Cell Type-Specific In Vitro Gene Expression Profiling of Stem Cell-Derived Neural Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction
2.2. Passaging and Maintenance of Cell Lines
2.3. Transfections and Lentivirus Production
2.4. FACS
2.5. Neuron Induction and Differentiation
2.6. Primary Mouse Astrocytes
2.7. hiPSC-MN and Primary Mouse Astrocyte Co-Cultures
2.8. Cortical-Enriched Organoid and Microglia Co-Cultures
2.9. Epitope-tagged RPL22 Immunoprecipitation
2.10. Western Blot
2.11. RNA Extraction
2.12. Reverse Transcription Digital PCR (RT-dPCR)
2.13. RNA Sequencing
2.14. RNA-Seq Analysis
3. Results
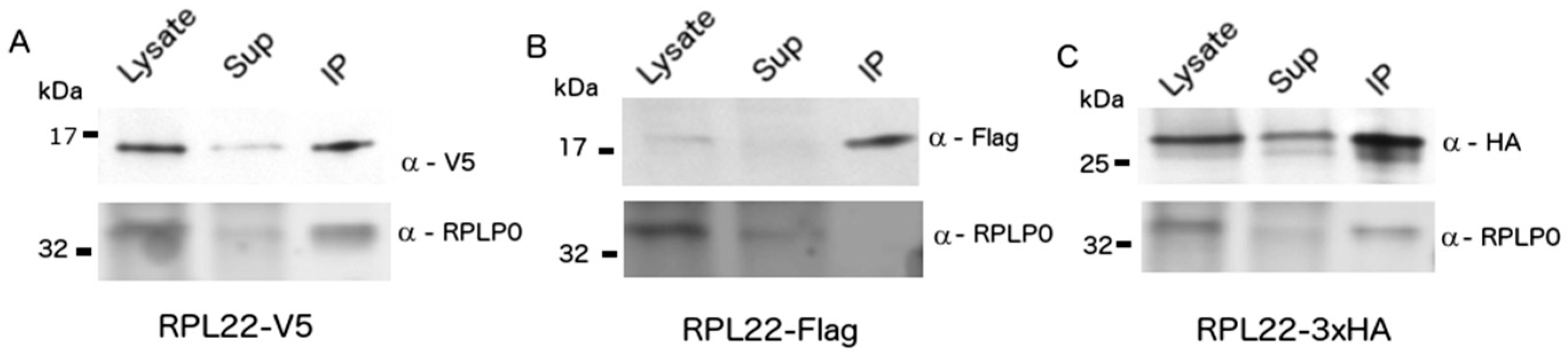
3.1. Construction of Epitope-Tagged RPL22
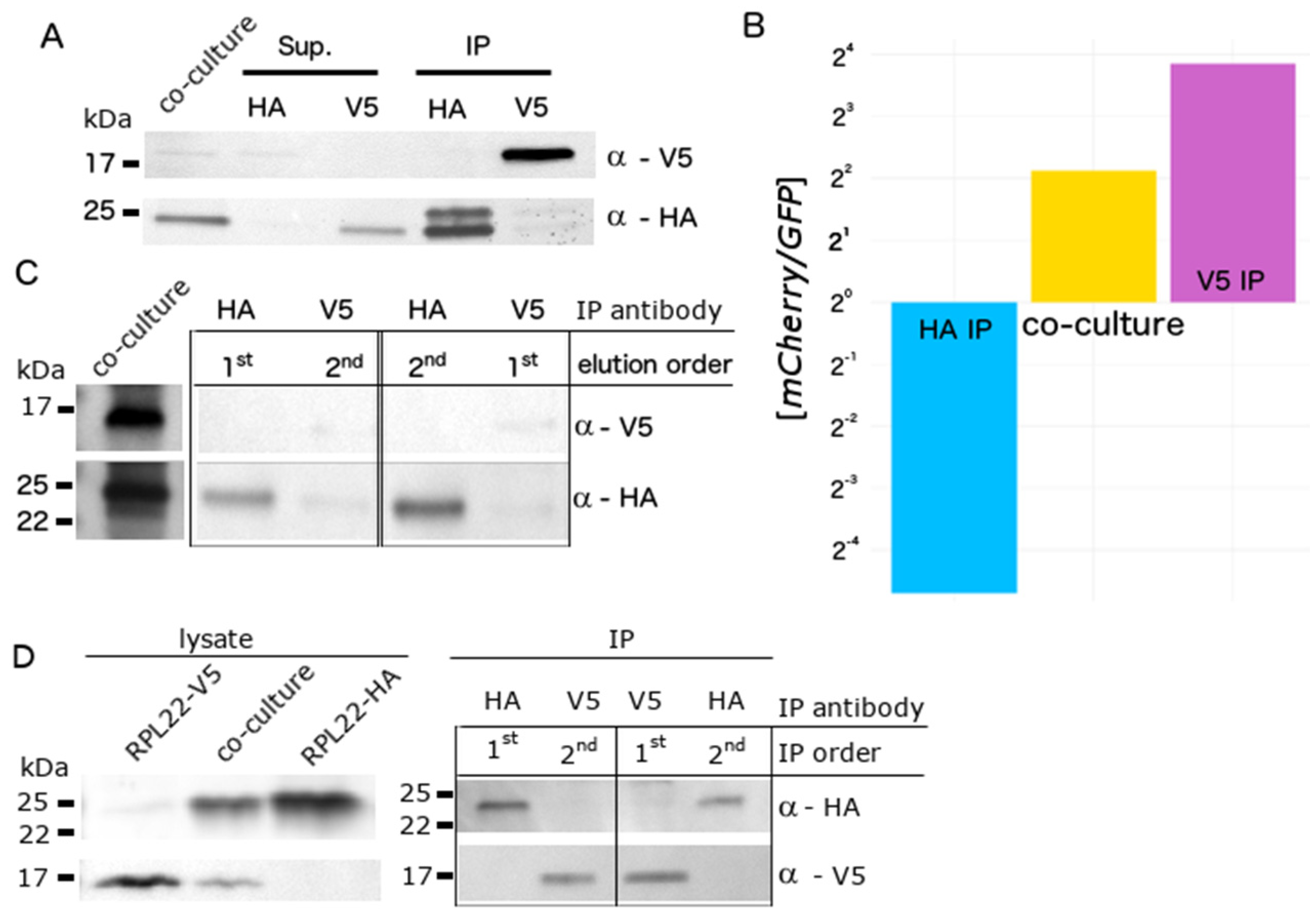
3.2. Biochemical Separation of Cell Type-Specific RNA in Co-Cultures Using RiboTags
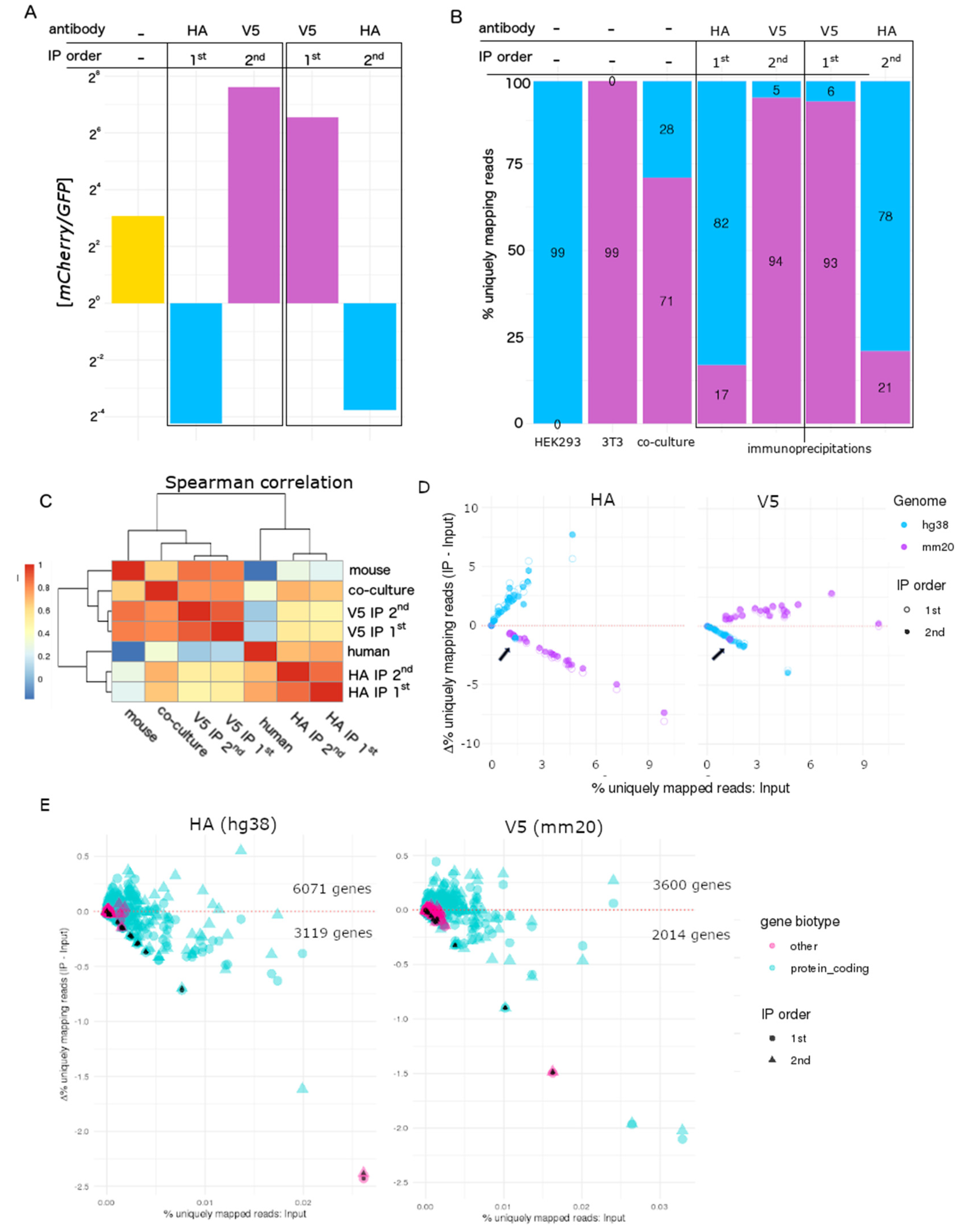
3.3. Transcriptome-Wide Cell Type-Specific Enrichment in Mixed Species Co-Cultures Using RiboTags
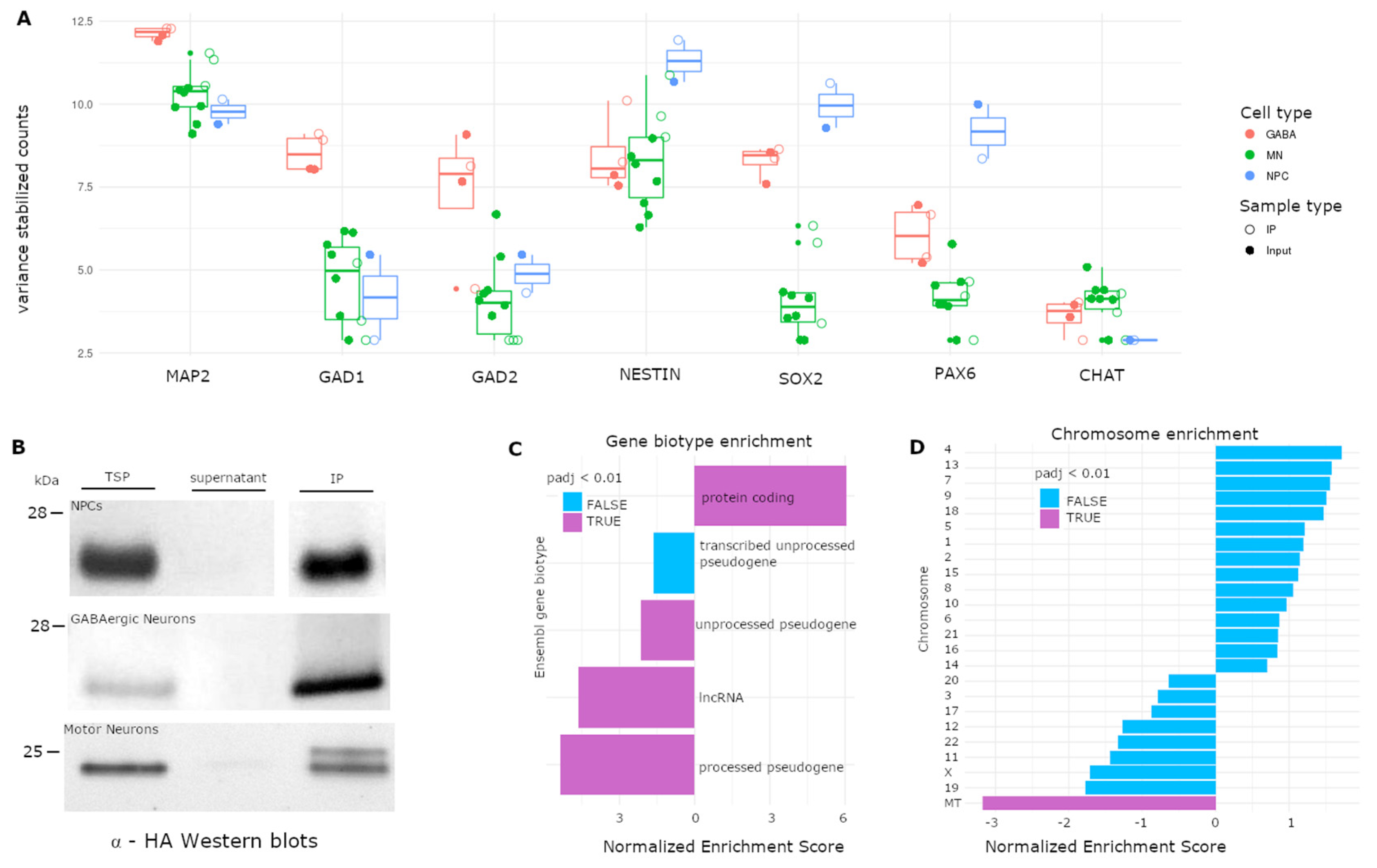
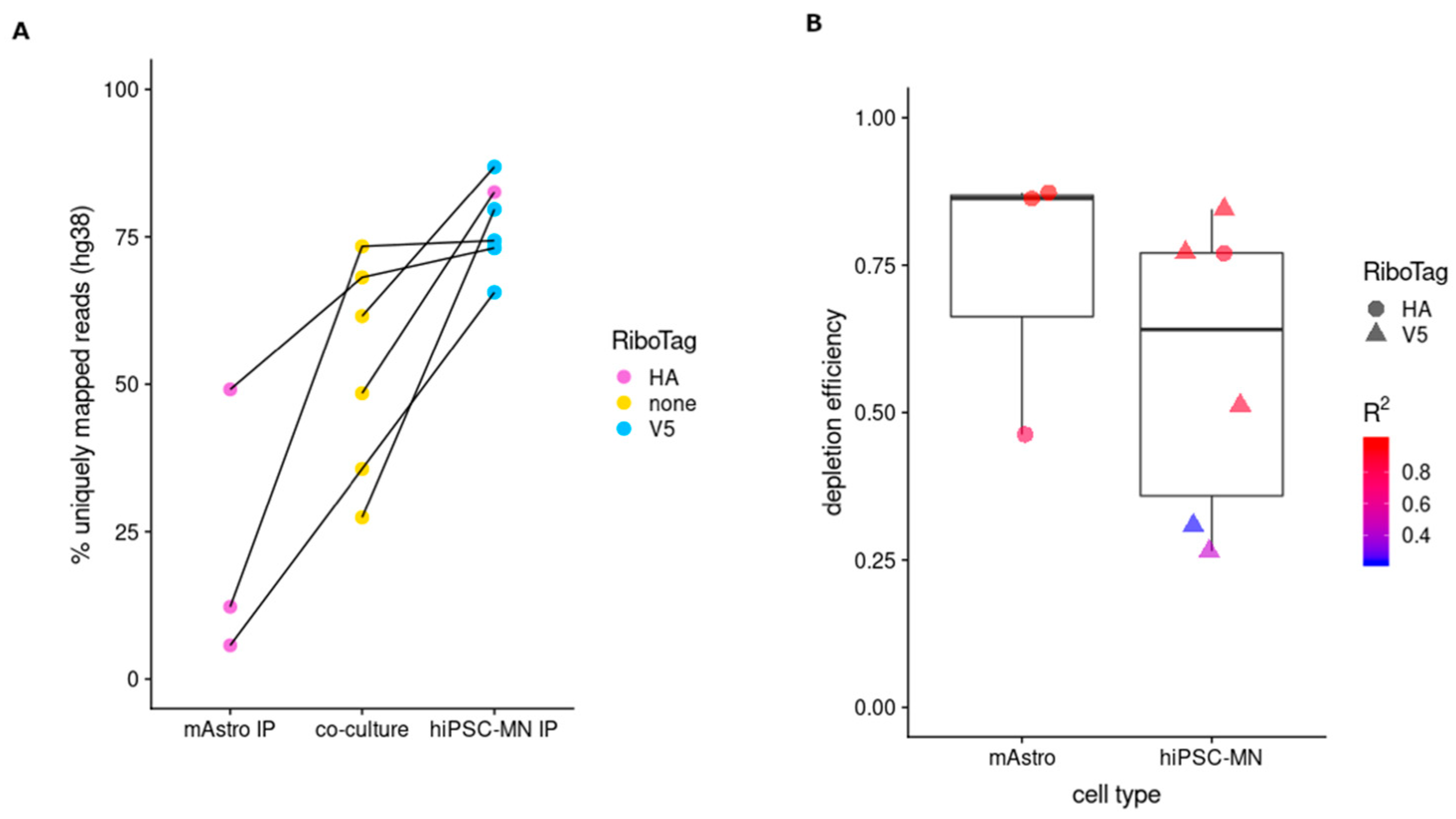
3.4. Application of RiboTag to hiPSC-Derived Neural Cell Types
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Horn, M.R.; Ruthazer, E.S. Glial regulation of synapse maturation and stabilization in the developing nervous system. Curr. Opin. Neurobiol. 2019, 54, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Sims, R.; van der Lee, S.J.; Naj, A.C.; Bellenguez, C.; Badarinarayan, N.; Jakobsdottir, J.; Kunkle, B.W.; Boland, A.; Raybould, R.; Bis, J.C.; et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 2017, 49, 1373–1384. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, D.M.; Gregory, J.; Brennand, K.J. The Importance of Non-neuronal Cell Types in hiPSC-Based Disease Modeling and Drug Screening. Front Cell Dev. Biol. 2017, 5, 117. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chao, J.; Shi, Y. Modeling neurological diseases using iPSC-derived neural cells: iPSC modeling of neurological diseases. Cell Tissue Res. 2018, 371, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N.; et al. