Emerin Phosphorylation during the Early Phase of the Oxidative Stress Response Influences Emerin–BAF Interaction and BAF Nuclear Localization
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures, Transfection and Treatments
2.2. Western Blotting and Immunoprecipitation
2.3. Immunofluorescence and Proximity Ligation Assay
2.4. Antibodies
3. Results
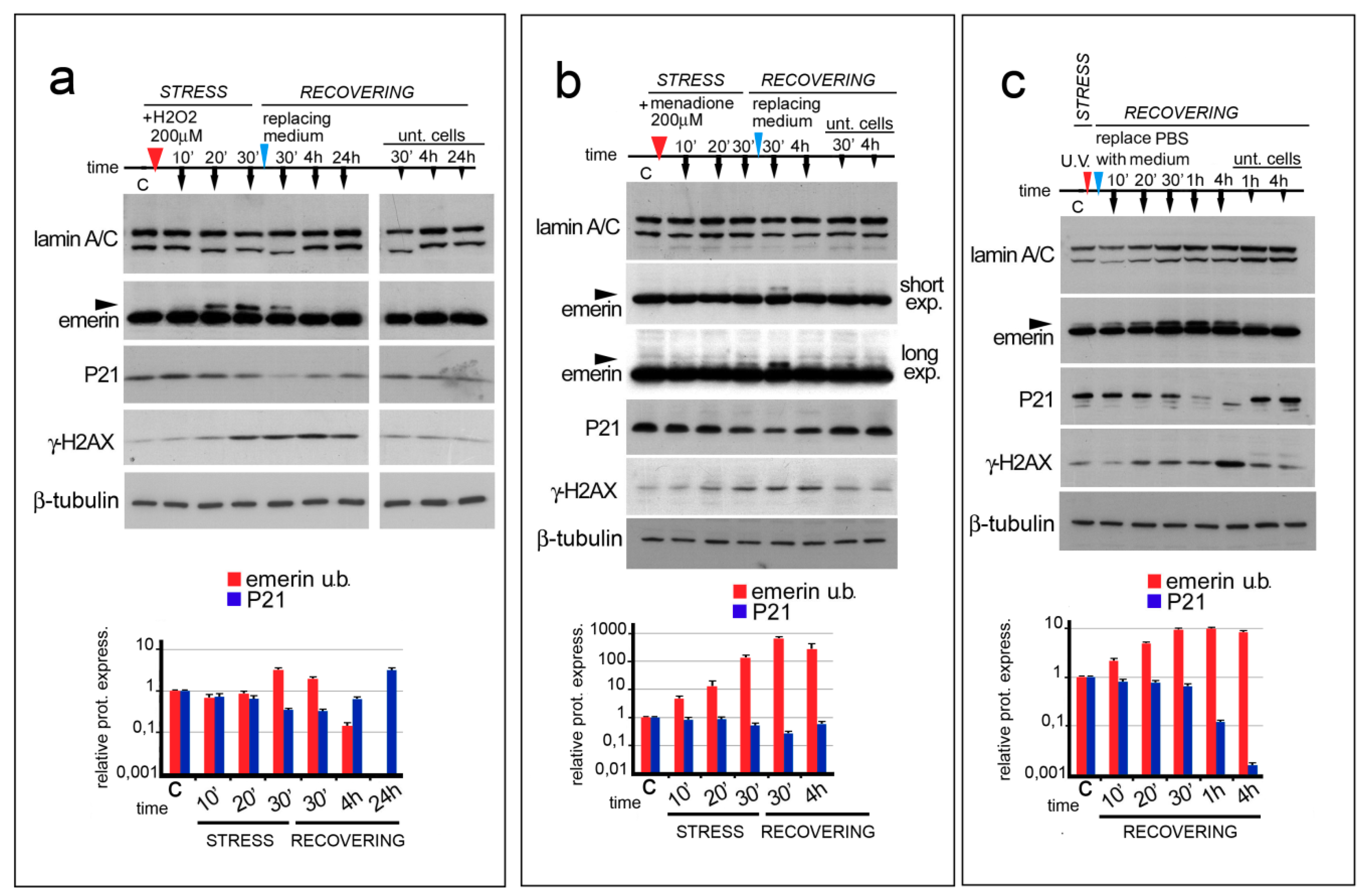
3.1. ROS Generating Agents Affect Emerin Molecular Weight
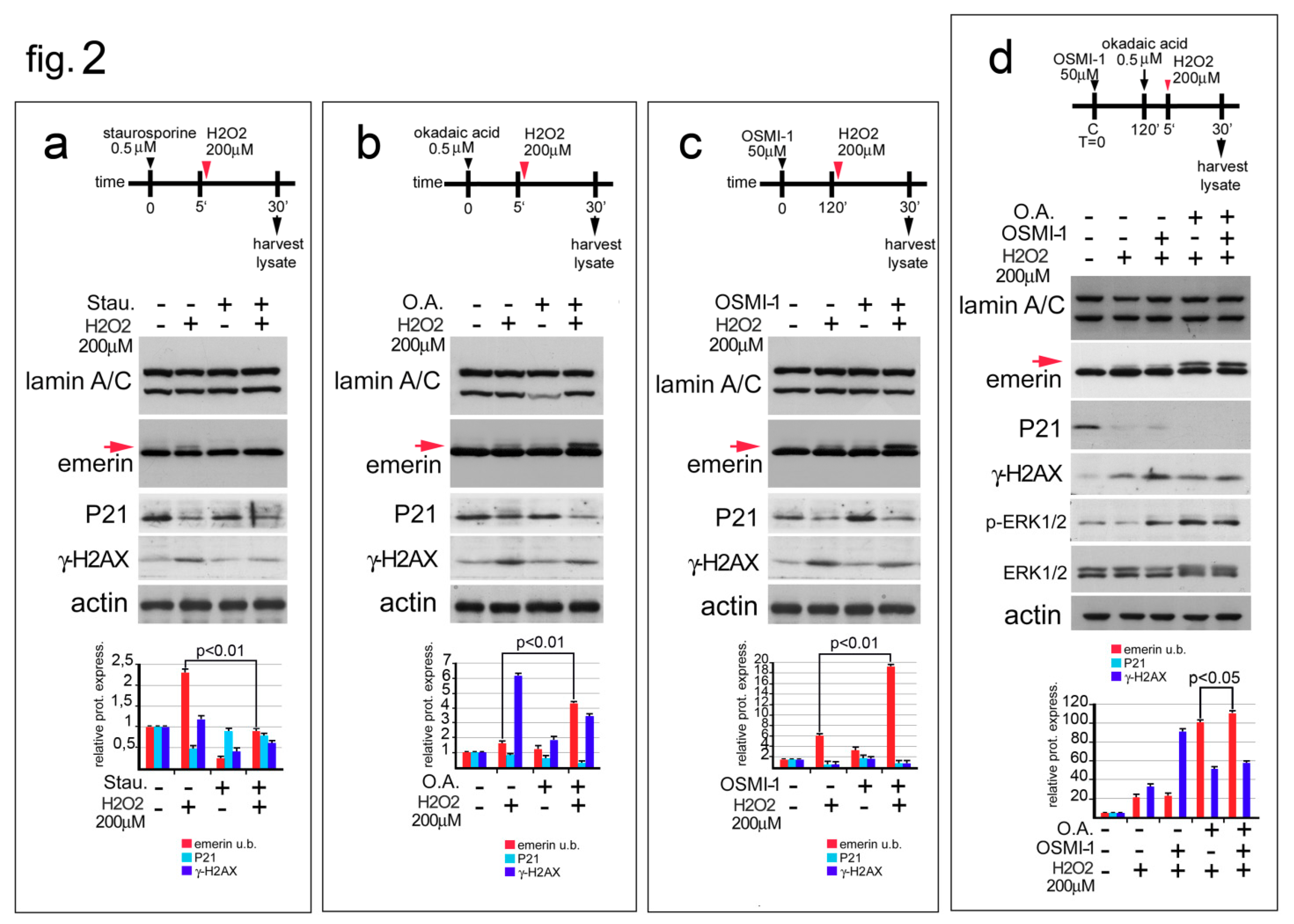
3.2. Emerin Phosphorylation Increase Molecular Weight during the Early Phase of the Oxidative Stress Response
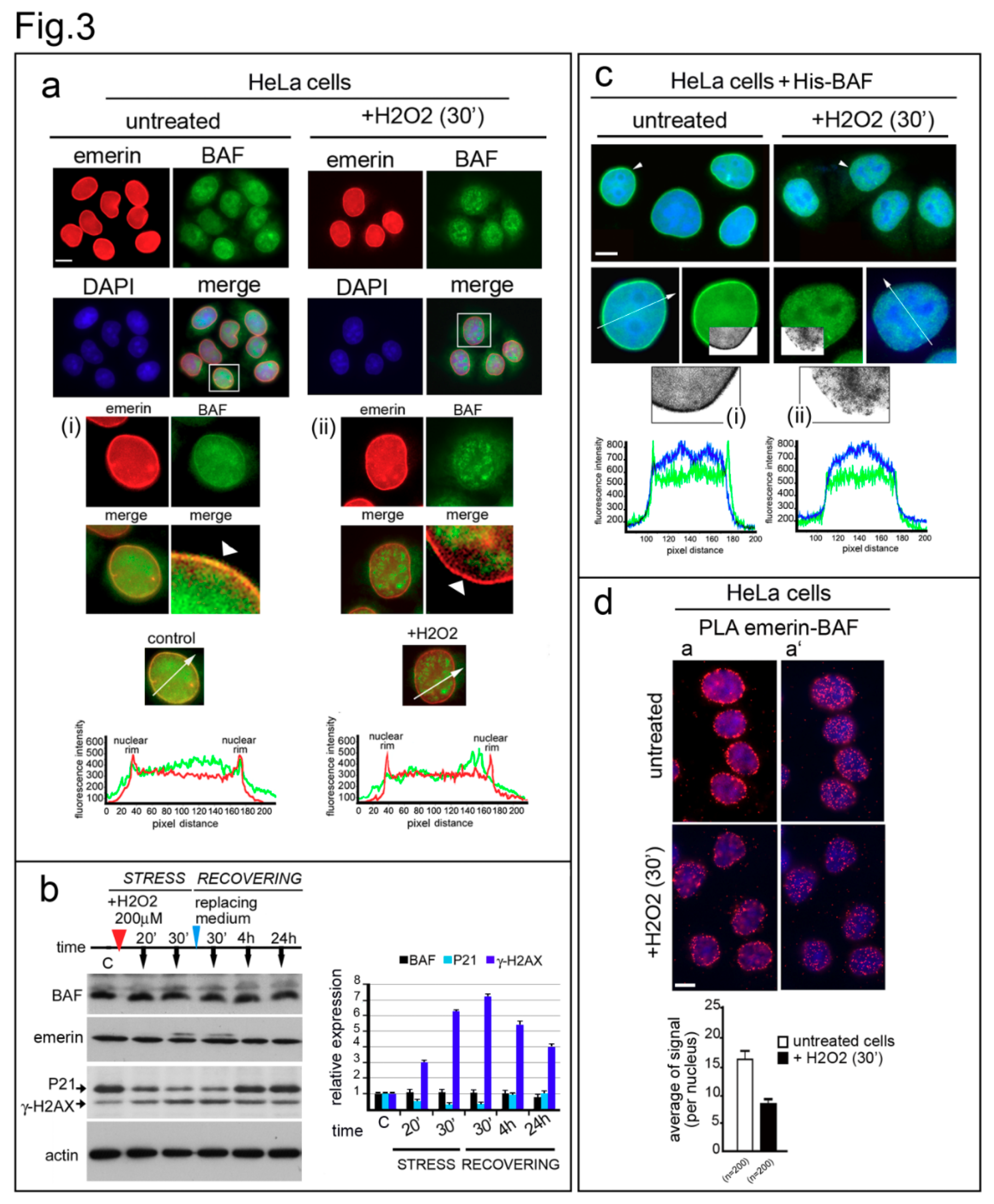
3.3. Emerin Interaction with BAF Is Affected during the Oxidative Stress Response
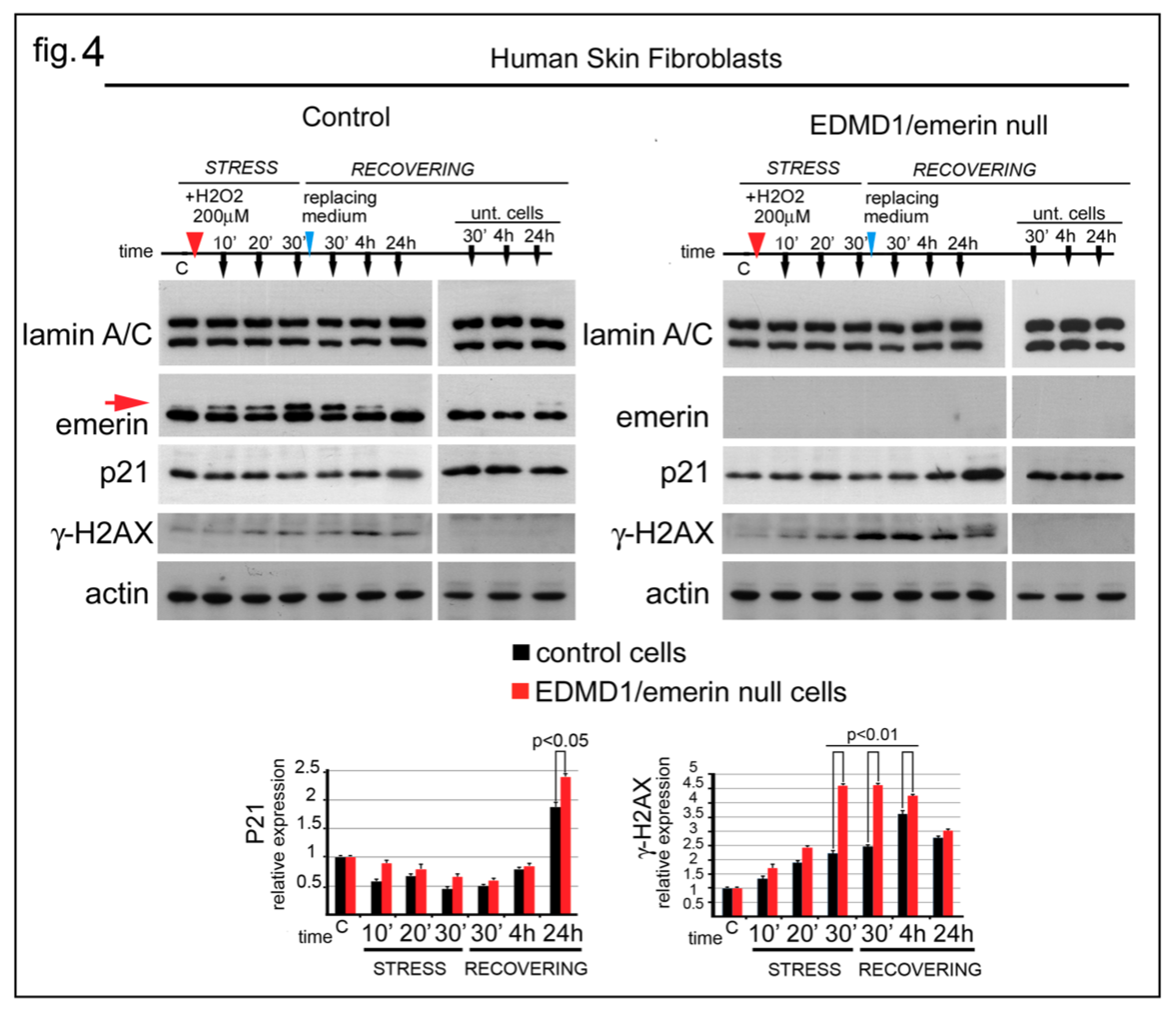
3.4. EDMD1 Cells Are More Sensitive to ROS-Induced DNA Damage
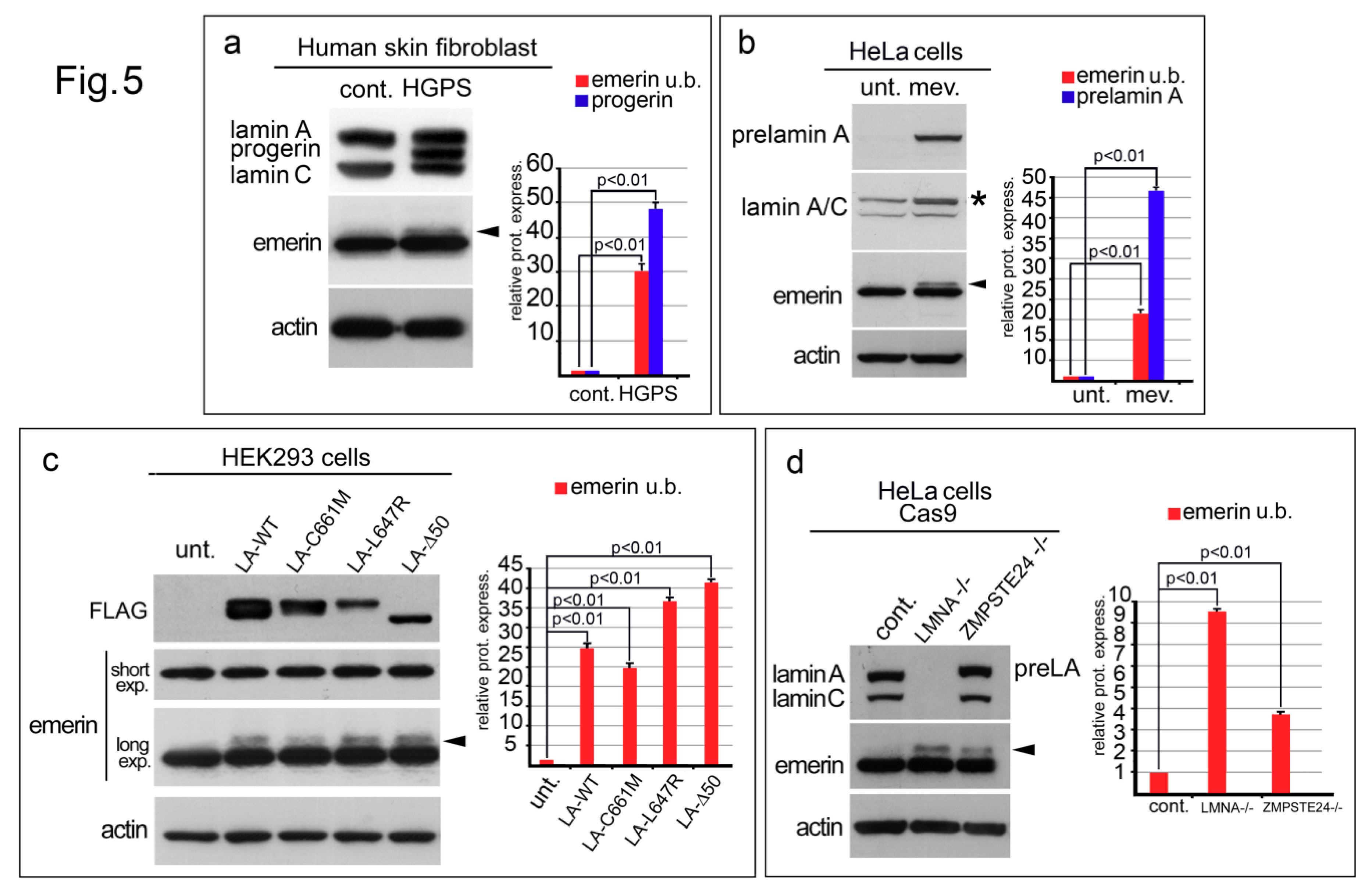
3.5. Emerin Is Also Affected in Various LaminA Deficiency Conditions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D.; et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757–772. [Google Scholar] [CrossRef]
- Cui, H.; Kong, Y.; Zhang, H. Oxidative Stress, Mitochondrial Dysfunction, and Aging. J. Signal Transduct. 2011, 2012. [Google Scholar] [CrossRef] [PubMed]
- Caron, M.; Auclair, M.; Donadille, B.; Béréziat, V.; Guerci, B.; Laville, M.; Narbonne, H.; Bodemer, C.; Lascols, O.; Capeau, J.; et al. Human lipodystrophies linked to mutations in A-type lamins and to HIV protease inhibitor therapy are both associated with prelamin A accumulation, oxidative stress and premature cellular senescence. Cell Death Differ. 2007, 14, 1759–1767. [Google Scholar] [CrossRef] [PubMed]
- Viteri, G.; Chung, Y.W.; Stadtman, E.R. Effect of progerin on the accumulation of oxidized proteins in fibroblasts from Hutchinson Gilford progeria patients. Mech. Ageing Dev. 2009, 131, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Peinado, J.R.; Quiros, P.M.; Pulido, M.R.; Mariño, G.; Martínez-Chantar, M.L.; Vázquez-Martínez, R.; Freije, J.M.P.; Lopez-Otin, C.; Malagón, M.M. Proteomic profiling of adipose tissue from Zmpste24−/− mice, a model of lipodystrophy and premature aging, reveals major changes in mitochondrial function and vimentin processing. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef]
