; Liu, H.; Li, W.; Zhang, C.; Qi, L.; Tuomilehto, J.; et al. Maternal Gestational Diabetes Is Associated with Offspring’s Hypertension. Am. J. Hypertens. 2019, 32, 335–342. [Google Scholar] [CrossRef]
- Wright, C.S.; Rifas-Shiman, S.L.; Rich-Edwards, J.W.; Taveras, E.M.; Gillman, M.W.; Oken, E. Intrauterine exposure to gestational diabetes, child adiposity, and blood pressure. Am. J. Hypertens. 2009, 22, 215–220. [Google Scholar] [CrossRef]
- Tam, W.H.; Ma, R.C.W.; Ozaki, R.; Li, A.M.; Chan, M.H.M.; Yuen, L.Y.; Lao, T.T.H.; Yang, X.; Ho, C.S.; Tutino, G.E.; et al. In Utero Exposure to Maternal Hyperglycemia Increases Childhood Cardiometabolic Risk in Offspring. Diabetes Care 2017, 40, 679–686. [Google Scholar] [CrossRef]
- Lowe, W.L., Jr.; Scholtens, D.M.; Kuang, A.; Linder, B.; Lawrence, J.M.; Lebenthal, Y.; McCance, D.; Hamilton, J.; Nodzenski, M.; Talbot, O.; et al. HAPO Follow-up Study Cooperative Research Group. Hyperglycemia and Adverse Pregnancy Outcome Follow-up Study (HAPO FUS): Maternal Gestational Diabetes Mellitus and Childhood Glucose Metabolism. Diabetes Care 2019, 42, 372–380. [Google Scholar] [CrossRef]
- Tsadok, M.A.; Friedlander, Y.; Paltiel, O.; Manor, O.; Meiner, V.; Hochner, H.; Sagy, Y.; Sharon, N.; Yazdgerdi, S.; Siscovick, D.; et al. Obesity and blood pressure in 17-year-old offspring of mothers with gestational diabetes: Insights from the Jerusalem Perinatal Study. Exp. Diabetes Res. 2011, 2011, 906154. [Google Scholar]
- Perng, W.; Hockett, C.W.; Sauder, K.A.; Dabelea, D. In utero exposure to gestational diabetes mellitus and cardiovascular risk factors in youth: A longitudinal analysis in the EPOCH cohort. Pediatr. Obes. 2020, e12611. [Google Scholar] [CrossRef]
- Leybovitz-Haleluya, N.; Wainstock, T.; Landau, D.; Sheiner, E. Maternal gestational diabetes mellitus and the risk of subsequent pediatric cardiovascular diseases of the offspring: A population-based cohort study with up to 18 years of follow up. Acta Diabetol. 2018, 55, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Arah, O.A.; Liew, Z.; Cnattingius, S.; Olsen, J.; Sørensen, H.T.; Qin, G.; Li, J. Maternal diabetes during pregnancy and early onset of cardiovascular disease in offspring: Population based cohort study with 40 years of follow-up. BMJ 2019, 367, l6398. [Google Scholar] [CrossRef]
- Franks, P.W.; Looker, H.C.; Kobes, S.; Touger, L.; Tataranni, P.A.; Hanson, R.L.; Knowler, W.C. Gestational glucose tolerance and risk of type 2 diabetes in young Pima Indian offspring. Diabetes 2006, 55, 460–465. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Clausen, T.D.; Mathiesen, E.R.; Hansen, T.; Pedersen, O.; Jensen, D.M.; Lauenborg, J.; Damm, P. High prevalence of type 2 diabetes and pre-diabetes in adult offspring of women with gestational diabetes mellitus or type 1 diabetes: The role of intrauterine hyperglycemia. Diabetes Care 2008, 31, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, J.M.; Garrett, A.J.; Schneider, L.A.; Hodyl, N.A.; Goldsworthy, M.R.; Coat, S.; Rowan, J.A.; Hague, W.M.; Pitcher, J.B. Reduced Cortical Excitability, Neuroplasticity, and Salivary Cortisol in 11-13-Year-Old Children Born to Women with Gestational Diabetes Mellitus. EBioMedicine 2018, 31, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Xiang, A.H.; Wang, X.; Martinez, M.P.; Getahun, D.; Page, K.A.; Buchanan, T.A.; Feldman, K. Maternal Gestational Diabetes Mellitus, Type 1 Diabetes, and Type 2 Diabetes During Pregnancy and Risk of ADHD in Offspring. Diabetes Care 2018, 41, 2502–2508. [Google Scholar] [CrossRef]
- Nomura, Y.; Marks, D.J.; Grossman, B.; Yoon, M.; Loudon, H.; Stone, J.; Halperin, J.M. Exposure to gestational diabetes mellitus and low socioeconomic status: Effects on neurocognitive development and risk of attention-deficit/hyperactivity disorder in offspring. Arch. Pediatr. Adolesc. Med. 2012, 166, 337–343. [Google Scholar]
- Xiang, A.H.; Wang, X.; Martinez, M.P.; Walthall, J.C.; Curry, E.S.; Page, K.; Buchanan, T.A.; Coleman, K.J.; Getahun, D. Association of maternal diabetes with autism in offspring. JAMA 2015, 313, 1425–1434. [Google Scholar] [CrossRef]
- Xu, G.; Jing, J.; Bowers, K.; Liu, B.; Bao, W. Maternal diabetes and the risk of autism spectrum disorders in the offspring: A systematic review and meta-analysis. J. Autism Dev. Disord. 2014, 44, 766–775. [Google Scholar] [CrossRef]
- Ornoy, A.; Wolf, A.; Ratzon, N.; Greenbaum, C.; Dulitzky, M. Neurodevelopmental outcome at early school age of children born to mothers with gestational diabetes. Arch. Dis. Child. Fetal Neonatal Ed. 1999, 81, F10–F14. [Google Scholar] [CrossRef] [PubMed]
- Nahum Sacks, K.; Friger, M.; Shoham-Vardi, I.; Abokaf, H.; Spiegel, E.; Sergienko, R.; Landau, D.; Sheiner, E. Prenatal exposure to gestational diabetes mellitus as an independent risk factor for long-term neuropsychiatric morbidity of the offspring. Am. J. Obstet. Gynecol. 2016, 215, 380e1–380e7. [Google Scholar] [CrossRef] [PubMed]
- Walter, E.; Tsumi, E.; Wainstock, T.; Spiegel, E.; Sheiner, E. Maternal gestational diabetes mellitus: Is it associated with long-term pediatric ophthalmic morbidity of the offspring? J. Maternal Fetal Neonatal Med. 2019, 32, 2529–2538. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.P.; Lin, J.; Chow, T.; Chung, J.; Wang, X.; Xiang, A.H. Maternal Gestational Diabetes and Type 2 Diabetes During Pregnancy and Risk of Childhood Asthma in Offspring. J. Pediatr. 2020, 219, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dong, X.; Wang, Z.; Wu, J. MicroRNA-1 in Cardiac Diseases and Cancers. Korean J. Physiol. Pharmacol. 2014, 18, 359–363. [Google Scholar] [CrossRef]
- Li, Y.Q.; Zhang, M.F.; Wen, H.Y.; Hu, C.L.; Liu, R.; Wei, H.Y.; Ai, C.M.; Wang, G.; Liao, X.X.; Li, X. Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics 2013, 68, 75–80. [Google Scholar] [CrossRef]
- Gasiulė, S.; Stankevičius, V.; Patamsytė, V.; Ražanskas, R.; Zukovas, G.; Kapustina, Z.; Zaliaduonytė, D.; Benetis, R.; Lesauskaitė, V.; Vilkaitis, G. Tissue-Specific miRNAs Regulate the Development of Thoracic Aortic Aneurysm: The Emerging Role of KLF4 Network. J. Clin. Med. 2019, 8, 1609. [Google Scholar] [CrossRef]
- Gerlinger-Romero, F.; Yonamine, C.Y.; Junior, D.C.; Esteves, J.V.; Machado, U.F. Dysregulation between TRIM63/FBXO32 expression and soleus muscle wasting in diabetic rats: Potential role of miR-1-3p, -29a/b-3p, and -133a/b-3p. Mol. Cell Biochem. 2017, 427, 187–199. [Google Scholar] [CrossRef]
- Kokkinopoulou, I.; Maratou, E.; Mitrou, P.; Boutati, E.; Sideris, D.C.; Fragoulis, E.G.; Christodoulou, M.I. Decreased expression of microRNAs targeting type-2 diabetes susceptibility genes in peripheral blood of patients and predisposed individuals. Endocrine 2019, 66, 226–239. [Google Scholar] [CrossRef]
- Hromadnikova, I.; Kotlabova, K.; Dvorakova, L.; Krofta, L. Evaluation of Vascular Endothelial Function in Young and Middle-Aged Women with Respect to a History of Pregnancy, Pregnancy-Related Complications, Classical Cardiovascular Risk Factors, and Epigenetics. Int. J. Mol. Sci. 2020, 21, 430. [Google Scholar] [CrossRef]
- Wang, X.; Shang, Y.; Dai, S.; Wu, W.; Yi, F.; Cheng, L. MicroRNA-16-5p aggravates myocardial infarction injury by targeting expression of insulin receptor substrates 1 and mediating myocardial apoptosis and angiogenesis. Curr. Neurovascular Res. 2020, 17, 11–17. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.F.; Neylon, A.; McGorrian, C.; Blake, G.J. miRNA-93-5p and other miRNAs as predictors of coronary artery disease and STEMI. Int. J. Cardiol. 2016, 224, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Vegter, E.L.; Schmitter, D.; Hagemeijer, Y.; Ovchinnikova, E.S.; van der Harst, P.; Teerlink, J.R.; O’Connor, C.M.; Metra, M.; Davison, B.A.; Bloomfield, D.; et al. Use of biomarkers to establish potential role and function of circulating microRNAs in acute heart failure. Int. J. Cardiol. 2016, 224, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Gacoń, J.; Badacz, R.; Stępień, E.; Karch, I.; Enguita, F.J.; Żmudka, K.; Przewłocki, T.; Kabłak-Ziembicka, A. Diagnostic and prognostic micro-RNAs in ischaemic stroke due to carotid artery stenosis and in acute coronary syndrome: A four-year prospective study. Kardiol. Polska 2018, 76, 362–369. [Google Scholar] [CrossRef]
- Duan, Y.R.; Chen, B.P.; Chen, F.; Yang, S.X.; Zhu, C.Y.; Ma, Y.L.; Li, Y.; Shi, J. Exosomal microRNA-16-5p from human urine-derived stem cells ameliorates diabetic nephropathy through protection of podocyte. J. Cell Mol. Med. 2019. [Google Scholar] [CrossRef]
- Assmann, T.S.; Recamonde-Mendoza, M.; Costa, A.R.; Puñales, M.; Tschiedel, B.; Canani, L.H.; Bauer, A.C.; Crispim, D. Circulating miRNAs in diabetic kidney disease: Case-control study and in silico analyses. Acta Diabetol. 2019, 56, 55–65. [Google Scholar] [CrossRef]
- Alicka, M.; Major, P.; Wysocki, M.; Marycz, K. Adipose-Derived Mesenchymal Stem Cells Isolated from Patients with Type 2 Diabetes Show Reduced “Stemness” through an Altered Secretome Profile, Impaired Anti-Oxidative Protection, and Mitochondrial Dynamics Deterioration. J. Clin. Med. 2019, 8, 765. [Google Scholar] [CrossRef]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef]
