Nanospheres Loaded with Curcumin Improve the Bioactivity of Umbilical Cord Blood-Mesenchymal Stem Cells via c-Src Activation during the Skin Wound Healing Process
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Culture of Human UCB-MSCs
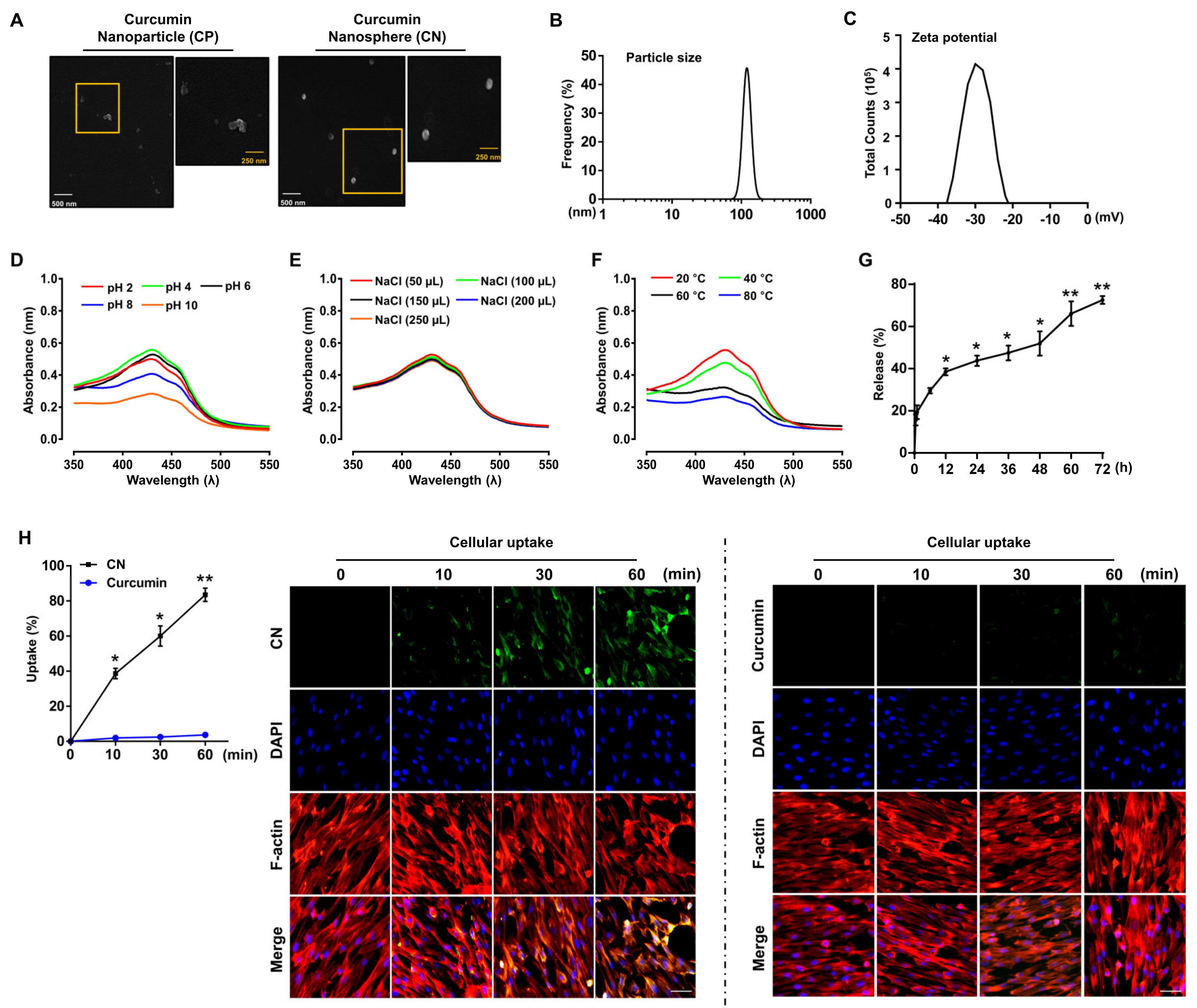
2.3. Preparation of the Curcumin Nanosphere (CN)
2.4. Field Emission Scanning Electron Microscope (FE-SEM) Measurements
2.5. Determination of the Particle Size and Zeta Potential
2.6. In Vitro Kinetic Release of Curcumin from CN
2.7. Determination of the Encapsulation Efficiency and Loading Capacity
2.8. Determination of CN Stability
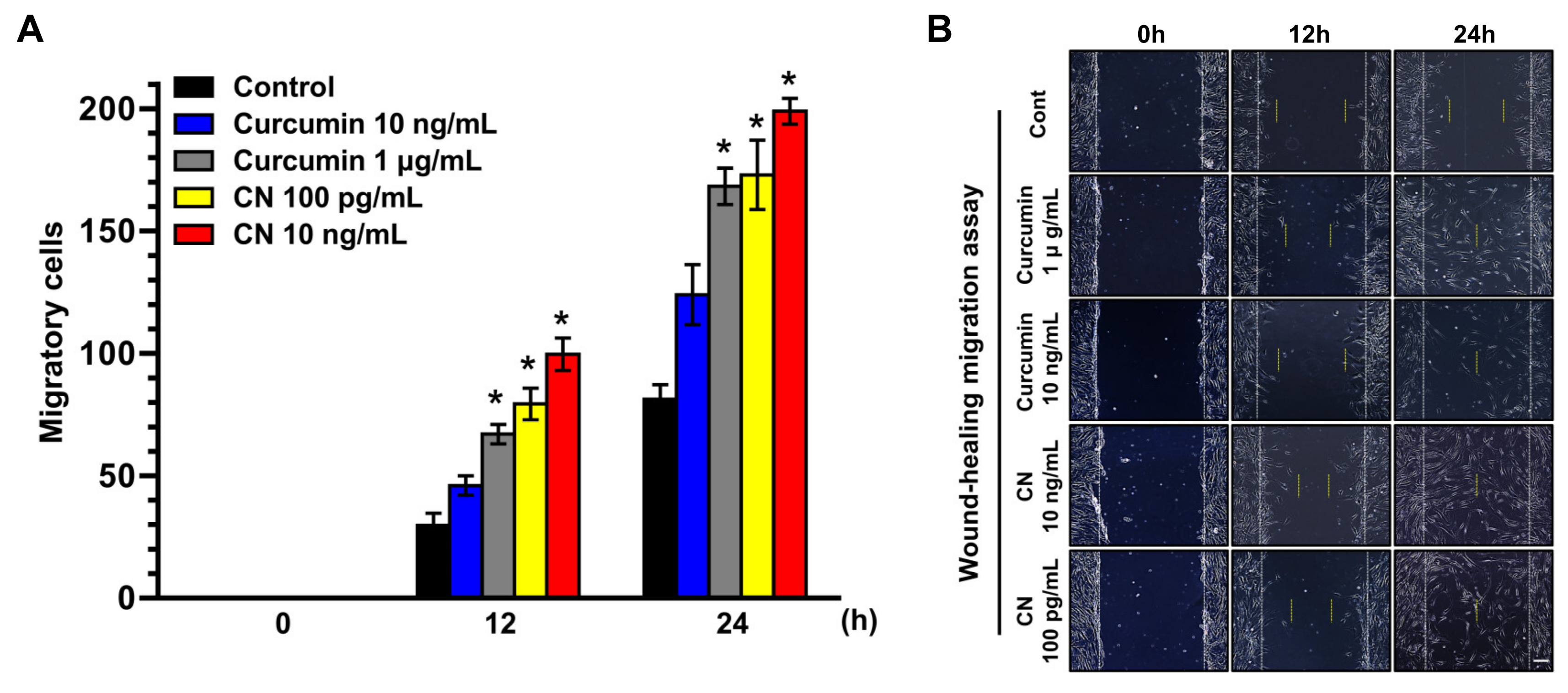
2.9. Wound-Healing Migration Assay
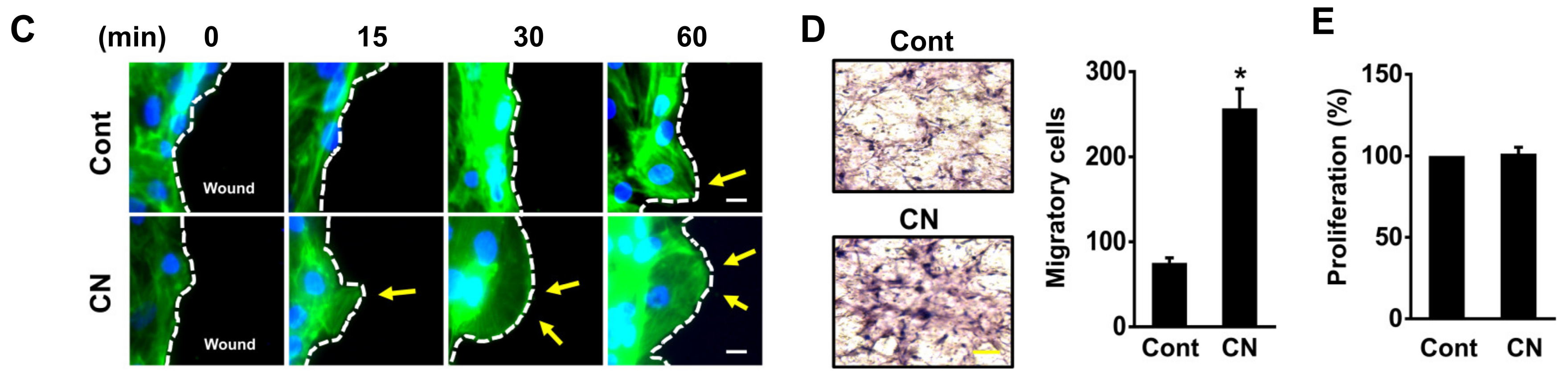
2.10. Lamellipodia Formation and Immunofluorescence Analysis
2.11. Cellular Uptake of CN
2.12. Transwell Migration Assay
2.13. Western Blot Analysis
2.14. Cell Proliferation Assay
2.15. Mouse Skin Wound Healing Model
2.16. Statistical Analysis
3. Results
3.1. Characterization of the Curcumin Nanosphere (CN)
3.2. The Effect of CN on Migration of Human UCB-MSCs
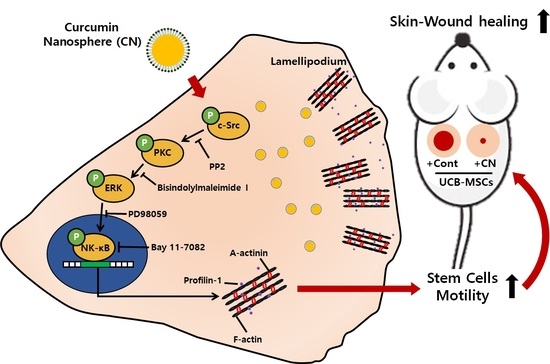
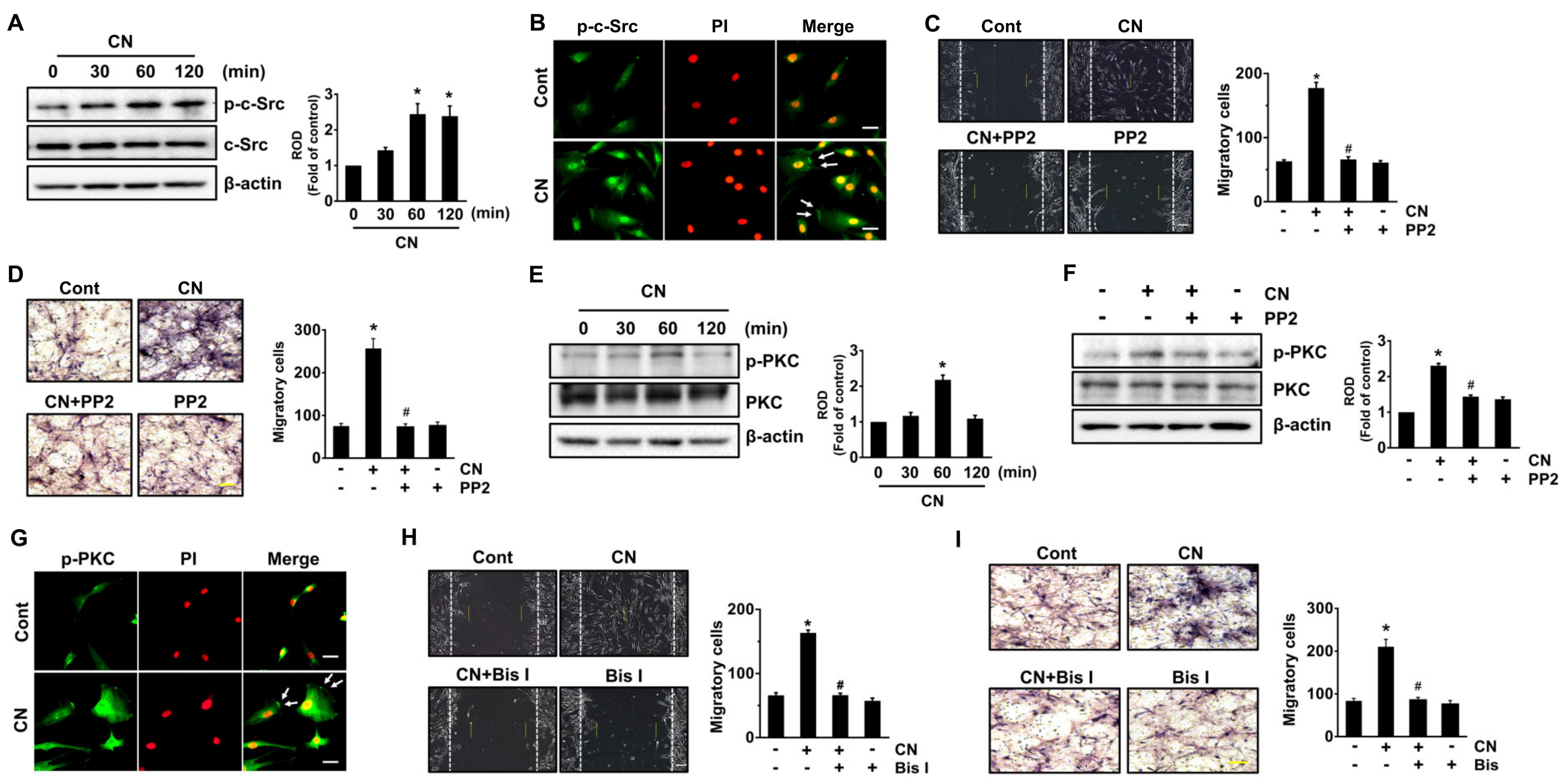
3.3. CN Regulates the Activation of c-Src and PKC
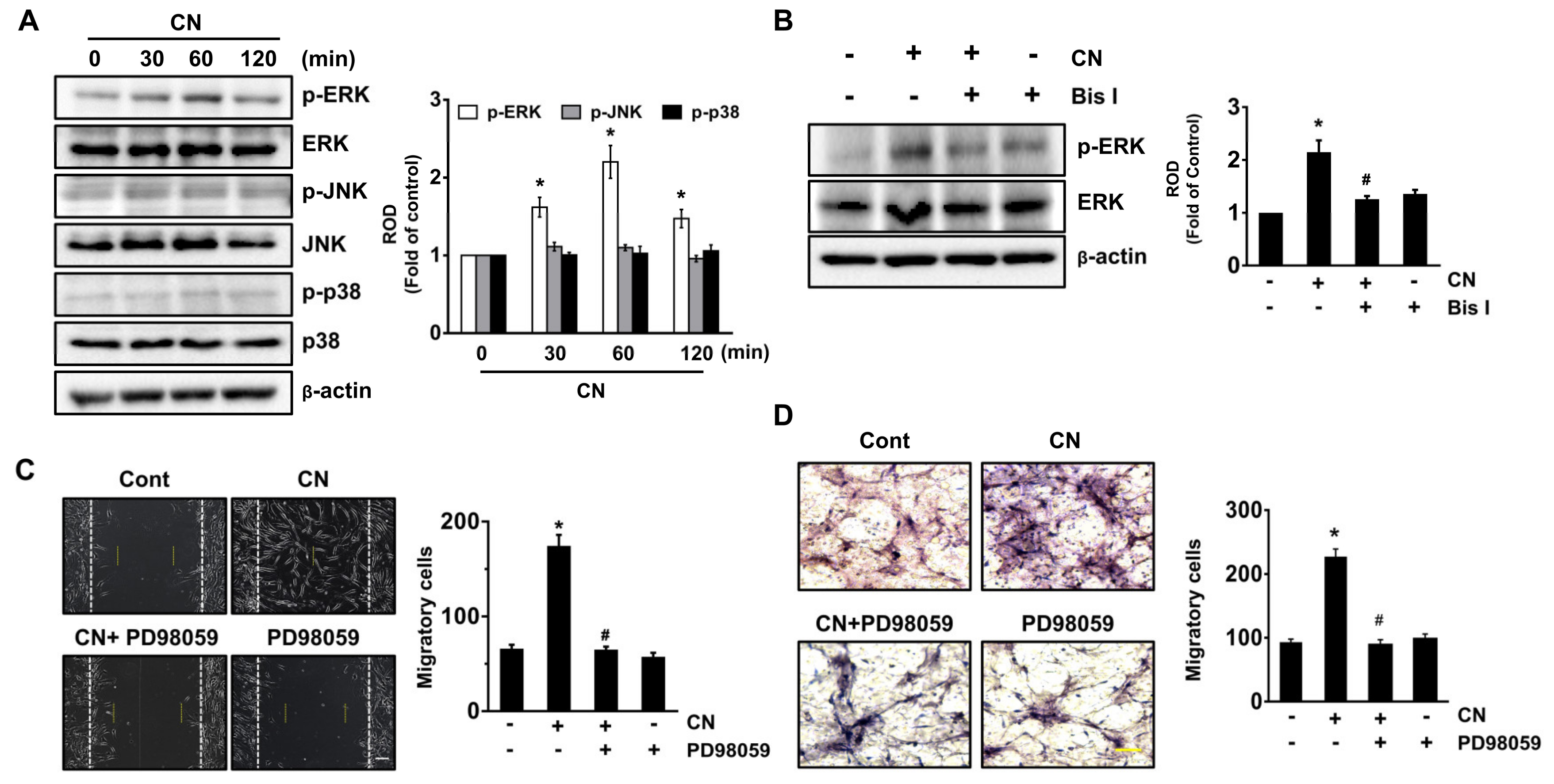
3.4. CN Uniquely Regulates the ERK Pathway Responsible for the Cell Migration
3.5. Regulatory Effect of CN on the Activation of NF-κB and the Cytoskeletal Reorganization
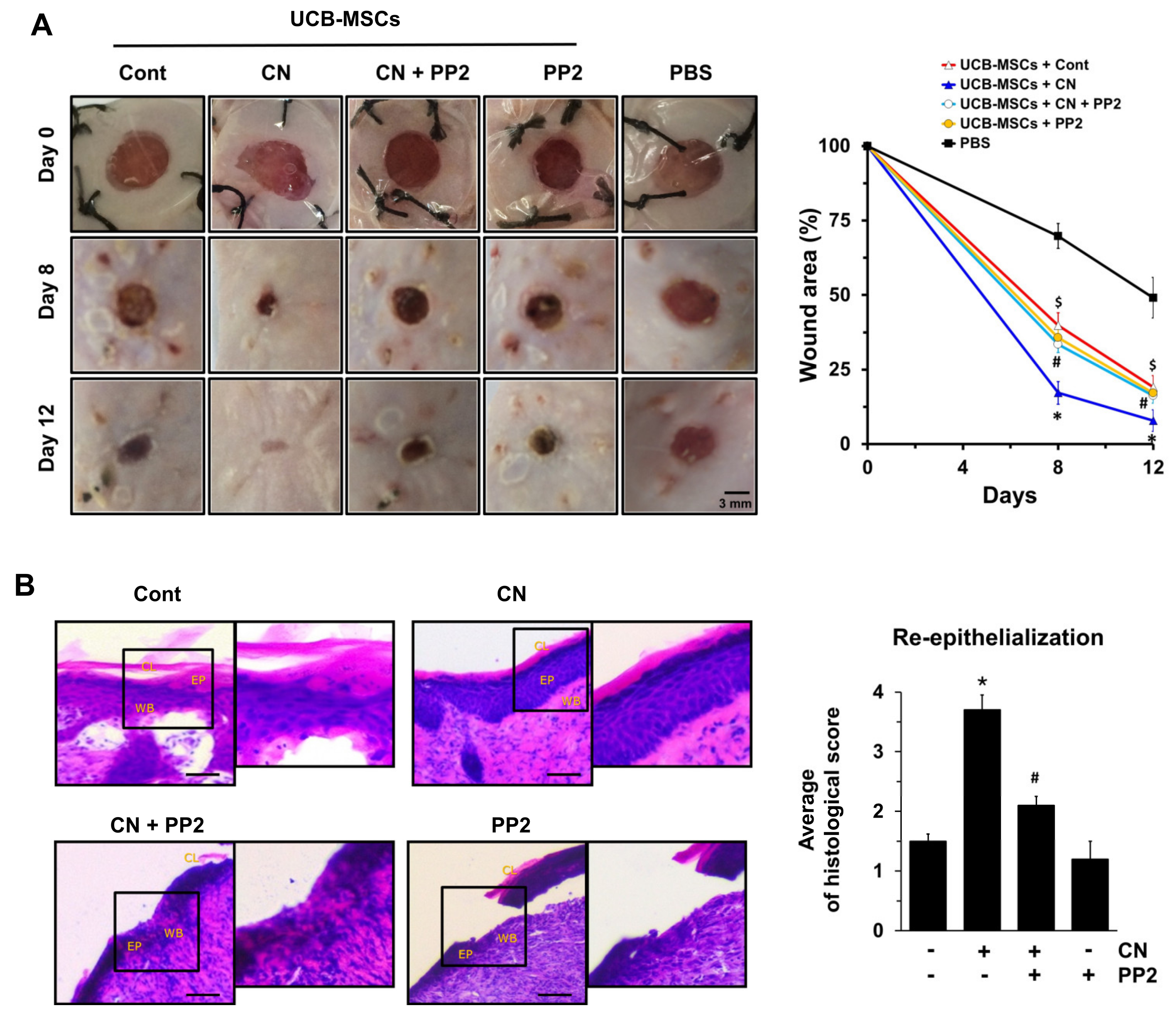
3.6. The Role of CN in Mouse Model for Skin Wound Healing
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Rorth, P. Collective cell migration. Annu. Rev. Cell Dev. Biol. 2009, 25, 407–429. [Google Scholar] [CrossRef]
- Rodero, M.P.; Khosrotehrani, K. Skin wound healing modulation by macrophages. Int. J. Clin. Exp. Pathol. 2010, 3, 643–653. [Google Scholar]
- Hocking, A.M.; Gibran, N.S. Mesenchymal stem cells: Paracrine signaling and differentiation during cutaneous wound repair. Exp. Cell Res. 2010, 316, 2213–2219. [Google Scholar] [CrossRef]
- Kim, S.W.; Zhang, H.Z.; Guo, L.; Kim, J.M.; Kim, M.H. Amniotic mesenchymal stem cells enhance wound healing in diabetic NOD/SCID mice through high angiogenic and engraftment capabilities. PLoS ONE 2012, 7, e41105. [Google Scholar] [CrossRef]
