Multi-Omics Analysis of Diabetic Heart Disease in the db/db Model Reveals Potential Targets for Treatment by a Longevity-Associated Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics
2.2. Viral Vector
2.3. In Vivo Protocol
2.4. Next-Generation RNA Sequencing
2.5. RNA Isolation and RT-qPCR
2.6. Protein Extraction and Western Blotting
2.7. Untargeted Metabolomics–Ultra-High-Performance Liquid Chromatography-Mass Spectrometry (UHPLC-MS)
2.8. Statistical Analysis
3. Results
3.1. Cardiac Lipid Metabolism is Altered in db/db Mouse Hearts with Further Changes Imparted by LAV-BPIFB4 Therapy
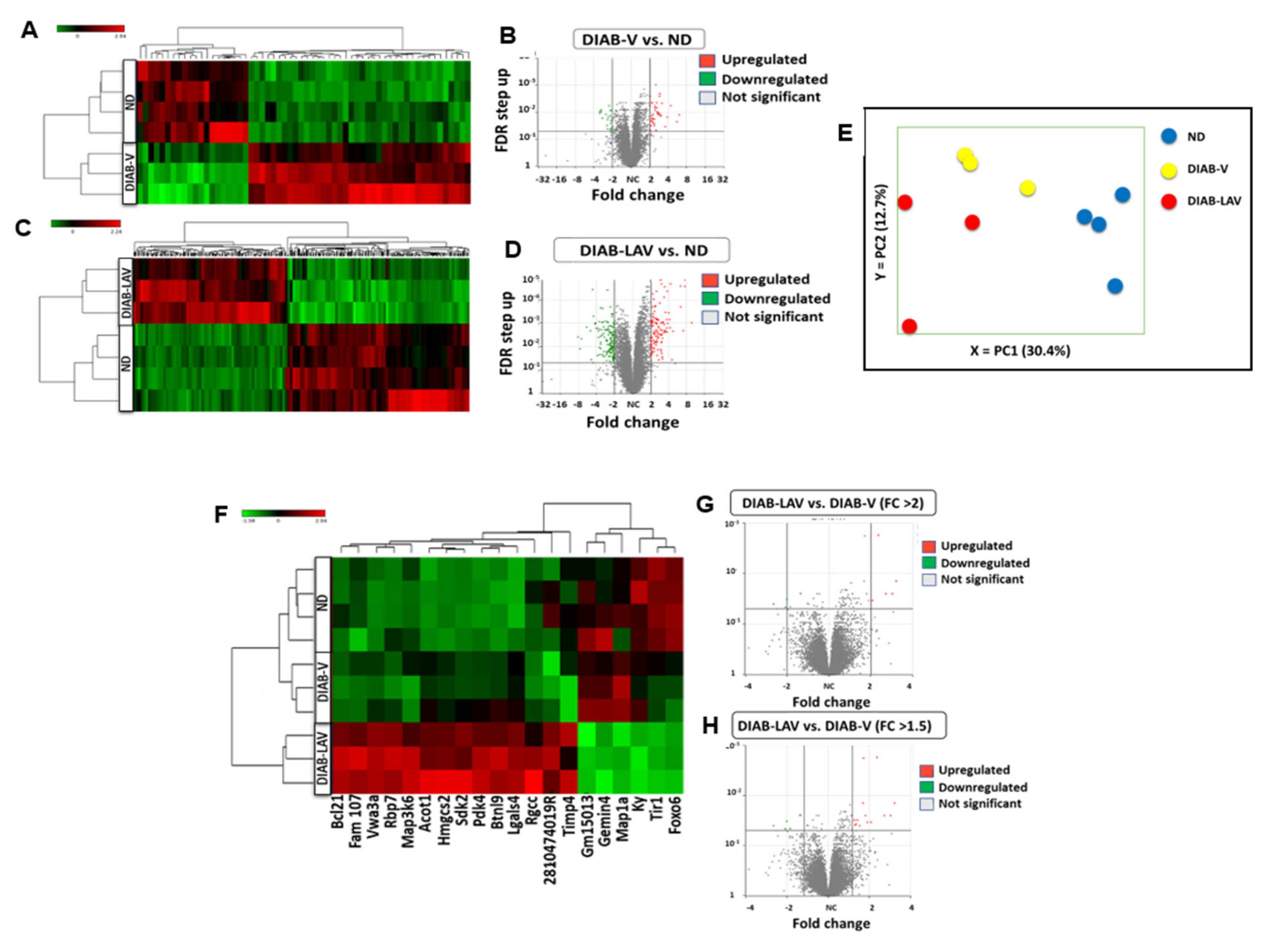
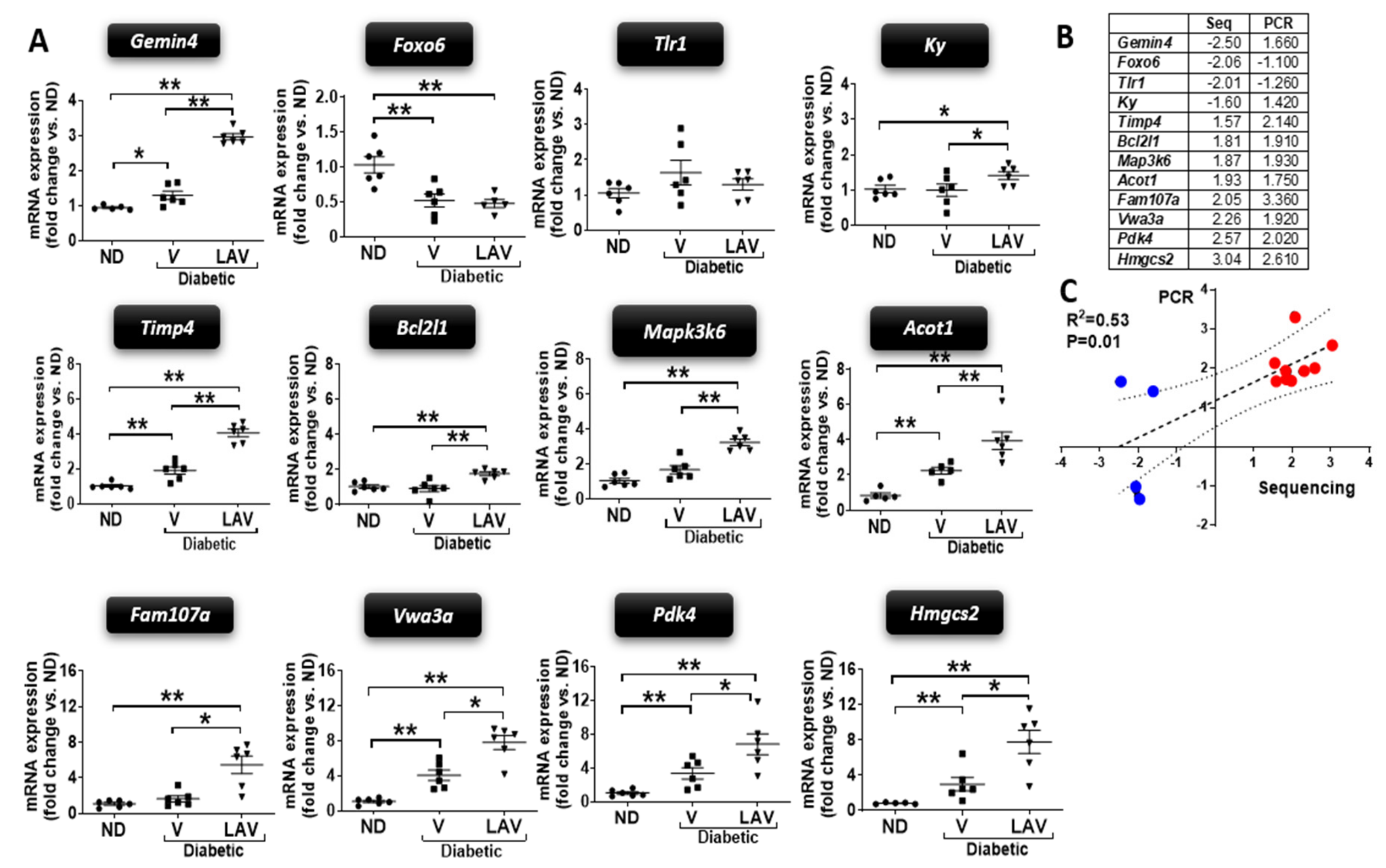
3.2. Changes in Cardiac Transcriptome of db/db Mice and Effect of LAV-BPIFB4 Therapy
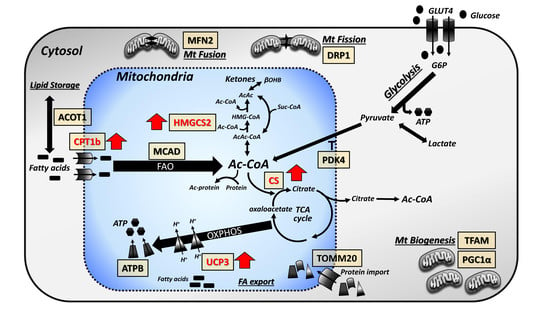
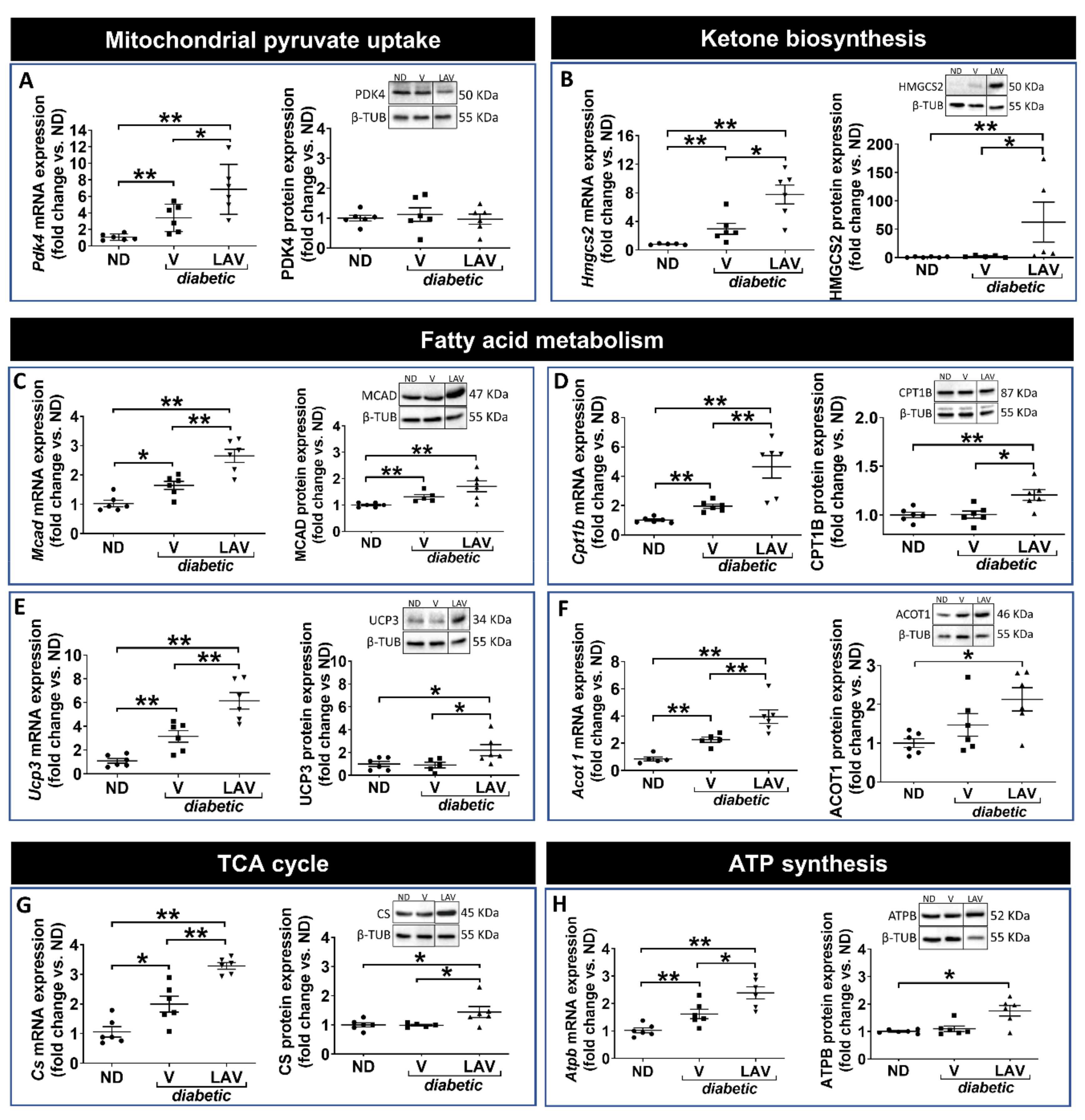
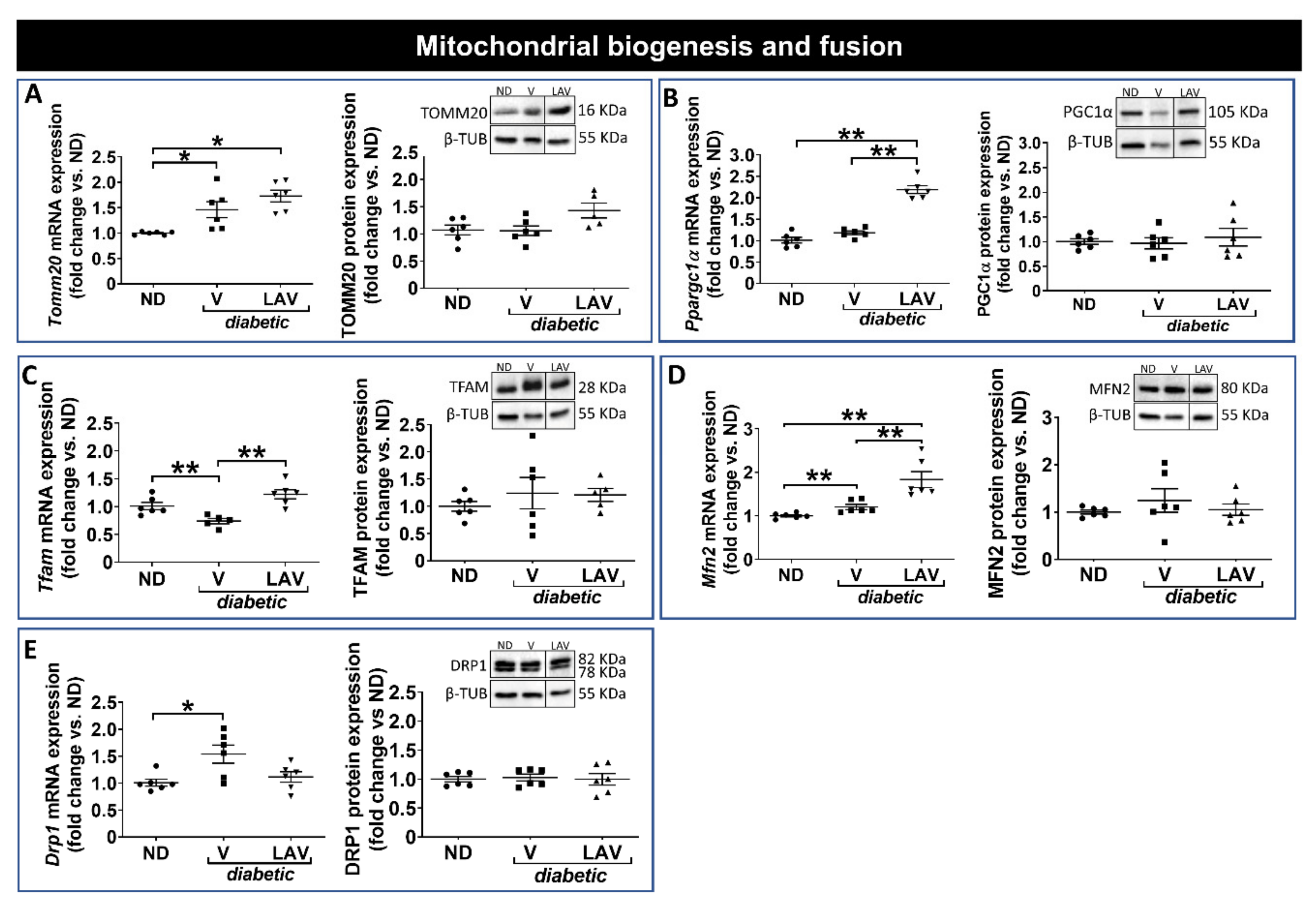
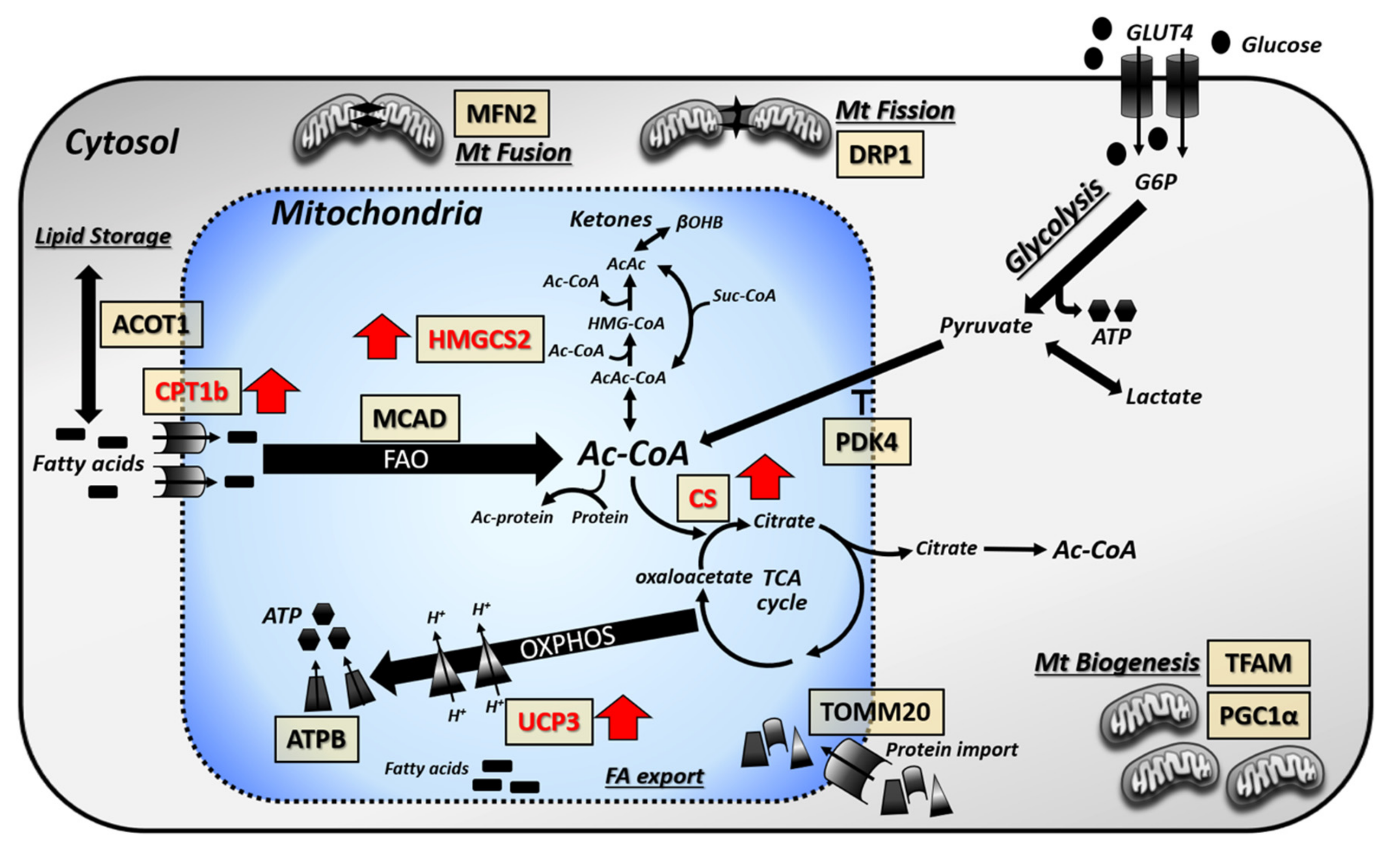
3.3. Focused Expression Analysis of Mitochondrial-Related Pathways
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jia, G.; Hill, M.A.; Sowers, J.R. Diabetic Cardiomyopathy. Circ. Res. 2018, 122, 624–638. [Google Scholar] [CrossRef] [PubMed]
- Lesnefsky, E.J.; Chen, Q.; Hoppel, C.L. Mitochondrial Metabolism in Aging Heart. Circ. Res. 2016, 118, 1593–1611. [Google Scholar] [CrossRef] [PubMed]
- Nichols, G.A.; Gullion, C.M.; Koro, C.E.; Ephross, S.A.; Brown, J.B. The incidence of congestive heart failure in type 2 diabetes: An update. Diabetes Care 2004, 27, 1879–1884. [Google Scholar] [CrossRef]