- Zhou, L.; Qi, R.Q.; Liu, M.; Xu, Y.P.; Li, G.; Weiland, M.; Kaplan, D.H.; Mi, Q.S. microRNA miR-17-92 cluster is highly expressed in epidermal Langerhans cells but not required for its development. Genes Immun. 2014, 15, 57–61. [Google Scholar] [CrossRef]
- Danielson, L.S.; Park, D.S.; Rotllan, N.; Chamorro-Jorganes, A.; Guijarro, M.V.; Fernandez-Hernando, C.; Fishman, G.I.; Phoon, C.K.; Hernando, E. Cardiovascular dysregulation of miR-17-92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2013, 27, 1460–1467. [Google Scholar] [CrossRef]
- Du, W.; Pan, Z.; Chen, X.; Wang, L.; Zhang, Y.; Li, S.; Liang, H.; Xu, C.; Zhang, Y.; Wu, Y.; et al. By targeting Stat3 microRNA-17-5p promotes cardiomyocyte apoptosis in response to ischemia followed by reperfusion. Cell Physiol. Biochem. 2014, 34, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Kaucsár, T.; Révész, C.; Godó, M.; Krenács, T.; Albert, M.; Szalay, C.I.; Rosivall, L.; Benyó, Z.; Bátkai, S.; Thum, T.; et al. Activation of the miR-17 family and miR-21 during murine kidney ischemia-reperfusion injury. Nucleic Acid Ther. 2013, 23, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Ellims, A.H.; Moore, X.L.; White, D.A.; Taylor, A.J.; Chin-Dusting, J.; Dart, A.M. Circulating microRNAs as biomarkers for diffuse myocardial fibrosis in patients with hypertrophic cardiomyopathy. J. Transl. Med. 2015, 13, 314. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Du, K.; Lu, X. Elevated expressions of serum miR-15a, miR-16, and miR-17-5p are associated with acute ischemic stroke. Int. J. Clin. Exp. Med. 2015, 8, 21071–21079. [Google Scholar] [PubMed]
- Chen, J.; Xu, L.; Hu, Q.; Yang, S.; Zhang, B.; Jiang, H. MiR-17-5p as circulating biomarkers for the severity of coronary atherosclerosis in coronary artery disease. Int. J. Cardiol. 2015, 197, 123–124. [Google Scholar] [CrossRef]
- Tian, L.; Song, Z.; Shao, W.; Du, W.W.; Zhao, L.R.; Zeng, K.; Yang, B.B.; Jin, T. Curcumin represses mouse 3T3-L1 cell adipogenic differentiation via inhibiting miR-17-5p and stimulating the Wnt signalling pathway effector Tcf7l2. Cell Death Dis. 2017, 8, e2559. [Google Scholar] [CrossRef]
- Chen, T.C.; Sung, M.L.; Kuo, H.C.; Chien, S.J.; Yen, C.K.; Chen, C.N. Differential regulation of human aortic smooth muscle cell proliferation by monocyte-derived macrophages from diabetic patients. PLoS ONE 2014, 9, e113752. [Google Scholar] [CrossRef]
- Mendell, J.T. miRiad roles for the miR-17-92 cluster in development and disease. Cell 2008, 133, 217–222. [Google Scholar] [CrossRef]
- Brock, M.; Samillan, V.J.; Trenkmann, M.; Schwarzwald, C.; Ulrich, S.; Gay, R.E.; Gassmann, M.; Ostergaard, L.; Gay, S.; Speich, R.; et al. AntagomiR directed against miR-20a restores functional BMPR2 signalling and prevents vascular remodelling in hypoxia-induced pulmonary hypertension. Eur. Heart J. 2014, 35, 3203–3211. [Google Scholar] [CrossRef]
- Platania, C.B.M.; Maisto, R.; Trotta, M.C.; D’Amico, M.; Rossi, S.; Gesualdo, C.; D’Amico, G.; Balta, C.; Herman, H.; Hermenean, A.; et al. Retinal and circulating miRNA expression patterns in diabetic retinopathy: An in silico and in vivo approach. Br. J. Pharmacol. 2019, 176, 2179–2194. [Google Scholar]
- Lareyre, F.; Clément, M.; Moratal, C.; Loyer, X.; Jean-Baptiste, E.; Hassen-Khodja, R.; Chinetti, G.; Mallat, Z.; Raffort, J. Differential micro-RNA expression in diabetic patients with abdominal aortic aneurysm. Biochimie 2019, 162, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.A.; Semus, H.M.; Montgomery, R.L.; Stack, C.; Latimer, P.A.; Lewton, S.M.; Lynch, J.M.; Hullinger, T.G.; Seto, A.G.; van Rooij, E. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur. J. Heart Fail. 2013, 15, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Flowers, E.; Aouizerat, B.E.; Abbasi, F.; Lamendola, C.; Grove, K.M.; Fukuoka, Y.; Reaven, G.M. Circulating microRNA-320a and microRNA-486 predict thiazolidinedione response: Moving towards precision health for diabetes prevention. Metabolism 2015, 64, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Katayama, M.; Wiklander, O.P.B.; Fritz, T.; Caidahl, K.; El-Andaloussi, S.; Zierath, J.R.; Krook, A. Circulating Exosomal miR-20b-5p Is Elevated in Type 2 Diabetes and Could Impair Insulin Action in Human Skeletal Muscle. Diabetes 2019, 68, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Chen, L.; Yan, C.; Zhou, W.; Endo, Y.; Liu, J.; Hu, L.; Hu, Y.; Mi, B.; Liu, G. Circulating Exosomal miR-20b-5p Inhibition Restores Wnt9b Signaling and Reverses Diabetes-Associated Impaired Wound Healing. Small 2020, 16, e1904044. [Google Scholar] [CrossRef]
- Zhu, K.; Hu, X.; Chen, H.; Li, F.; Yin, N.; Liu, A.L.; Shan, K.; Qin, Y.W.; Huang, X.; Chang, Q.; et al. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. EBioMedicine 2019, 49, 341–353. [Google Scholar] [CrossRef]
- Sekar, D.; Venugopal, B.; Sekar, P.; Ramalingam, K. Role of microRNA 21 in diabetes and associated/related diseases. Gene 2016, 582, 14–18. [Google Scholar] [CrossRef]
- Suárez, Y.; Fernández-Hernando, C.; Pober, J.S.; Sessa, W.C. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007, 100, 1164–1173. [Google Scholar] [CrossRef]
- Dong, S.; Ma, W.; Hao, B.; Hu, F.; Yan, L.; Yan, X.; Wang, Y.; Chen, Z.; Wang, Z. microRNA-21 promotes cardiac fibrosis and development of heart failure with preserved left ventricular ejection fraction by up-regulating Bcl-2. Int. J. Clin. Exp. Pathol. 2014, 7, 565–574. [Google Scholar]
- Zhang, J.; Xing, Q.; Zhou, X.; Li, J.; Li, Y.; Zhang, L.; Zhou, Q.; Tang, B. Circulating miRNA 21 is a promising biomarker for heart failure. Mol. Med. Rep. 2017, 16, 7766–7774. [Google Scholar] [CrossRef]
- Licholai, S.; Blaż, M.; Kapelak, B.; Sanak, M. Unbiased Profile of MicroRNA Expression in Ascending Aortic Aneurysm Tissue Appoints Molecular Pathways Contributing to the Pathology. Ann. Thorac. Surg. 2016, 102, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Kriegel, A.J.; Baker, M.A.; Liu, Y.; Liu, P.; Cowley, A.W., Jr.; Liang, M. Endogenous microRNAs in human microvascular endothelial cells regulate mRNAs encoded by hypertension-related genes. Hypertension 2015, 66, 793–799. [Google Scholar] [CrossRef]
- Velle-Forbord, T.; Eidlaug, M.; Debik, J.; Sæther, J.C.; Follestad, T.; Nauman, J.; Gigante, B.; Røsjø, H.; Omland, T.; Langaas, M.; et al. Circulating microRNAs as predictive biomarkers of myocardial infarction: Evidence from the HUNT study. Atherosclerosis 2019, 289, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Demirsoy, İ.H.; Ertural, D.Y.; Balci, Ş.; Çınkır, Ü.; Sezer, K.; Tamer, L.; Aras, N. Profiles of Circulating MiRNAs Following Metformin Treatment in Patients with Type 2 Diabetes. J. Med. Biochem. 2018, 37, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Spazzafumo, L.; Bonafè, M.; Recchioni, R.; Prattichizzo, F.; Marcheselli, F.; Micolucci, L.; Mensà, E.; Giuliani, A.; Santini, G.; et al. MiR-21-5p and miR-126a-3p levels in plasma and circulating angiogenic cells: Relationship with type 2 diabetes complications. Oncotarget 2015, 6, 35372–35382. [Google Scholar] [CrossRef]
- Assmann, T.S.; Recamonde-Mendoza, M.; De Souza, B.M.; Crispim, D. MicroRNA expression profiles and type 1 diabetes mellitus: Systematic review and bioinformatic analysis. Endocr. Connect. 2017, 6, 773–790. [Google Scholar] [CrossRef]
- Lakhter, A.J.; Pratt, R.E.; Moore, R.E.; Doucette, K.K.; Maier, B.F.; DiMeglio, L.A.; Sims, E.K. Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia 2018, 61, 1124–1134. [Google Scholar] [CrossRef]
- Grieco, G.E.; Cataldo, D.; Ceccarelli, E.; Nigi, L.; Catalano, G.; Brusco, N.; Mancarella, F.; Ventriglia, G.; Fondelli, C.; Guarino, E.; et al. Serum Levels of miR-148a and miR-21-5p Are Increased in Type 1 Diabetic Patients and Correlated with Markers of Bone Strength and Metabolism. Noncoding RNA 2018, 4, 37. [Google Scholar] [CrossRef]
- Gholaminejad, A.; Abdul Tehrani, H.; Gholami Fesharaki, M. Identification of candidate microRNA biomarkers in diabetic nephropathy: A meta-analysis of profiling studies. J. Nephrol. 2018, 31, 813–831. [Google Scholar] [CrossRef]
- Long, B.; Gan, T.Y.; Zhang, R.C.; Zhang, Y.H. miR-23a Regulates Cardiomyocyte Apoptosis by Targeting Manganese Superoxide Dismutase. Mol. Cells 2017, 40, 542–549. [Google Scholar] [CrossRef]
- Wang, S.; He, W.; Wang, C. MiR-23a Regulates the Vasculogenesis of Coronary Artery Disease by Targeting Epidermal Growth Factor Receptor. Cardiovasc. Ther. 2016, 34, 199–208. [Google Scholar] [CrossRef]
- Cong, X.; Li, Y.; Lu, N.; Dai, Y.; Zhang, H.; Zhao, X.; Liu, Y. Resveratrol attenuates the inflammatory reaction induced by ischemia/reperfusion in the rat heart. Mol. Med. Rep. 2014, 9, 2528–2532. [Google Scholar] [CrossRef] [PubMed]
- Černá, V.; Ostašov, P.; Pitule, P.; Moláček, J.; Třeška, V.; Pešta, M. The Expression Profile of MicroRNAs in Small and Large Abdominal Aortic Aneurysms. Cardiol. Res. Pract. 2019, 2019, 8645840. [Google Scholar] [CrossRef]