- Ojeh, N.; Pastar, I.; Tomic-Canic, M.; Stojadinovic, O. Stem cells in skin regeneration, wound healing, and their clinical applications. Int. J. Mol. Sci. 2015, 16, 25476–25501. [Google Scholar] [CrossRef] [PubMed]
- Braun, K.M.; Prowse, D.M. Distinct epidermal stem cell compartments are maintained by independent niche microenvironments. Stem Cell Rev. 2006, 2, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Yau, W.W.Y.; Rujitanaroj, P.O.; Lam, L.; Chew, S.Y. Directing stem cell fate by controlled RNA interference. Biomaterials 2012, 33, 2608–2628. [Google Scholar] [CrossRef]
- Vaca, P.; Berna, G.; Araujo, R.; Carneiro, E.M.; Bedoya, F.J.; Soria, B.; Martin, F. Nicotinamide induces differentiation of embryonic stem cells into insulin-secreting cells. Exp. Cell Res. 2008, 314, 969–974. [Google Scholar] [CrossRef]
- Madhyastha, R.; Madhyastha, H.; Nakajima, Y.; Omura, S.; Maruyama, M. Curcumin facilitates fibrinolysis and cellular migration during wound healing by modulating urokinase plasminogen activator expression. Pathophysiol. Haemost. Thromb. 2010, 37, 59–66. [Google Scholar] [CrossRef]
- Tejada, S.; Manayi, A.; Daglia, M.; Nabavi, S.F.; Sureda, A.; Hajheydari, Z.; Gortzi, O.; Pazoki-Toroudi, H.; Nabavi, S.M. Wound healing effects of curcumin: A short review. Curr. Pharm. Biotechnol. 2016, 17, 1002–1007. [Google Scholar] [CrossRef]
- Sahebkar, A. Curcuminoids for the management of hypertriglyceridaemia. Nat. Rev. Cardiol. 2014, 11, 123. [Google Scholar] [CrossRef]
- Goel, A.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin as “Curecumin”: From kitchen to clinic. Biochem. Pharmacol. 2008, 75, 787–809. [Google Scholar] [CrossRef] [PubMed]
- Sala de Oyanguren, F.J.; Rainey, N.E.; Moustapha, A.; Saric, A.; Sureau, F.; O’Connor, J.E.; Petit, P.X. Highlighting curcumin-induced crosstalk between autophagy and apoptosis as supported by its specific subcellular localization. Cells 2020, 9, 361. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Yan, R.; Xu, X.; Li, X.; Cao, L.; Gao, L.; Liu, J.; Zhou, X.; Yu, H.; Wang, X.; et al. Curcumin represses adipogenic differentiation of human bone marrow mesenchymal stem cells via inhibiting kruppel-like factor 15 expression. Acta Histochem. 2019, 121, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Dulbecco, P.; Savarino, V. Therapeutic potential of curcumin in digestive diseases. World J. Gastroenterol. 2013, 19, 9256–9270. [Google Scholar] [CrossRef]