- Huynh, K.; Bernardo, B.C.; McMullen, J.R.; Ritchie, R.H. Diabetic cardiomyopathy: Mechanisms and new treatment strategies targeting antioxidant signaling pathways. Pharmacol. Ther. 2014, 142, 375–415. [Google Scholar] [CrossRef] [PubMed]
- Motta, M.; Bennati, E.; Capri, M.; Ferlito, L.; Malaguarnera, M. Diabetes mellitus in the extreme longevity. Exp. Gerontol. 2008, 43, 102–105. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bennati, E.; Murphy, A.; Cambien, F.; Whitehead, A.; Archbold, G.; Young, I.S.; Rea, I.M. BELFAST Centenarians: A Case of Optimal Cardiovascular Risk? Curr. Pharm. Des. 2010, 16, 789–795. [Google Scholar] [CrossRef]
- Villa, F.; Carrizzo, A.; Spinelli, C.C.; Ferrario, A.; Malovini, A.; Maciag, A.; Damato, A.; Auricchio, A.; Spinetti, G.; Sangalli, E.; et al. Genetic Analysis Reveals a Longevity-Associated Protein Modulating Endothelial Function and Angiogenesis. Circ. Res. 2015, 117, 333–345. [Google Scholar] [CrossRef]
- Puca, A.A.; Carrizzo, A.; Spinelli, C.C.; Damato, A.; Ambrosio, M.; Villa, F.; Ferrario, A.; Maciag, A.; Fornai, F.; Lenzi, P.; et al. Single systemic transfer of a human gene associated with exceptional longevity halts the progression of atherosclerosis and inflammation in ApoE knockout mice through a CXCR4-mediated mechanism. Eur. Hear. J. 2019. [Google Scholar] [CrossRef]