- Lozano-Bartolomé, J.; Llauradó, G.; Portero-Otin, M.; Altuna-Coy, A.; Rojo-Martínez, G.; Vendrell, J.; Jorba, R.; Rodríguez-Gallego, E.; Chacón, M.R. Altered Expression of miR-181a-5p and miR-23a-3p Is Associated with Obesity and TNFα-Induced Insulin Resistance. J. Clin. Endocrinol. Metab. 2018, 103, 1447–1458. [Google Scholar] [CrossRef] [PubMed]
- Dolz, S.; Górriz, D.; Tembl, J.I.; Sánchez, D.; Fortea, G.; Parkhutik, V.; Lago, A. Circulating MicroRNAs as Novel Biomarkers of Stenosis Progression in Asymptomatic Carotid Stenosis. Stroke 2017, 48, 10–16. [Google Scholar] [CrossRef] [PubMed]
- De Gonzalo-Calvo, D.; Cenarro, A.; Garlaschelli, K.; Pellegatta, F.; Vilades, D.; Nasarre, L.; Camino-Lopez, S.; Crespo, J.; Carreras, F.; Leta, R.; et al. Translating the microRNA signature of microvesicles derived from human coronary artery smooth muscle cells in patients with familial hypercholesterolemia and coronary artery disease. J. Mol. Cell Cardiol. 2017, 106, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Gecys, D.; Tatarunas, V.; Veikutiene, A.; Lesauskaite, V. New potential modulators of CYP4F2 enzyme activity in angina pectoris: Hsa-miR-24-3p and hsa-miR-34a-5p. Biomarkers 2020, 25, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Onrat, S.T.; Onrat, E.; Ercan Onay, E.; Yalım, Z.; Avşar, A. The Genetic Determination of the Differentiation between Ischemic Dilated Cardiomyopathy and Idiopathic Dilated Cardiomyopathy. Genet. Test. Mol. Biomark. 2018, 22, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Qi, J.; Fan, B.Y.; Zhang, J.; Su, F.F.; Wang, H.T. MicroRNA-24-3p Attenuates Myocardial Ischemia/Reperfusion Injury by Suppressing RIPK1 Expression in Mice. Cell Physiol. Biochem. 2018, 51, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Lu, Z.; Lin, V.; May, A.; Shaw, D.H.; Wang, Z.; Che, B.; Tran, K.; Du, H.; Shaw, P.X. MicroRNA miR-24-3p Reduces Apoptosis and Regulates Keap1-Nrf2 Pathway in Mouse Cardiomyocytes Responding to Ischemia/Reperfusion Injury. Oxid. Med. Cell Longev. 2018, 2018, 7042105. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, Q.G. The role of miR-26 in tumors and normal tissues. Oncol. Lett. 2011, 2, 1019–1023. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Lin, S.; Lv, C. MiR-26a-5p regulates cardiac fibroblasts collagen expression by targeting ULK1. Sci. Rep. 2018, 8, 2104. [Google Scholar] [CrossRef] [PubMed]
- Bye, A.; Røsjø, H.; Nauman, J.; Silva, G.J.; Follestad, T.; Omland, T.; Wisløff, U. Circulating microRNAs predict future fatal myocardial infarction in healthy individuals - The HUNT study. J. Mol. Cell Cardiol. 2016, 97, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Hsu, A.; Chen, S.J.; Chang, Y.S.; Chen, H.C.; Chu, P.H. Systemic approach to identify serum microRNAs as potential biomarkers for acute myocardial infarction. Biomed. Res. Int. 2014, 2014, 418628. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Guo, S.; Zhang, G.; Liu, Y.; Bi, S.; Wang, X.; Lu, Q. miR-26a-5p protects against myocardial ischemia/reperfusion injury by regulating the PTEN/PI3K/AKT signaling pathway. Braz. J. Med. Biol. Res. 2020, 53, e9106. [Google Scholar] [CrossRef] [PubMed]
- Chouvarine, P.; Geldner, J.; Giagnorio, R.; Legchenko, E.; Bertram, H.; Hansmann, G. Trans-Right-Ventricle and Transpulmonary MicroRNA Gradients in Human Pulmonary Arterial Hypertension. Pediatr. Crit. Care Med. 2020, 21, 340–349. [Google Scholar] [CrossRef]
- Garavelli, S.; Bruzzaniti, S.; Tagliabue, E.; Prattichizzo, F.; Di Silvestre, D.; Perna, F.; La Sala, L.; Ceriello, A.; Mozzillo, E.; Fattorusso, V.; et al. Blood Co-Circulating Extracellular microRNAs and Immune Cell Subsets Associate with Type 1 Diabetes Severity. Int. J. Mol. Sci. 2020, 21, 477. [Google Scholar] [CrossRef]
- Ye, Y.; Hu, Z.; Lin, Y.; Zhang, C.; Perez-Polo, J.R. Downregulation of microRNA-29 by antisense inhibitors and a PPAR-gamma agonist protects against myocardial ischaemia-reperfusion injury. Cardiovasc. Res. 2010, 87, 535–544. [Google Scholar] [CrossRef]
- Moraes, L.N.; Fernandez, G.J.; Vechetti-Júnior, I.J.; Freire, P.P.; Souza, R.W.A.; Villacis, R.A.R.; Rogatto, S.R.; Reis, P.P.; Dal-Pai-Silva, M.; Carvalho, R.F. Integration of miRNA and mRNA expression profiles reveals microRNA-regulated networks during muscle wasting in cardiac cachexia. Sci. Rep. 2017, 7, 6998. [Google Scholar] [CrossRef]
- Zhao, Y.; Yuan, Y.; Qiu, C. Underexpression of CACNA1C Caused by Overexpression of microRNA-29a Underlies the Pathogenesis of Atrial Fibrillation. Med. Sci. Monit. 2016, 22, 2175–2181. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Xue, S.; Ding, H.; Wang, Y.; Qi, H.; Wang, Y.; Zhu, W.; Li, P. Clinical significance of circulating microRNAs as diagnostic biomarkers for coronary artery disease. J. Cell Mol. Med. 2020, 24, 1146–1150. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhu, J.; Han, W.; Jiang, X.; Xu, M.; Zhao, Y.; Dong, Q.; Pang, Z.; Guan, Q.; Gao, L.; et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: A clinical study. Acta Diabetol. 2011, 48, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Widlansky, M.E.; Jensen, D.M.; Wang, J.; Liu, Y.; Geurts, A.M.; Kriegel, A.J.; Liu, P.; Ying, R.; Zhang, G.; Casati, M.; et al. miR-29 contributes to normal endothelial function and can restore it in cardiometabolic disorders. EMBO Mol. Med. 2018, 10, e8046. [Google Scholar] [CrossRef] [PubMed]
- Bulent Vatan, M.; Kalaycı Yigin, A.; Akdemir, R.; Tarik Agac, M.; Akif Cakar, M.; Aksoy, M.; Tatli, E.; Kilic, H.; Gunduz, H.; Guzel, D.; et al. Altered Plasma MicroRNA Expression in Patients with Mitral Chordae Tendineae Rupture. J. Heart Valve Dis. 2016, 25, 580–588. [Google Scholar]
- Gumus, G.; Giray, D.; Bobusoglu, O.; Tamer, L.; Karpuz, D.; Hallioglu, O. MicroRNA values in children with rheumatic carditis: A preliminary study. Rheumatol. Int. 2018, 38, 1199–1205. [Google Scholar] [CrossRef]
- Rogg, E.M.; Abplanalp, W.T.; Bischof, C.; John, D.; Schulz, M.H.; Krishnan, J.; Fischer, A.; Poluzzi, C.; Schaefer, L.; Bonauer, A.; et al. Analysis of Cell Type-Specific Effects of MicroRNA-92a Provides Novel Insights into Target Regulation and Mechanism of Action. Circulation 2018, 138, 2545–2558. [Google Scholar] [CrossRef]
- Marques, F.Z.; Vizi, D.; Khammy, O.; Mariani, J.A.; Kaye, D.M. The transcardiac gradient of cardio-microRNAs in the failing heart. Eur. J. Heart Fail. 2016, 18, 1000–1008. [Google Scholar] [CrossRef]
- Liu, Y.; Li, Q.; Hosen, M.R.; Zietzer, A.; Flender, A.; Levermann, P.; Schmitz, T.; Frühwald, D.; Goody, P.; Nickenig, G.; et al. Atherosclerotic Conditions Promote the Packaging of Functional MicroRNA-92a-3p Into Endothelial Microvesicles. Circ. Res. 2019, 124, 575–587. [Google Scholar] [CrossRef]
- Wiese, C.B.; Zhong, J.; Xu, Z.Q.; Zhang, Y.; Ramirez Solano, M.A.; Zhu, W.; Linton, M.F.; Sheng, Q.; Kon, V.; Vickers, K.C. Dual inhibition of endothelial miR-92a-3p and miR-489-3p reduces renal injury-associated atherosclerosis. Atherosclerosis 2019, 282, 121–131. [Google Scholar] [CrossRef]
- Moncini, S.; Salvi, A.; Zuccotti, P.; Viero, G.; Quattrone, A.; Barlati, S.; De Petro, G.; Venturin, M.; Riva, P. The role of miR-103 and miR-107 in regulation of CDK5R1 expression and in cellular migration. PLoS ONE 2011, 6, e20038. [Google Scholar] [CrossRef]
- Huang, L.; Li, L.; Chen, X.; Zhang, H.; Shi, Z. MiR-103a targeting Piezo1 is involved in acute myocardial infarction through regulating endothelium function. Cardiol. J. 2016, 23, 556–562. [Google Scholar] [CrossRef]
- Deng, B.; Du, J.; Hu, R.; Wang, A.P.; Wu, W.H.; Hu, C.P.; Li, Y.J.; Li, X.H. MicroRNA-103/107 is involved in hypoxia-induced proliferation of pulmonary arterial smooth muscle cells by targeting HIF-1β. Life Sci. 2016, 147, 117–124. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Assmann, T.S.; Recamonde-Mendoza, M.; Puñales, M.; Tschiedel, B.; Canani, L.H.; Crispim, D. MicroRNA expression profile in plasma from type 1 diabetic patients: Case-control study and bioinformatic analysis. Diabetes Res. Clin. Pract. 2018, 141, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Shaham, L.; Binder, V.; Gefen, N.; Borkhardt, A.; Izraeli, S. MiR-125 in normal and malignant hematopoiesis. Leukemia 2012, 26, 2011–2018. [Google Scholar] [CrossRef] [PubMed]
- Tiedt, S.; Prestel, M.; Malik, R.; Schieferdecker, N.; Duering, M.; Kautzky, V.; Stoycheva, I.; Böck, J.; Northoff, B.H.; Klein, M.; et al. RNA-Seq Identifies Circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as Potential Biomarkers for Acute Ischemic Stroke. Circ. Res. 2017, 121, 970–980. [Google Scholar] [CrossRef]
- Jia, K.; Shi, P.; Han, X.; Chen, T.; Tang, H.; Wang, J. Diagnostic value of miR-30d-5p and miR-125b-5p in acute myocardial infarction. Mol. Med. Rep. 2016, 14, 184–194. [Google Scholar] [CrossRef]
- Bayoumi, A.S.; Park, K.M.; Wang, Y.; Teoh, J.P.; Aonuma, T.; Tang, Y.; Su, H.; Weintraub, N.L.; Kim, I.M. A carvedilol-responsive microRNA, miR-125b-5p protects the heart from acute myocardial infarction by repressing pro-apoptotic bak1 and klf13 in cardiomyocytes. J. Mol. Cell Cardiol. 2018, 114, 72–82. [Google Scholar] [CrossRef]
- Satake, E.; Pezzolesi, M.G.; Md Dom, Z.I.; Smiles, A.M.; Niewczas, M.A.; Krolewski, A.S. Circulating miRNA Profiles Associated with Hyperglycemia in Patients with Type 1 Diabetes. Diabetes 2018, 67, 1013–1023. [Google Scholar] [CrossRef]
