Genetic Control of Muscle Diversification and Homeostasis: Insights from Drosophila
Abstract
1. Introduction
1.1. General Overview
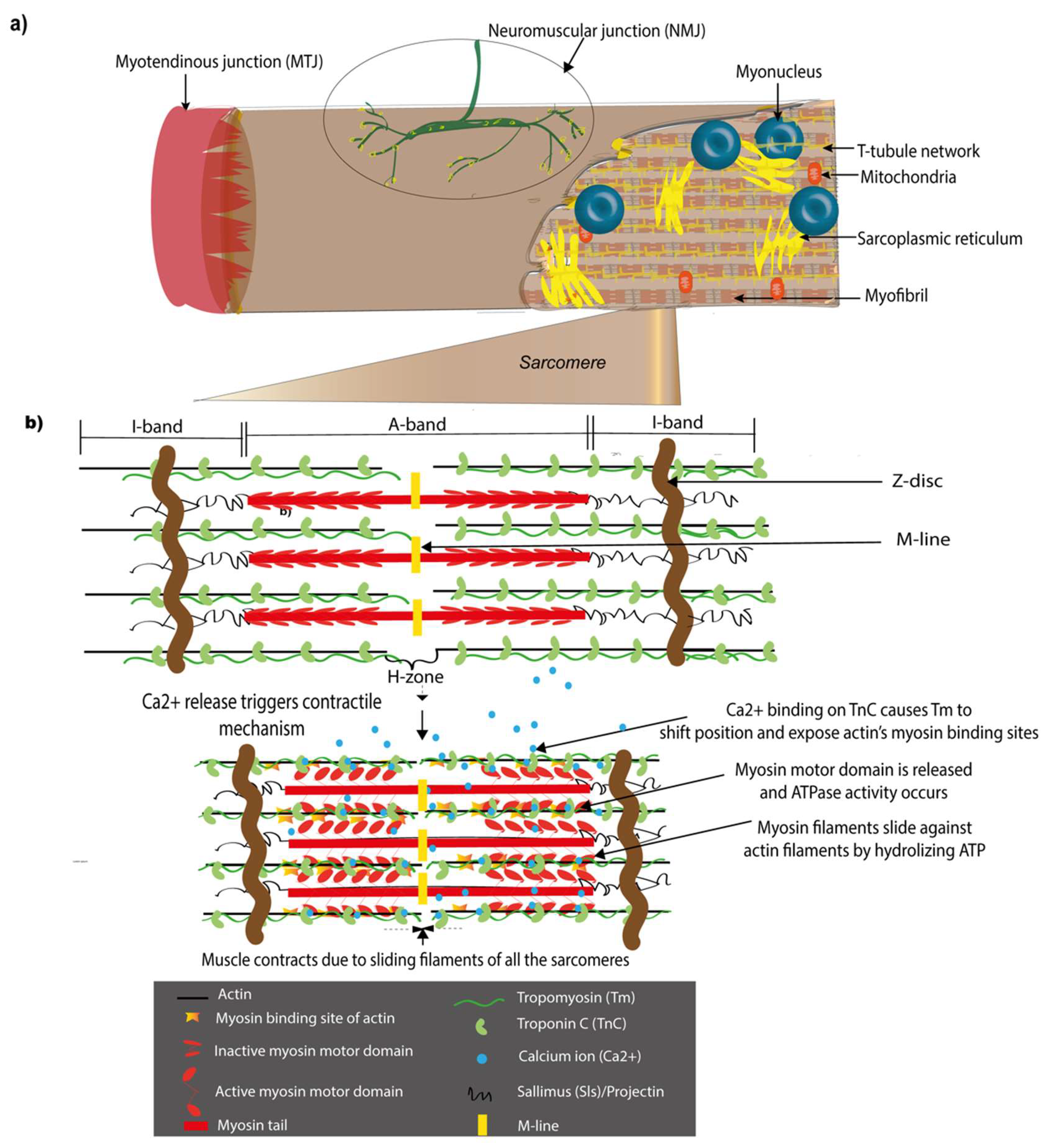
1.2. Major Structural Components of the Drosophila Muscle and Their Vertebrate Counterparts
1.2.1. Sarcomeres
1.2.2. Myotendinous Junctions (MTJs)
1.2.3. Neuromuscular Junctions (NMJs)
2. The Sarcomere and Molecular Mechanisms of Muscle Contraction
3. Muscle Diversification—On the Road to Muscle Homeostasis
- (a)
- (b)
- The two waves of myogenesis in Drosophila result in two homeostatic states, one in the larva and one in the adult. The larval homeostatic states are highly dynamic given the large growth spurt that occurs over the three larval instars. This might provide insights into mechanisms of muscle atrophy and hypertrophy. Forkhead box sub-group O (Foxo), for example, has been shown to inhibit larval muscle growth by repressing diminutive (myc) [62]. In mice, excess c-Myc has been shown to induce cardiac hypertrophy [63].
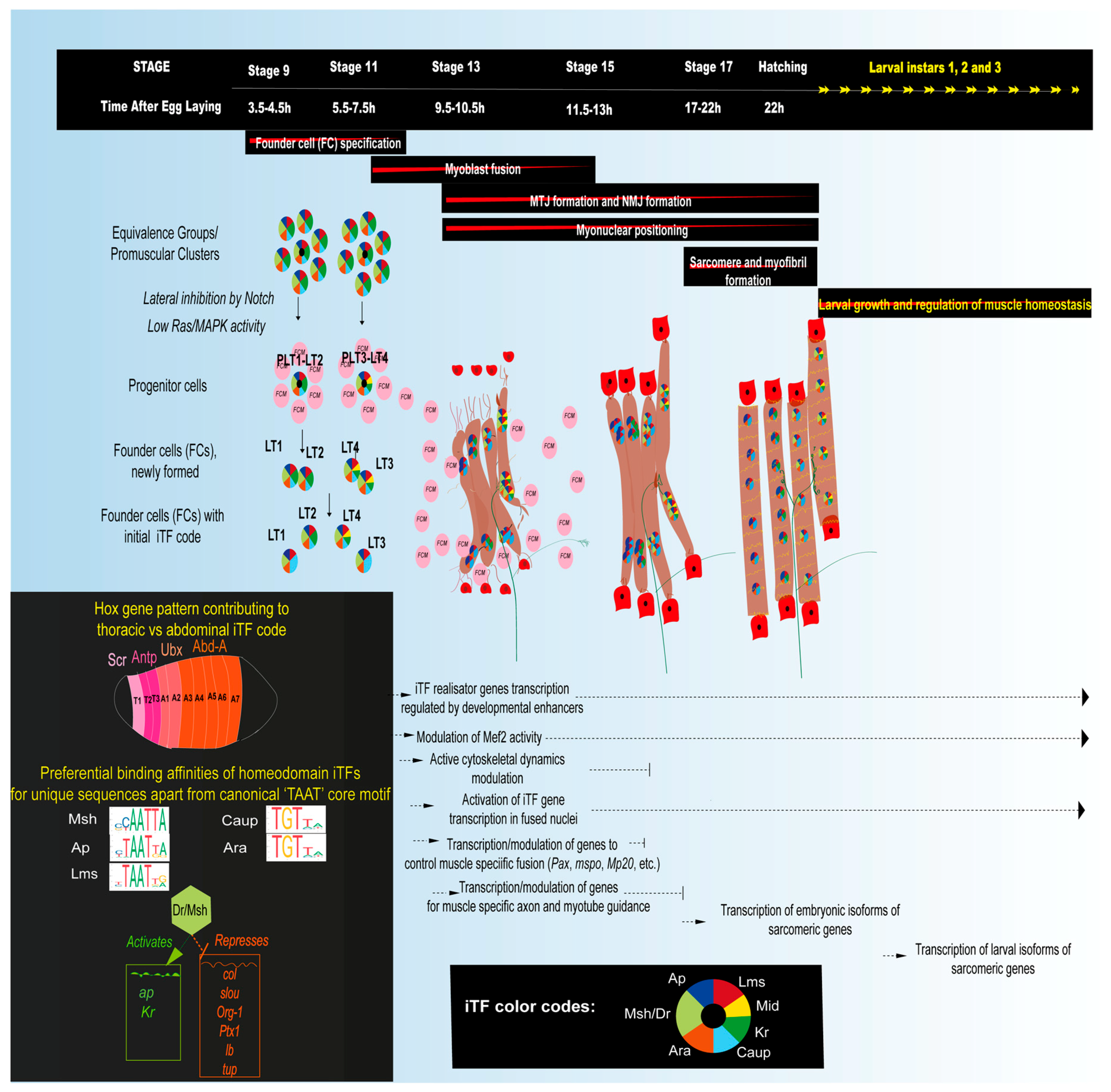
3.1. Embryonic Myogenesis of Larval Muscles
3.1.1. Muscle Diversification by the Specification of Muscle Founder Cells Expressing Identity Transcription Factors (iTFs)
3.1.2. The Role of iTFs
3.1.3. Mef2, a Key Muscle Differentiation Factor and Its Interactions with iTFs
3.1.4. Myoblast Fusion and Myonuclear Positioning
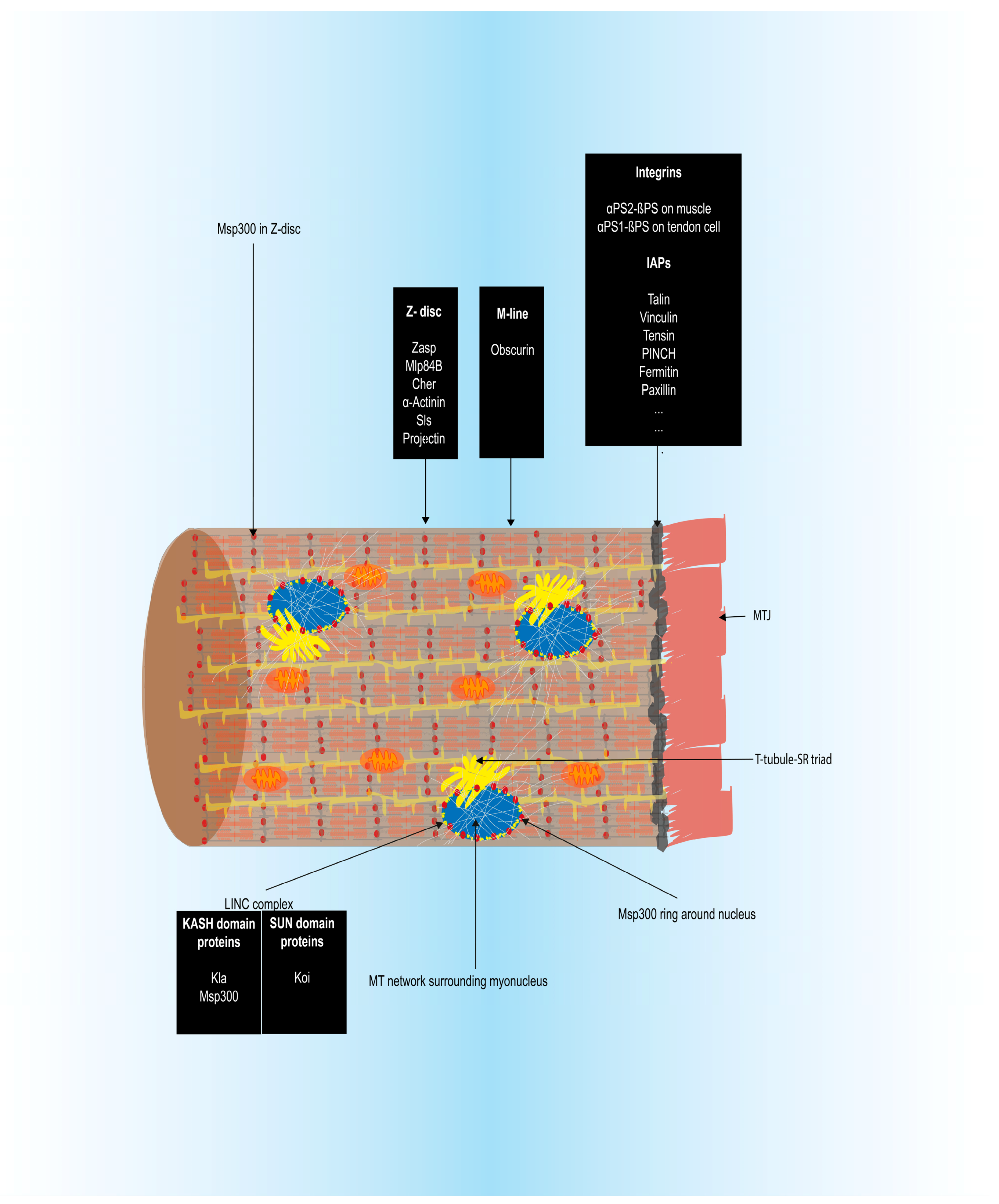
3.1.5. Myotendinous Junction (MTJ) Formation
3.1.6. Sarcomere Assembly and Myofibrillogenesis
3.1.7. Innervation and Neuromuscular Junction (NMJ) Formation
3.2. Pupal Myogenesis of Adult Muscles
3.2.1. Myoblast Pool Generation by Adult Muscle Precursors (AMPs) during Larval Stages
3.2.2. Histolysis of Larval Muscles, Adult iTF Code Refinement, and the Contribution of AMPs
3.2.3. MTJ Formation
3.2.4. Sarcomere Assembly
3.2.5. Innervation and NMJ Formation
3.2.6. Programmed Cell Death Following Eclosion of New Adults
4. The Maintenance of Muscle Homeostasis
4.1. Muscle Homeostasis under Normal Conditions
4.2. Re-Establishment of Muscle Homeostasis Following Muscle Injury
4.3. Muscle Homeostasis under Pathological Conditions
5. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Bate, M. The embryonic development of larval muscles in Drosophila. Development 1990, 110, 791. [Google Scholar] [PubMed]
- Abmayr, S.M.; Erickson, M.S.; Bour, B.A. Embryonic development of the larval body wall musculature of Drosophila melanogaster. Trends Genet. 1995, 11, 153–159. [Google Scholar] [CrossRef]
- Fyrberg, E. Study of contractile and cytoskeletal proteins using Drosophila genetics. Cell Motil. Cytoskelet. 1989, 14, 118–127. [Google Scholar] [CrossRef]
- Dobi, K.C.; Schulman, V.K.; Baylies, M.K. Specification of the somatic musculature in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 357–375. [Google Scholar] [CrossRef]
- Mukund, K.; Subramaniam, S. Skeletal muscle: A review of molecular structure and function, in health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1462. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M. Comparison of muscle development in Drosophila and vertebrates. In Muscle development in Drosophila; Sink, H., Ed.; Landes Bioscience/Springer: Georgetown, TX, USA; New York, NY, USA, 2006; pp. 169–203. [Google Scholar]
- Piccirillo, R.; Demontis, F.; Perrimon, N.; Goldberg, A.L. Mechanisms of muscle growth and atrophy in mammals and Drosophila. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2014, 243, 201–215. [Google Scholar] [CrossRef]
- Kohsaka, H.; Guertin, P.A.; Nose, A. Neural Circuits Underlying Fly Larval Locomotion. Curr. Pharm. Des. 2017, 23, 1722–1733. [Google Scholar] [CrossRef]
- Wolfe, R.R. The underappreciated role of muscle in health and disease. Am. J. Clin. Nutr. 2006, 84, 475–482. [Google Scholar] [CrossRef]
- Veratti, E. Investigations on the fine structure of striated muscle fiber read before the Reale Istituto Lombardo, 13 March 1902. J. Biophys. Biochem. Cytol. 1961, 10, 1–59. [Google Scholar] [CrossRef]
- Hanson, J.; Huxley, H.E. Structural Basis of the Cross-Striations in Muscle. Nature 1953, 172, 530–532. [Google Scholar] [CrossRef]
- Royuela, M.; Fraile, B.; Arenas, M.I.; Paniagua, R. Characterization of several invertebrate muscle cell types: A comparison with vertebrate muscles. Microsc. Res. Tech. 2000, 48, 107–115. [Google Scholar] [CrossRef]
- Littlefield, R.S.; Fowler, V.M. Thin filament length regulation in striated muscle sarcomeres: Pointed-end dynamics go beyond a nebulin ruler. Semin. Cell Dev. Biol. 2008, 19, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Lemke, S.B.; Schnorrer, F. Mechanical forces during muscle development. Mech. Dev. 2017, 144, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Geist, J.; Grogan, A.; Hu, L.-Y.R.; Kontrogianni-Konstantopoulos, A. Thick Filament Protein Network, Functions, and Disease Association. Compr. Physiol. 2018, 8, 631–709. [Google Scholar] [CrossRef] [PubMed]
- Hooper, S.L.; Thuma, J.B. Invertebrate Muscles: Muscle Specific Genes and Proteins. Physiol. Rev. 2005, 85, 1001–1060. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, R.; Zelzer, E.; Volk, T. Connecting muscles to tendons: Tendons and musculoskeletal development in flies and vertebrates. Dev. Camb. Engl. 2010, 137, 2807–2817. [Google Scholar] [CrossRef]
- Soler, C.; Laddada, L.; Jagla, K. Coordinated Development of Muscles and Tendon-Like Structures: Early Interactions in the Drosophila Leg. Front. Physiol. 2016, 7, 22. [Google Scholar] [CrossRef]
- Lemke, S.B.; Weidemann, T.; Cost, A.-L.; Grashoff, C.; Schnorrer, F. A small proportion of Talin molecules transmit forces at developing muscle attachments in vivo. PLoS Biol. 2019, 17, e3000057. [Google Scholar] [CrossRef]
- Richier, B.; Inoue, Y.; Dobramysl, U.; Friedlander, J.; Brown, N.H.; Gallop, J.L. Integrin signaling downregulates filopodia during muscle-tendon attachment. J. Cell Sci. 2018, 131, jcs217133. [Google Scholar] [CrossRef]
- Nawrotzki, R.; Willem, M.; Miosge, N.; Brinkmeier, H.; Mayer, U. Defective integrin switch and matrix composition at alpha 7-deficient myotendinous junctions precede the onset of muscular dystrophy in mice. Hum. Mol. Genet. 2003, 12, 483–495. [Google Scholar] [CrossRef]
- Marshall, J.L.; Chou, E.; Oh, J.; Kwok, A.; Burkin, D.J.; Crosbie-Watson, R.H. Dystrophin and utrophin expression require sarcospan: Loss of α7 integrin exacerbates a newly discovered muscle phenotype in sarcospan-null mice. Hum. Mol. Genet. 2012, 21, 4378–4393. [Google Scholar] [CrossRef] [PubMed]
- Broadie, K.; Bate, M. The Drosophila NMJ: A genetic model system for synapse formation and function. Semin. Dev. Biol. 1995, 6, 221–231. [Google Scholar] [CrossRef]
- Menon, K.P.; Carrillo, R.A.; Zinn, K. Development and plasticity of the Drosophila larval neuromuscular junction. WIREs Dev. Biol. 2013, 2, 647–670. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.P.; Littleton, J.T. Transmission, Development, and Plasticity of Synapses. Genetics 2015, 201, 345–375. [Google Scholar] [CrossRef]
- Lacin, H.; Truman, J.W. Lineage mapping identifies molecular and architectural similarities between the larval and adult Drosophila central nervous system. eLife 2016, 5, e13399. [Google Scholar] [CrossRef]
- Pérez-Moreno, J.J.; O’Kane, C.J. GAL4 Drivers Specific for Type Ib and Type Is Motor Neurons in Drosophila. G3 Genes Genomes Genet. 2019, 9, 453. [Google Scholar] [CrossRef]
- Dasari, S.; Cooper, R.L. Modulation of sensory–CNS–motor circuits by serotonin, octopamine, and dopamine in semi-intact Drosophila larva. Neurosci. Res. 2004, 48, 221–227. [Google Scholar] [CrossRef]
- Kohsaka, H.; Takasu, E.; Morimoto, T.; Nose, A. A Group of Segmental Premotor Interneurons Regulates the Speed of Axial Locomotion in Drosophila Larvae. Curr. Biol. 2014, 24, 2632–2642. [Google Scholar] [CrossRef]
- Babski, H.; Jovanic, T.; Surel, C.; Yoshikawa, S.; Zwart, M.F.; Valmier, J.; Thomas, J.B.; Enriquez, J.; Carroll, P.; Garcès, A. A GABAergic Maf-expressing interneuron subset regulates the speed of locomotion in Drosophila. Nat. Commun. 2019, 10, 4796. [Google Scholar] [CrossRef]
- Vigoreaux, J.O. Genetics of the Drosophila flight muscle myofibril: A window into the biology of complex systems. BioEssays 2001, 23, 1047–1063. [Google Scholar] [CrossRef]
- Campbell, K.B.; Chandra, M. Functions of stretch activation in heart muscle. J. Gen. Physiol. 2006, 127, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Huxley, H.E. Fifty years of muscle and the sliding filament hypothesis. Eur. J. Biochem. 2004, 271, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-H.; Clark, C.; Geisbrecht, E.R. Analysis of mitochondrial structure and function in the Drosophila larval musculature. Mitochondrion 2016, 26, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Auld, A.L.; Folker, E.S. Nucleus-dependent sarcomere assembly is mediated by the LINC complex. Mol. Biol. Cell 2016, 27, 2351–2359. [Google Scholar] [CrossRef] [PubMed]
- Al-Qusairi, L.; Laporte, J. T-tubule biogenesis and triad formation in skeletal muscle and implication in human diseases. Skelet. Muscle 2011, 1, 26. [Google Scholar] [CrossRef]