- Dang, Z.; Avolio, E.; Thomas, A.C.; Faulkner, A.; Beltrami, A.P.; Cervellin, C.; Carrizzo, A.; Maciag, A.; Gu, Y.; Ciaglia, E.; et al. Transfer of a human gene variant associated with exceptional longevity improves cardiac function in obese type-2 diabetic mice through induction of the SDF-1/CXCR4 signalling pathway. Eur. J. Heart Fail. 2020. [Google Scholar] [CrossRef] [PubMed]
- Aasum, E.; Hafstad, A.D.; Severson, D.L.; Larsen, T.S. Age-dependent changes in metabolism, contractile function, and ischemic sensitivity in hearts from db/db mice. Diabetes 2003, 52, 434–441. [Google Scholar] [CrossRef]
- Alex, L.; Russo, I.; Holoborodko, V.; Frangogiannis, N.G. Characterization of a mouse model of obesity-related fibrotic cardiomyopathy that recapitulates features of human heart failure with preserved ejection fraction. Am. J. Physiol. Circ. Physiol. 2018, 315, 934–949. [Google Scholar] [CrossRef] [PubMed]
- How, O.-J.; Aasum, E.; Severson, D.L.; Chan, W.A.; Essop, M.F.; Larsen, T.S.; Essop, M.F. Increased Myocardial Oxygen Consumption Reduces Cardiac Efficiency in Diabetic Mice. Diabetes 2006, 55, 466–473. [Google Scholar] [CrossRef] [PubMed]
- Kuo, T.H.; Moore, K.H.; Giacomelli, F.; Wiener, J. Defective Oxidative Metabolism of Heart Mitochondria from Genetically Diabetic Mice. Diabetes 1983, 32, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Demarco, V.G.; Ford, D.A.; Henriksen, E.J.; Aroor, A.R.; Johnson, M.S.; Habibi, J.; Ma, L.; Yang, M.; Albert, C.J.; Lally, J.W.; et al. Obesity-related alterations in cardiac lipid profile and nondipping blood pressure pattern during transition to diastolic dysfunction in male db/db mice. Endocrinology 2012, 154, 159–171. [Google Scholar] [CrossRef]