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Dezonne, R.S.; Sartore, R.C.; Nascimento, J.M.; Saia-Cereda, V.M.; Romão, L.F.; Alves-Leon, S.V.; de Souza, J.M.; Martins-de-Souza, D.; Rehen, S.K.; Gomes, F.C.A. Derivation of Functional Human Astrocytes from Cerebral Organoids. Sci. Rep. 2017, 7, 45091. [Google Scholar] [CrossRef]
- Marton, R.M.; Miura, Y.; Sloan, S.A.; Li, Q.; Revah, O.; Levy, R.J.; Huguenard, J.R.; Pașca, S.P. Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 2019, 22, 484–491. [Google Scholar] [CrossRef]
- Abud, E.M.; Ramirez, R.N.; Martinez, E.S.; Healy, L.M.; Nguyen, C.H.H.; Newman, S.A.; Yeromin, A.V.; Scarfone, V.M.; Marsh, S.E.; Fimbres, C.; et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 2017, 94, 278–293.e9. [Google Scholar] [CrossRef]
- Cakir, B.; Xiang, Y.; Tanaka, Y.; Kural, M.H.; Parent, M.; Kang, Y.-J.; Chapeton, K.; Patterson, B.; Yuan, Y.; He, C.-S.; et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods 2019, 16, 1169–1175. [Google Scholar] [CrossRef]
- Vatine, G.D.; Barrile, R.; Workman, M.J.; Sances, S.; Barriga, B.K.; Rahnama, M.; Barthakur, S.; Kasendra, M.; Lucchesi, C.; Kerns, J.; et al. Human iPSC-Derived Blood-Brain Barrier Chips Enable Disease Modeling and Personalized Medicine Applications. Cell Stem Cell 2019, 24, 995–1005.e6. [Google Scholar] [CrossRef]
- Lobo, M.K.; Karsten, S.L.; Gray, M.; Geschwind, D.H.; Yang, X.W. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat. Neurosci. 2006, 9, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Arlotta, P.; Molyneaux, B.J.; Chen, J.; Inoue, J.; Kominami, R.; Macklis, J.D. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 2005, 45, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Ramsköld, D.; Luo, S.; Wang, Y.