- Razzaq, A.; Robinson, I.M.; McMahon, H.T.; Skepper, J.N.; Su, Y.; Zelhof, A.C.; Jackson, A.P.; Gay, N.J.; O’Kane, C.J. Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 2001, 15, 2967–2979. [Google Scholar] [CrossRef]
- Ackermann, M.A.; Ziman, A.P.; Strong, J.; Zhang, Y.; Hartford, A.K.; Ward, C.W.; Randall, W.R.; Kontrogianni-Konstantopoulos, A.; Bloch, R.J. Integrity of the network sarcoplasmic reticulum in skeletal muscle requires small ankyrin 1. J. Cell Sci. 2011, 124, 3619–3630. [Google Scholar] [CrossRef]
- Maughan, D.W.; Godt, R.E. Equilibrium distribution of ions in a muscle fiber. Biophys. J. 1989, 56, 717–722. [Google Scholar] [CrossRef]
- Clausen, T. Na+-K+ Pump Stimulation Improves Contractility in Damaged Muscle Fibers. Ann. N. Y. Acad. Sci. 2006, 1066, 286–294. [Google Scholar] [CrossRef]
- Lebovitz, R.M.; Takeyasu, K.; Fambrough, D.M. Molecular characterization and expression of the (Na+ + K+)-ATPase alpha-subunit in Drosophila melanogaster. EMBO J. 1989, 8, 193–202. [Google Scholar] [CrossRef]
- Yatsenko, A.S.; Kucherenko, M.M.; Xie, Y.; Aweida, D.; Urlaub, H.; Scheibe, R.J.; Cohen, S.; Shcherbata, H.R. Profiling of the muscle-specific dystroglycan interactome reveals the role of Hippo signaling in muscular dystrophy and age-dependent muscle atrophy. BMC Med. 2020, 18, 8. [Google Scholar] [CrossRef] [PubMed]
- Huxley, H.; Hanson, J. Changes in the Cross-Striations of Muscle during Contraction and Stretch and their Structural Interpretation. Nature 1954, 173, 973–976. [Google Scholar] [CrossRef]
- Huxley, A.F.; Niedergerke, R. Structural Changes in Muscle During Contraction: Interference Microscopy of Living Muscle Fibres. Nature 1954, 173, 971–973. [Google Scholar] [CrossRef] [PubMed]
- Potter, J.D.; Sheng, Z.; Pan, B.-S.; Zhao, J. A Direct Regulatory Role for Troponin T and a Dual Role for Troponin C in the Ca2+ Regulation of Muscle Contraction. J. Biol. Chem. 1995, 270, 2557–2562. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.; Lakey, A.; Agianian, B.; Hutchings, A.; Butcher, G.W.; Labeit, S.; Leonard, K.; Bullard, B. Troponin C in different insect muscle types: Identification of two isoforms in Lethocerus, Drosophila and Anopheles that are specific to asynchronous flight muscle in the adult insect. Biochem. J. 2003, 371, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Vibert, P.; Craig, R.; Lehman, W. Steric-model for activation of muscle thin filaments 1 1 Edited by P.E. Wright. J. Mol. Biol. 1997, 266, 8–14. [Google Scholar] [CrossRef]
- Farman, G.P.; Miller, M.S.; Reedy, M.C.; Soto-Adames, F.N.; Vigoreaux, J.O.; Maughan, D.W.; Irving, T.C. Phosphorylation and the N-terminal extension of the regulatory light chain help orient and align the myosin heads in Drosophila flight muscle. J. Struct. Biol. 2009, 168, 240–249. [Google Scholar] [CrossRef]
- Beall, C.J.; Fyrberg, E. Muscle abnormalities in Drosophila melanogaster heldup mutants are caused by missing or aberrant troponin-I isoforms. J. Cell Biol. 1991, 114, 941–951. [Google Scholar] [CrossRef]
- Lee, K.H.; Sulbarán, G.; Yang, S.; Mun, J.Y.; Alamo, L.; Pinto, A.; Sato, O.; Ikebe, M.; Liu, X.; Korn, E.D.; et al. Interacting-heads motif has been conserved as a mechanism of myosin II inhibition since before the origin of animals. Proc. Natl. Acad. Sci. USA 2018, 115, E1991–E2000. [Google Scholar] [CrossRef]
- Jung, H.S.; Komatsu, S.; Ikebe, M.; Craig, R. Head-head and head-tail interaction: A general mechanism for switching off myosin II activity in cells. Mol. Biol. Cell 2008, 19, 3234–3242. [Google Scholar] [CrossRef]
- Green, H.J.; Griffiths, A.G.; Ylänne, J.; Brown, N.H. Novel functions for integrin-associated proteins revealed by analysis of myofibril attachment in Drosophila. eLife 2018, 7, e35783. [Google Scholar] [CrossRef] [PubMed]
- González-Morales, N.; Holenka, T.K.; Schöck, F. Filamin actin-binding and titin-binding fulfill distinct functions in Z-disc cohesion. PLoS Genet. 2017, 13, e1006880. [Google Scholar] [CrossRef] [PubMed]
- Liao, K.A.; González-Morales, N.; Schöck, F. Zasp52, a Core Z-disc Protein in Drosophila Indirect Flight Muscles, Interacts with α-Actinin via an Extended PDZ Domain. PLoS Genet. 2016, 12, e1006400. [Google Scholar] [CrossRef] [PubMed]
- Katzemich, A.; West, R.J.H.; Fukuzawa, A.; Sweeney, S.T.; Gautel, M.; Sparrow, J.; Bullard, B. Binding partners of the kinase domains in Drosophila obscurin and their effect on the structure of the flight muscle. J. Cell Sci. 2015, 128, 3386–3397. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.A.; Bland, J.M.; Beckerle, M.C. The Drosophila muscle LIM protein, Mlp84B, cooperates with D-titin to maintain muscle structural integrity. J. Cell Sci. 2007, 120, 2066. [Google Scholar] [CrossRef] [PubMed]
- Perkins, A.D.; Ellis, S.J.; Asghari, P.; Shamsian, A.; Moore, E.D.W.; Tanentzapf, G. Integrin-mediated adhesion maintains sarcomeric integrity. Dev. Biol. 2010, 338, 15–27. [Google Scholar] [CrossRef]
- Prill, K.; Dawson, J.F. Assembly and Maintenance of Sarcomere Thin Filaments and Associated Diseases. Int. J. Mol. Sci. 2020, 21, 542. [Google Scholar] [CrossRef]
- Ciglar, L.; Furlong, E.E. Conservation and divergence in developmental networks: A view from Drosophila myogenesis. Cell Differ. Cell Div. Growth Death 2009, 21, 754–760. [Google Scholar] [CrossRef]
- Karalaki, M.; Fili, S.; Philippou, A.; Koutsilieris, M. Muscle regeneration: Cellular and molecular events. Vivo Athens Greece 2009, 23, 779–796. [Google Scholar]
- Chaturvedi, D.; Reichert, H.; Gunage, R.D.; VijayRaghavan, K. Identification and functional characterization of muscle satellite cells in Drosophila. eLife 2017, 6, e30107. [Google Scholar] [CrossRef]
- Demontis, F.; Perrimon, N. Integration of Insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Dev. Camb. Engl. 2009, 136, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Mao, S.; Baumgarten, G.; Serrano, J.; Jordan, M.C.; Roos, K.P.; Fishbein, M.C.; MacLellan, W.R. Inducible activation of c-Myc in adult myocardium in vivo provokes cardiac myocyte hypertrophy and reactivation of DNA synthesis. Circ. Res. 2001, 89, 1122–1129. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.C. The Physical Mechanisms of Drosophila Gastrulation: Mesoderm and Endoderm Invagination. Genetics 2020, 214, 543. [Google Scholar] [CrossRef] [PubMed]
- Azpiazu, N.; Lawrence, P.A.; Vincent, J.P.; Frasch, M. Segmentation and specification of the Drosophila mesoderm. Genes Dev. 1996, 10, 3183–3194. [Google Scholar] [CrossRef]
- Baylies, M.K.; Bate, M. twist: A Myogenic Switch in Drosophila. Science 1996, 272, 1481. [Google Scholar] [CrossRef] [PubMed]
- Carmena, A.; Bate, M.; Jiménez, F. Lethal of scute, a proneural gene, participates in the specification of muscle progenitors during Drosophila embryogenesis. Genes Dev. 1995, 9, 2373–2383. [Google Scholar] [CrossRef]
- Carmena, A.; Buff, E.; Halfon, M.S.; Gisselbrecht, S.; Jiménez, F.; Baylies, M.K.; Michelson, A.M. Reciprocal Regulatory Interactions between the Notch and Ras Signaling Pathways in the Drosophila Embryonic Mesoderm. Dev. Biol. 2002, 244, 226–242. [Google Scholar] [CrossRef]
- Doe, C.Q.; Skeath, J.B. Neurogenesis in the insect central nervous system. Curr. Opin. Neurobiol. 1996, 6, 18–24. [Google Scholar] [CrossRef]
- Crews, S.T. Drosophila Embryonic CNS Development: Neurogenesis, Gliogenesis, Cell Fate, and Differentiation. Genetics 2019, 213, 1111. [Google Scholar] [CrossRef]
- Gomez Ruiz, M.; Bate, M. Segregation of myogenic lineages in Drosophila requires numb. Development 1997, 124, 4857. [Google Scholar]
- Liu, J.; Qian, L.; Wessells, R.J.; Bidet, Y.; Jagla, K.; Bodmer, R. Hedgehog and RAS pathways cooperate in the anterior–posterior specification and positioning of cardiac progenitor cells. Dev. Biol. 2006, 290, 373–385. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rushton, E.; Drysdale, R.; Abmayr, S.M.; Michelson, A.M.; Bate, M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development 1995, 121, 1979. [Google Scholar] [PubMed]
- Prokop, A.; Landgraf, M.; Rushton, E.; Broadie, K.; Bate, M. Presynaptic Development at the Drosophila Neuromuscular Junction: Assembly and Localization of Presynaptic Active Zones. Neuron 1996, 17, 617–626. [Google Scholar] [CrossRef]
- Halfon, M.S.; Carmena, A.; Gisselbrecht, S.; Sackerson, C.M.; Jiménez, F.; Baylies, M.K.; Michelson, A.M. Ras Pathway Specificity Is Determined by the Integration of Multiple Signal-Activated and Tissue-Restricted Transcription Factors. Cell 2000, 103, 63–74. [Google Scholar] [CrossRef]
- Cox, V.T.; Beckett, K.; Baylies, M.K. Delivery of wingless to the ventral mesoderm by the developing central nervous system ensures proper patterning of individual slouch-positive muscle progenitors. Dev. Biol. 2005, 287, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Dohrmann, C.; Azpiazu, N.; Frasch, M. A new Drosophila homeo box gene is expressed in mesodermal precursor cells of distinct muscles during embryogenesis. Genes Dev. 1990, 4, 2098–2111. [Google Scholar] [CrossRef]
- Bourgouin, C.; Lundgren, S.E.; Thomas, J.B. Apterous is a drosophila LIM domain gene required for the development of a subset of embryonic muscles. Neuron 1992, 9, 549–561. [Google Scholar] [CrossRef]
- Keller, C.A.; Grill, M.A.; Abmayr, S.M. A Role for nautilus in the Differentiation of Muscle Precursors. Dev. Biol. 1998, 202, 157–171. [Google Scholar] [CrossRef]