- Mora-Ortiz, M.; Ramos, P.N.; Oregioni, A.; Claus, S.P. NMR metabolomics identifies over 60 biomarkers associated with Type II Diabetes impairment in db/db mice. Metabolomics 2019, 15, 89. [Google Scholar] [CrossRef]
- Cefalu, W.T. Animal Models of Type 2 Diabetes: Clinical Presentation and Pathophysiological Relevance to the Human Condition. ILAR J. 2006, 47, 186–198. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Wu, H.; Southam, A.; Hines, A.; Viant, M.R. High-throughput tissue extraction protocol for NMR and MS-based metabolomics. Anal. Biochem. 2008, 372, 204–212. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.; Wedge, D.C.; Goodacre, R.; Kell, D.B.; Baker, P.N.; Kenny, L.C.; Mamas, M.A.; Neyses, L.; Dunn, W.B. Automated workflows for accurate mass-based putative metabolite identification in LC/MS-derived metabolomic datasets. Bioinformatics 2011, 27, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.A.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic Quotient Normalization as Robust Method to Account for Dilution of Complex Biological Mixtures. Application in1H NMR Metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef]
- Villa, F.; Malovini, A.; Carrizzo, A.; Spinelli, C.C.; Ferrario, A.; Maciag, A.; Madonna, M.; Bellazzi, R.; Milanesi, L.; Vecchione, C.; et al. Serum BPIFB4 levels classify health status in long-living individuals. Immun. Ageing 2015, 12, 27. [Google Scholar] [CrossRef]