- Mateos, J.; De La Fuente, A.; Lesende-Rodríguez, I.; Pernas, P.F.; Arufe, M.C.; Blanco, F.J.; Blanco, F.J. Lamin A deregulation in human mesenchymal stem cells promotes an impairment in their chondrogenic potential and imbalance in their response to oxidative stress. Stem Cell Res. 2013, 11, 1137–1148. [Google Scholar] [CrossRef]
- Maraldi, N.M.; Squarzoni, S.; Sabatelli, P.; Capanni, C.; Mattioli, E.; Ognibene, A.; Lattanzi, G. Laminopathies: Involvement of structural nuclear proteins in the pathogenesis of an increasing number of human diseases. J. Cell. Physiol. 2005, 203, 319–327. [Google Scholar] [CrossRef]
- Burke, B.; Stewart, C.L. The nuclear lamins: Flexibility in function. Nat. Rev. Mol. Cell Biol. 2012, 14, 13–24. [Google Scholar] [CrossRef]
- Maraldi, N.M.; Mattioli, E.; Lattanzi, G.; Columbaro, M.; Capanni, C.; Camozzi, D.; Squarzoni, S.; Manzoli, F.A. Prelamin A processing and heterochromatin dynamics in laminopathies. Adv. Enzym. Regul. 2007, 47, 154–167. [Google Scholar] [CrossRef]
- De Sandre-Giovannoli, A. Lamin a Truncation in Hutchinson-Gilford Progeria. Science 2003, 300, 2055. [Google Scholar] [CrossRef]
- Eriksson, M.; Brown, W.T.; Gordon, L.B.; Glynn, M.W.; Singer, J.; Scott, L.; Erdos, M.R.; Robbins, C.M.; Moses, T.Y.; Berglund, P.; et al. Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 2003, 423, 293–298. [Google Scholar] [CrossRef]
- Navarro, C.; Cadiñanos, J.; De Sandre-Giovannoli, A.; Bernard, R.; Courrier, S.; Boccaccio, I.; Boyer, A.; Kleijer, W.J.; Wagner, A.; Giuliano, F.; et al. Loss of ZMPSTE24 (FACE-1) causes autosomal recessive restrictive dermopathy and accumulation of Lamin A precursors. Hum. Mol. Genet. 2005, 14, 1503–1513. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, A.K.; Fryns, J.-P.; Auchus, R.J.; Garg, A. Zinc metalloproteinase, ZMPSTE24, is mutated in mandibuloacral dysplasia. Hum. Mol. Genet. 2003, 12, 1995–2001. [Google Scholar] [CrossRef] [PubMed]