- Knirr, S.; Azpiazu, N.; Frasch, M. The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development 1999, 126, 4525. [Google Scholar]
- Boukhatmi, H.; Frendo, J.L.; Enriquez, J.; Crozatier, M.; Dubois, L.; Vincent, A. Tup/Islet1 integrates time and position to specify muscle identity in Drosophila. Development 2012, 139, 3572. [Google Scholar] [CrossRef]
- Crozatier, M.; Vincent, A. Requirement for the Drosophila COE transcription factor Collier in formation of an embryonic muscle: Transcriptional response to notch signalling. Development 1999, 126, 1495. [Google Scholar] [PubMed]
- Busser, B.W.; Shokri, L.; Jaeger, S.A.; Gisselbrecht, S.S.; Singhania, A.; Berger, M.F.; Zhou, B.; Bulyk, M.L.; Michelson, A.M. Molecular mechanism underlying the regulatory specificity of a Drosophila homeodomain protein that specifies myoblast identity. Dev. Camb. Engl. 2012, 139, 1164–1174. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Busser, B.W.; Gisselbrecht, S.S.; Shokri, L.; Tansey, T.R.; Gamble, C.E.; Bulyk, M.L.; Michelson, A.M. Contribution of distinct homeodomain DNA binding specificities to Drosophila embryonic mesodermal cell-specific gene expression programs. PLoS ONE 2013, 8, e69385. [Google Scholar] [CrossRef] [PubMed]
- Dubois, L.; Frendo, J.-L.; Chanut-Delalande, H.; Crozatier, M.; Vincent, A. Genetic dissection of the Transcription Factor code controlling serial specification of muscle identities in Drosophila. eLife 2016, 5, e14979. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Bell, J.B.; Simmonds, A.J. Vestigial is required during late-stage muscle differentiation in Drosophila melanogaster embryos. Mol. Biol. Cell 2010, 21, 3304–3316. [Google Scholar] [CrossRef][Green Version]
- Carrasco-Rando, M.; Tutor, A.S.; Prieto-Sánchez, S.; González-Pérez, E.; Barrios, N.; Letizia, A.; Martín, P.; Campuzano, S.; Ruiz-Gómez, M. Drosophila araucan and caupolican integrate intrinsic and signalling inputs for the acquisition by muscle progenitors of the lateral transverse fate. PLoS Genet. 2011, 7, e1002186. [Google Scholar] [CrossRef]
- Enriquez, J.; de Taffin, M.; Crozatier, M.; Vincent, A.; Dubois, L. Combinatorial coding of Drosophila muscle shape by Collier and Nautilus. Dev. Biol. 2012, 363, 27–39. [Google Scholar] [CrossRef]
- Nose, A.; Isshiki, T.; Takeichi, M. Regional specification of muscle progenitors in Drosophila: The role of the msh homeobox gene. Development 1998, 125, 215. [Google Scholar]
- Lord, P.C.W.; Lin, M.-H.; Hales, K.H.; Storti, R.V. Normal Expression and the Effects of Ectopic Expression of the Drosophila muscle segment homeobox (msh) Gene Suggest a Role in Differentiation and Patterning of Embryonic Muscles. Dev. Biol. 1995, 171, 627–640. [Google Scholar] [CrossRef][Green Version]
- Knirr, S.; Frasch, M. Molecular Integration of Inductive and Mesoderm-Intrinsic Inputs Governs even-skipped Enhancer Activity in a Subset of Pericardial and Dorsal Muscle Progenitors. Dev. Biol. 2001, 238, 13–26. [Google Scholar] [CrossRef]
- Fujioka, M.; Wessells, R.J.; Han, Z.; Liu, J.; Fitzgerald, K.; Yusibova, G.L.; Zamora, M.; Ruiz-Lozano, P.; Bodmer, R.; Jaynes, J.B. Embryonic even skipped-dependent muscle and heart cell fates are required for normal adult activity, heart function, and lifespan. Circ. Res. 2005, 97, 1108–1114. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Gomez, M.; Romani, S.; Hartmann, C.; Jackle, H.; Bate, M. Specific muscle identities are regulated by Kruppel during Drosophila embryogenesis. Development 1997, 124, 3407. [Google Scholar] [PubMed]
- Jagla, T.; Bellard, F.; Lutz, Y.; Dretzen, G.; Bellard, M.; Jagla, K. ladybird determines cell fate decisions during diversification of Drosophila somatic muscles. Development 1998, 125, 3699. [Google Scholar] [PubMed]
- Müller, D.; Jagla, T.; Bodart, L.M.; Jährling, N.; Dodt, H.-U.; Jagla, K.; Frasch, M. Regulation and functions of the lms homeobox gene during development of embryonic lateral transverse muscles and direct flight muscles in Drosophila. PLoS ONE 2010, 5, e14323. [Google Scholar] [CrossRef]
- Kumar, R.P.; Dobi, K.C.; Baylies, M.K.; Abmayr, S.M. Muscle cell fate choice requires the T-box transcription factor midline in Drosophila. Genetics 2015, 199, 777–791. [Google Scholar] [CrossRef][Green Version]
- Corbin, V.; Michelson, A.M.; Abmayr, S.M.; Neel, V.; Alcamo, E.; Maniatis, T.; Young, M.W. A role for the Drosophila neurogenic genes in mesoderm differentiation. Cell 1991, 67, 311–323. [Google Scholar] [CrossRef]
- Schaub, C.; Nagaso, H.; Jin, H.; Frasch, M. Org-1, the Drosophila ortholog of Tbx1, is a direct activator of known identity genes during muscle specification. Development 2012, 139, 1001. [Google Scholar] [CrossRef]
- Duan, H.; Zhang, C.; Chen, J.; Sink, H.; Frei, E.; Noll, M. A key role of Pox meso in somatic myogenesis of Drosophila. Development 2007, 134, 3985. [Google Scholar] [CrossRef]
- Vorbrüggen, G.; Constien, R.; Zilian, O.; Wimmer, E.A.; Dowe, G.; Taubert, H.; Noll, M.; Jäckle, H. Embryonic expression and characterization of a Ptx1 homolog in Drosophila. Mech. Dev. 1997, 68, 139–147. [Google Scholar] [CrossRef]
- Drysdale, R.; Rushton, E.; Bate, M. Genes required for embryonic muscle development in Drosophila melanogaster A survey of the X chromosome. Rouxs Arch. Dev. Biol. Off. Organ EDBO 1993, 202, 276–295. [Google Scholar] [CrossRef]
- Liu, Y.-H.; Jakobsen, J.S.; Valentin, G.; Amarantos, I.; Gilmour, D.T.; Furlong, E.E.M. A Systematic Analysis of Tinman Function Reveals Eya and JAK-STAT Signaling as Essential Regulators of Muscle Development. Dev. Cell 2009, 16, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Clark, I.B.N.; Boyd, J.; Hamilton, G.; Finnegan, D.J.; Jarman, A.P. D-six4 plays a key role in patterning cell identities deriving from the Drosophila mesoderm. Dev. Biol. 2006, 294, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Jagla, T.; Bidet, Y.; Da Ponte, J.P.; Dastugue, B.; Jagla, K. Cross-repressive interactions of identity genes are essential for proper specification of cardiac and muscular fates in Drosophila. Development 2002, 129, 1037. [Google Scholar] [PubMed]
- Dobi, K.C.; Halfon, M.S.; Baylies, M.K. Whole-Genome Analysis of Muscle Founder Cells Implicates the Chromatin Regulator Sin3A in Muscle Identity. Cell Rep. 2014, 8, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Capovilla, M.; Kambris, Z.; Botas, J. Direct regulation of the muscle-identity gene apterous by a Hox protein in the somatic mesoderm. Development 2001, 128, 1221. [Google Scholar] [PubMed]
- Michelson, A.M. Muscle pattern diversification in Drosophila is determined by the autonomous function of homeotic genes in the embryonic mesoderm. Development 1994, 120, 755. [Google Scholar]
- Enriquez, J.; Boukhatmi, H.; Dubois, L.; Philippakis, A.A.; Bulyk, M.L.; Michelson, A.M.; Crozatier, M.; Vincent, A. Multi-step control of muscle diversity by Hox proteins in the Drosophila embryo. Dev. Camb. Engl. 2010, 137, 457–466. [Google Scholar] [CrossRef][Green Version]
- Domsch, K.; Carnesecchi, J.; Disela, V.; Friedrich, J.; Trost, N.; Ermakova, O.; Polychronidou, M.; Lohmann, I. The Hox transcription factor Ubx stabilizes lineage commitment by suppressing cellular plasticity in Drosophila. eLife 2019, 8, e42675. [Google Scholar] [CrossRef]
- Hessinger, C.; Technau, G.M.; Rogulja-Ortmann, A. The Drosophila Hox gene Ultrabithorax acts in both muscles and motoneurons to orchestrate formation of specific neuromuscular connections. Development 2017, 144, 139. [Google Scholar] [CrossRef]
- Junion, G.; Bataillé, L.; Jagla, T.; Da Ponte, J.P.; Tapin, R.; Jagla, K. Genome-wide view of cell fate specification: Ladybird acts at multiple levels during diversification of muscle and heart precursors. Genes Dev. 2007, 21, 3163–3180. [Google Scholar] [CrossRef]
- Bataillé, L.; Delon, I.; Da Ponte, J.P.; Brown, N.H.; Jagla, K. Downstream of Identity Genes: Muscle-Type-Specific Regulation of the Fusion Process. Dev. Cell 2010, 19, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Black, B.L.; Olson, E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (Mef2) proteins. Annu. Rev. Cell Dev. Biol. 1998, 14, 167–196. [Google Scholar] [CrossRef] [PubMed]
- Pon, J.R.; Marra, M.A. MEF2 transcription factors: Developmental regulators and emerging cancer genes. Oncotarget 2016, 7, 2297–2312. [Google Scholar] [CrossRef]
- Taylor, M.V.; Hughes, S.M. Mef2 and the skeletal muscle differentiation program. Skelet. Muscle Dev. 30th Anniv. MyoD 2017, 72, 33–44. [Google Scholar] [CrossRef]
- Bour, B.A.; O’Brien, M.A.; Lockwood, W.L.; Goldstein, E.S.; Bodmer, R.; Taghert, P.H.; Abmayr, S.M.; Nguyen, H.T. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995, 9, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Ranganayakulu, G.; Zhao, B.; Dokidis, A.; Molkentin, J.D.; Olson, E.N.; Schulz, R.A. A Series of Mutations in the D-MEF2 Transcription Factor Reveal Multiple Functions in Larval and Adult Myogenesis in Drosophila. Dev. Biol. 1995, 171, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Elgar, S.J.; Han, J.; Taylor, M.V. mef2 activity levels differentially affect gene expression during Drosophila muscle development. Proc. Natl. Acad. Sci. USA 2008, 105, 918–923. [Google Scholar] [CrossRef]
