CDK12: A Potent Target and Biomarker for Human Cancer Therapy
Abstract
1. Introduction
2. Cyclin Dependent Kinase 12 (CDK12): Gene, Structure and Expression
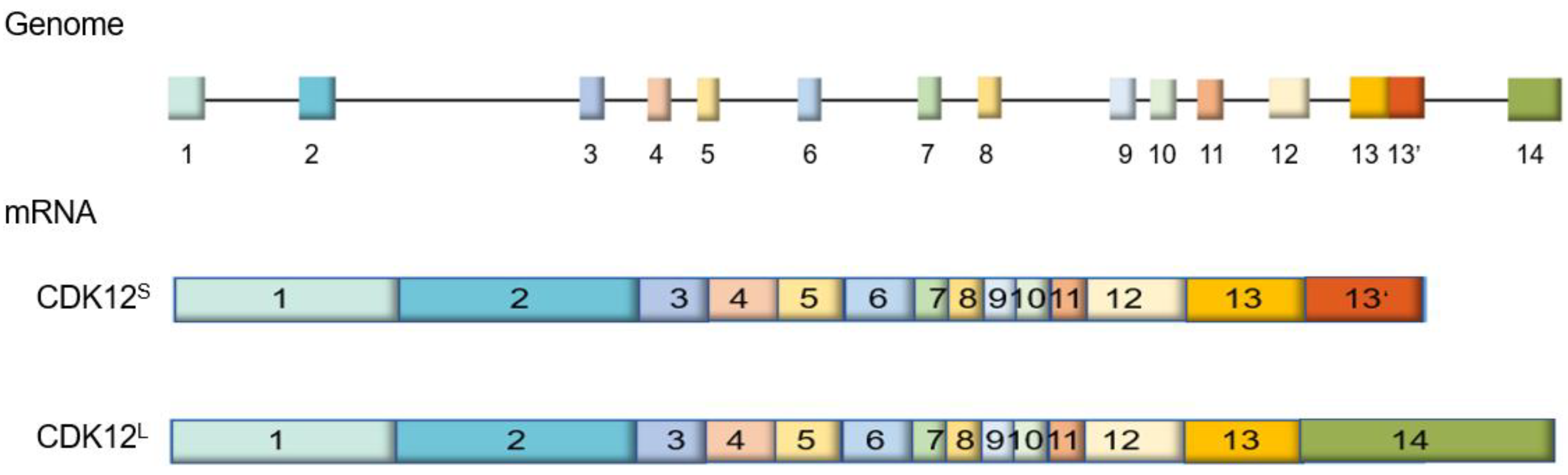
2.1. Gene and Isoforms of CDK12
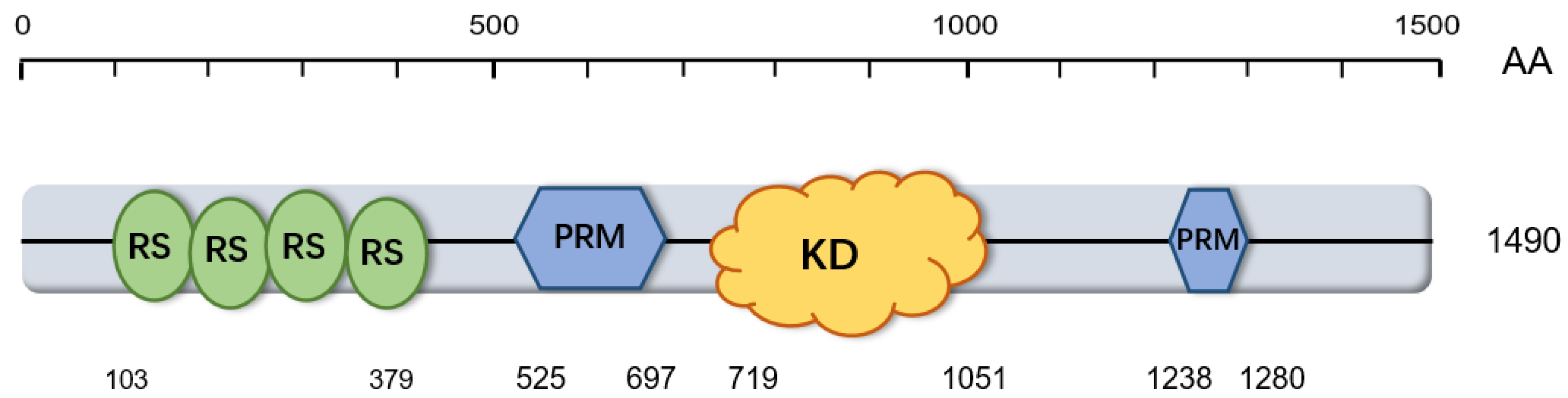
2.2. Structure of CDK12
2.3. CDK12 Expression
3. CDK12′s Biological Functions
3.1. CDK12 in Gene Transcription
3.2. CDK12 in RNA Splicing
3.3. CDK12 in Translation
3.4. CDK12 in Cell Cycle
3.5. CDK12 in Cell Proliferation
3.6. CDK12 in DNA Damage Response (DDR)
4. CDK12 and Human Cancer
4.1. CDK12 in Breast Cancer
4.2. CDK12 in Ovarian Cancer
4.3. CDK12 in Prostate Cancer
4.4. CDK12 in Gastric Cancer
5. CDK12 as a Potential Target and Biomarker for Cancer Therapy
6. Conclusion and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lim, S.; Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P. Genetic control of cell size at cell division in yeast. Nature 1975, 256, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J. Bacteriol. 1973, 115, 966–974. [Google Scholar] [CrossRef]
- Loyer, P.; Trembley, J.H.; Katona, R.; Kidd, V.J.; Lahti, J.M. Role of CDK/cyclin complexes in transcription and RNA splicing. Cell. Signal. 2005, 17, 1033–1051. [Google Scholar] [CrossRef]
- Paculova, H.; Kohoutek, J. The emerging roles of CDK12 in tumorigenesis. Cell Div. 2017, 12, 7. [Google Scholar] [CrossRef]
- Cheng, S.W.; Kuzyk, M.A.; Moradian, A.; Ichu, T.A.; Chang, V.C.; Tien, J.F.; Vollett, S.E.; Griffith, M.; Marra, M.A.; Morin, G.B. Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Mol. Cell. Biol. 2012, 32, 4691–4704. [Google Scholar] [CrossRef]
- Chirackal Manavalan, A.P.; Pilarova, K.; Kluge, M.; Bartholomeeusen, K.; Rajecky, M.; Oppelt, J.; Khirsariya, P.; Paruch, K.; Krejci, L.; Friedel, C.C.; et al. CDK12 controls G1/S progression by regulating RNAPII processivity at core DNA replication genes. EMBO Rep. 2019, 20, e47592. [Google Scholar] [CrossRef]
- Krajewska, M.; Dries, R.; Grassetti, A.V.; Dust, S.; Gao, Y.; Huang, H.; Sharma, B.; Day, D.S.; Kwiatkowski, N.; Pomaville, M.; et al. CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat. Commun. 2019, 10, 1757. [Google Scholar] [CrossRef]
- Dubbury, S.J.; Boutz, P.L.; Sharp, P.A. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 2018, 564, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Kwiatkowski, N.; Olson, C.M.; Dixon-Clarke, S.E.; Abraham, B.J.; Greifenberg, A.K.; Ficarro, S.B.; Elkins, J.M.; Liang, Y.; Hannett, N.M.; et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 2016, 12, 876–884. [Google Scholar] [CrossRef] [PubMed]
- Blazek, D.; Kohoutek, J.; Bartholomeeusen, K.; Johansen, E.; Hulinkova, P.; Luo, Z.; Cimermancic, P.; Ule, J.; Peterlin, B.M. The Cyclin K/CDK12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011, 25, 2158–2172. [Google Scholar] [CrossRef]
- Choi, S.H.; Martinez, T.F.; Kim, S.; Donaldson, C.; Shokhirev, M.N.; Saghatelian, A.; Jones, K.A. CDK12 phosphorylates 4E-BP1 to enable mTORC1-dependent translation and mitotic genome stability. Genes Dev. 2019, 33, 418–435. [Google Scholar] [CrossRef] [PubMed]