- Zhang, L.; Ussher, J.R.; Oka, T.; Cadete, V.J.; Wagg, C.; Lopaschuk, G.D. Cardiac diacylglycerol accumulation in high fat-fed mice is associated with impaired insulin-stimulated glucose oxidation. Cardiovasc. Res. 2010, 89, 148–156. [Google Scholar] [CrossRef]
- Park, T.-S.; Hu, Y.; Noh, H.-L.; Drosatos, K.; Okajima, K.; Buchanan, J.; Tuinei, J.; Homma, S.; Jiang, X.-C.; Abel, E.D.; et al. Ceramide is a cardiotoxin in lipotoxic cardiomyopathy. J. Lipid Res. 2008, 49, 2101–2112. [Google Scholar] [CrossRef]
- Schulze, P.; Drosatos, K.; Goldberg, I.J. Lipid Use and Misuse by the Heart. Circ. Res. 2016, 118, 1736–1751. [Google Scholar] [CrossRef]
- Law, B.A.; Liao, X.; Moore, K.S.; Southard, A.; Roddy, P.; Ji, R.; Szulc, Z.; Bielawska, A.; Schulze, P.C.; Cowart, L.A.; et al. Lipotoxic very-long-chain ceramides cause mitochondrial dysfunction, oxidative stress, and cell death in cardiomyocytes. FASEB J. 2018, 32, 1403–1416. [Google Scholar] [CrossRef]
- Han, X.; Yang, J.; Cheng, H.; Yang, K.; Abendschein, D.R.; Gross, R.W. Shotgun Lipidomics Identifies Cardiolipin Depletion in Diabetic Myocardium Linking Altered Substrate Utilization with Mitochondrial Dysfunction. Biochemestry 2005, 44, 16684–16694. [Google Scholar] [CrossRef]
- Han, X.; Yang, J.; Yang, K.; Zhongdan, Z.; Abendschein, S.D.R.; Gross, S.R.W.; Zhao, Z. Alterations in Myocardial Cardiolipin Content and Composition Occur at the Very Earliest Stages of Diabetes: A Shotgun Lipidomics Study†. Biochemistry 2007, 46, 6417–6428. [Google Scholar] [CrossRef] [PubMed]