-C.; Li, R.; Deng, Q.; Faridani, O.R.; Daniels, G.A.; Khrebtukova, I.; Loring, J.F.; Laurent, L.C.; et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012, 30, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Bonner, R.F.; Emmert-Buck, M.; Cole, K.; Pohida, T.; Chuaqui, R.; Goldstein, S.; Liotta, L.A. Laser capture microdissection: Molecular analysis of tissue. Science 1997, 278, 1481–1483. [Google Scholar] [CrossRef] [PubMed]
- Emmert-Buck, M.R.; Bonner, R.F.; Smith, P.D.; Chuaqui, R.F.; Zhuang, Z.; Goldstein, S.R.; Weiss, R.A.; Liotta, L.A. Laser capture microdissection. Science 1996, 274, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Rossner, M.J.; Hirrlinger, J.; Wichert, S.P.; Boehm, C.; Newrzella, D.; Hiemisch, H.; Eisenhardt, G.; Stuenkel, C.; von Ahsen, O.; Nave, K.-A. Global transcriptome analysis of genetically identified neurons in the adult cortex. J. Neurosci. 2006, 26, 9956–9966. [Google Scholar] [CrossRef]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef]
- Mimitou, E.P.; Cheng, A.; Montalbano, A.; Hao, S.; Stoeckius, M.; Legut, M.; Roush, T.; Herrera, A.; Papalexi, E.; Ouyang, Z.; et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 2019, 16, 409–412. [Google Scholar] [CrossRef]
- Coenen, C.; Liedtke, S.; Kogler, G. RNA Amplification Protocol Leads to Biased Polymerase Chain Reaction Results Especially for Low-Copy Transcripts of Human Bone Marrow-Derived Stromal Cells. PLoS ONE 2015, 10, e0141070. [Google Scholar] [CrossRef][Green Version]
- Okaty, B.W.; Sugino, K.; Nelson, S.B. Cell type-specific transcriptomics in the brain. J. Neurosci. 2011, 31, 6939–6943. [Google Scholar] [CrossRef] [PubMed]
- Cobos, F.A.; Vandesompele, J.; Mestdagh, P.; De Preter, K. Computational deconvolution of transcriptomics data from mixed cell populations. Bioinformatics 2018, 34, 1969–1979. [Google Scholar] [CrossRef] [PubMed]
- Sturm, G.; Finotello, F.; Petitprez, F.; Zhang, J.D.; Baumbach, J.; Fridman, W.H.; List, M.; Aneichyk, T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 2019, 35, i436–i445. [Google Scholar] [CrossRef] [PubMed]