- Cunha, P.M.F.; Sandmann, T.; Gustafson, E.H.; Ciglar, L.; Eichenlaub, M.P.; Furlong, E.E.M. Combinatorial binding leads to diverse regulatory responses: Lmd is a tissue-specific modulator of Mef2 activity. PLoS Genet. 2010, 6, e1001014. [Google Scholar] [CrossRef]
- Junion, G.; Jagla, T.; Duplant, S.; Tapin, R.; Da Ponte, J.-P.; Jagla, K. Mapping Dmef2-binding regulatory modules by using a ChIP-enriched in silico targets approach. Proc. Natl. Acad. Sci. USA 2005, 102, 18479. [Google Scholar] [CrossRef]
- Tanaka, K.K.K.; Bryantsev, A.L.; Cripps, R.M. Myocyte Enhancer Factor 2 and Chorion Factor 2 Collaborate in Activation of the Myogenic Program in Drosophila. Mol. Cell. Biol. 2008, 28, 1616. [Google Scholar] [CrossRef]
- García-Zaragoza, E.; Mas, J.A.; Vivar, J.; Arredondo, J.J.; Cervera, M. CF2 activity and enhancer integration are required for proper muscle gene expression in Drosophila. Mech. Dev. 2008, 125, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Cripps, R.M.; Lovato, T.L.; Olson, E.N. Positive autoregulation of the Myocyte enhancer factor-2 myogenic control gene during somatic muscle development in Drosophila. Dev. Biol. 2004, 267, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Xu, X. Drosophila mef2 Expression during Mesoderm Development Is Controlled by a Complex Array ofcis-Acting Regulatory Modules. Dev. Biol. 1998, 204, 550–566. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Liang, S.; Zhao, Y.; Han, Z. miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Dev. Camb. Engl. 2012, 139, 3543–3552. [Google Scholar] [CrossRef] [PubMed]
- Cripps, R.M.; Black, B.L.; Zhao, B.; Lien, C.L.; Schulz, R.A.; Olson, E.N. The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 1998, 12, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Nowak, S.J.; Aihara, H.; Gonzalez, K.; Nibu, Y.; Baylies, M.K. Akirin links twist-regulated transcription with the Brahma chromatin remodeling complex during embryogenesis. PLoS Genet. 2012, 8, e1002547. [Google Scholar] [CrossRef]
- Wang, F.; Minakhina, S.; Tran, H.; Changela, N.; Kramer, J.; Steward, R. Tet protein function during Drosophila development. PLoS ONE 2018, 13, e0190367. [Google Scholar] [CrossRef]
- Deng, H.; Hughes, S.C.; Bell, J.B.; Simmonds, A.J. Alternative requirements for Vestigial, Scalloped, and Dmef2 during muscle differentiation in Drosophila melanogaster. Mol. Biol. Cell 2009, 20, 256–269. [Google Scholar] [CrossRef][Green Version]
- Haralalka, S.; Abmayr, S.M. Myoblast fusion in Drosophila. Exp. Cell Res. 2010, 316, 3007–3013. [Google Scholar] [CrossRef]
- Deng, S.; Azevedo, M.; Baylies, M. Acting on identity: Myoblast fusion and the formation of the syncytial muscle fiber. Semin. Cell Dev. Biol. 2017, 72, 45–55. [Google Scholar] [CrossRef]
- Lee, D.M.; Chen, E.H. Drosophila Myoblast Fusion: Invasion and Resistance for the Ultimate Union. Annu. Rev. Genet. 2019, 53, 67–91. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Gómez, M.; Coutts, N.; Price, A.; Taylor, M.V.; Bate, M. Drosophila Dumbfounded: A Myoblast Attractant Essential for Fusion. Cell 2000, 102, 189–198. [Google Scholar] [CrossRef]
- Strünkelnberg, M.; Bonengel, B.; Moda, L.M.; Hertenstein, A.; de Couet, H.G.; Ramos, R.G.P.; Fischbach, K.-F. rst and its paralogue kirre act redundantly during embryonic muscle development in Drosophila. Development 2001, 128, 4229. [Google Scholar]
- Bour, B.A.; Chakravarti, M.; West, J.M.; Abmayr, S.M. Drosophila SNS, a member of the immunoglobulin superfamily that is essential for myoblast fusion. Genes Dev. 2000, 14, 1498–1511. [Google Scholar] [PubMed]
- Artero, R.D.; Castanon, I.; Baylies, M.K. The immunoglobulin-like protein Hibris functions as a dose-dependent regulator of myoblast fusion and is differentially controlled by Ras and Notch signaling. Development 2001, 128, 4251. [Google Scholar]
- Bataillé, L.; Boukhatmi, H.; Frendo, J.-L.; Vincent, A. Dynamics of transcriptional (re)-programming of syncytial nuclei in developing muscles. BMC Biol. 2017, 15. [Google Scholar] [CrossRef]
- Azevedo, M.; Baylies, M.K. Getting into Position: Nuclear Movement in Muscle Cells. Trends Cell Biol. 2020, 30, 303–316. [Google Scholar] [CrossRef]
- Roman, W.; Gomes, E.R. Nuclear positioning in skeletal muscle. SI Nucl. Position. 2018, 82, 51–56. [Google Scholar] [CrossRef]
- Cadot, B.; Gache, V.; Gomes, E.R. Moving and positioning the nucleus in skeletal muscle - one step at a time. Nucl. Austin Tex 2015, 6, 373–381. [Google Scholar] [CrossRef]
- Starr, D.A.; Fridolfsson, H.N. Interactions Between Nuclei and the Cytoskeleton Are Mediated by SUN-KASH Nuclear-Envelope Bridges. Annu. Rev. Cell Dev. Biol. 2010, 26, 421–444. [Google Scholar] [CrossRef]
- Folker, E.S.; Baylies, M.K. Nuclear positioning in muscle development and disease. Front. Physiol. 2013, 4, 363. [Google Scholar] [CrossRef] [PubMed]
- Volk, T. Singling out Drosophila tendon cells: A dialogue between two distinct cell types. Trends Genet. 1999, 15, 448–453. [Google Scholar] [CrossRef]
- Schnorrer, F.; Dickson, B.J. Muscle Building: Mechanisms of Myotube Guidance and Attachment Site Selection. Dev. Cell 2004, 7, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Schilling, T.F. Tendon development and musculoskeletal assembly: Emerging roles for the extracellular matrix. Dev. Camb. Engl. 2015, 142, 4191–4204. [Google Scholar] [CrossRef]
- Lejard, V.; Blais, F.; Guerquin, M.-J.; Bonnet, A.; Bonnin, M.-A.; Havis, E.; Malbouyres, M.; Bidaud, C.B.; Maro, G.; Gilardi-Hebenstreit, P.; et al. EGR1 and EGR2 involvement in vertebrate tendon differentiation. J. Biol. Chem. 2011, 286, 5855–5867. [Google Scholar] [CrossRef]
- Volohonsky, G.; Edenfeld, G.; Klämbt, C.; Volk, T. Muscle-dependent maturation of tendon cells is induced by post-transcriptional regulation of stripeA. Development 2007, 134, 347. [Google Scholar] [CrossRef]
- Kramer, S.G.; Kidd, T.; Simpson, J.H.; Goodman, C.S. Switching Repulsion to Attraction: Changing Responses to Slit During Transition in Mesoderm Migration. Science 2001, 292, 737. [Google Scholar] [CrossRef]
- Callahan, C.A.; Bonkovsky, J.L.; Scully, A.L.; Thomas, J.B. derailed is required for muscle attachment site selection in Drosophila. Development 1996, 122, 2761. [Google Scholar]
- Schnorrer, F.; Kalchhauser, I.; Dickson, B.J. The Transmembrane Protein Kon-tiki Couples to Dgrip to Mediate Myotube Targeting in Drosophila. Dev. Cell 2007, 12, 751–766. [Google Scholar] [CrossRef]
- Estrada, B.; Gisselbrecht, S.S.; Michelson, A.M. The transmembrane protein Perdido interacts with Grip and integrins to mediate myotube projection and attachment in the Drosophila embryo. Development 2007, 134, 4469. [Google Scholar] [CrossRef]
- Swan, L.E.; Schmidt, M.; Schwarz, T.; Ponimaskin, E.; Prange, U.; Boeckers, T.; Thomas, U.; Sigrist, S.J. Complex interaction of Drosophila GRIP PDZ domains and Echinoid during muscle morphogenesis. EMBO J. 2006, 25, 3640–3651. [Google Scholar] [CrossRef] [PubMed]
- Chanana, B.; Graf, R.; Koledachkina, T.; Pflanz, R.; Vorbrüggen, G. αPS2 integrin-mediated muscle attachment in Drosophila requires the ECM protein Thrombospondin. Mech. Dev. 2007, 124, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Katzemich, A.; Long, J.Y.; Panneton, V.; Fisher, L.A.B.; Hipfner, D.; Schöck, F. Slik phosphorylation of Talin T152 is crucial for proper Talin recruitment and maintenance of muscle attachment in Drosophila. Development 2019, 146, dev176339. [Google Scholar] [CrossRef] [PubMed]
- Pines, M.; Das, R.; Ellis, S.J.; Morin, A.; Czerniecki, S.; Yuan, L.; Klose, M.; Coombs, D.; Tanentzapf, G. Mechanical force regulates integrin turnover in Drosophila in vivo. Nat. Cell Biol. 2012, 14, 935–943. [Google Scholar] [CrossRef]
- Sparrow, J.C.; Schöck, F. The initial steps of myofibril assembly: Integrins pave the way. Nat. Rev. Mol. Cell Biol. 2009, 10, 293–298. [Google Scholar] [CrossRef]
- Gautel, M.; Djinović-Carugo, K. The sarcomeric cytoskeleton: From molecules to motion. J. Exp. Biol. 2016, 219, 135. [Google Scholar] [CrossRef]
- Rhee, D.; Sanger, J.M.; Sanger, J.W. The premyofibril: Evidence for its role in myofibrillogenesis. Cell Motil. Cytoskeleton 1994, 28, 1–24. [Google Scholar] [CrossRef]
- Jirka, C.; Pak, J.H.; Grosgogeat, C.A.; Marchetii, M.M.; Gupta, V.A. Dysregulation of NRAP degradation by KLHL41 contributes to pathophysiology in nemaline myopathy. Hum. Mol. Genet. 2019, 28, 2549–2560. [Google Scholar] [CrossRef]
- Antin, P.B.; Tokunaka, S.; Nachmias, V.T.; Holtzer, H. Role of stress fiber-like structures in assembling nascent myofibrils in myosheets recovering from exposure to ethyl methanesulfonate. J. Cell Biol. 1986, 102, 1464–1479. [Google Scholar] [CrossRef]
- Epstein, H.; Fischman, D. Molecular analysis of protein assembly in muscle development. Science 1991, 251, 1039–1044. [Google Scholar] [CrossRef]
- Rui, Y.; Bai, J.; Perrimon, N. Sarcomere formation occurs by the assembly of multiple latent protein complexes. PLoS Genet. 2010, 6, e1001208. [Google Scholar] [CrossRef] [PubMed]