- Dudek, J.; Hartmann, M.; Rehling, P. The role of mitochondrial cardiolipin in heart function and its implication in cardiac disease. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Vecchini, A.; Del Rosso, F.; Binaglia, L.; Dhalla, N.S.; Panagia, V. Molecular Defects in Sarcolemmal Glycerophospholipid Subclasses in Diabetic Cardiomyopathy. J. Mol. Cell. Cardiol. 2000, 32, 1061–1074. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Zhang, R.; Liang, Y.; Shi, J.; Li, J.; Shang, F.; Mao, X.; Sun, J. Changes of myocardial lipidomics profiling in a rat model of diabetic cardiomyopathy using UPLC/Q-TOF/MS analysis. Diabetol. Metab. Syndr. 2017, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yao, M.; Wang, R.; Shi, Y.; Hou, L.; Hou, Z.; Lian, K.; Zhang, N.; Wang, Y.; Li, W.; et al. Profile of cardiac lipid metabolism in STZ-induced diabetic mice. Lipids Heal. Dis. 2018, 17, 231. [Google Scholar] [CrossRef]
- Messina-Graham, S.; Broxmeyer, H.E. SDF-1/CXCL12 modulates mitochondrial respiration of immature blood cells in a bi-phasic manner. Blood Cells Mol. Dis. 2016, 58, 13–18. [Google Scholar] [CrossRef]
- Yu, B.-B.; Zhi, H.; Zhang, X.-Y.; Liang, J.-W.; He, J.; Su, C.; Xia, W.-H.; Zhang, G.-X.; Tao, J. Mitochondrial dysfunction-mediated decline in angiogenic capacity of endothelial progenitor cells is associated with capillary rarefaction in patients with hypertension via downregulation of CXCR4/JAK2/SIRT5 signaling. E BioMed. 2019, 42, 64–75. [Google Scholar] [CrossRef]