- Sanz, E.; Yang, L.; Su, T.; Morris, D.R.; McKnight, G.S.; Amieux, P.S. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. USA 2009, 106, 13939–13944. [Google Scholar] [CrossRef]
- Doyle, J.P.; Dougherty, J.D.; Heiman, M.; Schmidt, E.F.; Stevens, T.R.; Ma, G.; Bupp, S.; Shrestha, P.; Shah, R.D.; Doughty, M.L.; et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 2008, 135, 749–762. [Google Scholar] [CrossRef]
- Heiman, M.; Schaefer, A.; Gong, S.; Peterson, J.D.; Day, M.; Ramsey, K.E.; Suárez-Fariñas, M.; Schwarz, C.; Stephan, D.A.; Surmeier, D.J.; et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 2008, 135, 738–748. [Google Scholar] [CrossRef]
- Nguyen, T.-M.; Schreiner, D.; Xiao, L.; Traunmüller, L.; Bornmann, C.; Scheiffele, P. An alternative splicing switch shapes neurexin repertoires in principal neurons versus interneurons in the mouse hippocampus. Elife 2016, 5, e22757. [Google Scholar] [CrossRef]
- Brichta, L.; Shin, W.; Jackson-Lewis, V.; Blesa, J.; Yap, E.-L.; Walker, Z.; Zhang, J.; Roussarie, J.-P.; Alvarez, M.J.; Califano, A.; et al. Identification of neurodegenerative factors using translatome-regulatory network analysis. Nat. Neurosci. 2015, 18, 1325–1333. [Google Scholar] [CrossRef]
- Haimon, Z.; Volaski, A.; Orthgiess, J.; Boura-Halfon, S.; Varol, D.; Shemer, A.; Yona, S.; Zuckerman, B.; David, E.; Chappell-Maor, L.; et al. Re-evaluating microglia expression profiles using RiboTag and cell isolation strategies. Nat. Immunol. 2018, 19, 636–644. [Google Scholar] [CrossRef]
- Michalovicz, L.T.; Kelly, K.A.; Vashishtha, S.; Ben-Hamo, R.; Efroni, S.; Miller, J.V.; Locker, A.R.; Sullivan, K.; Broderick, G.; Miller, D.B.; et al. Astrocyte-specific transcriptome analysis using the ALDH1L1 bacTRAP mouse reveals novel biomarkers of astrogliosis in response to neurotoxicity. J. Neurochem. 2019, 150, 420–440. [Google Scholar] [CrossRef]