- Lui, G.Y.L.; Grandori, C.; Kemp, C.J. CDK12: An emerging therapeutic target for cancer. J. Clin. Pathol. 2018, 71, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Lei, T.; Zhang, P.; Zhang, X.; Xiao, X.; Zhang, J.; Qiu, T.; Dai, Q.; Zhang, Y.; Min, L.; Li, Q.; et al. Cyclin K regulates prereplicative complex assembly to promote mammalian cell proliferation. Nat. Commun. 2018, 9, 1876. [Google Scholar] [CrossRef]
- Chila, R.; Guffanti, F.; Damia, G. Role and therapeutic potential of CDK12 in human cancers. Cancer Treat. Rev. 2016, 50, 83–88. [Google Scholar] [CrossRef]
- Pilarova, K.; Herudek, J.; Blazek, D. CDK12: Cellular functions and therapeutic potential of versatile player in cancer. NAR Cancer 2020, 2. [Google Scholar] [CrossRef]
- Peng, F.; Yang, C.; Kong, Y.; Huang, X.; Chen, Y.; Zhou, Y.; Xie, X.; Liu, P. CDK12 Promotes Breast Cancer Progression and Maintains Stemness by Activating c-myc/beta -catenin Signaling. Curr. Cancer Drug Targets 2020, 20, 156–165. [Google Scholar] [CrossRef]
- Tien, J.F.; Mazloomian, A.; Cheng, S.G.; Hughes, C.S.; Chow, C.C.T.; Canapi, L.T.; Oloumi, A.; Trigo-Gonzalez, G.; Bashashati, A.; Xu, J.; et al. CDK12 regulates alternative last exon mRNA splicing and promotes breast cancer cell invasion. Nucleic Acids Res. 2017, 45, 6698–6716. [Google Scholar] [CrossRef]
- Ekumi, K.M.; Paculova, H.; Lenasi, T.; Pospichalova, V.; Bosken, C.A.; Rybarikova, J.; Bryja, V.; Geyer, M.; Blazek, D.; Barboric, M. Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the CDK12/CycK complex. Nucleic Acids Res. 2015, 43, 2575–2589. [Google Scholar] [CrossRef] [PubMed]
- Ko, T.K.; Kelly, E.; Pines, J. CrkRS: A novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J. Cell Sci. 2001, 114, 2591–2603. [Google Scholar] [PubMed]
- Chen, H.H.; Wang, Y.C.; Fann, M.J. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol. Cel. Biol. 2006, 26, 2736–2745. [Google Scholar] [CrossRef] [PubMed]
- Bartkowiak, B.; Liu, P.; Phatnani, H.P.; Fuda, N.J.; Cooper, J.J.; Price, D.H.; Adelman, K.; Lis, J.T.; Greenleaf, A.L. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010, 24, 2303–2316. [Google Scholar] [CrossRef]
- Dai, Q.; Lei, T.; Zhao, C.; Zhong, J.; Tang, Y.Z.; Chen, B.; Yang, J.; Li, C.; Wang, S.; Song, X.; et al. Cyclin K-containing kinase complexes maintain self-renewal in murine embryonic stem cells. J. Biol. Chem. 2012, 287, 25344–25352. [Google Scholar] [CrossRef]
- Kohoutek, J.; Blazek, D. Cyclin K goes with CDK12 and Cdk13. Cell Div. 2012, 7, 12. [Google Scholar] [CrossRef]
- Greifenberg, A.K.; Hönig, D.; Pilarova, K.; Düster, R.; Bartholomeeusen, K.; Bösken, C.A.; Anand, K.; Blazek, D.; Geyer, M. Structural and Functional Analysis of the Cdk13/Cyclin K Complex. Cell Rep. 2016, 14, 320–331. [Google Scholar] [CrossRef]
- Greenleaf, A.L. Human CDK12 and CDK13, multi-tasking CTD kinases for the new millenium. Transcription 2019, 10, 91–110. [Google Scholar] [CrossRef]