- Filesi, I.; Gullotta, F.; Lattanzi, G.; D’Apice, M.R.; Capanni, C.; Nardone, A.M.; Columbaro, M.; Scarano, G.; Mattioli, E.; Sabatelli, P.; et al. Alterations of nuclear envelope and chromatin organization in mandibuloacral dysplasia, a rare form of laminopathy. Physiol. Genom. 2005, 23, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Sieprath, T.; Corne, T.; Nooteboom, M.; Grootaert, C.; Rajkovic, A.; Buysschaert, B.; Robijns, J.; Broers, J.L.; Ramaekers, F.C.; Koopman, W.J.; et al. Sustained accumulation of prelamin A and depletion of lamin A/C both cause oxidative stress and mitochondrial dysfunction but induce different cell fates. Nucleus 2015, 6, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Muchir, A.; Van Engelen, B.G.; Lammens, M.; Mislow, J.M.; McNally, E.; Schwartz, K.; Bonne, G. Nuclear envelope alterations in fibroblasts from LGMD1B patients carrying nonsense Y259X heterozygous or homozygous mutation in lamin A/C gene. Exp. Cell Res. 2003, 291, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Bione, S.; Maestrini, E.; Rivella, S.; Mancini, M.; Regis, S.; Romeo, G.; Toniolo, D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 1994, 8, 323–327. [Google Scholar] [CrossRef]
- Samson, C.; Petitalot, A.; Celli, F.; Herrada, I.; Ropars, V.; Le Du, M.-H.; Nhiri, N.; Jacquet, E.; Arteni, A.-A.; Buendia, B.; et al. Structural analysis of the ternary complex between lamin A/C, BAF and emerin identifies an interface disrupted in autosomal recessive progeroid diseases. Nucleic Acids Res. 2018, 46, 10460–10473. [Google Scholar] [CrossRef]
- Wu, D.; Flannery, A.R.; Cai, H.; Ko, E.; Cao, K. Nuclear localization signal deletion mutants of lamin A and progerin reveal insights into lamin A processing and emerin targeting. Nucleus 2014, 5, 66–74. [Google Scholar] [CrossRef][Green Version]