- Kim, J.Y.; Lee, Y.M.; Kim, D.W.; Min, T.; Lee, S.J. Nanosphere loaded with curcumin inhibits the gastrointestinal cell death signaling pathway induced by the foodborne pathogen vibrio vulnificus. Cells 2020, 9, 631. [Google Scholar] [CrossRef] [PubMed]
- Park, J.B.; Lee, Y.M.; Park, M.K.; Min, T.; Lee, S.J. Effects of anti-ecotoxicological curcumin nanospheres on feed efficiency and fecal odor in mice. J. Environ. Sci. Int. 2019, 28, 183–189. [Google Scholar] [CrossRef]
- Qiao, C.; Xu, W.; Zhu, W.; Hu, J.; Qian, H.; Yin, Q.; Jiang, R.; Yan, Y.; Mao, F.; Yang, H.; et al. Human mesenchymal stem cells isolated from the umbilical cord. Cell Biol. Int. 2008, 32, 8–15. [Google Scholar] [CrossRef]
- Le Blanc, K.; Tammik, L.; Sundberg, B.; Haynesworth, S.E.; Ringden, O. Mesenchymal stem cells inhibit and stimulate mixed lymphocyte cultures and mitogenic responses independently of the major histocompatibility complex. Scand. J. Immunol. 2003, 57, 11–20. [Google Scholar] [CrossRef]
- Yang, S.E.; Ha, C.W.; Jung, M.; Jin, H.J.; Lee, M.; Song, H.; Choi, S.; Oh, W.; Yang, Y.S. Mesenchymal stem/progenitor cells developed in cultures from UC blood. Cytotherapy 2004, 6, 476–486. [Google Scholar] [CrossRef]
- Sensebe, L.; Krampera, M.; Schrezenmeier, H.; Bourin, P.; Giordano, R. Mesenchymal stem cells for clinical application. Vox Sang. 2010, 98, 93–107. [Google Scholar] [CrossRef] [PubMed]
- Wakitani, S.; Nawata, M.; Tensho, K.; Okabe, T.; Machida, H.; Ohgushi, H. Repair of articular cartilage defects in the patello-femoral joint with autologous bone marrow mesenchymal cell transplantation: Three case reports involving nine defects in five knees. J. Tissue Eng. Regen. Med. 2007, 1, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Peled, A.; Petit, I.; Kollet, O.; Magid, M.; Ponomaryov, T.; Byk, T.; Nagler, A.; Ben-Hur, H.; Many, A.; Shultz, L.; et al. Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science 1999, 283, 845–848. [Google Scholar] [CrossRef] [PubMed]