- Valcárcel, J.; Green, M.R. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci. 1996, 21, 296–301. [Google Scholar] [CrossRef]
- Kay, B.K.; Williamson, M.P.; Sudol, M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2000, 14, 231–241. [Google Scholar] [CrossRef]
- Ball, L.J.; Kuhne, R.; Schneider-Mergener, J.; Oschkinat, H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angewandte Chem. 2005, 44, 2852–2869. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, T.; Li, L.; Li, S.S. The SH3 domain—A family of versatile peptide- and protein-recognition module. Front. Biosci. J. Virtual Library 2008, 13, 4938–4952. [Google Scholar] [CrossRef] [PubMed]
- Mayer, B.J. SH3 domains: Complexity in moderation. J. Cell Sci. 2001, 114, 1253–1263. [Google Scholar] [PubMed]
- Ponna, S.K.; Myllykoski, M.; Boeckers, T.M.; Kursula, P. Structure of an unconventional SH3 domain from the postsynaptic density protein Shank3 at ultrahigh resolution. Biochem. Biophys. Res. Commun. 2017, 490, 806–812. [Google Scholar] [CrossRef]
- Bedford, M.T.; Chan, D.C.; Leder, P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997, 16, 2376–2383. [Google Scholar] [CrossRef]
- Castillo, J.; Knol, J.C.; Korver, C.M.; Piersma, S.R.; Pham, T.V.; de Goeij-de Haas, R.R.; van Pelt, A.M.M.; Jimenez, C.R.; Jansen, B.J.H. Human Testis Phosphoproteome Reveals Kinases as Potential Targets in Spermatogenesis and Testicular Cancer. Mol. Cell. Proteom. MCP 2019, 18, S132–S144. [Google Scholar] [CrossRef] [PubMed]
- Bosken, C.A.; Farnung, L.; Hintermair, C.; Merzel Schachter, M.; Vogel-Bachmayr, K.; Blazek, D.; Anand, K.; Fisher, R.P.; Eick, D.; Geyer, M. The structure and substrate specificity of human CDK12/Cyclin K. Nat. Commun. 2014, 5, 3505. [Google Scholar] [CrossRef]
- Fuda, N.J.; Ardehali, M.B.; Lis, J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 2009, 461, 186–192. [Google Scholar] [CrossRef]
- Egloff, S.; Murphy, S. Cracking the RNA polymerase II CTD code. Trends Genet. TIG 2008, 24, 280–288. [Google Scholar] [CrossRef]
- Eick, D.; Geyer, M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013, 113, 8456–8490. [Google Scholar] [CrossRef]
- Davidson, L.; Muniz, L.; West, S. 3’ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014, 28, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Yang, W.; Ni, T.; Tang, Z.; Nakadai, T.; Zhu, J.; Roeder, R.G. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 2015, 350, 1383–1386. [Google Scholar] [CrossRef]
- Edwards, M.C.; Wong, C.; Elledge, S.J. Human cyclin K, a novel RNA polymerase II-associated cyclin possessing both carboxy-terminal domain kinase and Cdk-activating kinase activity. Mol. Cell. Biol. 1998, 18, 4291–4300. [Google Scholar] [CrossRef] [PubMed]