- Zhu, Y.; Lyapichev, K.; Lee, D.H.; Motti, D.; Ferraro, N.M.; Zhang, Y.; Yahn, S.; Soderblom, C.; Zha, J.; Bethea, J.R.; et al. Macrophage Transcriptional Profile Identifies Lipid Catabolic Pathways That Can Be Therapeutically Targeted after Spinal Cord Injury. J. Neurosci. 2017, 37, 2362–2376. [Google Scholar] [CrossRef] [PubMed]
- Southern, J.A.; Young, D.F.; Heaney, F.; Baumgärtner, W.K.; Randall, R.E. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 1991, 72, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Field, J.; Nikawa, J.; Broek, D.; MacDonald, B.; Rodgers, L.; Wilson, I.A.; Lerner, R.A.; Wigler, M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 1988, 8, 2159–2165. [Google Scholar] [CrossRef] [PubMed]
- Hopp, T.P.; Prickett, K.S.; Price, V.L.; Libby, R.T.; March, C.J.; Pat Cerretti, D.; Urdal, D.L.; Conlon, P.J. A Short Polypeptide Marker Sequence Useful for Recombinant Protein Identification and Purification. Biotechnology 1988, 6, 1204–1210. [Google Scholar] [CrossRef]
- Topol, A.; Zhu, S.; Hartley, B.J.; English, J.; Hauberg, M.E.; Tran, N.; Rittenhouse, C.A.; Simone, A.; Ruderfer, D.M.; Johnson, J.; et al. Dysregulation of miRNA-9 in a Subset of Schizophrenia Patient-Derived Neural Progenitor Cells. Cell Rep. 2017, 20, 2525. [Google Scholar] [CrossRef]
- Lee, I.S.; Carvalho, C.M.B.; Douvaras, P.; Ho, S.-M.; Hartley, B.J.; Zuccherato, L.W.; Ladran, I.G.; Siegel, A.J.; McCarthy, S.; Malhotra, D.; et al. Characterization of molecular and cellular phenotypes associated with a heterozygous CNTNAP2 deletion using patient-derived hiPSC neural cells. NPJ Schizophr. 2015, 1, 15019. [Google Scholar] [CrossRef]
- Ho, S.-M.; Hartley, B.J.; Tcw, J.; Beaumont, M.; Stafford, K.; Slesinger, P.A.; Brennand, K.J. Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells. Methods 2016, 101, 113–124. [Google Scholar] [CrossRef]