- Cai, W.-F.; Kang, K.; Huang, W.; Liang, J.-L.; Feng, Y.-L.; Liu, G.-S.; Chang, D.-H.; Wen, Z.-L.; Paul, C.; Xu, M.; et al. CXCR4 attenuates cardiomyocytes mitochondrial dysfunction to resist ischaemia-reperfusion injury. J. Cell. Mol. Med. 2015, 19, 1825–1835. [Google Scholar] [CrossRef]
- Buchanan, J.; Mazumder, P.K.; Hu, P.; Chakrabarti, G.; Roberts, M.W.; Yun, U.J.; Cooksey, R.C.; Litwin, S.E.; Abel, E.D. Reduced Cardiac Efficiency and Altered Substrate Metabolism Precedes the Onset of Hyperglycemia and Contractile Dysfunction in Two Mouse Models of Insulin Resistance and Obesity. Endocrinology 2005, 146, 5341–5349. [Google Scholar] [CrossRef]
- Boudina, S.; Sena, S.; Theobald, H.; Sheng, X.; Wright, J.; Hu, X.X.; Aziz, S.; Johnson, J.I.; Bugger, H.; Zaha, V.G.; et al. Mitochondrial Energetics in the Heart in Obesity-Related Diabetes: Direct Evidence for Increased Uncoupled Respiration and Activation of Uncoupling Proteins. Diabetes 2007, 56, 2457–2466. [Google Scholar] [CrossRef]
- Dabkowski, E.R.; Baseler, W.A.; Williamson, C.L.; Powell, M.; Razunguzwa, T.T.; Frisbee, J.C.; Hollander, J.M. Mitochondrial dysfunction in the type 2 diabetic heart is associated with alterations in spatially distinct mitochondrial proteomes. Am. J. Physiol. Circ. Physiol. 2010, 299, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Kesherwani, V.; Shahshahan, H.R.; Mishra, P.K. Cardiac transcriptome profiling of diabetic Akita mice using microarray and next generation sequencing. PLoS ONE 2017, 12, e0182828. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faulkner, A.; Dang, Z.; Avolio, E.; Thomas, A.C.; Batstone, T.; Lloyd, G.R.; Weber, R.J.; Najdekr, L.; Jankevics, A.; Dunn, W.B.; et al. Multi-Omics Analysis of Diabetic Heart Disease in the db/db Model Reveals Potential Targets for Treatment by a Longevity-Associated Gene. Cells 2020, 9, 1283. https://doi.org/10.3390/cells9051283
Faulkner A, Dang Z, Avolio E, Thomas AC, Batstone T, Lloyd GR, Weber RJ, Najdekr L, Jankevics A, Dunn WB, et al. Multi-Omics Analysis of Diabetic Heart Disease in the db/db Model Reveals Potential Targets for Treatment by a Longevity-Associated Gene. Cells. 2020; 9(5):1283. https://doi.org/10.3390/cells9051283
Chicago/Turabian StyleFaulkner, Ashton, Zexu Dang, Elisa Avolio, Anita C Thomas, Thomas Batstone, Gavin R Lloyd, Ralf JM Weber, Lukáš Najdekr, Andris Jankevics, Warwick B Dunn, and et al. 2020. "Multi-Omics Analysis of Diabetic Heart Disease in the db/db Model Reveals Potential Targets for Treatment by a Longevity-Associated Gene" Cells 9, no. 5: 1283. https://doi.org/10.3390/cells9051283
APA StyleFaulkner, A., Dang, Z., Avolio, E., Thomas, A. C., Batstone, T., Lloyd, G. R., Weber, R. J., Najdekr, L., Jankevics, A., Dunn, W. B., Spinetti, G., Vecchione, C., Puca, A. A., & Madeddu, P. (2020). Multi-Omics Analysis of Diabetic Heart Disease in the db/db Model Reveals Potential Targets for Treatment by a Longevity-Associated Gene. Cells, 9(5), 1283. https://doi.org/10.3390/cells9051283