- Son, B.R.; Marquez-Curtis, L.A.; Kucia, M.; Wysoczynski, M.; Turner, A.R.; Ratajczak, J.; Ratajczak, M.Z.; Janowska-Wieczorek, A. Migration of bone marrow and cord blood mesenchymal stem cells in vitro is regulated by stromal-derived factor-1-CXCR4 and hepatocyte growth factor-c-met axes and involves matrix metalloproteinases. Stem Cells 2006, 24, 1254–1264. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Jeon, E.S.; Lee, J.S.; Cho, M.; Suh, D.S.; Chang, C.L.; Kim, J.H. Lysophosphatidic acid in malignant ascites stimulates migration of human mesenchymal stem cells. J. Cell. Biochem. 2008, 104, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Han, B.; Cai, S.; Lei, Y.; Sun, T.; Sheng, Z. Migration of bone marrow-derived mesenchymal stem cells induced by tumor necrosis factor-α and its possible role in wound healing. Wound Repair Regen. 2009, 17, 185–191. [Google Scholar] [CrossRef]
- Kang, Y.Y.; Jung, H.; Yu, G.; Chong, Y.; Mok, H. Complexation of curcumin with 2-aminoethyl diphenyl borate and implications for spatiotemporal fluorescence monitoring. Int. J. Pharm. 2016, 515, 669–676. [Google Scholar] [CrossRef]
- Kishibe, M.; Bando, Y.; Tanaka, T.; Ishida-Yamamoto, A.; Iizuka, H.; Yoshida, S. Kallikrein-related peptidase 8-dependent skin wound healing is associated with upregulation of kallikrein-related peptidase 6 and PAR2. J. Investig. Dermatol. 2012, 132, 1717–1724. [Google Scholar] [CrossRef]
- Lee, K.B.; Choi, J.; Cho, S.B.; Chung, J.Y.; Moon, E.S.; Kim, N.S.; Han, H.J. Topical embryonic stem cells enhance wound healing in diabetic rats. J. Bone. Miner. Res. 2011, 29, 1554–1562. [Google Scholar] [CrossRef]
- Rajabi, M.A.; Rajabi, F. The effect of estrogen on wound healing in rats. Pak. J. Med. Sci. 2007, 23, 349–352. [Google Scholar]
- Altavilla, D.; Galeano, M.; Bitto, A.; Minutoli, L.; Squadrito, G.; Seminara, P.; Venuti, F.S.; Torre, V.; Calo, M.; Colonna, M.; et al. Lipid peroxidation inhibition by raxofelast improves angiogenesis and wound healing in experimental burn wounds. Shock 2005, 24, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Galeano, M.; Altavilla, D.; Cucinotta, D.; Russo, G.T.; Calo, M.; Bitto, A.; Marini, H.; Marini, R.; Adamo, E.B.; Seminara, P.; et al. Recombinant human erythropoietin stimulates angiogenesis and wound healing in the genetically diabetic mouse. Diabetes 2004, 53, 2509–2517. [Google Scholar] [CrossRef] [PubMed]