- Weitkunat, M.; Kaya-Çopur, A.; Grill, S.W.; Schnorrer, F. Tension and Force-Resistant Attachment Are Essential for Myofibrillogenesis in Drosophila Flight Muscle. Curr. Biol. 2014, 24, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Orfanos, Z.; Leonard, K.; Elliott, C.; Katzemich, A.; Bullard, B.; Sparrow, J. Sallimus and the Dynamics of Sarcomere Assembly in Drosophila Flight Muscles. J. Mol. Biol. 2015, 427, 2151–2158. [Google Scholar] [CrossRef] [PubMed]
- Röper, K.; Mao, Y.; Brown, N.H. Contribution of sequence variation in Drosophila actins to their incorporation into actin-based structures in vivo. J. Cell Sci. 2005, 118, 3937. [Google Scholar] [CrossRef] [PubMed]
- Bulgakova, N.A.; Wellmann, J.; Brown, N.H. Diverse integrin adhesion stoichiometries caused by varied actomyosin activity. Open Biol. 2017, 7, 160250. [Google Scholar] [CrossRef] [PubMed]
- Zappia, M.P.; Frolov, M.V. E2F function in muscle growth is necessary and sufficient for viability in Drosophila. Nat. Commun. 2016, 7, 10509. [Google Scholar] [CrossRef] [PubMed]
- Molnár, I.; Migh, E.; Szikora, S.; Kalmár, T.; Végh, A.G.; Deák, F.; Barkó, S.; Bugyi, B.; Orfanos, Z.; Kovács, J.; et al. DAAM is required for thin filament formation and Sarcomerogenesis during muscle development in Drosophila. PLoS Genet. 2014, 10, e1004166. [Google Scholar] [CrossRef]
- Bai, J.; Hartwig, J.H.; Perrimon, N. SALS, a WH2-Domain-Containing Protein, Promotes Sarcomeric Actin Filament Elongation from Pointed Ends during Drosophila Muscle Growth. Dev. Cell 2007, 13, 828–842. [Google Scholar] [CrossRef]
- Mardahl-Dumesnil, M.; Fowler, V.M. Thin filaments elongate from their pointed ends during myofibril assembly in Drosophila indirect flight muscle. J. Cell Biol. 2001, 155, 1043–1053. [Google Scholar] [CrossRef]
- Bloor, J.W.; Kiehart, D.P. zipper Nonmuscle Myosin-II Functions Downstream of PS2 Integrin in Drosophila Myogenesis and Is Necessary for Myofibril Formation. Dev. Biol. 2001, 239, 215–228. [Google Scholar] [CrossRef]
- Volk, T.; Fessler, L.I.; Fessler, J.H. A role for integrin in the formation of sarcomeric cytoarchitecture. Cell 1990, 63, 525–536. [Google Scholar] [CrossRef]
- Kreiskôther, N.; Reichert, N.; Buttgereit, D.; Hertenstein, A.; Fischbach, K.-F.; Renkawitz-Pohl, R. Drosophila Rolling pebbles colocalises and putatively interacts with alpha-Actinin and the Sls isoform Zormin in the Z-discs of the sarcomere and with Dumbfounded/Kirre, alpha-Actinin and Zormin in the terminal Z-discs. J. Muscle Res. Cell Motil. 2006, 27, 93. [Google Scholar] [CrossRef] [PubMed]
- Kelemen-Valkony, I.; Kiss, M.; Csiha, J.; Kiss, A.; Bircher, U.; Szidonya, J.; Maróy, P.; Juhász, G.; Komonyi, O.; Csiszár, K.; et al. Drosophila basement membrane collagen col4a1 mutations cause severe myopathy. Matrix Biol. 2012, 31, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Katzemich, A.; Liao, K.A.; Czerniecki, S.; Schöck, F. Alp/Enigma family proteins cooperate in Z-disc formation and myofibril assembly. PLoS Genet. 2013, 9, e1003342. [Google Scholar] [CrossRef]
- Crisp, S.J.; Evers, J.F.; Bate, M. Endogenous patterns of activity are required for the maturation of a motor network. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 10445–10450. [Google Scholar] [CrossRef]
- Broadie, K.; Bate, M. Development of the embryonic neuromuscular synapse of Drosophila melanogaster. J. Neurosci. 1993, 13, 144. [Google Scholar] [CrossRef]
- Keshishian, H.; Broadie, K.; Chiba, A.; Bate, M. The Drosophila Neuromuscular Junction: A Model System for Studying Synaptic Development and Function. Annu. Rev. Neurosci. 1996, 19, 545–575. [Google Scholar] [CrossRef]
- Ruiz-Cañada, C.; Budnik, V. Introduction on The Use of The Drosophila Embryonic/Larval Neuromuscular Junction as A Model System to Study Synapse Development and Function, and A Brief Summary of Pathfinding and Target Recognition. In International Review of Neurobiology; Academic Press: New York, NY, USA, 2006; pp. 1–31. ISBN 0074-7742. [Google Scholar]
- Landgraf, M.; Bossing, T.; Technau, G.M.; Bate, M. The origin, location, and projections of the embryonic abdominal motorneurons of Drosophila. J. Neurosci. Off. J. Soc. Neurosci. 1997, 17, 9642–9655. [Google Scholar] [CrossRef]
- Schmid, A.; Chiba, A.; Doe, C.Q. Clonal analysis of Drosophila embryonic neuroblasts: Neural cell types, axon projections and muscle targets. Development 1999, 126, 4653. [Google Scholar]
- Landgraf, M.; Jeffrey, V.; Fujioka, M.; Jaynes, J.B.; Bate, M. Embryonic Origins of a Motor System: Motor Dendrites Form a Myotopic Map in Drosophila. PLoS Biol. 2003, 1, e41. [Google Scholar] [CrossRef]
- Ritzenthaler, S.; Suzuki, E.; Chiba, A. Postsynaptic filopodia in muscle cells interact with innervating motoneuron axons. Nat. Neurosci. 2000, 3, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Nose, A. Generation of neuromuscular specificity in Drosophila: Novel mechanisms revealed by new technologies. Front. Mol. Neurosci. 2012, 5, 62. [Google Scholar] [CrossRef] [PubMed]
- Nose, A.; Umeda, T.; Takeichi, M. Neuromuscular target recognition by a homophilic interaction of connectin cell adhesion molecules in Drosophila. Development 1997, 124, 1433. [Google Scholar] [PubMed]
- Patel, N.H.; Ball, E.E.; Goodman, C.S. Changing role of even-skipped during the evolution of insect pattern formation. Nature 1992, 357, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Labrador, J.P.; O’Keefe, D.; Yoshikawa, S.; McKinnon, R.D.; Thomas, J.B.; Bashaw, G.J. The Homeobox Transcription Factor Even-skipped Regulates Netrin-Receptor Expression to Control Dorsal Motor-Axon Projections in Drosophila. Curr. Biol. 2005, 15, 1413–1419. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Featherstone, D.E. Discs-large (DLG) is clustered by presynaptic innervation and regulates postsynaptic glutamate receptor subunit composition in Drosophila. BMC Biol. 2005, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, A.; Kobler, O.; Kittel, R.J.; Wichmann, C.; Sierralta, J.; Sigrist, S.J.; Gundelfinger, E.D.; Knust, E.; Thomas, U. A perisynaptic ménage à trois between Dlg, DLin-7, and Metro controls proper organization of Drosophila synaptic junctions. J. Neurosci. Off. J. Soc. Neurosci. 2010, 30, 5811–5824. [Google Scholar] [CrossRef]
- Broadie, K.; Bate, M. Innervation directs receptor synthesis and localization in Drosophila embryo synaptogenesis. Nature 1993, 361, 350–353. [Google Scholar] [CrossRef]
- Kim, M.D.; Wen, Y.; Jan, Y.-N. Patterning and organization of motor neuron dendrites in the Drosophila larva. Dev. Biol. 2009, 336, 213–221. [Google Scholar] [CrossRef]
- Syed, A.; Lukacsovich, T.; Pomeroy, M.; Bardwell, A.J.; Decker, G.T.; Waymire, K.G.; Purcell, J.; Huang, W.; Gui, J.; Padilla, E.M.; et al. Miles to go (mtgo) encodes FNDC3 proteins that interact with the chaperonin subunit CCT3 and are required for NMJ branching and growth in Drosophila. Dev. Biol. 2019, 445, 37–53. [Google Scholar] [CrossRef]
- Beumer, K.J.; Rohrbough, J.; Prokop, A.; Broadie, K. A role for PS integrins in morphological growth and synaptic function at the postembryonic neuromuscular junction of Drosophila. Development 1999, 126, 5833. [Google Scholar] [PubMed]
- Vonhoff, F.; Keshishian, H. In Vivo Calcium Signaling during Synaptic Refinement at the Drosophila Neuromuscular Junction. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 5511–5526. [Google Scholar] [CrossRef]
- DePew, A.T.; Aimino, M.A.; Mosca, T.J. The Tenets of Teneurin: Conserved Mechanisms Regulate Diverse Developmental Processes in the Drosophila Nervous System. Front. Neurosci. 2019, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Bate, M.; Rushton, E.; Currie, D.A. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development 1991, 113, 79. [Google Scholar] [PubMed]
- Figeac, N.; Jagla, T.; Aradhya, R.; Da Ponte, J.P.; Jagla, K. Drosophila adult muscle precursors form a network of interconnected cells and are specified by the rhomboid-triggered EGF pathway. Development 2010, 137, 1965. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, G.; Zmojdzian, M.; Da Ponte, J.P.; Junion, G.; Jagla, K. Drosophila adult muscle precursor cells contribute to motor axon pathfinding and proper innervation of embryonic muscles. Development 2020, 147, dev183004. [Google Scholar] [CrossRef]
- Gunage, R.D.; Dhanyasi, N.; Reichert, H.; VijayRaghavan, K. Drosophila adult muscle development and regeneration. Skelet. Muscle Dev. 30th Anniv. MyoD 2017, 72, 56–66. [Google Scholar] [CrossRef]
- Laurichesse, Q.; Soler, C. Muscle development: A view from adult myogenesis in Drosophila. Semin. Cell Dev. Biol. 2020. [Google Scholar] [CrossRef]
- Oas, S.T.; Bryantsev, A.L.; Cripps, R.M. Arrest is a regulator of fiber-specific alternative splicing in the indirect flight muscles of Drosophila. J. Cell Biol. 2014, 206, 895–908. [Google Scholar] [CrossRef]
- DeAguero, A.A.; Castillo, L.; Oas, S.T.; Kiani, K.; Bryantsev, A.L.; Cripps, R.M. Regulation of fiber-specific actin expression by the Drosophila SRF ortholog Blistered. Dev. Camb. Engl. 2019, 146, dev164129. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C. Fiber Types in Mammalian Skeletal Muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef] [PubMed]
- Hastings, G.A.; Emerson, C.P., Jr. Myosin functional domains encoded by alternative exons are expressed in specific thoracic muscles of Drosophila. J. Cell Biol. 1991, 114, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Figeac, N.; Daczewska, M.; Marcelle, C.; Jagla, K. Muscle stem cells and model systems for their investigation. Dev. Dyn. 2007, 236, 3332–3342. [Google Scholar] [CrossRef] [PubMed]