- Capanni, C.; Del Coco, R.; Mattioli, E.; Camozzi, D.; Columbaro, M.; Schena, E.; Merlini, L.; Squarzoni, S.; Maraldi, N.M.; Lattanzi, G. Emerin-prelamin A interplay in human fibroblasts. Biol. Cell 2009, 101, 541–554. [Google Scholar] [CrossRef]
- Jamin, A.; Wiebe, M. Barrier to Autointegration Factor (BANF1): Interwoven roles in nuclear structure, genome integrity, innate immunity, stress responses and progeria. Curr. Opin. Cell Biol. 2015, 34, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, L.; Wilson, K.L. Barrier-to-Autointegration Factor Phosphorylation on Ser-4 Regulates Emerin Binding to Lamin a In Vitro and Emerin Localization In Vivo. Mol. Biol. Cell 2006, 17, 1154–1163. [Google Scholar] [CrossRef] [PubMed]
- Berk, J.M.; Simon, D.N.; Jenkins-Houk, C.R.; Westerbeck, J.W.; Grønning-Wang, L.M.; Carlson, C.R.; Wilson, K.L. The molecular basis of emerin-emerin and emerin-BAF interactions. J. Cell Sci. 2014, 127, 3956–3969. [Google Scholar] [CrossRef] [PubMed]
- Berk, J.M.; Maitra, S.; Dawdy, A.W.; Shabanowitz, J.; Hunt, D.F.; Wilson, K.L. O-linked β-n-acetylglucosamine (o-glcnac) regulates emerin binding to barrier to autointegration factor (baf) in a chromatin- and lamin b-enriched “niche”. J. Biol. Chem. 2013, 288, 30192–30209. [Google Scholar] [CrossRef]
- Capanni, C.; Cenni, V.; Haraguchi, T.; Squarzoni, S.; Schüchner, S.; Ogris, E.; Novelli, G.; Maraldi, N.; Lattanzi, G. Lamin A precursor induces barrier-to-autointegration factor nuclear localization. Cell Cycle 2010, 9, 2600–2610. [Google Scholar] [CrossRef]
- Loi, M.; Cenni, V.; Duchi, S.; Squarzoni, S.; Lopez-Otin, C.; Foisner, R.; Lattanzi, G.; Capanni, C. Barrier-to-Autointegration Factor (BAF) involvement in prelamin A-related chromatin organization changes. Oncotarget 2015, 7, 15662–15677. [Google Scholar] [CrossRef]
- Park, C.H.; Ryu, H.G.; Kim, S.-H.; Lee, D.; Song, H.; Kim, K.-T. Presumed pseudokinase VRK3 functions as a BAF kinase. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1853, 1738–1748. [Google Scholar] [CrossRef]