- Tiscornia, G.; Singer, O.; Verma, I.M. Production and purification of lentiviral vectors. Nat. Protoc. 2006, 1, 241–245. [Google Scholar] [CrossRef]
- Maury, Y.; Côme, J.; Piskorowski, R.A.; Salah-Mohellibi, N.; Chevaleyre, V.; Peschanski, M.; Martinat, C.; Nedelec, S. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 2015, 33, 89–96. [Google Scholar] [CrossRef]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef]
- O’Doherty, U.; Swiggard, W.J.; Malim, M.H. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 2000, 74, 10074–10080. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Chanda, S.; Marro, S.; Ng, Y.-H.; Janas, J.A.; Haag, D.; Ang, C.E.; Tang, Y.; Flores, Q.; Mall, M.; et al. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods 2017, 14, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Barretto, N.; Zhang, H.; Powell, S.K.; Fernando, M.B.; Zhang, S.; Flaherty, E.K.; Ho, S.-M.; Slesinger, P.A.; Duan, J.; Brennand, K.J. ASCL1- and DLX2-induced GABAergic neurons from hiPSC-derived NPCs. J. Neurosci. Methods 2020, 334, 108548. [Google Scholar] [CrossRef] [PubMed]
- Harder, J.M.; Braine, C.E.; Williams, P.A.; Zhu, X.; MacNicoll, K.H.; Sousa, G.L.; Buchanan, R.A.; Smith, R.S.; Libby, R.T.; Howell, G.R.; et al. Early immune responses are independent of RGC dysfunction in glaucoma with complement component C3 being protective. Proc. Natl. Acad. Sci. USA 2017, 114, E3839–E3848. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-J.; Elahi, L.S.; Pașca, A.M.; Marton, R.M.; Gordon, A.; Revah, O.; Miura, Y.; Walczak, E.M.; Holdgate, G.M.; Fan, H.C.; et al. Reliability of human cortical organoid generation. Nat. Methods 2019, 16, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Karch, C.M.; Kao, A.W.; Karydas, A.; Onanuga, K.; Martinez, R.; Argouarch, A.; Wang, C.; Huang, C.; Sohn, P.D.; Bowles, K.R.; et al. A Comprehensive Resource for Induced Pluripotent Stem Cells from Patients with Primary Tauopathies. Stem Cell Rep. 2019, 13, 939–955. [Google Scholar] [CrossRef]