- Eifler, T.T.; Shao, W.; Bartholomeeusen, K.; Fujinaga, K.; Jäger, S.; Johnson, J.R.; Luo, Z.; Krogan, N.J.; Peterlin, B.M. Cyclin-dependent kinase 12 increases 3’ end processing of growth factor-induced c-FOS transcripts. Mol. Cell. Biol. 2015, 35, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.; Thuma, L.; Klambt, C. The regulation of glial-specific splicing of Neurexin IV requires HOW and CDK12 activity. Development 2012, 139, 1765–1776. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.; Gao, X.; Gilmore, J.M.; Florens, L.; Washburn, M.P.; Smith, E.; Shilatifard, A. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol. Cell. Biol. 2015, 35, 928–938. [Google Scholar] [CrossRef]
- Bertoli, C.; Skotheim, J.M.; de Bruin, R.A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef]
- Gutgemann, I.; Lehman, N.L.; Jackson, P.K.; Longacre, T.A. Emi1 protein accumulation implicates misregulation of the anaphase promoting complex/cyclosome pathway in ovarian clear cell carcinoma. Mod. Pathol. 2008, 21, 445–454. [Google Scholar] [CrossRef]
- Messner, D.J.; Kowdley, K.V. Neoplastic transformation of rat liver epithelial cells is enhanced by non-transferrin-bound iron. BMC Gastroenterol. 2008, 8, 2. [Google Scholar] [CrossRef]
- Perez-Posada, A.; Dudin, O.; Ocana-Pallares, E.; Ruiz-Trillo, I.; Ondracka, A. Cell cycle transcriptomics of Capsaspora provides insights into the evolution of cyclin-CDK machinery. PLoS Genet. 2020, 16, e1008584. [Google Scholar] [CrossRef]
- Geng, M.; Yang, Y.; Cao, X.; Dang, L.; Zhang, T.; Zhang, L. Targeting CDK12-mediated transcription regulation in anaplastic thyroid carcinoma. Biochem. Biophys. Res. Commun. 2019, 520, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.R.; Juan, H.C.; Wong, Y.H.; Tsai, J.W.; Fann, M.J. CDK12 Regulates Neurogenesis and Late-Arising Neuronal Migration in the Developing Cerebral Cortex. Cerebral Cortex 2017, 27, 2289–2302. [Google Scholar] [CrossRef] [PubMed]
- Schecher, S.; Walter, B.; Falkenstein, M.; Macher-Goeppinger, S.; Stenzel, P.; Krümpelmann, K.; Hadaschik, B.; Perner, S.; Kristiansen, G.; Duensing, S.; et al. Cyclin K dependent regulation of Aurora B affects apoptosis and proliferation by induction of mitotic catastrophe in prostate cancer. Int. J. Cancer 2017, 141, 1643–1653. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, F.; Shi, X.; Liu, X.; Yang, H.; Zhang, Z. Identification and Characterization of the Cyclin-Dependent Kinases Gene Family in Silkworm, Bombyx mori. DNA Cell Biol. 2016, 35, 13–23. [Google Scholar] [CrossRef]
- Blow, J.J.; Gillespie, P.J. Replication licensing and cancer--a fatal entanglement? Nat. Rev. Cancer 2008, 8, 799–806. [Google Scholar] [CrossRef]
- Wang, C.; Wang, H.; Lieftink, C.; du Chatinier, A.; Gao, D.; Jin, G.; Jin, H.; Beijersbergen, R.L.; Qin, W.; Bernards, R. CDK12 inhibition mediates DNA damage and is synergistic with sorafenib treatment in hepatocellular carcinoma. Gut 2019. [Google Scholar] [CrossRef]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
- Chen, B.; Zhang, G.; Wei, G.; Wang, Y.; Guo, L.; Lin, J.; Li, K.; Mok, H.; Cao, L.; Ren, C.; et al. Heterogeneity of genomic profile in patients with HER2-positive breast cancer. Endocr. Related Cancer 2020, 27, 153–162. [Google Scholar] [CrossRef]