- Beira, J.V.; Paro, R. The legacy of Drosophila imaginal discs. Chromosoma 2016, 125, 573–592. [Google Scholar] [CrossRef] [PubMed]
- Aradhya, R.; Zmojdzian, M.; Da Ponte, J.P.; Jagla, K. Muscle niche-driven Insulin-Notch-Myc cascade reactivates dormant Adult Muscle Precursors in Drosophila. eLife 2015, 4, e08497. [Google Scholar] [CrossRef]
- Tavi, P.; Korhonen, T.; Hänninen, S.L.; Bruton, J.D.; Lööf, S.; Simon, A.; Westerblad, H. Myogenic skeletal muscle satellite cells communicate by tunnelling nanotubes. J. Cell. Physiol. 2010, 223, 376–383. [Google Scholar] [CrossRef]
- Gunage, R.D.; Reichert, H.; VijayRaghavan, K. Identification of a new stem cell population that generates Drosophila flight muscles. eLife 2014, 3, e03126. [Google Scholar] [CrossRef]
- Vishal, K.; Brooks, D.S.; Bawa, S.; Gameros, S.; Stetsiv, M.; Geisbrecht, E.R. Adult Muscle Formation Requires Drosophila Moleskin for Proliferation of Wing Disc-Associated Muscle Precursors. Genetics 2017, 206, 199. [Google Scholar] [CrossRef]
- Bernard, F.; Dutriaux, A.; Silber, J.; Lalouette, A. Notch pathway repression by vestigial is required to promote indirect flight muscle differentiation in Drosophila melanogaster. Dev. Biol. 2006, 295, 164–177. [Google Scholar] [CrossRef]
- Dutta, D.; Umashankar, M.; Lewis, E.B.; Rodrigues, V.; VijayRaghavan, K. Hox Genes Regulate Muscle Founder Cell Pattern Autonomously and Regulate Morphogenesis Through Motor Neurons. J. Neurogenet. 2010, 24, 95–108. [Google Scholar] [CrossRef]
- Sudarsan, V.; Anant, S.; Guptan, P.; VijayRaghavan, K.; Skaer, H. Myoblast Diversification and Ectodermal Signaling in Drosophila. Dev. Cell 2001, 1, 829–839. [Google Scholar] [CrossRef]
- Maqbool, T.; Soler, C.; Jagla, T.; Daczewska, M.; Lodha, N.; Palliyil, S.; VijayRaghavan, K.; Jagla, K. Shaping leg muscles in Drosophila: Role of ladybird, a conserved regulator of appendicular myogenesis. PLoS ONE 2006, 1, e122. [Google Scholar] [CrossRef] [PubMed]
- Ghazi, A.; Anant, S.; Vijay Raghavan, K. Apterous mediates development of direct flight muscles autonomously and indirect flight muscles through epidermal cues. Development 2000, 127, 5309. [Google Scholar] [PubMed]
- Dutta, D.; Shaw, S.; Maqbool, T.; Pandya, H.; VijayRaghavan, K. Drosophila Heartless Acts with Heartbroken/Dof in Muscle Founder Differentiation. PLoS Biol. 2005, 3, e337. [Google Scholar] [CrossRef]
- Dutta, D.; Anant, S.; Ruiz-Gomez, M.; Bate, M.; VijayRaghavan, K. Founder myoblasts and fibre number during adult myogenesis in Drosophila. Development 2004, 131, 3761. [Google Scholar] [CrossRef]
- Bryantsev, A.L.; Duong, S.; Brunetti, T.M.; Chechenova, M.B.; Lovato, T.L.; Nelson, C.; Shaw, E.; Uhl, J.D.; Gebelein, B.; Cripps, R.M. Extradenticle and homothorax control adult muscle fiber identity in Drosophila. Dev. Cell 2012, 23, 664–673. [Google Scholar] [CrossRef] [PubMed]
- Schönbauer, C.; Distler, J.; Jährling, N.; Radolf, M.; Dodt, H.-U.; Frasch, M.; Schnorrer, F. Spalt mediates an evolutionarily conserved switch to fibrillar muscle fate in insects. Nature 2011, 479, 406–409. [Google Scholar] [CrossRef]
- DeSimone, S.; Coelho, C.; Roy, S.; VijayRaghavan, K.; White, K. ERECT WING, the Drosophila member of a family of DNA binding proteins is required in imaginal myoblasts for flight muscle development. Development 1996, 122, 31. [Google Scholar]
- Fernandes, J.; Bate, M.; Vijayraghavan, K. Development of the indirect flight muscles of Drosophila. Development 1991, 113, 67. [Google Scholar]
- Kuleesha, Y.; Puah, W.C.; Wasser, M. Live imaging of muscle histolysis in Drosophila metamorphosis. BMC Dev. Biol. 2016, 16, 12. [Google Scholar] [CrossRef]
- Fernandes, J.J.; Keshishian, H. Motoneurons regulate myoblast proliferation and patterning in Drosophila. Dev. Biol. 2005, 277, 493–505. [Google Scholar] [CrossRef]
- Currie, D.A.; Bate, M. The development of adult abdominal muscles in Drosophila: Myoblasts express twist and are associated with nerves. Development 1991, 113, 91. [Google Scholar] [PubMed]
- Rivlin, P.K.; Schneiderman, A.M.; Booker, R. Imaginal Pioneers Prefigure the Formation of Adult Thoracic Muscles in Drosophila melanogaster. Dev. Biol. 2000, 222, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.; Gildor, B.; Shilo, B.-Z.; VijayRaghavan, K.; Schejter, E.D. The actin nucleator WASp is required for myoblast fusion during adult Drosophila myogenesis. Dev. Camb. Engl. 2011, 138, 2347–2357. [Google Scholar] [CrossRef] [PubMed]
- Ghazi, A.; Paul, L.; VijayRaghavan, K. Prepattern genes and signaling molecules regulate stripe expression to specify Drosophila flight muscle attachment sites. Mech. Dev. 2003, 120, 519–528. [Google Scholar] [CrossRef]
- Laddada, L.; Jagla, K.; Soler, C. Odd-skipped and Stripe act downstream of Notch to promote the morphogenesis of long appendicular tendons in Drosophila. Biol. Open 2019, 8, bio038760. [Google Scholar] [CrossRef] [PubMed]
- Weitkunat, M.; Brasse, M.; Bausch, A.R.; Schnorrer, F. Mechanical tension and spontaneous muscle twitching precede the formation of cross-striated muscle in vivo. Dev. Camb. Engl. 2017, 144, 1261–1272. [Google Scholar] [CrossRef] [PubMed]
- Spletter, M.L.; Schnorrer, F. Transcriptional regulation and alternative splicing cooperate in muscle fiber-type specification in flies and mammals. Dev. Biol. 2014, 321, 90–98. [Google Scholar] [CrossRef]
- Reedy, M.C.; Beall, C. Ultrastructure of Developing Flight Muscle in Drosophila. I. Assembly of Myofibrils. Dev. Biol. 1993, 160, 443–465. [Google Scholar] [CrossRef]
- Spletter, M.L.; Barz, C.; Yeroslaviz, A.; Zhang, X.; Lemke, S.B.; Bonnard, A.; Brunner, E.; Cardone, G.; Basler, K.; Habermann, B.H.; et al. A transcriptomics resource reveals a transcriptional transition during ordered sarcomere morphogenesis in flight muscle. eLife 2018, 7, e34058. [Google Scholar] [CrossRef]
- Shwartz, A.; Dhanyasi, N.; Schejter, E.D.; Shilo, B.-Z. The Drosophila formin Fhos is a primary mediator of sarcomeric thin-filament array assembly. eLife 2016, 5, e16540. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, I.; Schöck, F. The nebulin repeat protein Lasp regulates I-band architecture and filament spacing in myofibrils. J. Cell Biol. 2014, 206, 559–572. [Google Scholar] [CrossRef]
- Qiu, F.; Brendel, S.; Cunha, P.M.F.; Astola, N.; Song, B.; Furlong, E.E.M.; Leonard, K.R.; Bullard, B. Myofilin, a protein in the thick filaments of insect muscle. J. Cell Sci. 2005, 118, 1527. [Google Scholar] [CrossRef] [PubMed]
- Ball, E.; Karlik, C.C.; Beall, C.J.; Saville, D.L.; Sparrow, J.C.; Bullard, B.; Fyrberg, E.A. Arthrin, a myofibrillar protein of insect flight muscle, is an actin-ubiquitin conjugate. Cell 1987, 51, 221–228. [Google Scholar] [CrossRef]
- Becker, K.D.; O’Donnell, P.T.; Heitz, J.M.; Vito, M.; Bernstein, S.I. Analysis of Drosophila paramyosin: Identification of a novel isoform which is restricted to a subset of adult muscles. J. Cell Biol. 1992, 116, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Maroto, M.; Arredondo, J.J.; Román, M.S.; Marco, R.; Cervera, M. Analysis of the Paramyosin/Miniparamyosin Gene: Miniparamyosin is an independently transcribed, distinct paramyosin isoform, widely distributed in invertebrates. J. Biol. Chem. 1995, 270, 4375–4382. [Google Scholar] [CrossRef]
- Vigoreaux, J.O. Alterations in flightin phosphorylation inDrosophila flight muscles are associated with myofibrillar defects engendered by actin and myosin heavy-chain mutant alleles. Biochem. Genet. 1994, 32, 301–314. [Google Scholar] [CrossRef]
- Reedy, M.C.; Bullard, B.; Vigoreaux, J.O. Flightin is essential for thick filament assembly and sarcomere stability in Drosophila flight muscles. J. Cell Biol. 2000, 151, 1483–1500. [Google Scholar] [CrossRef]
- Craig, R.; Woodhead, J.L. Structure and function of myosin filaments. Curr. Opin. Struct. Biol. 2006, 16, 204–212. [Google Scholar] [CrossRef]
- Orfanos, Z.; Sparrow, J.C. Myosin isoform switching during assembly of the Drosophila flight muscle thick filament lattice. J. Cell Sci. 2013, 126, 139. [Google Scholar] [CrossRef]
- Contompasis, J.L.; Nyland, L.R.; Maughan, D.W.; Vigoreaux, J.O. Flightin Is Necessary for Length Determination, Structural Integrity, and Large Bending Stiffness of Insect Flight Muscle Thick Filaments. J. Mol. Biol. 2010, 395, 340–348. [Google Scholar] [CrossRef]
- Sparrow, J.; Reedy, M.; Ball, E.; Kyrtatas, V.; Molloy, J.; Durston, J.; Hennessey, E.; White, D. Functional and ultrastructural effects of a missense mutation in the indirect flight muscle-specific actin gene of Drosophila melanogaster. J. Mol. Biol. 1991, 222, 963–982. [Google Scholar] [CrossRef]
- Burkart, C.; Qiu, F.; Brendel, S.; Benes, V.; Hååg, P.; Labeit, S.; Leonard, K.; Bullard, B. Modular Proteins from the Drosophila sallimus (sls) Gene and their Expression in Muscles with Different Extensibility. J. Mol. Biol. 2007, 367, 953–969. [Google Scholar] [CrossRef]
- González-Morales, N.; Xiao, Y.S.; Schilling, M.A.; Marescal, O.; Liao, K.A.; Schöck, F. Myofibril diameter is set by a finely tuned mechanism of protein oligomerization in Drosophila. eLife 2019, 8, e50496. [Google Scholar] [CrossRef]