- Petrie, R.J.; Doyle, A.D.; Yamada, K.M. Random versus directionally persistent cell migration. Nat. Rev. Mol. Cell Biol. 2009, 10, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Fogh, B.S.; Multhaupt, H.A.; Couchman, J.R. Protein kinase C, focal adhesions and the regulation of cell migration. J. Histochem. Cytochem. 2014, 62, 172–184. [Google Scholar] [CrossRef]
- Arias-Salgado, E.G.; Lizano, S.; Sarkar, S.; Brugge, J.S.; Ginsberg, M.H.; Shattil, S.J. Src kinase activation by direct interaction with the integrin β cytoplasmic domain. Proc. Natl. Acad. Sci. USA 2003, 100, 13298–13302. [Google Scholar] [CrossRef]
- Deng, R.; Li, F.; Wu, H.; Wang, W.Y.; Dai, L.; Zhang, Z.R.; Fu, J. Anti-inflammatory mechanism of geniposide: Inhibiting the hyperpermeability of fibroblast-like synoviocytes via the RhoA/p38 MAPK/NF-κB/F-actin signal pathway. Front. Pharmacol. 2018, 9, 105. [Google Scholar] [CrossRef]
- Ibrahim, N.; Wong, S.K.; Mohamed, I.N.; Mohamed, N.; Chin, K.Y.; Ima-Nirwana, S.; Shuid, A.N. Wound healing properties of selected natural products. Int. J. Environ. Res. Public Health 2018, 15, 2360. [Google Scholar] [CrossRef]
- Liu, C.H.; Chang, F.Y. Development and characterization of eucalyptol microemulsions for topic delivery of curcumin. Chem. Pharm. Bull. 2011, 59, 172–178. [Google Scholar] [CrossRef]
- Gera, M.; Sharma, N.; Ghosh, M.; Huynh, D.L.; Lee, S.J.; Min, T.; Kwon, T.; Jeong, D.K. Nanoformulations of curcumin: An emerging paradigm for improved remedial application. Oncotarget 2017, 8, 66680–66698. [Google Scholar] [CrossRef]
- Frohlich, E.; Roblegg, E. Models for oral uptake of nanoparticles in consumer products. Toxicology 2012, 291, 10–17. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential—What they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, K.; Pan, J.; Liu, B.; Feng, S.-S.J.B. Folic acid conjugated nanoparticles of mixed lipid monolayer shell and biodegradable polymer core for targeted delivery of Docetaxel. Biomaterials 2010, 31, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Ziello, J.E.; Huang, Y.; Jovin, I.S.J.M.M. Cellular endocytosis and gene delivery. Mol. Med. 2010, 16, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Rejman, J.; Oberle, V.; Zuhorn, I.S.; Hoekstra, D.J.B.j. Size-dependent internalization of particles via the pathways of clathrin-and caveolae-mediated endocytosis. Biochem. J. 2004, 377, 159–169. [Google Scholar] [CrossRef]