- Mattioli, E.; Andrenacci, D.; Mai, A.; Valente, S.; Robijns, J.; De Vos, W.H.; Capanni, C.; Lattanzi, G. Statins and Histone Deacetylase Inhibitors Affect Lamin A/C - Histone Deacetylase 2 Interaction in Human Cells. Front. Cell Dev. Biol. 2019, 7, 6. [Google Scholar] [CrossRef]
- Houthaeve, G.; Xiong, R.; Robijns, J.; Luyckx, B.; Beulque, Y.; Brans, T.; Campsteijn, C.; Samal, S.K.; Stremersch, S.; De Smedt, S.C.; et al. Targeted Perturbation of Nuclear Envelope Integrity with Vapor Nanobubble-Mediated Photoporation. ACS Nano 2018, 12, 7791–7802. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.; Wright, J.; Agarwala, V.; A Scott, D.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Ye, B.; Hou, N.; Xiao, L.; Xu, Y.; Xu, H.; Li, F. Dynamic monitoring of oxidative DNA double-strand break and repair in cardiomyocytes. Cardiovasc. Pathol. 2015, 25, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Zlatanou, A.; Despras, E.; Braz-Petta, T.; Boubakour-Azzouz, I.; Pouvelle, C.; Stewart, G.S.; Nakajima, S.; Yasui, A.; Ishchenko, A.A.; Kannouche, P.L. The hMsh2-hMsh6 Complex Acts in Concert with Monoubiquitinated PCNA and Pol η in Response to Oxidative DNA Damage in Human Cells. Mol. Cell 2011, 43, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Meoz, R.; Jiang, J.; Lazarus, M.B.; Orman, M.; Janetzko, J.; Fan, C.; Duveau, D.Y.; Tan, Z.-W.; Thomas, C.J.; Walker, S. A Small Molecule That Inhibits OGT Activity in Cells. ACS Chem. Biol. 2015, 10, 1392–1397. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Douki, T.; Ravanat, J.-L. Oxidatively Generated Damage to Cellular DNA by UVB and UVA Radiation, Photochem. Photobiol. 2014, 91, 140–155. [Google Scholar] [CrossRef] [PubMed]
- Slupphaug, G.; Kavli, B.; E Krokan, H. The interacting pathways for prevention and repair of oxidative DNA damage. Mutat. Res. Mol. Mech. Mutagen. 2003, 531, 231–251. [Google Scholar] [CrossRef]