- Choi, H.J.; Jin, S.; Cho, H.; Won, H.Y.; An, H.W.; Jeong, G.Y.; Park, Y.U.; Kim, H.Y.; Park, M.K.; Son, T.; et al. CDK12 drives breast tumor initiation and trastuzumab resistance via WNT and IRS1-ErbB-PI3K signaling. EMBO Rep. 2019, 20, e48058. [Google Scholar] [CrossRef]
- Al-Mahmood, S.; Sapiezynski, J.; Garbuzenko, O.B.; Minko, T. Metastatic and triple-negative breast cancer: Challenges and treatment options. Drug Deliv. Transl. Res. 2018, 8, 1483–1507. [Google Scholar] [CrossRef]
- Naidoo, K.; Wai, P.T.; Maguire, S.L.; Daley, F.; Haider, S.; Kriplani, D.; Campbell, J.; Mirza, H.; Grigoriadis, A.; Tutt, A.; et al. Evaluation of CDK12 Protein Expression as a Potential Novel Biomarker for DNA Damage Response-Targeted Therapies in Breast Cancer. Mol. Cancer Ther. 2018, 17, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Koonings, P.P.; Campbell, K.; Mishell, D.R., Jr.; Grimes, D.A. Relative frequency of primary ovarian neoplasms: A 10-year review. Obstet. Gynecol. 1989, 74, 921–926. [Google Scholar] [CrossRef]
- Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; DiSaia, P.; Gabra, H.; Glenn, P.; et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Sokol, E.S.; Pavlick, D.; Frampton, G.M.; Ross, J.S.; Miller, V.A.; Ali, S.M.; Lotan, T.L.; Pardoll, D.M.; Chung, J.H.; Antonarakis, E.S. Pan-Cancer Analysis of CDK12 Loss-of-Function Alterations and Their Association with the Focal Tandem-Duplicator Phenotype. Oncologist 2019, 24, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Kanakkanthara, A.; Kurmi, K.; Ekstrom, T.L.; Hou, X.; Purfeerst, E.R.; Heinzen, E.P.; Correia, C.; Huntoon, C.J.; O’Brien, D.; Wahner Hendrickson, A.E.; et al. BRCA1 Deficiency Upregulates NNMT, Which Reprograms Metabolism and Sensitizes Ovarian Cancer Cells to Mitochondrial Metabolic Targeting Agents. Cancer Res. 2019, 79, 5920–5929. [Google Scholar] [CrossRef] [PubMed]
- Bogdanova, N.V.; Schurmann, P.; Valova, Y.; Dubrowinskaja, N.; Turmanov, N.; Yugay, T.; Essimsiitova, Z.; Mingazheva, E.; Prokofyeva, D.; Bermisheva, M.; et al. A Splice Site Variant of CDK12 and Breast Cancer in Three Eurasian Populations. Front. Oncol. 2019, 9, 493. [Google Scholar] [CrossRef]
- Zeng, M.; Kwiatkowski, N.P.; Zhang, T.; Nabet, B.; Xu, M.; Liang, Y.; Quan, C.; Wang, J.; Hao, M.; Palakurthi, S.; et al. Targeting MYC dependency in ovarian cancer through inhibition of CDK7 and CDK12/13. eLife 2018, 7. [Google Scholar] [CrossRef]
- Reimers, M.A.; Yip, S.M.; Zhang, L.; Cieslik, M.; Dhawan, M.; Montgomery, B.; Wyatt, A.W.; Chi, K.N.; Small, E.J.; Chinnaiyan, A.M.; et al. Clinical Outcomes in Cyclin-dependent Kinase 12 Mutant Advanced Prostate Cancer. Eur. Urol. 2020, 77, 333–341. [Google Scholar] [CrossRef]
- Wu, Y.M.; Cieślik, M.; Lonigro, R.J.; Vats, P.; Reimers, M.A.; Cao, X.; Ning, Y.; Wang, L.; Kunju, L.P.; de Sarkar, N.; et al. Inactivation of CDK12 Delineates a Distinct Immunogenic Class of Advanced Prostate Cancer. Cell 2018, 173, 1770–1782. [Google Scholar] [CrossRef]