- Samandari, N.; Mirza, A.H.; Kaur, S.; Hougaard, P.; Nielsen, L.B.; Fredheim, S.; Mortensen, H.B.; Pociot, F. Influence of Disease Duration on Circulating Levels of miRNAs in Children and Adolescents with New Onset Type 1 Diabetes. Noncoding RNA 2018, 4, 35. [Google Scholar] [CrossRef]
- Yu, C.Y.; Yang, C.Y.; Rui, Z.L. MicroRNA-125b-5p improves pancreatic β-cell function through inhibiting JNK signaling pathway by targeting DACT1 in mice with type 2 diabetes mellitus. Life Sci. 2019, 224, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.J.; Zhao, Z.F.; Kang, X.J.; Wang, H.J.; Zhao, J.; Pu, X.M. MicroRNA-126-3p suppresses cell proliferation by targeting PIK3R2 in Kaposi’s sarcoma cells. Oncotarget 2016, 7, 36614–36621. [Google Scholar] [CrossRef] [PubMed]
- Matsha, T.E.; Kengne, A.P.; Hector, S.; Mbu, D.L.; Yako, Y.Y.; Erasmus, R.T. MicroRNA profiling and their pathways in South African individuals with prediabetes and newly diagnosed type 2 diabetes mellitus. Oncotarget 2018, 9, 30485–30498. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lan, X.; Wu, L.; Wu, N.; Chen, Q.; Li, Y.; Du, X.; Wei, C.; Feng, L.; Li, Y.; Osoro, E.K.; et al. Long Noncoding RNA lnc-HC Regulates PPARγ-Mediated Hepatic Lipid Metabolism through miR-130b-3p. Mol. Ther. Nucleic Acids 2019, 18, 954–965. [Google Scholar] [CrossRef]
- Zhang, J.; Jazii, F.R.; Haghighi, M.M.; Alvares, D.; Liu, L.; Khosraviani, N.; Adeli, K. miR-130b is a potent stimulator of hepatic very-low-density lipoprotein assembly and secretion via marked induction of microsomal triglyceride transfer protein. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E262–E275. [Google Scholar] [CrossRef]
- Li, P.; Zhang, Q.; Wu, X.; Yang, X.; Zhang, Y.; Li, Y.; Jiang, F. Circulating microRNAs serve as novel biological markers for intracranial aneurysms. J. Am. Heart Assoc. 2014, 3, e000972. [Google Scholar] [CrossRef]
- Tian, C.; Li, Z.; Yang, Z.; Huang, Q.; Liu, J.; Hong, B. Plasma MicroRNA-16 Is a Biomarker for Diagnosis, Stratification, and Prognosis of Hyperacute Cerebral Infarction. PLoS ONE 2016, 11, e0166688. [Google Scholar] [CrossRef]
- Prabu, P.; Rome, S.; Sathishkumar, C.; Aravind, S.; Mahalingam, B.; Shanthirani, C.S.; Gastebois, C.; Villard, A.; Mohan, V.; Balasubramanyam, M. Circulating MiRNAs of ‘Asian Indian Phenotype’ Identified in Subjects with Impaired Glucose Tolerance and Patients with Type 2 Diabetes. PLoS ONE 2015, 10, e0128372. [Google Scholar] [CrossRef]
- Feng, T.; Li, K.; Zheng, P.; Wang, Y.; Lv, Y.; Shen, L.; Chen, Y.; Xue, Z.; Li, B.; Jin, L.; et al. Weighted Gene Coexpression Network Analysis Identified MicroRNA Coexpression Modules and Related Pathways in Type 2 Diabetes Mellitus. Oxid. Med. Cell Longev. 2019, 2019, 9567641. [Google Scholar] [CrossRef]
- Liang, H.W.; Yang, X.; Wen, D.Y.; Gao, L.; Zhang, X.Y.; Ye, Z.H.; Luo, J.; Li, Z.Y.; He, Y.; Pang, Y.Y.; et al. Utility of miR 133a 3p as a diagnostic indicator for hepatocellular carcinoma: An investigation combined with GEO, TCGA, meta analysis and bioinformatics. Mol. Med. Rep. 2018, 17, 1469–1484. [Google Scholar] [CrossRef]
- Van Rooij, E.; Olson, E.N. MicroRNAs: Powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Investig. 2007, 117, 2369–2376. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, R.; Lin, F.; Zhang, S.; Zhang, G.; Hu, S.; Zheng, Z. MicroRNA: Novel regulators involved in the remodeling and reverse remodeling of the heart. Cardiology 2009, 113, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Kukreja, R.C.; Yin, C.; Salloum, F.N. MicroRNAs: New players in cardiac injury and protection. Mol. Pharmacol. 2011, 80, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Duisters, R.F.; Tijsen, A.J.; Schroen, B.; Leenders, J.J.; Lentink, V.; van der Made, I.; Herias, V.; van Leeuwen, R.E.; Schellings, M.W.; Barenbrug, P.; et al. miR-133 and miR-30 regulate connective tissue growth factor: Implications for a role of microRNAs in myocardial matrix remodeling. Circ. Res. 2009, 104, 170–178. [Google Scholar] [CrossRef]
- Liu, W.; Ling, S.; Sun, W.; Liu, T.; Li, Y.; Zhong, G.; Zhao, D.; Zhang, P.; Song, J.; Jin, X.; et al. Circulating microRNAs correlated with the level of coronary artery calcification in symptomatic patients. Sci. Rep. 2015, 5, 16099. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, M.; He, H.; Chen, J.; Zeng, H.; Li, J.; Duan, R. MicroRNA/mRNA profiling and regulatory network of intracranial aneurysm. BMC Med. Genom. 2013, 6, 36. [Google Scholar] [CrossRef]
- Liu, H.; Xiong, W.; Liu, F.; Lin, F.; He, J.; Liu, C.; Lin, Y.; Dong, S. Significant role and mechanism of microRNA-143-3p/KLLN axis in the development of coronary heart disease. Am. J. Transl. Res. 2019, 11, 3610–3619. [Google Scholar]
- Li, C.; Li, J.; Xue, K.; Zhang, J.; Wang, C.; Zhang, Q.; Chen, X.; Gao, C.; Yu, X.; Sun, L. MicroRNA-143-3p promotes human cardiac fibrosis via targeting sprouty3 after myocardial infarction. J. Mol. Cell Cardiol. 2019, 129, 281–292. [Google Scholar] [CrossRef]
- Yu, B.; Zhao, Y.; Zhang, H.; Xie, D.; Nie, W.; Shi, K. Inhibition of microRNA-143-3p attenuates myocardial hypertrophy by inhibiting inflammatory response. Cell Biol. Int. 2018, 42, 1584–1593. [Google Scholar] [CrossRef]
- Jiao, M.; You, H.Z.; Yang, X.Y.; Yuan, H.; Li, Y.L.; Liu, W.X.; Jin, M.; Du, J. Circulating microRNA signature for the diagnosis of childhood dilated cardiomyopathy. Sci. Rep. 2018, 8, 724. [Google Scholar] [CrossRef]
- Deng, L.; Blanco, F.J.; Stevens, H.; Lu, R.; Caudrillier, A.; McBride, M.; McClure, J.D.; Grant, J.; Thomas, M.; Frid, M.; et al. MicroRNA-143 Activation Regulates Smooth Muscle and Endothelial Cell Crosstalk in Pulmonary Arterial Hypertension. Circ. Res. 2015, 117, 870–883. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Tian, C.; Sun, L.; Cao, F.; Meng, Z. The lncRNA TUG1/miR-145-5p/FGF10 regulates proliferation and migration in VSMCs of hypertension. Biochem. Biophys. Res. Commun. 2018, 501, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Niu, X.; Xiao, Y.; Lin, K.; Chen, X. MiRNA expression profiles in healthy OSAHS and OSAHS with arterial hypertension: Potential diagnostic and early warning markers. Respir. Res. 2018, 19, 194. [Google Scholar] [CrossRef] [PubMed]
- Toro, R.; Blasco-Turrión, S.; Morales-Ponce, F.J.; Gonzalez, P.; Martínez-Camblor, P.; López-Granados, A.; Brugada, R.; Campuzano, O.; Pérez-Serra, A.; Rosa Longobardo, F.; et al. Plasma microRNAs as biomarkers for Lamin A/C-related dilated cardiomyopathy. J. Mol. Med. 2018, 96, 845–856. [Google Scholar] [CrossRef]
- Yuan, M.; Zhang, L.; You, F.; Zhou, J.; Ma, Y.; Yang, F.; Tao, L. MiR-145-5p regulates hypoxia-induced inflammatory response and apoptosis in cardiomyocytes by targeting CD40. Mol. Cell Biochem. 2017, 431, 123–131. [Google Scholar] [CrossRef]
- Wu, G.; Tan, J.; Li, J.; Sun, X.; Du, L.; Tao, S. miRNA-145-5p induces apoptosis after ischemia-reperfusion by targeting dual specificity phosphatase 6. J. Cell Physiol. 2019, 234, 16281–16289. [Google Scholar] [CrossRef]
- Xie, X.; Peng, L.; Zhu, J.; Zhou, Y.; Li, L.; Chen, Y.; Yu, S.; Zhao, Y. miR-145-5p/Nurr1/TNF-α Signaling-Induced Microglia Activation Regulates Neuron Injury of Acute Cerebral Ischemic/Reperfusion in Rats. Front. Mol. Neurosci. 2017, 10, 383. [Google Scholar] [CrossRef]
- Nunez Lopez, Y.O.; Retnakaran, R.; Zinman, B.; Pratley, R.E.; Seyhan, A.A. Predicting and understanding the response to short-term intensive insulin therapy in people with early type 2 diabetes. Mol. Metab. 2019, 20, 63–78. [Google Scholar] [CrossRef]
- Zhang, J.; Cui, C.; Xu, H. Downregulation of miR-145-5p elevates retinal ganglion cell survival to delay diabetic retinopathy progress by targeting FGF5. Biosci. Biotechnol. Biochem. 2019, 83, 1655–1662. [Google Scholar] [CrossRef]
- Mona Zamanian, A.; Rezaei-Tavirani, M.; Rezaei-Tavirani, M.; Robati, R.M. Gestational Diabetes Mellitus Regulatory Network Identifies hsa-miR-145-5p and hsa-miR-875-5p as Potential Biomarkers. Int. J. Endocrinol. Metab. 2019, 17, e86640. [Google Scholar]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Paterson, M.R.; Kriegel, A.J. MiR-146a/b: A family with shared seeds and different roots. Physiol. Genom. 2017, 49, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ye, Z.H.; Liang, H.W.; Ren, F.H.; Li, P.; Dang, Y.W.; Chen, G. Down-regulation of miR-146a-5p and its potential targets in hepatocellular carcinoma validated by a TCGA- and GEO-based study. FEBS Open Bio 2017, 7, 504–521. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ha, T.; Liu, L.; Zou, J.; Zhang, X.; Kalbfleisch, J.; Gao, X.; Williams, D.; Li, C. Increased expression of microRNA-146a decreases myocardial ischaemia/reperfusion injury. Cardiovasc. Res. 2013, 97, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.; Ji, Y.; Zhang, C.; Guo, X.; Zhang, Y.; Jia, S.; Ma, W.; Fan, Y.; Wang, C. Circulating MiR-146a May be a Potential Biomarker of Coronary Heart Disease in Patients with Subclinical Hypothyroidism. Cell Physiol. Biochem. 2018, 45, 226–236. [Google Scholar] [CrossRef]