- Moldovan, G.-L.; Pfander, B.; Jentsch, S. PCNA, the Maestro of the Replication Fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef]
- Tifft, K.E.; Bradbury, K.A.; Wilson, K.L. Tyrosine phosphorylation of nuclear-membrane protein emerin by Src, Abl and other kinases. J. Cell Sci. 2009, 122, 3780–3790. [Google Scholar] [CrossRef]
- Wheeler, M.A.; Warley, A.; Roberts, R.; Ehler, E.; Ellis, J.A. Identification of an emerin–β-catenin complex in the heart important for intercalated disc architecture and β-catenin localisation. Cell. Mol. Life Sci. 2009, 67, 781–796. [Google Scholar] [CrossRef]
- Cartegni, L.; Di Barletta, M.R.; Barresi, R.; Squarzoni, S.; Sabatelli, P.; Maraldi, N.; Mora, M.; Di Blasi, C.; Cornelio, F.; Merlini, L.; et al. Heart-specific localization of emerin: New insights into Emery-Dreifuss muscular dystrophy. Hum. Mol. Genet. 1997, 6, 2257–2264. [Google Scholar] [CrossRef]
- De Queiroz, R.M.; Madan, R.; Chien, J.; Dias, W.; Slawson, C. Changes in O-Linked N-Acetylglucosamine (O-GlcNAc) Homeostasis Activate the p53 Pathway in Ovarian Cancer Cells. J. Biol. Chem. 2016, 291, 18897–18914. [Google Scholar] [CrossRef]
- Kim, M.Y.; Kim, Y.S.; Kim, M.; Choi, M.Y.; Roh, G.S.; Lee, D.H.; Kim, H.J.; Kang, S.S.; Cho, G.J.; Shin, J.K.; et al. Metformin inhibits cervical cancer cell proliferation via decreased ampk o-glcnacylation. Anim. Cells Syst. 2019, 23, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Cenni, V.; Capanni, C.; Mattioli, E.; Schena, E.; Squarzoni, S.; Bacalini, M.G.; Garagnani, P.; Salvioli, S.; Franceschi, C.; Lattanzi, G. Lamin A involvement in ageing processes. Ageing Res. Rev. 2020, 101073. [Google Scholar] [CrossRef] [PubMed]
- De Oca, R.M.; Andreassen, P.R.; Wilson, K.L. Barrier-to-Autointegration Factor influences specific histone modifications. Nucleus 2011, 2, 580–590. [Google Scholar] [CrossRef] [PubMed]
- De Oca, R.M.; Shoemaker, C.J.; Gucek, M.; Cole, R.N.; Wilson, K.L. Barrier-to-Autointegration Factor Proteome Reveals Chromatin-Regulatory Partners. PLoS ONE 2009, 4, e7050. [Google Scholar] [CrossRef]