- Mateo, J.; Seed, G.; Bertan, C.; Rescigno, P.; Dolling, D.; Figueiredo, I.; Miranda, S.; Nava Rodrigues, D.; Gurel, B.; Clarke, M.; et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J. Clin. Investig. 2020. [Google Scholar] [CrossRef]
- Quigley, D.A.; Dang, H.X.; Zhao, S.G.; Lloyd, P.; Aggarwal, R.; Alumkal, J.J.; Foye, A.; Kothari, V.; Perry, M.D.; Bailey, A.M.; et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 2018, 174, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.R.; Ha, G.; Hoff, A.M.; Wala, J.A.; Carrot-Zhang, J.; Whelan, C.W.; Haradhvala, N.J.; Freeman, S.S.; Reed, S.C.; Rhoades, J.; et al. Structural Alterations Driving Castration-Resistant Prostate Cancer Revealed by Linked-Read Genome Sequencing. Cell 2018, 174, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Niu, L.; Xiao, Z.; Zheng, C.; Shen, Y.; Shi, Y.; Han, X. Whole-genome sequencing of prostate cancer reveals novel mutation-driven processes and molecular subgroups. Life Sci. 2019. [Google Scholar] [CrossRef]
- Zhou, C.; Feng, X.; Yuan, F.; Ji, J.; Shi, M.; Yu, Y.; Zhu, Z.; Zhang, J. Difference of molecular alterations in HER2-positive and HER2-negative gastric cancers by whole-genome sequencing analysis. Cancer Manag. Res. 2018, 10, 3945–3954. [Google Scholar] [CrossRef]
- Ji, J.; Zhou, C.; Wu, J.; Cai, Q.; Shi, M.; Zhang, H.; Yu, Y.; Zhu, Z.; Zhang, J. Expression pattern of CDK12 protein in gastric cancer and its positive correlation with CD8(+) cell density and CCL12 expression. Int. J. Med. Sci. 2019, 16, 1142–1148. [Google Scholar] [CrossRef] [PubMed]
- Quereda, V.; Bayle, S.; Vena, F.; Frydman, S.M.; Monastyrskyi, A.; Roush, W.R.; Duckett, D.R. Therapeutic Targeting of CDK12/CDK13 in Triple-Negative Breast Cancer. Cancer Cell 2019, 36, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.; Quigley, D.A.; Robinson, T.M.; Feng, F.Y.; Ashworth, A. Transcription-Associated Cyclin-Dependent Kinases as Targets and Biomarkers for Cancer Therapy. Cancer Discov. 2020, 10, 351–370. [Google Scholar] [CrossRef]
- Schiewer, M.J.; Goodwin, J.F.; Han, S.; Brenner, J.C.; Augello, M.A.; Dean, J.L.; Liu, F.; Planck, J.L.; Ravindranathan, P.; Chinnaiyan, A.M.; et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012, 2, 1134–1149. [Google Scholar] [CrossRef] [PubMed]
- Paculova, H.; Kramara, J.; Simeckova, S.; Fedr, R.; Soucek, K.; Hylse, O.; Paruch, K.; Svoboda, M.; Mistrik, M.; Kohoutek, J. BRCA1 or CDK12 loss sensitizes cells to CHK1 inhibitors. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317727479. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]
- Johnson, S.F.; Cruz, C.; Greifenberg, A.K.; Dust, S.; Stover, D.G.; Chi, D.; Primack, B.; Cao, S.; Bernhardy, A.J.; Coulson, R.; et al. CDK12 Inhibition Reverses De Novo and Acquired PARP Inhibitor Resistance in BRCA Wild-Type and Mutated Models of Triple-Negative Breast Cancer. Cell Rep. 2016, 17, 2367–2381. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.; Eastman, A. The cancer therapeutic potential of Chk1 inhibitors: How mechanistic studies impact on clinical trial design. Br. J. Clin. Pharmacol. 