- Li, S.H.; Chen, L.; Pang, X.M.; Su, S.Y.; Zhou, X.; Chen, C.Y.; Huang, L.G.; Li, J.P.; Liu, J.L. Decreased miR-146a expression in acute ischemic stroke directly targets the Fbxl10 mRNA and is involved in modulating apoptosis. Neurochem. Int. 2017, 107, 156–167. [Google Scholar] [CrossRef]
- Barberio, M.D.; Kasselman, L.J.; Playford, M.P.; Epstein, S.B.; Renna, H.A.; Goldberg, M.; DeLeon, J.; Voloshyna, I.; Barlev, A.; Salama, M.; et al. Cholesterol efflux alterations in adolescent obesity: Role of adipose-derived extracellular vesical microRNAs. J. Transl. Med. 2019, 17, 232. [Google Scholar] [CrossRef]
- Gaudet, A.D.; Fonken, L.K.; Gushchina, L.V.; Aubrecht, T.G.; Maurya, S.K.; Periasamy, M.; Nelson, R.J.; Popovich, P.G. miR-155 Deletion in Female Mice Prevents Diet-Induced Obesity. Sci. Rep. 2016, 6, 22862. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, S.Y.; Yang, C.Q.; Ma, B.M.; Jiang, D. MiR-155-5p inhibits the proliferation and migration of VSMCs and HUVECs in atherosclerosis by targeting AKT1. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2223–2233. [Google Scholar]
- Zhu, M.; Wei, Y.; Geißler, C.; Abschlag, K.; Corbalán Campos, J.; Hristov, M.; Möllmann, J.; Lehrke, M.; Karshovska, E.; Schober, A. Hyperlipidemia-Induced MicroRNA-155-5p Improves β-Cell Function by Targeting Mafb. Diabetes 2017, 66, 3072–3084. [Google Scholar] [CrossRef]
- Li, S.; Lee, C.; Song, J.; Lu, C.; Liu, J.; Cui, Y.; Liang, H.; Cao, C.; Zhang, F.; Chen, H. Circulating microRNAs as potential biomarkers for coronary plaque rupture. Oncotarget 2017, 8, 48145–48156. [Google Scholar] [CrossRef] [PubMed]
- Mukai, N.; Nakayama, Y.; Murakami, S.; Tanahashi, T.; Sessler, D.I.; Ishii, S.; Ogawa, S.; Tokuhira, N.; Mizobe, T.; Sawa, T.; et al. Potential contribution of erythrocyte microRNA to secondary erythrocytosis and thrombocytopenia in congenital heart disease. Pediatr. Res. 2018, 83, 866–873. [Google Scholar] [CrossRef] [PubMed]
- Klimczak, D.; Kuch, M.; Pilecki, T.; Żochowska, D.; Wirkowska, A.; Pączek, L. Plasma microRNA-155-5p is increased among patients with chronic kidney disease and nocturnal hypertension. J. Am. Soc. Hypertens. 2017, 11, 831–841.e4. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, L.; Ding, W.; Cai, S.; Zhao, Q. Ablation alleviates atrial fibrillation by regulating the signaling pathways of endothelial nitric oxide synthase/nitric oxide via miR-155-5p and miR-24-3p. J. Cell Biochem. 2019, 120, 4451–4462. [Google Scholar] [CrossRef]
- Sun, X.; Sit, A.; Feinberg, M.W. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc. Med. 2014, 24, 105–112. [Google Scholar] [CrossRef]
- Hulsmans, M.; Sinnaeve, P.; Van der Schueren, B.; Mathieu, C.; Janssens, S.; Holvoet, P. Decreased miR-181a expression in monocytes of obese patients is associated with the occurrence of metabolic syndrome and coronary artery disease. J. Clin. Endocrinol. Metab. 2012, 97, E1213–E1218. [Google Scholar] [CrossRef]
- Du, X.; Yang, Y.; Xu, C.; Peng, Z.; Zhang, M.; Lei, L.; Gao, W.; Dong, Y.; Shi, Z.; Sun, X.; et al. Upregulation of miR-181a impairs hepatic glucose and lipid homeostasis. Oncotarget 2017, 8, 91362–91378. [Google Scholar] [CrossRef]
- Wu, J.; Fan, C.L.; Ma, L.J.; Liu, T.; Wang, C.; Song, J.X.; Lv, Q.S.; Pan, H.; Zhang, C.N.; Wang, J.J. Distinctive expression signatures of serum microRNAs in ischaemic stroke and transient ischaemic attack patients. Thromb. Haemost. 2017, 117, 992–1001. [Google Scholar]
- Zhu, J.; Yao, K.; Wang, Q.; Guo, J.; Shi, H.; Ma, L.; Liu, H.; Gao, W.; Zou, Y.; Ge, J. Circulating miR-181a as a Potential Novel Biomarker for Diagnosis of Acute Myocardial Infarction. Cell Physiol. Biochem. 2016, 40, 1591–1602. [Google Scholar] [CrossRef]
- Nabih, E.S.; Andrawes, N.G. The Association Between Circulating Levels of miRNA-181a and Pancreatic Beta Cells Dysfunction via SMAD7 in Type 1 Diabetic Children and Adolescents. J. Clin. Lab. Anal. 2016, 30, 727–731. [Google Scholar] [CrossRef]
- He, J.F.; Luo, Y.M.; Wan, X.H.; Jiang, D. Biogenesis of MiRNA-195 and its role in biogenesis, the cell cycle, and apoptosis. J. Biochem. Mol. Toxicol. 2011, 25, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Liu, N.; Williams, A.H.; McAnally, J.; Gerard, R.D.; Richardson, J.A.; Olson, E.N. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA 2006, 103, 18255–18260. [Google Scholar] [CrossRef] [PubMed]
- You, X.Y.; Huang, J.H.; Liu, B.; Liu, S.J.; Zhong, Y.; Liu, S.M. HMGA1 is a new target of miR-195 involving isoprenaline-induced cardiomyocyte hypertrophy. Biochemistry 2014, 79, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Attia, R.; Mayr, U.; Gomes, R.S.; Phinikaridou, A.; Yin, X.; Langley, S.R.; Willeit, P.; Lu, R.; Fanshawe, B.; et al. Role of miR-195 in aortic aneurysmal disease. Circ. Res. 2014, 115, 857–866. [Google Scholar] [CrossRef]
- Du, J.; Zheng, R.; Xiao, F.; Zhang, S.; He, K.; Zhang, J.; Shao, Y. Downregulated MicroRNA-195 in the Bicuspid Aortic Valve Promotes Calcification of Valve Interstitial Cells via Targeting SMAD7. Cell Physiol. Biochem. 2017, 44, 884–896. [Google Scholar] [CrossRef]
- Collares, C.V.; Evangelista, A.F.; Xavier, D.J.; Rassi, D.M.; Arns, T.; Foss-Freitas, M.C.; Foss, M.C.; Puthier, D.; Sakamoto-Hojo, E.T.; Passos, G.A.; et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res. Notes 2013, 6, 491. [Google Scholar] [CrossRef]
- Massaro, J.D.; Polli, C.D.; Costa, E.; Silva, M.; Alves, C.C.; Passos, G.A.; Sakamoto-Hojo, E.T.; Rodrigues de Holanda Miranda, W.; Bispo Cezar, N.J.; Rassi, D.M.; et al. Post-transcriptional markers associated with clinical complications in Type 1 and Type 2 diabetes mellitus. Mol. Cell Endocrinol. 2019, 490, 1–14. [Google Scholar] [CrossRef]
- Li, M.; Luan, L.; Liu, Q.; Liu, Y.; Lan, X.; Li, Z.; Liu, W. MiRNA-199a-5p Protects Against Cerebral Ischemic Injury by Down-Regulating DDR1 in Rats. World Neurosurg. 2019, 131, e486–e494. [Google Scholar] [CrossRef]
- Yan, M.; Yang, S.; Meng, F.; Zhao, Z.; Tian, Z.; Yang, P. MicroRNA 199a-5p induces apoptosis by targeting JunB. Sci. Rep. 2018, 8, 6699. [Google Scholar] [CrossRef]
- Lynch, S.M.; Ward, M.; McNulty, H.; Angel, C.Z.; Horigan, G.; Strain, J.J.; Purvis, J.; Tackett, M.; McKenna, D.J. Serum levels of miR-199a-5p correlates with blood pressure in premature cardiovascular disease patients homozygous for the MTHFR 677C > T polymorphism. Genom. 2020, 112, 669–676. [Google Scholar] [CrossRef]
- Tian, X.; Yu, C.; Shi, L.; Li, D.; Chen, X.; Xia, D.; Zhou, J.; Xu, W.; Ma, C.; Gu, L.; et al. MicroRNA-199a-5p aggravates primary hypertension by damaging vascular endothelial cells through inhibition of autophagy and promotion of apoptosis. Exp. Ther. Med. 2018, 16, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Pang, B.; Xiao, Y.; Zhou, S.; He, B.; Zhang, F.; Liu, W.; Peng, H.; Li, P. The protective microRNA-199a-5p-mediated unfolded protein response in hypoxic cardiomyocytes is regulated by STAT3 pathway. J. Physiol. Biochem. 2019, 75, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, G.; Zhang, H.; Wang, J. MiRNA-199a-5p influences pulmonary artery hypertension via downregulating Smad3. Biochem. Biophys. Res. Commun. 2016, 473, 859–866. [Google Scholar] [CrossRef]
- Wang, J.; Yu, G. A Systems Biology Approach to Characterize Biomarkers for Blood Stasis Syndrome of Unstable Angina Patients by Integrating MicroRNA and Messenger RNA Expression Profiling. Evid. Based Complement. Altern. Med. 2013, 2013, 510208. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Gu, T.; Shi, E.; Wang, Y.; Fang, Q.; Wang, C. Dysregulation of renal microRNA expression after deep hypothermic circulatory arrest in rats. Eur J. Cardiothorac. Surg. 2016, 49, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Hirt, M.N.; Werner, T.; Indenbirken, D.; Alawi, M.; Demin, P.; Kunze, A.C.; Stenzig, J.; Starbatty, J.; Hansen, A.; Fiedler, J.; et al. Deciphering the microRNA signature of pathological cardiac hypertrophy by engineered heart tissue- and sequencing-technology. J. Mol. Cell Cardiol. 2015, 81, 1–9. [Google Scholar] [CrossRef]
- Aguado-Fraile, E.; Ramos, E.; Conde, E.; Rodríguez, M.; Martín-Gómez, L.; Lietor, A.; Candela, Á.; Ponte, B.; Liaño, F.; García-Bermejo, M.L. A Pilot Study Identifying a Set of microRNAs As Precise Diagnostic Biomarkers of Acute Kidney Injury. PLoS ONE 2015, 10, e0127175. [Google Scholar] [CrossRef]
- Ma, H.; Chen, P.; Sang, C.; Huang, D.; Geng, Q.; Wang, L. Modulation of apoptosis-related microRNAs following myocardial infarction in fat-1 transgenic mice vs wild-type mice. J. Cell Mol. Med. 2018, 22, 5698–5707. [Google Scholar] [CrossRef]
- Qiao, X.R.; Wang, L.; Liu, M.; Tian, Y.; Chen, T. MiR-210-3p attenuates lipid accumulation and inflammation in atherosclerosis by repressing IGF2. Biosci. Biotechnol. Biochem. 2020, 84, 321–329. [Google Scholar] [CrossRef]
- Derda, A.A.; Pfanne, A.; Bwangär, C.; Schimmel, K.; Kennel, P.J.; Xiao, K.; Schulze, P.C.; Bauersachs, J.; Thum, T. Blood-based microRNA profiling in patients with cardiac amyloidosis. PLoS ONE 2018, 13, e0204235. [Google Scholar] [CrossRef]