- Truman, J.W.; Schuppe, H.; Shepherd, D.; Williams, D.W. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development 2004, 131, 5167. [Google Scholar] [CrossRef]
- Brierley, D.J.; Blanc, E.; Reddy, O.V.; Vijayraghavan, K.; Williams, D.W. Dendritic targeting in the leg neuropil of Drosophila: The role of midline signalling molecules in generating a myotopic map. PLoS Biol. 2009, 7, e1000199. [Google Scholar] [CrossRef]
- Enriquez, J.; Venkatasubramanian, L.; Baek, M.; Peterson, M.; Aghayeva, U.; Mann, R.S. Specification of Individual Adult Motor Neuron Morphologies by Combinatorial Transcription Factor Codes. Neuron 2015, 86, 955–970. [Google Scholar] [CrossRef]
- Baek, M.; Mann, R.S. Lineage and Birth Date Specify Motor Neuron Targeting and Dendritic Architecture in Adult Drosophila. J. Neurosci. 2009, 29, 6904. [Google Scholar] [CrossRef]
- Fernandes, J.J.; Keshishian, H. Nerve-muscle interactions during flight muscle development in Drosophila. Development 1998, 125, 1769. [Google Scholar]
- Fernandes, J.; VijayRaghavan, K. The development of indirect flight muscle innervation in Drosophila melanogaster. Development 1993, 118, 215. [Google Scholar]
- Rival, T.; Soustelle, L.; Cattaert, D.; Strambi, C.; Iché, M.; Birman, S. Physiological requirement for the glutamate transporter dEAAT1 at the adult Drosophila neuromuscular junction. J. Neurobiol. 2006, 66, 1061–1074. [Google Scholar] [CrossRef]
- Kimura, K.I.; Truman, J.W. Postmetamorphic cell death in the nervous and muscular systems of Drosophila melanogaster. J. Neurosci. Off. J. Soc. Neurosci. 1990, 10, 403–411. [Google Scholar] [CrossRef]
- Soler, C.; Taylor, M.V. The Him gene inhibits the development of Drosophila flight muscles during metamorphosis. Mech. Dev. 2009, 126, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Soler, C.; Han, J.; Taylor, M.V. The conserved transcription factor Mef2 has multiple roles in adult Drosophila musculature formation. Development 2012, 139, 1270. [Google Scholar] [CrossRef]
- Zhang, S.; Bernstein, S.I. Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech. Dev. 2001, 101, 35–45. [Google Scholar] [CrossRef]
- Swank, D.M.; Bartoo, M.L.; Knowles, A.F.; Iliffe, C.; Bernstein, S.I.; Molloy, J.E.; Sparrow, J.C. Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J. Biol. Chem. 2001, 276, 15117–15124. [Google Scholar] [CrossRef]
- Karlik, C.C.; Fyrberg, E.A. Two Drosophila melanogaster tropomyosin genes: Structural and functional aspects. Mol. Cell. Biol. 1986, 6, 1965. [Google Scholar] [CrossRef]
- Mateos, J.; Herranz, R.; Domingo, A.; Sparrow, J.; Marco, R. The structural role of high molecular weight tropomyosins in dipteran indirect flight muscle and the effect of phosphorylation. J. Muscle Res. Cell Motil. 2006, 27, 189–201. [Google Scholar] [CrossRef]
- Katzemich, A.; Long, J.Y.; Jani, K.; Lee, B.R.; Schöck, F. Muscle type-specific expression of Zasp52 isoforms in Drosophila. Gene Expr. Patterns 2011, 11, 484–490. [Google Scholar] [CrossRef]
- Katzemich, A.; Kreisköther, N.; Alexandrovich, A.; Elliott, C.; Schöck, F.; Leonard, K.; Sparrow, J.; Bullard, B. The function of the M-line protein obscurin in controlling the symmetry of the sarcomere in the flight muscle of Drosophila. J. Cell Sci. 2012, 125, 3367. [Google Scholar] [CrossRef]
- Marín, M.-C.; Rodríguez, J.-R.; Ferrús, A. Transcription of Drosophila Troponin I Gene Is Regulated by Two Conserved, Functionally Identical, Synergistic Elements. Mol. Biol. Cell 2004, 15, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Mas, J.-A.; García-Zaragoza, E.; Cervera, M. Two Functionally Identical Modular Enhancers in Drosophila Troponin T Gene Establish the Correct Protein Levels in Different Muscle Types. Mol. Biol. Cell 2004, 15, 1931–1945. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gremke, L.; Lord, P.C.W.; Sabacan, L.; Lin, S.-C.; Wohlwill, A.; Storti, R.V. Coordinate Regulation of Drosophila Tropomyosin Gene Expression Is Controlled by Multiple Muscle-Type-Specific Positive and Negative Enhancer Elements. Dev. Biol. 1993, 159, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.H.; Nguyen, H.T.; Dybala, C.; Storti, R.V. Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression. Proc. Natl. Acad. Sci. USA 1996, 93, 4623. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.L.; Wohlwill, A.; Dzitoeva, S.; Lin, M.-H.; Holbrook, S.; Storti, R.V. The Drosophila PAR Domain Protein 1 (Pdp1) Gene Encodes Multiple Differentially Expressed mRNAs and Proteins through the Use of Multiple Enhancers and Promoters. Dev. Biol. 2000, 224, 401–414. [Google Scholar] [CrossRef]
- Vigoreaux, J.O.; Saide, J.D.; Pardue, M.L. Structurally different Drosophila striated muscles utilize distinct variants of Z-band-associated proteins. J. Muscle Res. Cell Motil. 1991, 12, 340–354. [Google Scholar] [CrossRef]
- Zhao, C.; Swank, D.M. The Drosophila indirect flight muscle myosin heavy chain isoform is insufficient to transform the jump muscle into a highly stretch-activated muscle type. Am. J. Physiol. Cell Physiol. 2017, 312, C111–C118. [Google Scholar] [CrossRef]
- Vigoreaux, J.O.; Perry, L.M. Multiple isoelectric variants of flightin in Drosophila stretch-activated muscles are generated by temporally regulated phosphorylations. J. Muscle Res. Cell Motil. 1994, 15, 607–616. [Google Scholar] [CrossRef]
- Daley, J.; Southgate, R.; Ayme-Southgate, A. Structure of the Drosophila projectin protein: Isoforms and implication for projectin filament assembly11Edited by M. F. Moody. J. Mol. Biol. 1998, 279, 201–210. [Google Scholar] [CrossRef]
- Glasheen, B.M.; Ramanath, S.; Patel, M.; Sheppard, D.; Puthawala, J.T.; Riley, L.A.; Swank, D.M. Five Alternative Myosin Converter Domains Influence Muscle Power, Stretch Activation, and Kinetics. Biophys. J. 2018, 114, 1142–1152. [Google Scholar] [CrossRef]
- Belozerov, V.E.; Ratkovic, S.; McNeill, H.; Hilliker, A.J.; McDermott, J.C. In vivo interaction proteomics reveal a novel p38 mitogen-activated protein kinase/Rack1 pathway regulating proteostasis in Drosophila muscle. Mol. Cell. Biol. 2014, 34, 474–484. [Google Scholar] [CrossRef] [PubMed]
- Haas, K.F.; Woodruff, E., 3rd; Broadie, K. Proteasome function is required to maintain muscle cellular architecture. Biol. Cell 2007, 99, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Voza, F.; Ezzeddine, N.; Frasch, M. Drosophila mind bomb2 is required for maintaining muscle integrity and survival. J. Cell Biol. 2007, 179, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Valdez, C.; Scroggs, R.; Chassen, R.; Reiter, L.T. Variation in Dube3a expression affects neurotransmission at the Drosophila neuromuscular junction. Biol. Open 2015, 4, 776–782. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marco-Ferreres, R.; Arredondo, J.J.; Fraile, B.; Cervera, M. Overexpression of troponin T in Drosophila muscles causes a decrease in the levels of thin-filament proteins. Biochem. J. 2005, 386, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Firdaus, H.; Mohan, J.; Naz, S.; Arathi, P.; Ramesh, S.R.; Nongthomba, U. A cis-regulatory mutation in troponin-I of Drosophila reveals the importance of proper stoichiometry of structural proteins during muscle assembly. Genetics 2015, 200, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Wójtowicz, I.; Jabłońska, J.; Zmojdzian, M.; Taghli-Lamallem, O.; Renaud, Y.; Junion, G.; Daczewska, M.; Huelsmann, S.; Jagla, K.; Jagla, T. Drosophila small heat shock protein CryAB ensures structural integrity of developing muscles, and proper muscle and heart performance. Development 2015, 142, 994. [Google Scholar] [CrossRef]
- Jabłońska, J.; Dubińska-Magiera, M.; Jagla, T.; Jagla, K.; Daczewska, M. Drosophila Hsp67Bc hot-spot variants alter muscle structure and function. Cell. Mol. Life Sci. CMLS 2018, 75, 4341–4356. [Google Scholar] [CrossRef]
- Wang, S.; Reuveny, A.; Volk, T. Nesprin provides elastic properties to muscle nuclei by cooperating with spectraplakin and EB1. J. Cell Biol. 2015, 209, 529–538. [Google Scholar] [CrossRef]
- Wang, S.; Volk, T. Composite biopolymer scaffolds shape muscle nucleus: Insights and perspectives from Drosophila. Bioarchitecture 2015, 5, 35–43. [Google Scholar] [CrossRef][Green Version]
- Lorber, D.; Rotkopf, R.; Volk, T. In vivo imaging of myonuclei during spontaneous muscle contraction reveals non-uniform nuclear mechanical dynamics in Nesprin/klar mutants. bioRxiv 2019, 643015. [Google Scholar] [CrossRef]
- Elhanany-Tamir, H.; Yu, Y.V.; Shnayder, M.; Jain, A.; Welte, M.; Volk, T. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J. Cell Biol. 2012, 198, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Morel, V.; Lepicard, S.N.; Rey, A.; Parmentier, M.-L.; Schaeffer, L. Drosophila Nesprin-1 controls glutamate receptor density at neuromuscular junctions. Cell. Mol. Life Sci. 2014, 71, 3363–3379. [Google Scholar] [CrossRef] [PubMed]
- Goel, P.; Dufour Bergeron, D.; Böhme, M.A.; Nunnelly, L.; Lehmann, M.; Buser, C.; Walter, A.M.; Sigrist, S.J.; Dickman, D. Homeostatic scaling of active zone scaffolds maintains global synaptic strength. J. Cell Biol. 2019, 218, 1706–1724. [Google Scholar] [CrossRef] [PubMed]