- Bar, D.Z.; Davidovich, M.; Lamm, A.; Zer, H.; Wilson, K.L.; Gruenbaum, Y. BAF-1 mobility is regulated by environmental stresses. Mol. Biol. Cell 2014, 25, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Guilluy, C.; Osborne, L.D.; Van Landeghem, L.; Sharek, L.; Superfine, R.; Garcia-Mata, R.; Burridge, K. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nature 2014, 16, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Essawy, N.; Samson, C.; Petitalot, A.; Moog, S.; Bigot, A.; Herrada, I.; Marcelot, A.; Arteni, A.-A.; Coirault, C.; Zinn-Justin, S. An Emerin LEM-Domain Mutation Impairs Cell Response to Mechanical Stress. Cells 2019, 8, 570. [Google Scholar] [CrossRef]
- Savio, M.; Coppa, T.; Cazzalini, O.; Perucca, P.; Necchi, D.; Nardo, T.; Stivala, L.A.; Prosperi, E. Degradation of p21CDKN1A after DNA damage is independent of type of lesion, and is not required for DNA repair. DNA Repair 2009, 8, 778–785. [Google Scholar] [CrossRef]
- Gioia, U.; Francia, S.; Cabrini, M.; Brambillasca, S.; Michelini, F.; Jones-Weinert, C.W.; Daddadifagagna, F. Pharmacological boost of DNA damage response and repair by enhanced biogenesis of DNA damage response RNAs. Sci. Rep. 2019, 9, 6460. [Google Scholar] [CrossRef]
- Moser, B.; Basílio, J.; Gotzmann, J.; Brachner, A.; Foisner, R. Comparative Interactome Analysis of Emerin, MAN1 and LEM2 Reveals a Unique Role for LEM2 in Nucleotide Excision Repair. Cells 2020, 9, 463. [Google Scholar] [CrossRef]
- El-Mahdy, M.A.; Zhu, Q.; Wang, Q.-E.; Wani, G.; Prætorius-Ibba, M.; Wani, A.A. Cullin 4A-mediated Proteolysis of DDB2 Protein at DNA Damage Sites Regulatesin VivoLesion Recognition by XPC. J. Biol. Chem. 2006, 281, 13404–13411. [Google Scholar] [CrossRef]
- Sugasawa, K.; Okuda, Y.; Saijo, M.; Nishi, R.; Matsuda, N.; Chu, G.; Mori, T.; Iwai, S.; Tanaka, K.; Tanaka, K.; et al. UV-Induced Ubiquitylation of XPC Protein Mediated by UV-DDB-Ubiquitin Ligase Complex. Cell 2005, 121, 387–400. [Google Scholar] [CrossRef]