- Verjans, R.; Peters, T.; Beaumont, F.J.; van Leeuwen, R.; van Herwaarden, T.; Verhesen, W.; Munts, C.; Bijnen, M.; Henkens, M.; Diez, J.; et al. MicroRNA-221/222 Family Counteracts Myocardial Fibrosis in Pressure Overload-Induced Heart Failure. Hypertension 2018, 71, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Li, R.; Maimaitijiang, A.; Liu, R.; Yan, F.; Hu, H.; Gao, X.; Shi, H. miR-221-3p inhibits oxidized low-density lipoprotein induced oxidative stress and apoptosis via targeting a disintegrin and metalloprotease-22. J. Cell Biochem. 2019, 120, 6304–6314. [Google Scholar] [CrossRef] [PubMed]
- Pereira-da-Silva, T.; Coutinho Cruz, M.; Carrusca, C.; Cruz Ferreira, R.; Napoleão, P.; Mota Carmo, M. Circulating microRNA profiles in different arterial territories of stable atherosclerotic disease: A systematic review. Am. J. Cardiovasc. Dis. 2018, 8, 1–13. [Google Scholar] [PubMed]
- Coffey, S.; Williams, M.J.; Phillips, L.V.; Galvin, I.F.; Bunton, R.W.; Jones, G.T. Integrated microRNA and messenger RNA analysis in aortic stenosis. Sci. Rep. 2016, 6, 36904. [Google Scholar] [CrossRef]
- Coskunpinar, E.; Cakmak, H.A.; Kalkan, A.K.; Tiryakioglu, N.O.; Erturk, M.; Ongen, Z. Circulating miR-221-3p as a novel marker for early prediction of acute myocardial infarction. Gene 2016, 591, 90–96. [Google Scholar] [CrossRef]
- Sørensen, S.S.; Nygaard, A.B.; Nielsen, M.Y.; Jensen, K.; Christensen, T. miRNA expression profiles in cerebrospinal fluid and blood of patients with acute ischemic stroke. Transl. Stroke Res. 2014, 5, 711–718. [Google Scholar] [CrossRef]
- Gusar, V.A.; Timofeeva, A.V.; Zhanin, I.S.; Shram, S.I.; Pinelis, V.G. Estimation of Time-Dependent microRNA Expression Patterns in Brain Tissue, Leukocytes, and Blood Plasma of Rats under Photochemically Induced Focal Cerebral Ischemia. Mol. Biol. 2017, 51, 683–695. [Google Scholar] [CrossRef]
- Nie, X.; Chen, Y.; Tan, J.; Dai, Y.; Mao, W.; Qin, G.; Ye, S.; Sun, J.; Yang, Z.; Chen, J. MicroRNA-221-3p promotes pulmonary artery smooth muscle cells proliferation by targeting AXIN2 during pulmonary arterial hypertension. Vascul. Pharmacol. 2019, 116, 24–35. [Google Scholar] [CrossRef]
- Villard, A.; Marchand, L.; Thivolet, C.; Rome, S. Diagnostic Value of Cell-free Circulating MicroRNAs for Obesity and Type 2 Diabetes: A Meta-analysis. J. Mol. Biomark. Diagn. 2015, 6, 251. [Google Scholar] [CrossRef]
- Wang, L.; Xu, L.; Xu, M.; Liu, G.; Xing, J.; Sun, C.; Ding, H. Obesity-Associated MiR-342-3p Promotes Adipogenesis of Mesenchymal Stem Cells by Suppressing CtBP2 and Releasing C/EBPα from CtBP2 Binding. Cell Physiol. Biochem. 2015, 35, 2285–2298. [Google Scholar] [CrossRef]
- Hezova, R.; Slaby, O.; Faltejskova, P.; Mikulkova, Z.; Buresova, I.; Raja, K.R.; Hodek, J.; Ovesna, J.; Michalek, J. microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol. 2010, 260, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Matboli, M.; Bekhet, M.M. Clinical verification of a novel urinary microRNA panal: 133b, -342 and -30 as biomarkers for diabetic nephropathy identified by bioinformatics analysis. Biomed. Pharmacother. 2016, 83, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Cui, Y.; Fan, L.; Mu, X.; Hua, Y. T2DM inhibition of endothelial miR-342-3p facilitates angiogenic dysfunction via repression of FGF11 signaling. Biochem. Biophys. Res. Commun. 2018, 503, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Khalyfa, A.; Kheirandish-Gozal, L.; Bhattacharjee, R.; Khalyfa, A.A.; Gozal, D. Circulating microRNAs as Potential Biomarkers of Endothelial Dysfunction in Obese Children. Chest 2016, 149, 786–800. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, M. MicroRNA-499-5p: A therapeutic target in the context of cardiovascular disease. Ann. Transl. Med. 2016, 4, 539. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, B.; Zhang, W.; Sun, L. Effect of miR-499a-5p on damage of cardiomyocyte induced by hypoxia-reoxygenation via downregulating CD38 protein. J. Cell Biochem. 2020, 121, 996–1004. [Google Scholar] [CrossRef]
- Neshati, V.; Mollazadeh, S.; Fazly Bazzaz, B.S.; de Vries, A.A.F.; Mojarrad, M.; Naderi-Meshkin, H.; Neshati, Z.; Mirahmadi, M.; Kerachian, M.A. MicroRNA-499a-5p Promotes Differentiation of Human Bone Marrow-Derived Mesenchymal Stem Cells to Cardiomyocytes. Appl. Biochem. Biotechnol. 2018, 186, 245–255. [Google Scholar] [CrossRef]
- Boštjančič, E.; Zidar, N.; Glavač, D. MicroRNAs and cardiac sarcoplasmic reticulum calcium ATPase-2 in human myocardial infarction: Expression and bioinformatic analysis. BMC Genom. 2012, 13, 552. [Google Scholar] [CrossRef]
- Salinas, J.; Lin, H.; Aparico, H.J.; Huan, T.; Liu, C.; Rong, J.; Beiser, A.; Himali, J.J.; Freedman, J.E.; Larson, M.G.; et al. Whole blood microRNA expression associated with stroke: Results from the Framingham Heart Study. PLoS ONE 2019, 14, e0219261. [Google Scholar] [CrossRef]
- Baldeón Rojas, L.; Weigelt, K.; de Wit, H.; Ozcan, B.; van Oudenaren, A.; Sempértegui, F.; Sijbrands, E.; Grosse, L.; Van Zonneveld, A.J.; Drexhage, H.A.; et al. Study on inflammation-related genes and microRNAs, with special emphasis on the vascular repair factor HGF and miR-574-3p, in monocytes and serum of patients with T2D. Diabetol. Metab. Syndr. 2016, 8, 6. [Google Scholar] [CrossRef]
- Houshmand-Oeregaard, A.; Schrölkamp, M.; Kelstrup, L.; Hansen, N.S.; Hjort, L.; Thuesen, A.C.B.; Broholm, C.; Mathiesen, E.R.; Clausen, T.D.; Vaag, A.; et al. Increased Expression of microRNA-15a and microRNA-15b in Skeletal Muscle from Adult Offspring of Women with Diabetes in Pregnancy. Hum. Mol. Genet. 2018, 27, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- International Association of Diabetes and Pregnancy Study Groups Consensus Panel; Metzger, B.E.; Gabbe, S.G.; Persson, B.; Buchanan, T.A.; Catalano, P.A.; Damm, P.; Dyer, A.R.; Leiva, A.D.; Hod, M.; et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care 2010, 33, 676–682. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus (Position Statement). Diabetes Care 2009, 32, S62–S67. [Google Scholar] [CrossRef] [PubMed]
- Metzger, B.E.; Coustan, D.R. Summary and recommendations of the Fourth International Workshop-Conference on Gestational Diabetes Mellitus. The Organizing Committee. Diabetes Care 1998, 21, B161–B167. [Google Scholar] [PubMed]
- Hromadnikova, I.; Kotlabova, K.; Dvorakova, L.; Krofta, L.; Sirc, J. Postnatal Expression Profile of microRNAs Associated with Cardiovascular and Cerebrovascular Diseases in Children at the Age of 3 to 11 Years in Relation to Previous Occurrence of Pregnancy-Related Complications. Int. J. Mol. Sci. 2019, 20, 654. [Google Scholar] [CrossRef] [PubMed]
- National High Blood Pressure Education Program Working Group on High Blood Pressure in Children and Adolescents The fourth report on the diagnosis, evaluation, and treatment of high blood pressure in children and adolescents. Pediatrics 2004, 114, 555–576. [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk—database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef]
- Hromadnikova, I. Postnatální Epigenetický Profil Kardiovaskulárních MikroRNA U Dětí Narozených Z Komplikovaných Gravidit-Nové Biomarkery Kardiovaskulárního Rizika. CZ Patent 308102, 31 October 2018. Issued 20 November 2019. Industrial Property Office, Czech Republic. [Google Scholar]
- Hromadnikova, I. A Method of Predicting a Risk of Cardiovascular Disease for a Child Born from a Complicated Pregnancy. PCT International Application No. PCT/CZ2019/050050, 30 October 2019. Industrial Property Office, Czech Republic. [Google Scholar]
- Rivera, R.M.; Bennett, L.B. Epigenetics in Humans: An Overview. Curr. Opin. Endocrinol. Diabetes Obes. 2010, 17, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Saetrom, P.; Snøve, O., Jr.; Rossi, J.J. Epigenetics and microRNAs. Pediatr. Res. 2007, 61, 17R–23R. [Google Scholar] [CrossRef] [PubMed]
- Tost, J. Epigenetics; Caister Academic Press: Norfolk, UK, 2008; pp. 1–404. [Google Scholar]
- Li, E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 2002, 3, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Sinkkonen, L.; Hugenschmidt, T.; Berninger, P.; Gaidatzis, D.; Mohn, F.; Artus-Revel, C.G.; Zavolan, M.; Svoboda, P.; Filipowicz, W. MicroRNAs Control De Novo DNA Methylation Through Regulation of Transcriptional Repressors in Mouse Embryonic Stem Cells. Nat. Struct. Mol. Biol. 2008, 15, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Flemr, M.; Stein, P.; Berninger, P.; Malik, R.; Zavolan, M.; Svoboda, P.; Schultz, R.M. MicroRNA Activity Is Suppressed in Mouse Oocytes. Curr. Biol. 2010, 20, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Suh, N.; Baehner, L.; Moltzahn, F.; Melton, C.; Shenoy, A.; Chen, J.; Blelloch, R. MicroRNA Function Is Globally Suppressed in Mouse Oocytes and Early Embryos. Curr. Biol. 2010, 20, 271–277. [Google Scholar] [CrossRef]
- Gu, Y.; Sun, J.; Groome, L.J.; Wang, Y. Differential miRNA expression profiles between the first and third trimester human placentas. Am. J. Physiol. Endocrinol. Metab. 2013, 304, E836–E843. [Google Scholar] [CrossRef]
- Maccani, M.A.; Padbury, J.F.; Lester, B.M.; Knopik, V.S.; Marsit, C.J. Placental miRNA expression profiles are associated with measures of infant neurobehavioral outcomes. Pediatr. Res. 2013, 74, 272–278. [Google Scholar] [CrossRef]