- Goel, P.; Dickman, D. Distinct homeostatic modulations stabilize reduced postsynaptic receptivity in response to presynaptic DLK signaling. Nat. Commun. 2018, 9, 1856. [Google Scholar] [CrossRef]
- Ziegler, A.B.; Augustin, H.; Clark, N.L.; Berthelot-Grosjean, M.; Simonnet, M.M.; Steinert, J.R.; Geillon, F.; Manière, G.; Featherstone, D.E.; Grosjean, Y. The Amino Acid Transporter JhI-21 Coevolves with Glutamate Receptors, Impacts NMJ Physiology, and Influences Locomotor Activity in Drosophila Larvae. Sci. Rep. 2016, 6, 19692. [Google Scholar] [CrossRef]
- Hong, H.; Zhao, K.; Huang, S.; Huang, S.; Yao, A.; Jiang, Y.; Sigrist, S.; Zhao, L.; Zhang, Y.Q. Structural Remodeling of Active Zones Is Associated with Synaptic Homeostasis. J. Neurosci. 2020, 40, 2817. [Google Scholar] [CrossRef]
- James, T.D.; Zwiefelhofer, D.J.; Frank, C.A. Maintenance of homeostatic plasticity at the Drosophila neuromuscular synapse requires continuous IP3-directed signaling. eLife 2019, 8, e39643. [Google Scholar] [CrossRef]
- Mourikis, P.; Gopalakrishnan, S.; Sambasivan, R.; Tajbakhsh, S. Cell-autonomous Notch activity maintains the temporal specification potential of skeletal muscle stem cells. Dev. Camb. Engl. 2012, 139, 4536–4548. [Google Scholar] [CrossRef]
- Boukhatmi, H.; Bray, S. A population of adult satellite-like cells in Drosophila is maintained through a switch in RNA-isoforms. eLife 2018, 7, e35954. [Google Scholar] [CrossRef]
- Siles, L.; Ninfali, C.; Cortés, M.; Darling, D.S.; Postigo, A. ZEB1 protects skeletal muscle from damage and is required for its regeneration. Nat. Commun. 2019, 10, 1364. [Google Scholar] [CrossRef] [PubMed]
- Kuang, S.; Kuroda, K.; Le Grand, F.; Rudnicki, M.A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 2007, 129, 999–1010. [Google Scholar] [CrossRef] [PubMed]
- de Haro, M.; Al-Ramahi, I.; De Gouyon, B.; Ukani, L.; Rosa, A.; Faustino, N.A.; Ashizawa, T.; Cooper, T.A.; Botas, J. MBNL1 and CUGBP1 modify expanded CUG-induced toxicity in a Drosophila model of myotonic dystrophy type 1. Hum. Mol. Genet. 2006, 15, 2138–2145. [Google Scholar] [CrossRef] [PubMed]
- Picchio, L.; Plantie, E.; Renaud, Y.; Poovthumkadavil, P.; Jagla, K. Novel Drosophila model of myotonic dystrophy type 1: Phenotypic characterization and genome-wide view of altered gene expression. Hum. Mol. Genet. 2013, 22, 2795–2810. [Google Scholar] [CrossRef]
- Yatsenko, A.S.; Shcherbata, H.R. Drosophila miR-9a Targets the ECM Receptor Dystroglycan to Canalize Myotendinous Junction Formation. Dev. Cell 2014, 28, 335–348. [Google Scholar] [CrossRef]
- Kucherenko, M.M.; Marrone, A.K.; Rishko, V.M.; Magliarelli, H.d.F.; Shcherbata, H.R. Stress and muscular dystrophy: A genetic screen for Dystroglycan and Dystrophin interactors in Drosophila identifies cellular stress response components. Dev. Biol. 2011, 352, 228–242. [Google Scholar] [CrossRef]
- Dialynas, G.; Shrestha, O.K.; Ponce, J.M.; Zwerger, M.; Thiemann, D.A.; Young, G.H.; Moore, S.A.; Yu, L.; Lammerding, J.; Wallrath, L.L. Myopathic lamin mutations cause reductive stress and activate the nrf2/keap-1 pathway. PLoS Genet. 2015, 11, e1005231. [Google Scholar] [CrossRef]
- Chandran, S.; Suggs, J.A.; Wang, B.J.; Han, A.; Bhide, S.; Cryderman, D.E.; Moore, S.A.; Bernstein, S.I.; Wallrath, L.L.; Melkani, G.C. Suppression of myopathic lamin mutations by muscle-specific activation of AMPK and modulation of downstream signaling. Hum. Mol. Genet. 2019, 28, 351–371. [Google Scholar] [CrossRef]
- Jungbluth, H.; Gautel, M. Pathogenic mechanisms in centronuclear myopathies. Front. Aging Neurosci. 2014, 6, 339. [Google Scholar] [CrossRef]
- Schulman, V.K.; Folker, E.S.; Rosen, J.N.; Baylies, M.K. Syd/JIP3 and JNK signaling are required for myonuclear positioning and muscle function. PLoS Genet. 2014, 10, e1004880. [Google Scholar] [CrossRef]
- Rosen, J.N.; Azevedo, M.; Soffar, D.B.; Boyko, V.P.; Brendel, M.B.; Schulman, V.K.; Baylies, M.K. The Drosophila Ninein homologue Bsg25D cooperates with Ensconsin in myonuclear positioning. J. Cell Biol. 2019, 218, 524–540. [Google Scholar] [CrossRef] [PubMed]
- Naimi, B.; Harrison, A.; Cummins, M.; Nongthomba, U.; Clark, S.; Canal, I.; Ferrus, A.; Sparrow, J.C. A Tropomyosin-2 Mutation Suppresses a Troponin I Myopathy inDrosophila. Mol. Biol. Cell 2001, 12, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Dahl-Halvarsson, M.; Olive, M.; Pokrzywa, M.; Ejeskär, K.; Palmer, R.H.; Uv, A.E.; Tajsharghi, H. Drosophila model of myosin myopathy rescued by overexpression of a TRIM-protein family member. Proc. Natl. Acad. Sci. USA 2018, 115, E6566–E6575. [Google Scholar] [CrossRef] [PubMed]
- Ugur, B.; Chen, K.; Bellen, H.J. Drosophila tools and assays for the study of human diseases. Dis. Model. Mech. 2016, 9, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Day, K.; Shefer, G.; Shearer, A.; Yablonka-Reuveni, Z. The depletion of skeletal muscle satellite cells with age is concomitant with reduced capacity of single progenitors to produce reserve progeny. Dev. Biol. 2010, 340, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Wen, Y.; Kuroda, K.; Hannon, K.; Rudnicki, M.A.; Kuang, S. Notch signaling deficiency underlies age-dependent depletion of satellite cells in muscular dystrophy. Dis. Models Mech. 2014, 7, 997. [Google Scholar] [CrossRef]
- Letsou, A.; Bohmann, D. Small flies—Big discoveries: Nearly a century of Drosophila genetics and development. Dev. Dyn. 2005, 232, 526–528. [Google Scholar] [CrossRef]
- Bellen, H.J.; Tong, C.; Tsuda, H. 100 years of Drosophila research and its impact on vertebrate neuroscience: A history lesson for the future. Nat. Rev. Neurosci. 2010, 11, 514–522. [Google Scholar] [CrossRef]
| iTF | Human Orthologs | FCs Expressing iTF 1 | References | Embryonic Somatic Muscle Pattern |
|---|---|---|---|---|
| Apterous (Ap) | LHX | LT1, LT2, LT3, LT4, VA2, VA3 | [78] | External muscles are represented in dark brown, intermediate muscles in a medium shade of brown, and internal muscles in fuchsia. |
| Araucan (Ara) | IRX | LT1, LT2, LT3, LT4, SBM, DT1-DO3 | [87] | |
| Caupolican (Caup) | IRX | LT1, LT2, LT3, LT4, SBM, DT1-DO3 | [87] | |
| Collier (Col)/Knot (Kn) | EBF | DA2,DA3-DO5,DT1-DO3, LL1-DO4 | [82,85,88] | |
| Drop (Dr)/Muscle segment homeobox (Msh) | MSX | DO1, DO2, LT1-LT2, LT3-LT4, VA2, VA3 | [89,90] | |
| Even-skipped (Eve) | EVX | DA1, DO2 | [91,92] | |
| Krüppel (Kr) | KLF | DA1, DO1, LT1-LT2, LT3-LT4, LL1,VA1-VA2, DO2, VL3, VO2, VO5 | [87,92,93] | |
| Ladybird (Lb) | LBX | SBM | [94] | |
| Lateral muscles scarcer (Lms) | - | LT1-LT2, LT3-LT4 | [95] | |
| Midline (Mid) | TBX20 | LT3-LT4, LO1, VA1-VA2 | [96] | |
| Nautilus (Nau) | MYOD | DO1, DA2, DA3-DO5, DO3, LL1- DO4, LO1, VA1 | [79,85,88,97] | |
| Optomotor-blind-related-1 (Org-1) | TBX1 | LO1, VT1, SBM | [98] | |
| Pox meso (Poxm) | PAX | DT1-DO3, VA1-VA2, VA3 | [99] | |
| Ptx1 | PITX | Ventral muscles | [100] | |
| Runt | DO2, VA3, VO4 | [92,101] | ||
| Slouch (Slou)/S59 | NKX1 | DT1-DO3, VA1-VA2, VA3, VT1, LO1 | [77,80,87] | |
| Scalloped (Sd) | TEF-1 | All FCs transiently, maintained in VL1, VL2, VL3, VL4 | [86] | |
| Vestigial (Vg) | VGLL | DA1-DA2, DA3, LL1, VL1, VL2, VL3, VL4 | [86] | |
| Tailup (Tup) | ISL | DA1, DA2, DO1, DO2 | [81] | |
| Eyes absent (Eya) | Differential temporal expression in multiple FCs | [85,102] | ||
| Six4 | SIX | Differential temporal expression in multiple FCs | [102,103] | |
| Sine occulis (So) | SIX | DA2, DA3-DO5, LL1-DO4 | [85] | |
| No ocelli (Noc) | ZNF | DA3-DO5 | [85] | |
| ETS-domain lacking (Edl) | - | DA2, DA3 | [85] |
| Adult iTF | Human Orthologs | Adult Myoblast Expression | Embryonic iTF Function 1 | References | Adult Flight and Leg Muscle Pattern |
|---|---|---|---|---|---|
| Vestigial (Vg) | VGLL | IFM | DA1-DA2, DA3, LL1, VL1, VL2, VL3, VL4 | [211] | Indirect flight muscles (IFM) are shown in shades of red and the direct flight muscles (DFM) in dark brown. Among the leg muscles, only the tergal depressor of trochanter (TDT) muscles are highlighted in olive green. Other leg muscles are in a light shade of green. |
| Extradenticle (Exd) | PBX | IFM | [218] | ||
| Homeothorax (Hth) | MEIS | IFM | [218] | ||
| Spalt major (Salm) | SALL | IFM | [219] | ||
| Erect wing (Ewg) | NRF1 | IFM | [220] | ||
| Cut (Ct) | DFM | [213,214] | |||
| Lateral muscles scarcer (Lms) | -- | DFM | LT1-LT2, LT3-LT4 | [95] | |
| Apterous (Ap) | LHX | DFM | LT1, LT2, LT3, LT4, VA2, VA3 | [215] | |
| Ladybird (Lb) | LBX | Leg muscles | SBM | [214] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poovathumkadavil, P.; Jagla, K. Genetic Control of Muscle Diversification and Homeostasis: Insights from Drosophila. Cells 2020, 9, 1543. https://doi.org/10.3390/cells9061543
Poovathumkadavil P, Jagla K. Genetic Control of Muscle Diversification and Homeostasis: Insights from Drosophila. Cells. 2020; 9(6):1543. https://doi.org/10.3390/cells9061543
Chicago/Turabian StylePoovathumkadavil, Preethi, and Krzysztof Jagla. 2020. "Genetic Control of Muscle Diversification and Homeostasis: Insights from Drosophila" Cells 9, no. 6: 1543. https://doi.org/10.3390/cells9061543
APA StylePoovathumkadavil, P., & Jagla, K. (2020). Genetic Control of Muscle Diversification and Homeostasis: Insights from Drosophila. Cells, 9(6), 1543. https://doi.org/10.3390/cells9061543