- Villanueva-Flores, F.; Castro-Lugo, A.; Ramírez, O.T.; Palomares, L.A.J.N. Understanding cellular interactions with nanomaterials: Towards a rational design of medical nanodevices. Nanotechnology 2020, 31, 132002. [Google Scholar] [CrossRef]
- Chen, K.; Cai, H.; Zhang, H.; Zhu, H.; Gu, Z.; Gong, Q.; Luo, K. Stimuli-responsive polymer-doxorubicin conjugate: Antitumor mechanism and potential as nano-prodrug. Acta Biomater. 2019, 84, 339–355. [Google Scholar] [CrossRef]
- Duan, Z.; Cai, H.; Zhang, H.; Chen, K.; Li, N.; Xu, Z.; Gong, Q.; Luo, K. PEGylated multistimuli-responsive dendritic prodrug-based nanoscale system for enhanced anticancer activity. ACS Appl. Mater. Interfaces 2018, 10, 35770–35783. [Google Scholar] [CrossRef]
- Yameen, B.; Choi, W.I.; Vilos, C.; Swami, A.; Shi, J.; Farokhzad, O.C. Insight into nanoparticle cellular uptake and intracellular targeting. J. Control. Release 2014, 190, 485–499. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Ruan, S.B.; Zhang, Q.Y.; He, Q.; Gao, H.L. Lapatinib-incorporated lipoprotein-like nanoparticles: Preparation and a proposed breast cancer-targeting mechanism. Acta Pharmacol. Sin. 2014, 35, 846–852. [Google Scholar] [CrossRef]
- Lee, S.S.; Lee, S.J.; Lee, S.H.; Ryu, J.M.; Lim, H.S.; Kim, J.S.; Song, E.J.; Jung, Y.H.; Lee, H.J.; Kim, C.H.; et al. Netrin-1-induced stem cell bioactivity contributes to the regeneration of injured tissues via the lipid raft-dependent integrin α6β4 signaling pathway. Sci. Rep. 2016, 6, 37526. [Google Scholar] [CrossRef]
- Lee, S.J.; Jung, Y.H.; Oh, S.Y.; Yong, M.S.; Ryu, J.M.; Han, H.J. Netrin-1 induces MMP-12-dependent E-cadherin degradation via the distinct activation of PKCα and FAK/Fyn in promoting mesenchymal stem cell motility. Stem Cells Dev. 2014, 23, 1870–1882. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K. FAK-Dependent Cell motility and cell elongation. Cells 2020, 9, 192. [Google Scholar] [CrossRef] [PubMed]
- Burridge, K. Focal adhesions: A personal perspective on a half century of progress. FEBS J. 2017, 284, 3355–3361. [Google Scholar] [CrossRef] [PubMed]
- Kaul, S.; Blackford, J.A., Jr.; Cho, S.; Simons, S.S., Jr. Ubc9 is a novel modulator of the induction properties of glucocorticoid receptors. J. Biol. Chem. 2002, 277, 12541–12549. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Na, S.I.; Heo, J.S.; Kim, M.H.; Kim, Y.H.; Lee, M.Y.; Kim, S.H.; Lee, Y.J.; Han, H.J. Arachidonic acid release by H2O2 mediated proliferation of mouse embryonic stem cells: Involvement of Ca2+/PKC and MAPKs-induced EGFR transactivation. J. Cell. Biochem. 2009, 106, 787–797. [Google Scholar] [CrossRef]