- Obrig, T.G.; Culp, W.J.; McKeehan, W.L.; Hardesty, B. The mechanism by which cycloheximide and related glutarimide antibiotics inhibit peptide synthesis on reticulocyte ribosomes. J. Biol. Chem. 1971, 246, 174–181. [Google Scholar]
- Krueger, F. Trim Galore: A Wrapper Tool around Cutadapt and FastQC to Consistently Apply Quality and Adapter Trimming to FastQ Files, With Some Extra Functionality for MspI-digested RRBS-type (Reduced Representation Bisufite-Seq) Libraries. 2012. Available online: http://www.Bioinformatics.Babraham.AC.UK/projects/trim_galore/ (accessed on 28 April 2016).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zalenski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Korotkevich, G.; Sukhov, V.; Sergushichev, A. Fast gene set enrichment analysis. bioRxiv 2019, 060012. [Google Scholar] [CrossRef]
- Lesiak, A.J.; Brodsky, M.; Neumaier, J.F. RiboTag is a flexible tool for measuring the translational state of targeted cells in heterogeneous cell cultures. Biotechniques 2015, 58, 308–317. [Google Scholar] [CrossRef] [PubMed]
| Sample | IP Order | Species | On/Off Target | Slope | R2 | p-Value |
|---|---|---|---|---|---|---|
| V5-1 (3T3) | 1 | human | off target | −0.8 | 0.99 | p < 0.001 |
| 1 | mouse | on target | 0.12 | 0.08 | p < 0.10 | |
| V5-2 (3T3) | 2 | human | off target | −0.85 | 0.99 | p < 0.001 |
| 2 | mouse | on target | 0.14 | 0.12 | p < 0.06 | |
| HA-1 (HEK293) | 1 | human | on target | 1.26 | 0.59 | p < 0.001 |
| 1 | mouse | off target | −0.81 | 0.99 | p < 0.001 | |
| HA-2 (HEK293) | 2 | human | on target | 1.56 | 0.74 | p < 0.001 |
| 2 | mouse | off target | −0.74 | 0.99 | p < 0.001 |
| Library | Input Library | MN Differentiation | IP Cell Type | RiboTag | Species | Slope | R Squared | p Value | RNA Yield (ng) |
|---|---|---|---|---|---|---|---|---|---|
| 5 | 2 | 20190603 | mAstro | HA | mouse | −0.46 | 0.82 | 0.001 | 593.6 |