- Soni, K.; Choudhary, A.; Patowary, A.; Singh, A.R.; Bhatia, S.; Sivasubbu, S.; Chandrasekaran, S.; Pillai, B. miR-34 Is Maternally Inherited in Drosophila Melanogaster and Danio Rerio. Nucleic Acids Res. 2013, 41, 4470–4480. [Google Scholar] [CrossRef]
- Babenko, O.; Kovalchuka, I.; Metz, G.A. Epigenetic programming of neurodegenerative diseases by an adverse environment. Brain Res. 2015, 1444, 96–111. [Google Scholar] [CrossRef]
- Rodgers, A.B.; Morgan, C.P.; Leu, N.A.; Bale, T.L. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc. Natl. Acad. Sci. USA 2015, 112, 13699–13704. [Google Scholar] [CrossRef] [PubMed]
- Fraser, R.; Lin, C.J. Epigenetic reprogramming of the zygote in mice and men: On your marks, get set, go! Reproduction 2016, 152, R211–R222. [Google Scholar] [CrossRef] [PubMed]
- Pang, T.Y.C.; Short, A.K.; Bredy, T.W.; Hannan, A.J. Transgenerational paternal transmission of acquired traits: Stress-induced modification of the sperm regulatory transcriptome and offspring phenotypes. Curr. Opin. Behav. Sci. 2017, 14, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Yeshurun, S.; Hannan, A.J. Transgenerational epigenetic influences of paternal environmental exposures on brain function and predisposition to psychiatric disorders. Mol. Psychiatry 2019, 24, 536–548. [Google Scholar] [CrossRef] [PubMed]
- Lacal, I.; Ventura, R. Epigenetic Inheritance: Concepts, Mechanisms and Perspectives. Front. Mol. Neurosci. 2018, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Mathers, J.C.; Strathdee, G.; Relton, C.L. Induction of epigenetic alterations by dietary and other environmental factors. Adv. Genet. 2010, 71, 3–39. [Google Scholar]
- Christensen, B.C.; Marsit, C.J. Epigenomics in environmental health. Front. Genet. 2011, 2, 84. [Google Scholar] [CrossRef]
- Talikka, M.; Sierro, N.; Ivanov, N.V.; Chaudhary, N.; Peck, M.J.; Hoeng, J.; Coggins, C.R.E.; Peitsch, C.M. Genomic Impact of Cigarette Smoke, With Application to Three Smoking-Related Diseases. Crit. Rev. Toxicol. 2012, 42, 877–889. [Google Scholar] [CrossRef]
- Cabib, S.; Puglisi-Allegra, S. The mesoaccumbens dopamine in coping with stress. Neurosci. Biobehav. Rev. 2012, 36, 79–89. [Google Scholar] [CrossRef]
- Van Otterdijk, S.D.; Mathers, J.C.; Strathdee, G. Do age-related changes in DNA methylation play a role in the development of age-related diseases? Biochem. Soc. Trans. 2013, 41, 803–807. [Google Scholar] [CrossRef]
- Jung, M.; Pfeifer, G.P. Aging and DNA methylation. BMC Biol 2015, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Deans, C.; Maggert, K.A. What Do You Mean, “Epigenetic”? Genetics 2015, 199, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Zoghbi, H.Y.; Beaudet, A.L. Epigenetics and Human Disease. Cold Spring Harb. Perspect. Biol. 2016, 8, a019497. [Google Scholar] [CrossRef] [PubMed]
- Tobias, D.K.; Chavarro, J.E.; Williams, M.A.; Buck Louis, G.M.; Hu, F.B.; Rich-Edwards, J.; Missmer, S.A.; Zhang, C. History of Infertility and Risk of Gestational Diabetes Mellitus: A Prospective Analysis of 40,773 Pregnancies. Am. J. Epidemiol. 2013, 178, 1219–1225. [Google Scholar] [CrossRef]
- Kouhkan, A.; Khamseh, M.E.; Pirjani, R.; Moini, A.; Arabipoor, A.; Maroufizadeh, S.; Hosseini, R.; Baradaran, H.R. Obstetric and perinatal outcomes of singleton pregnancies conceived via assisted reproductive technology complicated by gestational diabetes mellitus: A prospective cohort study. BMC Pregnancy Childbirth 2018, 18, 495. [Google Scholar] [CrossRef]
- Mohammadi, M.; Morasae, E.K.; Maroufizadeh, S.; Almasi-Hashiani, A.; Navid, B.; Amini, P.; Omani-Samani, R.; Alizadeh, A. Assisted reproductive technology and the risk of gestational diabetes mellitus: A systemic review and meta-analysis. Middle East Fertil. Soc. J. 2020, 25, 6. [Google Scholar] [CrossRef]
| miRBase ID | Gene Location on Chromosome | Role in the Pathogenesis of Diabetes Mellitus and Cardiovascular/Cerebrovascular Diseases |
|---|---|---|
| hsa-miR-1-3p | 20q13.3 [46] 18q11.2 | Acute myocardial infarction, heart ischemia, post-myocardial infarction complications [47], thoracic aortic aneurysm [48], diabetes mellitus [49,50], vascular endothelial dysfunction [51]. |
| hsa-miR-16-5p | 13q14.2 | Myocardial infarction [52,53], heart failure [54], acute coronary syndrome, cerebral ischaemic events [55], gestational diabetes mellitus [9,13,20], diabetes mellitus [56,57,58]. |
| hsa-miR-17-5p | 13q31.3 [59,60] | Cardiac development [61], ischemia/reperfusion-induced cardiac injury [62], kidney ischemia-reperfusion injury [63], diffuse myocardial fibrosis in hypertrophic cardiomyopathy [64], acute ischemic stroke [65], coronary artery disease [66], adipogenic differentiation [67], gestational diabetes mellitus [9,13], diabetes mellitus [58,68]. |
| hsa-miR-20a-5p | 13q31.3 [69] | Pulmonary hypertension [70], gestational diabetes mellitus [9,13,17], diabetic retinopathy [71], diabetes with abdominal aortic aneurysm [72]. |
| hsa-miR-20b-5p | Xq26.2 [69] | Hypertension-induced heart failure [73], insulin resistance [74], T2DM [75,76], diabetic retinopathy [77]. |
| hsa-miR-21-5p | 17q23.2 [78] | Homeostasis of the cardiovascular system [79], cardiac fibrosis and heart failure [80,81], thoracic aortic aneurysm [48], ascending aortic aneurysm [82], regulation of hypertension-related genes [83], myocardial infarction [84], insulin resistance [74], T2DM [85], T2DM with major cardiovascular events [86], T1DM [87,88,89], diabetic nephropathy [90]. |
| hsa-miR-23a-3p | 19p13.12 | Heart failure [91], coronary artery disease [92], cerebral ischemia-reperfusion [93], vascular endothelial dysfunction [51], small and large abdominal aortic aneurysm [94], obesity and insulin resistance [95]. |
| hsa-miR-24-3p | 19p13.12 | Asymptomatic carotid stenosis [96], familial hypercholesterolemia and coronary artery disease [97], angina pectoris [98], ischemic dilated cardiomyopathy [99], small and large abdominal aortic aneurysm [94], myocardial ischemia/reperfusion [100,101], diabetes mellitus [50,58,62,64]. |
| hsa-miR-26a-5p | 3p22.2 [102] 12q14.1 | Heart failure, cardiac hypertrophy [103], myocardial infarction [84,104,105], ischemia/reperfusion injury [106], pulmonary arterial hypertension [107], T1DM [108], diabetic nephropathy [90]. |
| hsa-miR-29a-3p | 7q32.3 | Ischemia/reperfusion-induced cardiac injury [109], cardiac cachexia, heart failure [110], atrial fibrillation [111], diffuse myocardial fibrosis in hypertrophic cardiomyopathy [64], coronary artery disease [112], pulmonary arterial hypertension [107], gestational diabetes mellitus [12], diabetes mellitus [49,57,113,114]. |
| hsa-miR-92a-3p | 13q31.3 Xq26.2 | Mitral chordae tendineae rupture [115], children with rheumatic carditis [116], myocardial infarction [117], heart failure [118], coronary artery disease [119], renal injury-associated atherosclerosis [120]. |
| hsa-miR-100-5p | 11q24.1 | Failing human heart, idiopathic dilated cardiomyopathy, ischemic cardiomyopathy [99], regulation of hypertension-related genes [83], T1DM [87]. |
| hsa-miR-103a-3p | 5q34 [121] 20p13 | Hypertension [122], hypoxia-induced pulmonary hypertension [123], myocardial ischemia/reperfusion injury, acute myocardial infarction [124], ischemic dilated cardiomyopathy [99], obesity, regulation of insulin sensitivity [125], T1DM [126]. |
| hsa-miR-125b-5p | 11q24.1 [126] 21q21.1 | Acute ischemic stroke [127], acute myocardial infarction [128,129], ischemic dilated cardiomyopathy [99], ascending aortic aneurysm [82], gestational diabetes mellitus [19], T1DM [130,131], T2DM [132]. |
| hsa-miR-126-3p | 9q34.3 [133] | Acute myocardial infarction [105], thoracic aortic aneurysm [48], T2DM [86,134], T2DM with major cardiovascular events [86], gestational diabetes mellitus [10]. |
| hsa-miR-130b-3p | 22q11.21 | Hypertriglyceridemia [135,136], intracranial aneurysms [137], hyperacute cerebral infarction [138], T2DM [85,139,140], gestational diabetes mellitus [10]. |
| hsa-miR-133a-3p | 18q11.2 [141] 20q13.33 | Heart failure [142], myocardial fibrosis in hypertrophic cardiomyopathy [64,143], arrhythmogenesis in the hypertrophic and failing hearts [144,145], coronary artery calcification [146], thoracic aortic aneurysm [48], ascending aortic aneurysm [82], diabetes mellitus [49,50]. |
| hsa-miR-143-3p | 5q33 | Intracranial aneurysms [147], coronary heart disease [148], myocardial infarction [149], myocardial hypertrophy [150], dilated cardiomyopathy [151], pulmonary arterial hypertension [152], acute ischemic stroke [127], ascending aortic aneurysm [82]. |
| hsa-miR-145-5p | 5q33 | Hypertension [153,154], dilated cardiomyopathy [155], myocardial infarction [156,157], stroke [157], acute cerebral ischemic/reperfusion [158], T2DM [58,159], T1DM [85], diabetic retinopathy [160], gestational diabetes mellitus [161]. |
| hsa-miR-146a-5p | 5q33.3 [162,163] | Angiogenesis [164], hypoxia, ischemia/reperfusion-induced cardiac injury [165], myocardial infarction [53], coronary atherosclerosis, coronary heart disease in patients with subclinical hypothyroidism [166], thoracic aortic aneurysm [48], acute ischemic stroke, acute cerebral ischemia [167], T2DM [58,85], T1DM [108], diabetic nephropathy [90]. |