- Etienne-Manneville, S.; Hall, A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCζ. Cell 2001, 106, 489–498. [Google Scholar] [CrossRef]
- Laudanna, C.; Sorio, C.; Tecchio, C.; Butcher, E.C.; Bonora, A.; Bassi, C.; Scarpa, A. Motility analysis of pancreatic adenocarcinoma cells reveals a role for the atypical ζ isoform of protein kinase C in cancer cell movement. Lab. Investig. 2003, 83, 1155–1163. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Forde, S.; Tye, B.J.; Newey, S.E.; Roubelakis, M.; Smythe, J.; McGuckin, C.P.; Pettengell, R.; Watt, S.M. Endolyn (CD164) modulates the CXCL12-mediated migration of umbilical cord blood CD133+ cells. Blood 2007, 109, 1825–1833. [Google Scholar] [CrossRef]
- Yun, S.P.; Lee, S.J.; Oh, S.Y.; Jung, Y.H.; Ryu, J.M.; Suh, H.N.; Kim, M.O.; Oh, K.B.; Han, H.J. Reactive oxygen species induce MMP12-dependent degradation of collagen 5 and fibronectin to promote the motility of human umbilical cord-derived mesenchymal stem cells. Br. J. Pharmacol. 2014, 171, 3283–3297. [Google Scholar] [CrossRef]
- Yamini, B. NF-κB, mesenchymal differentiation and glioblastoma. Cells 2018, 7, 125. [Google Scholar] [CrossRef]
- Lidke, D.S.; Huang, F.; Post, J.N.; Rieger, B.; Wilsbacher, J.; Thomas, J.L.; Pouyssegur, J.; Jovin, T.M.; Lenormand, P. ERK nuclear translocation is dimerization-independent but controlled by the rate of phosphorylation. J. Biol. Chem. 2010, 285, 3092–3102. [Google Scholar] [CrossRef] [PubMed]
- Menke, A.; Giehl, K. Regulation of adherens junctions by Rho GTPases and p120-catenin. Arch. Biochem. Biophys. 2012, 524, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Konakahara, S.; Ohashi, K.; Mizuno, K.; Itoh, K.; Tsuji, T. CD29 integrin- and LIMK1/cofilin-mediated actin reorganization regulates the migration of haematopoietic progenitor cells underneath bone marrow stromal cells. Genes Cells 2004, 9, 345–358. [Google Scholar] [CrossRef]
- Araki, N.; Hatae, T.; Yamada, T.; Hirohashi, S. Actinin-4 is preferentially involved in circular ruffling and macropinocytosis in mouse macrophages: Analysis by fluorescence ratio imaging. J. Cell. Sci. 2000, 113, 3329–3340. [Google Scholar] [PubMed]