2013, 76, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.X.; Janetka, J.W.; Piwnica-Worms, H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol. Med. 2011, 17, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.X.; Cai, S.; Li, S.; Ryan, C.E.; Guo, Z.; Schaiff, W.T.; Lin, L.; Hoog, J.; Goiffon, R.J.; Prat, A.; et al. Targeting Chk1 in p53-deficient triple-negative breast cancer is therapeutically beneficial in human-in-mouse tumor models. J. Clin. Investig. 2012, 122, 1541–1552. [Google Scholar] [CrossRef]
- Guzi, T.J.; Paruch, K.; Dwyer, M.P.; Labroli, M.; Shanahan, F.; Davis, N.; Taricani, L.; Wiswell, D.; Seghezzi, W.; Penaflor, E.; et al. Targeting the replication checkpoint using SCH 900776, a potent and functionally selective CHK1 inhibitor identified via high content screening. Mol. Cancer Ther. 2011, 10, 591–602. [Google Scholar] [CrossRef]
- Iniguez, A.B.; Stolte, B.; Wang, E.J.; Conway, A.S.; Alexe, G.; Dharia, N.V.; Kwiatkowski, N.; Zhang, T.; Abraham, B.J.; Mora, J.; et al. EWS/FLI Confers Tumor Cell Synthetic Lethality to CDK12 Inhibition in Ewing Sarcoma. Cancer cell 2018, 33, 202–216. [Google Scholar] [CrossRef]
- Delehouzé, C.; Godl, K.; Loaëc, N.; Bruyère, C.; Desban, N.; Oumata, N.; Galons, H.; Roumeliotis, T.I.; Giannopoulou, E.G.; Grenet, J.; et al. CDK/CK1 inhibitors roscovitine and CR8 downregulate amplified MYCN in neuroblastoma cells. Oncogene 2014, 33, 5675–5687. [Google Scholar] [CrossRef]
- Riggi, N.; Knoechel, B.; Gillespie, S.M.; Rheinbay, E.; Boulay, G.; Suva, M.L.; Rossetti, N.E.; Boonseng, W.E.; Oksuz, O.; Cook, E.B.; et al. EWS-FLI1 utilizes divergent chromatin remodeling mechanisms to directly activate or repress enhancer elements in Ewing sarcoma. Cancer Cell 2014, 26, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Menghi, F.; Barthel, F.P.; Yadav, V.; Tang, M.; Ji, B.; Tang, Z.; Carter, G.W.; Ruan, Y.; Scully, R.; Verhaak, R.G.W.; et al. The Tandem Duplicator Phenotype Is a Prevalent Genome-Wide Cancer Configuration Driven by Distinct Gene Mutations. Cancer Cell 2018, 34, 197–210. [Google Scholar] [CrossRef]
- Kwiatkowski, N.; Zhang, T.; Rahl, P.B.; Abraham, B.J.; Reddy, J.; Ficarro, S.B.; Dastur, A.; Amzallag, A.; Ramaswamy, S.; Tesar, B.; et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 2014, 511, 616–620. [Google Scholar] [CrossRef]
- Asghar, U.; Witkiewicz, A.K.; Turner, N.C.; Knudsen, E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 2015, 14, 130–146. [Google Scholar] [CrossRef] [PubMed]
- Novotna, E.; Bukum, N.; Hofman, J.; Flaxova, M.; Kouklikova, E.; Louvarova, D.; Wsol, V. Aldo-keto reductase 1C3 (AKR1C3): A missing piece of the puzzle in the dinaciclib interaction profile. Arch. Toxicol. 2018, 92, 2845–2857. [Google Scholar] [CrossRef] [PubMed]
- Parry, D.; Guzi, T.; Shanahan, F.; Davis, N.; Prabhavalkar, D.; Wiswell, D.; Seghezzi, W.; Paruch, K.; Dwyer, M.P.; Doll, R.; et al. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol. Cancer Ther. 2010, 9, 2344–2353. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.S.J.; Sharko, A.C.; Munie, J.; Kassler, S.; Melendez, T.; Lim, C.U.; Broude, E.V. CDK7 Inhibition is Effective in all the Subtypes of Breast Cancer: Determinants of Response and Synergy with EGFR Inhibition. Cells 2020, 9, 638. [Google Scholar] [CrossRef] [PubMed]