| hsa-miR-155-5p | 21q21.3 | Thoracic aortic aneurysm [48], type 1 diabetes [125], gestational diabetes mellitus [20], adolescent obesity [168], diet-induced obesity and obesity resistance [168], atherosclerosis [170], hyperlipidemia—associated endotoxemia [171], coronary plaque rupture [172], children with cyanotic heart disease [173], chronic kidney disease and nocturnal hypertension [174], atrial fibrillation [175]. |
| hsa-miR-181a-5p | 1q32.1 [176] 9q33.3 | Regulation of hypertension-related genes [65], atherosclerosis [176], metabolic syndrome, coronary artery disease [177], non-alcoholic fatty liver disease [178], ischaemic stroke, transient ischaemic attack, acute myocardial infarction [179,180], obesity and insulin resistance [95,176,177], T1DM [85,181], T2DM [176,180]. |
| hsa-miR-195-5p | 17p13.1 [182] | Cardiac hypertrophy, heart failure [183,184], abdominal aortic aneurysms [185], aortic stenosis [186], T2DM [159], gestational diabetes mellitus [18]. |
| hsa-miR-199a-5p | 1q24.3 19p13.2 | T1DM, T2DM, gestational diabetes mellitus [187], diabetic retinopathy [188], cerebral ischemic injury [189], heart failure [190], hypertension [191,192], congenital heart disease [193], pulmonary artery hypertension [194], unstable angina [195], hypoxia in myocardium [193], acute kidney injury [196]. |
| hsa-miR-210-3p | 11p15.5 | Cardiac hypertrophy [197], acute kidney injury [198], myocardial infarction [199], atherosclerosis [200]. |
| hsa-miR-221-3p | Xp11.3 | Asymptomatic carotid stenosis [96], cardiac amyloidosis [201], heart failure [202], atherosclerosis [203,204], aortic stenosis [205], acute myocardial infarction [206], acute ischemic stroke [207], focal cerebral ischemia [208], pulmonary artery hypertension [209], obesity [210]. |
| hsa-miR-342-3p | 14q32.2 | Cardiac amyloidosis [201], obesity [211], T1DM [85,187,212], T2DM [187,213,214], gestational diabetes mellitus [187], endothelial dysfunction [215]. |
| hsa-miR-499a-5p | 20q11.22 | Myocardial infarction [53,216], hypoxia [217], cardiac regeneration [218], vascular endothelial dysfunction [51]. |
| hsa-miR-574-3p | 4p14 | Myocardial infarction [219], coronary artery disease [136], cardiac amyloidosis [201], stroke [220], T2DM [140,221]. |
| Normal Pregnancies Normal Clinical Findings (n = 48) | Normal Pregnancies Abnormal Clinical Findings (n = 37) | Gestational Diabetes Mellitus (GDM) Normal Clinical Findings (n = 61) | GDM Abnormal Clinical Findings (n= 57) | p-Value1 | p-Value2 | p-Value3 | |
|---|---|---|---|---|---|---|---|
| Children at follow-up | |||||||
| Age (years) | 5 (3–11) | 5 (3–11) | 5 (3–10) | 5 (3–9) | 0.228 | 0.980 | 0.358 |
| Height (cm) | 115.5 (98–144.5) | 118.0 (100–153) | 114.0 (99–143.5) | 113.5 (98–153) | 0.332 | 0.546 | 0.274 |
| Weight (kg) | 20.8 (14–37) | 22.3 (14.7–40.8) | 19.5 (14.4–37.4) | 19.6 (15–47.1) | 0.104 | 0.604 | 0.565 |
| BMI (kg/m2) | 15.41 (13.22–18.09) | 15.80 (13.3–20) | 15.20 (13.53–18.09) | 15.58 (12.97–20.08) | 0.067 | 0.976 | 0.166 |
| Systolic BP (mmHg) | 98 (84–115) | 104 (89–123) | 99 (82–113) | 101 (87–125) | <0.001 | 0.864 | 0.027 |
| Diastolic BP (mmHg) | 60 (38–68) | 64.0 (43–81) | 60 (47–67) | 61 (49–79) | 0.002 | 0.795 | 0.008 |
| Heart rate (n/min) | 90 (67–110) | 90.0 (51–120) | 96 (64–118) | 98 (78–122) | 0.954 | 0.022 | <0.001 |
| During Gestation | |||||||
| Maternal age at delivery (years) | 31.5 (21–40) | 32 (25–46) | 34 (27–42) | 33 (27–45) | 0.233 | 0.009 | 0.046 |
| GA at delivery (weeks) | 39.86 (37.71–41.57) | 39.86 (37.86–41.86) | 39.72 (37.43–41.28) | 39.43 (37.00–41.14) | 0.852 | 0.111 | 0.020 |
| Fetal birth weight (g) | 3425 (2730–4220) | 3280 (2530–4450) | 3500 (2700-4330) | 3420 (2770–4400) | 0.989 | 0.157 | 0.252 |
| Mode of delivery | 0.698 | <0.001 | 0.001 | ||||
| Vaginal | 44 (91.67%) | 33 (89.19%) | 38 (62.30%) | 37 (64.91%) | |||
| CS | 4 (8.33%) | 4 (10.81%) | 23 (37.70%) | 20 (35.09%) | |||
| Fetal sex | 0.346 | 0.771 | 0.273 | ||||
| Boy | 27 (56.25%) | 17 (45.95%) | 36 (59.02%) | 38 (66.67%) | |||
| Girl | 21 (43.75%) | 20 (54.05%) | 25 (40.98%) | 19 (33.33%) | |||
| Primiparity | 0.069 | 0.181 | 0.091 | ||||
| Yes | 29 (60.42%) | 15 (40.54%) | 29 (47.54%) | 25 (43.86%) | |||
| No | 19 (39.58%) | 22 (59.46%) | 32 (52.46%) | 32 (56.14%) | |||
| Birth order of index pregnancy | 0.058 | 0.080 | 0.294 | ||||
| 1st | 26 (54.17%) | 11 (29.73%) | 20 (32.79%) | 22 (38.60%) | |||
| 2nd | 16 (33.33%) | 14 (37.84%) | 23 (37.70%) | 22 (38.60%) | |||
| 3rd | 4 (8.33%) | 10 (27.03%) | 10 (16.39%) | 6 (10.52%) | |||
| 4th+ | 2 (4.17%) | 2 (5.40%) | 8 (13.11%) | 7 (12.28%) | |||
| Infertility treatment | 0.852 | 0.014 | 0.084 | ||||
| Yes | 1 (2.08%) | 1 (2.70%) | 10 (16.39%) | 6 (10.53%) | |||
| No | 47 (97.92%) | 36 (97.30%) | 51 (83.61%) | 51 (89.47%) | |||
| Maternal BMI (kg/m2) | |||||||
| Prepregnancy BMI | 21.88 (14.77–30.3) | 21.22 (17.37–28.04) | 22.02 (16.3–30.85) | 22.64 (17.53–30.49) | 1.000 | 1.000 | 0.567 |
| BMI < 18.5 | 4 (8.33%) | 5 (13.51%) | 5 (8.20%) | 1 (1.75%) | - | - | - |
| BMI 18.5–24.9 | 38 (79.17%) | 31 (83.78%) | 43 (70.49%) | 37 (64.91%) | |||
| BMI 25.0–29.9 | 5 (10.42%) | 1 (2.70%) | 11 (18.03%) | 17 (29.82%) | |||
| BMI > 30 | 1 (2.08%) | 0 (0%) | 2 (3.28%) | 2 (3.51%) | |||
| BMI at admission for delivery | 26.17 (20.88–34.82) | 26.45 (20.72–33.17) | 25.97 (19.84–36.85) | 27.53 (20.18–36.73) | 1.000 | 1.000 | 1.000 |
| Total gestational weight gain (GWG) (kg) | 14.5 (8–25.5) | 14.5 (8–21) | 10 (2–21) | 11 (3–26) | 1.000 | <0.001 | 0.008 |
| BMI at follow-up | 23.26 (17.7–39.08) | 22.11 (18.17–29.17) | 23.82 (17.39–32.14) | 23.42 (17.39–34.37) | 0.414 | 1.000 | 1.000 |
| Serum Metabolic Biochemical Parameters of Mothers During the Third Trimester of Gestation | |||||||
| HbA1c | - | - | 32 (26–42) | 32.5 (26–40) | - | - | - |
| Creatinine (μmol/L) | 56.0 (44.0–70.0) | 52.5 (43.0–83.0) | 56.0 (40.0–78.0) | 55.0 (36.0–83.0) | 1.000 | 1.000 | 1.000 |
| Uric acid (μmol/L) | 286.5 (200.0–377.0) | 294.5 (221.0–345.0) | 298 (180.0–419.0) | 289 (157.0–471.0) | 1.000 | 1.000 | 1.000 |
| Total bilirubin (μmol/L) | 4.0 (3.0–7.0) | 4.0 (0.18–12.0) | 6.0 (2.70–20.0) | 6.0 (3.0–20.0) | 1.000 | 0.497 | 0.108 |
| ALT (μkat/L) | 0.22 (0.11–2.4) | 0.17 (0.07–0.36) | 0.23 (0.11–1.09) | 0.22 (0.11–0.46) | 1.000 | 1.000 | 1.000 |
| AST (μkat/L) | 0.38 (0.25–2.36) | 0.47 (0.22–2.29) | 0.38 (0.22–1.46) | 0.40 (0.24–0.84) | 1.000 | 1.000 | 1.000 |
| ALP (μkat/L) | 2.12 (1.86–4.66) | 2.51 (1.71–3.52) | 2.67 (1.35–6.15) | 2.36 (1.46–5.32) | 1.000 | 1.000 | 1.000 |
| Cholesterol (mmol/L) | 7.50 (6.30–11.40) | 9.2 (6.90–10.5)) | 7.71 (4.47–9.5) | 6.93 (6.13–9.10) | 1.000 | 1.000 | 1.000 |
| Triglyceride (mmol/L) | 3.10 (3.00–3.20) | 2.9 (2.7–3.3) | 3.05 (1.80–4.80) | 3.35 (2.50–8.90) | 1.000 | 1.000 | 1.000 |
| Total protein (g/L) | 63.7 (48.7–70.8) | 64,35 (47.2–68.9) | 60.3 (37.7–70.8) | 61.6 (43.8–67.9%) | 1.000 | 1.000 | 1.000 |
| Albumin (g/L) | 37.35 (28.9–40.6) | 36.9 (27.9–41.6) | 36.5 (4.40–42.1) | 36.2(26.3–40.9) | 1.000 | 1.000 | 1.000 |
| Blood glucose (mmol/L) | 4.7 (4.2–5.1) | 4.5 (4.3–5.8) | 4.65 (4.1–8.0) | 4.4 (3.8–5.6) | 1.000 | 1.000 | 1.000 |
| Normal Pregnancy | GDM Pregnancy | OR (95% CI) | p-Value | |
|---|---|---|---|---|
| Overweight/obese | 8/85 (9.41%) | 6/118 (5.08%) | 0.515 (0.172–1.545) | 0.237 |
| Prehypertension/ hypertension | 15/85 (17.65%) | 19/118 (16.10%) | 0.896 (0.426–1.883) | 0.771 |
| Valve problems or heart defects | 17/85 (20.0%) | 42/118 (35.59%) | 2.240 (1.167–4.300) | 0.015 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hromadnikova, I.; Kotlabova, K.; Dvorakova, L.; Krofta, L.; Sirc, J. Substantially Altered Expression Profile of Diabetes/Cardiovascular/Cerebrovascular Disease Associated microRNAs in Children Descending from Pregnancy Complicated by Gestational Diabetes Mellitus—One of Several Possible Reasons for an Increased Cardiovascular Risk. Cells 2020, 9, 1557. https://doi.org/10.3390/cells9061557
Hromadnikova I, Kotlabova K, Dvorakova L, Krofta L, Sirc J. Substantially Altered Expression Profile of Diabetes/Cardiovascular/Cerebrovascular Disease Associated microRNAs in Children Descending from Pregnancy Complicated by Gestational Diabetes Mellitus—One of Several Possible Reasons for an Increased Cardiovascular Risk. Cells. 2020; 9(6):1557. https://doi.org/10.3390/cells9061557
Chicago/Turabian StyleHromadnikova, Ilona, Katerina Kotlabova, Lenka Dvorakova, Ladislav Krofta, and Jan Sirc. 2020. "Substantially Altered Expression Profile of Diabetes/Cardiovascular/Cerebrovascular Disease Associated microRNAs in Children Descending from Pregnancy Complicated by Gestational Diabetes Mellitus—One of Several Possible Reasons for an Increased Cardiovascular Risk" Cells 9, no. 6: 1557. https://doi.org/10.3390/cells9061557
APA StyleHromadnikova, I., Kotlabova, K., Dvorakova, L., Krofta, L., & Sirc, J. (2020). Substantially Altered Expression Profile of Diabetes/Cardiovascular/Cerebrovascular Disease Associated microRNAs in Children Descending from Pregnancy Complicated by Gestational Diabetes Mellitus—One of Several Possible Reasons for an Increased Cardiovascular Risk. Cells, 9(6), 1557. https://doi.org/10.3390/cells9061557