- Lee, S.J.; Jung, Y.H.; Oh, S.Y.; Yun, S.P.; Han, H.J. Melatonin enhances the human mesenchymal stem cells motility via melatonin receptor 2 coupling with Gαq in skin wound healing. J. Pineal. Res. 2014, 57, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.W.; Heo, S.C.; Jeong, G.O.; Yoon, J.W.; Mo, W.M.; Lee, M.J.; Jang, I.H.; Kwon, S.M.; Lee, J.S.; Kim, J.H. Tumor necrosis factor-α-activated mesenchymal stem cells promote endothelial progenitor cell homing and angiogenesis. Biochim. Biophys. Acta 2013, 1832, 2136–2144. [Google Scholar] [CrossRef] [PubMed]

| Score | Re-Epithelialization |
|---|---|
| 0 | Absence of epithelial proliferation in > 70% of the tissue |
| 1 | Poor epidermal organization in > 60% of the tissue |
| 2 | Incomplete epidermal organization in > 40% of the tissue |
| 3 | Moderate epithelial proliferation in > 60% of the tissue |
| 4 | Complete epidermal remodeling in > 80% of the tissue |
| Encapsulation Efficiency (%) | Loading Capacity (%) | |
|---|---|---|
| CN | 87.1 ± 0.1 | 9.0 ± 0.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.-W.; Choi, C.-H.; Park, J.P.; Lee, S.-J. Nanospheres Loaded with Curcumin Improve the Bioactivity of Umbilical Cord Blood-Mesenchymal Stem Cells via c-Src Activation during the Skin Wound Healing Process. Cells 2020, 9, 1467. https://doi.org/10.3390/cells9061467
Kim D-W, Choi C-H, Park JP, Lee S-J. Nanospheres Loaded with Curcumin Improve the Bioactivity of Umbilical Cord Blood-Mesenchymal Stem Cells via c-Src Activation during the Skin Wound Healing Process. Cells. 2020; 9(6):1467. https://doi.org/10.3390/cells9061467
Chicago/Turabian StyleKim, Do-Wan, Chang-Hyung Choi, Jong Pil Park, and Sei-Jung Lee. 2020. "Nanospheres Loaded with Curcumin Improve the Bioactivity of Umbilical Cord Blood-Mesenchymal Stem Cells via c-Src Activation during the Skin Wound Healing Process" Cells 9, no. 6: 1467. https://doi.org/10.3390/cells9061467
APA StyleKim, D.-W., Choi, C.-H., Park, J. P., & Lee, S.-J. (2020). Nanospheres Loaded with Curcumin Improve the Bioactivity of Umbilical Cord Blood-Mesenchymal Stem Cells via c-Src Activation during the Skin Wound Healing Process. Cells, 9(6), 1467. https://doi.org/10.3390/cells9061467