| 8 | 2 | 20190603 | MN | V5 | human | −0.26 | 0.42 | 0.001 | <1 |
| 12 | 10 | 20190528 | mAstro | HA | mouse | −0.87 | 0.99 | 0.001 | 158.2 |
| 14 | 10 | 20190528 | MN | V5 | human | −0.31 | 0.23 | 0.017 | 422.8 |
| 20 | 17 | 20190417 | mAstro | HA | mouse | −0.86 | 0.99 | 0.001 | 431.2 |
| 18 | 17 | 20190417 | MN | V5 | human | −0.51 | 0.91 | 0.001 | <1 |
| 28 | 21 | 20190610 | MN | V5 | human | −0.77 | 0.96 | 0.001 | 187.6 |
| 30 | 23 | 20190610 | MN | HA | human | −0.77 | 0.94 | 0.001 | <1 |
| 31 | 25 | 20190617 | MN | V5 | human | −0.84 | 0.96 | 0.001 | <1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gregory, J.A.; Hoelzli, E.; Abdelaal, R.; Braine, C.; Cuevas, M.; Halpern, M.; Barretto, N.; Schrode, N.; Akbalik, G.; Kang, K.; et al. Cell Type-Specific In Vitro Gene Expression Profiling of Stem Cell-Derived Neural Models. Cells 2020, 9, 1406. https://doi.org/10.3390/cells9061406
Gregory JA, Hoelzli E, Abdelaal R, Braine C, Cuevas M, Halpern M, Barretto N, Schrode N, Akbalik G, Kang K, et al. Cell Type-Specific In Vitro Gene Expression Profiling of Stem Cell-Derived Neural Models. Cells. 2020; 9(6):1406. https://doi.org/10.3390/cells9061406
Chicago/Turabian StyleGregory, James A., Emily Hoelzli, Rawan Abdelaal, Catherine Braine, Miguel Cuevas, Madeline Halpern, Natalie Barretto, Nadine Schrode, Güney Akbalik, Kristy Kang, and et al. 2020. "Cell Type-Specific In Vitro Gene Expression Profiling of Stem Cell-Derived Neural Models" Cells 9, no. 6: 1406. https://doi.org/10.3390/cells9061406
APA StyleGregory, J. A., Hoelzli, E., Abdelaal, R., Braine, C., Cuevas, M., Halpern, M., Barretto, N., Schrode, N., Akbalik, G., Kang, K., Cheng, E., Bowles, K., Lotz, S., Goderie, S., Karch, C. M., Temple, S., Goate, A., Brennand, K. J., & Phatnani, H. (2020). Cell Type-Specific In Vitro Gene Expression Profiling of Stem Cell-Derived Neural Models. Cells, 9(6), 1406. https://doi.org/10.3390/cells9061406