- Christensen, C.L.; Kwiatkowski, N.; Abraham, B.J.; Carretero, J.; Al-Shahrour, F.; Zhang, T.; Chipumuro, E.; Herter-Sprie, G.S.; Akbay, E.A.; Altabef, A.; et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer Cell 2014, 26, 909–922. [Google Scholar] [CrossRef]
- Bayles, I.; Krajewska, M.; Pontius, W.D.; Saiakhova, A.; Morrow, J.J.; Bartels, C.; Lu, J.; Faber, Z.J.; Fedorov, Y.; Hong, E.S.; et al. Ex vivo screen identifies CDK12 as a metastatic vulnerability in osteosarcoma. J. Clin. Investig. 2019, 129, 4377–4392. [Google Scholar] [CrossRef]
| Cancer Type | CDK12′s Function | Mechanism | References |
|---|---|---|---|
| Breast cancer (HER2 1 -positive breast cancer) | Tumor promoter | Overexpression of CDK12 regulates the splicing of ATM 5 and DNAJB6-L 6 and actives WNT 7 and IRS1-ErbB-PI3K 8 signaling | [20,59] |
| Breast cancer (TNBC 2) | Tumor suppressor | Loss of CDK12 leads to downregulation of DDR 9 genes | [61,76] |
| Ovarian cancer (HGSOC 3) | Tumor suppressor | Loss of CDK12 leads to downregulation of DDR 9 genes | [21,63,65] |
| Prostate cancer (mCRPC 4) | Tumor suppressor | Loss of CDK12 leads to downregulation of DDR 9 genes | [68,69] |
| Gastric cancer | Tumor promoter | Overexpression of CDK12 actives the CDK12/CCL21 10 pathway | [75] |
| Treatment | Function | Cancer Type | References |
|---|---|---|---|
| Dinaciclib | Inhibition of multiple CDKs including CDK12 | Breast cancer and metastatic osteosarcoma | [59,96] |
| THZ1 | Inhibition of CDK7/12 | Ovarian cancer and neuroblastoma | [67,87] |
| THZ531 | Inhibition of CDK12/13 | Breast cancer, hepatocellular carcinoma and metastatic osteosarcoma | [56,59,96] |
| SR-4835 | Inhibition of CDK12/13 | TNBC 3 (use with PARP inhibitors) | [76] |
| PARP 1 inhibitors + CDK12 inhibitors | Synthetic lethality | TNBC 3, ovarian cancer and Ewing sarcoma | [76,81,86] |
| CHK1 2 inhibitors | Synthetic lethality | Ovarian cancer | [79] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, S.; Hu, L.; Wu, Z.; Chen, Z.; Liu, S.; Xu, X.; Qian, A. CDK12: A Potent Target and Biomarker for Human Cancer Therapy. Cells 2020, 9, 1483. https://doi.org/10.3390/cells9061483
Liang S, Hu L, Wu Z, Chen Z, Liu S, Xu X, Qian A. CDK12: A Potent Target and Biomarker for Human Cancer Therapy. Cells. 2020; 9(6):1483. https://doi.org/10.3390/cells9061483
Chicago/Turabian StyleLiang, Shujing, Lifang Hu, Zixiang Wu, Zhihao Chen, Shuyu Liu, Xia Xu, and Airong Qian. 2020. "CDK12: A Potent Target and Biomarker for Human Cancer Therapy" Cells 9, no. 6: 1483. https://doi.org/10.3390/cells9061483
APA StyleLiang, S., Hu, L., Wu, Z., Chen, Z., Liu, S., Xu, X., & Qian, A. (2020). CDK12: A Potent Target and Biomarker for Human Cancer Therapy. Cells, 9(6), 1483. https://doi.org/10.3390/cells9061483