- Bolderson, E.; Burgess, J.T.; Li, J.; Gandhi, N.S.; Boucher, D.; Croft, L.V.; Beard, S.; Plowman, J.J.; Suraweera, A.; Adams, M.N.; et al. Barrier-to-autointegration factor 1 (Banf1) regulates poly [ADP-ribose] polymerase 1 (PARP1) activity following oxidative DNA damage. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Capanni, C.; Del Coco, R.; Squarzoni, S.; Columbaro, M.; Mattioli, E.; Camozzi, D.; Rocchi, A.; Scotlandi, K.; Maraldi, N.; Foisner, R.; et al. Prelamin A is involved in early steps of muscle differentiation. Exp. Cell Res. 2008, 314, 3628–3637. [Google Scholar] [CrossRef]
- Malińska, D.; Kudin, A.P.; Bejtka, M.; Kunz, W.S. Changes in mitochondrial reactive oxygen species synthesis during differentiation of skeletal muscle cells. Mitochondrion 2012, 12, 144–148. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cenni, V.; Squarzoni, S.; Loi, M.; Mattioli, E.; Lattanzi, G.; Capanni, C. Emerin Phosphorylation during the Early Phase of the Oxidative Stress Response Influences Emerin–BAF Interaction and BAF Nuclear Localization. Cells 2020, 9, 1415. https://doi.org/10.3390/cells9061415
Cenni V, Squarzoni S, Loi M, Mattioli E, Lattanzi G, Capanni C. Emerin Phosphorylation during the Early Phase of the Oxidative Stress Response Influences Emerin–BAF Interaction and BAF Nuclear Localization. Cells. 2020; 9(6):1415. https://doi.org/10.3390/cells9061415
Chicago/Turabian StyleCenni, Vittoria, Stefano Squarzoni, Manuela Loi, Elisabetta Mattioli, Giovanna Lattanzi, and Cristina Capanni. 2020. "Emerin Phosphorylation during the Early Phase of the Oxidative Stress Response Influences Emerin–BAF Interaction and BAF Nuclear Localization" Cells 9, no. 6: 1415. https://doi.org/10.3390/cells9061415
APA StyleCenni, V., Squarzoni, S., Loi, M., Mattioli, E., Lattanzi, G., & Capanni, C. (2020). Emerin Phosphorylation during the Early Phase of the Oxidative Stress Response Influences Emerin–BAF Interaction and BAF Nuclear Localization. Cells, 9(6), 1415. https://doi.org/10.3390/cells9061415





