Abstract
Laminopathies are causally associated with mutations on the Lamin A/C gene (LMNA). To date, more than 400 mutations in LMNA have been reported in patients. These mutations are widely distributed throughout the entire gene and are associated with a wide range of phenotypes. Unfortunately, little is known about the mechanisms underlying the effect of the majority of these mutations. This is the case of more than 40 mutations that are located at exon 4. Using CRISPR/Cas9 technology, we generated a collection of Lmna exon 4 mutants in mouse C2C12 myoblasts. These cell models included different types of exon 4 deletions and the presence of R249W mutation, one of the human variants associated with a severe type of laminopathy, LMNA-associated congenital muscular dystrophy (L-CMD). We characterized these clones by measuring their nuclear circularity, myogenic differentiation capacity in 2D and 3D conditions, DNA damage, and levels of p-ERK and p-AKT (phosphorylated Mitogen-Activated Protein Kinase 1/3 and AKT serine/threonine kinase 1). Our results indicated that Lmna exon 4 mutants showed abnormal nuclear morphology. In addition, levels and/or subcellular localization of different members of the lamin and LINC (LInker of Nucleoskeleton and Cytoskeleton) complex were altered in all these mutants. Whereas no significant differences were observed for ERK and AKT activities, the accumulation of DNA damage was associated to the Lmna p.R249W mutant myoblasts. Finally, significant myogenic differentiation defects were detected in the Lmna exon 4 mutants. These results have key implications in the development of future therapeutic strategies for the treatment of laminopathies.
1. Introduction
Lamins are nuclear intermediate filaments proteins that form a meshwork underlying the inner side of the nuclear membrane [1]. Nuclear lamins have been associated with a wide spectrum of cellular functions including structural support, mechanosensing, chromatin organization, transcription regulation, replication, nuclear assembly, and nuclear pore complex activity [2,3,4,5]. Laminopathies are a group of human rare diseases mainly associated with different mutations on the Lamin A/C gene (LMNA). Lamin A/C proteins are main components of the lamin. Laminopathies include at least 15 different diseases that are divided in 4 different categories depending on the affected tissues: striated muscle, adipose tissue, peripheral nerves, or multiple tissues [6]. This last group includes Hutchinson–Gilford progeria syndrome (HGPS), which is characterized by aging-like symptoms emerging in childhood.
The LMNA gene contains 12 exons and codes for 2 different proteins, Lamin A and C. Both are members of the intermediate filament group of proteins, which are characterized for the presence of a short N-terminal domain, a central domain composed of four α-helical domains that are separated by three linker regions and a globular C-terminal region. To date, more than 400 mutations in LMNA have been reported in patients (http://www.umd.be/LMNA/). These mutations are widely distributed throughout the entire gene. Moreover, there is no association between the gene location of these mutations and the wide range of phenotypes included in this group of rare diseases. Two main hypotheses have been postulated to explain the development and progression of laminopathies [7]. One of them assumes that LMNA mutations provoke structural and mechanical changes that are responsible for the development of the disease, whereas the other relies on functional abnormalities induced by changes in gene expression patterns and differentiation programs. Unfortunately, little is known about the exact mechanisms underlying the effects induced by the majority of the laminopathy-causing LMNA mutations.
No curative treatment is currently available for any type of laminopathy. A number of pre-clinical studies have explored the therapeutic potential of different compounds that modulate the Mechanistic Target Of Rapamycin Kinase (mTOR) pathway [8,9], the mitogen-activated protein kinase (MAPK) cascade [10,11], and the epigenetic regulator N-Acetyltransferase 10 (NAT10) [12], among others. Gene therapy approaches are being studied as well. In this regard, the most promising results have been obtained for HGPS by introducing frameshift mutations in the LMNA gene using CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR associated protein 9) [13,14]. Besides these advances, we are still far from a cure for the majority of the laminopathies. More information is therefore needed about the molecular and cellular defects induced for each disease-associated LMNA mutation in order to obtain the therapeutic strategies that better fit each case.
Here, we explored whether mutations in Lmna exon 4 affect the biology and function of mouse myoblasts. This exon codes for a linker domain located between the two central α-helical domains of Lamins A and C (Figure S1). Although up to 47 mutations in LMNA exon 4 have been reported with different disorders linked to this gene (http://www.umd.be/LMNA/), little is known about the cellular and molecular changes induced by them that are causally related with laminopathy development and progression. Thus, although LMNA p.R249W is the most frequent mutation for LMNA-associated congenital muscular dystrophy, a severe type of laminopathy [15], and has been found to be related to abnormal nuclear morphology and myogenic differentiation, mislocalization of Lamin B, altered sense of microenvironment stiffness, and DNA damage [16,17,18,19], the causal connections between this mutation and the disease remain mainly unknown. On the other hand, because LMNA exons 3 and 5 are in frame, exon 4 skipping might be considered as a therapeutic strategy for all those laminopathies associated with LMNA exon 4 mutations. Although it has been reported that exon skipping could be successfully used for the treatment of diseases associated with LMNA mutations in exon 5 [20], the potential of this approach has not been tested yet for LMNA exon 4.
Using mouse C2C12 myoblasts and CRISPR/Cas9, we generated a collection of Lmna exon 4 mutants that were classified as Lmna-null, Lmna-R249W, or carriers of Lmna exon4 in-frame deletions. The study of these mutants allowed us to find critical information on the importance of Lmna exon 4 in myoblast biology, specifically in the integrity of the nucleus, the protein levels, and sub-cellular location of nuclear envelope proteins and the capacity to differentiate to myogenic fibers. Given the fact that LMNA mutations are causally associated with several rare diseases, this work provides new and valuable information for a better understanding and future treatment of these diseases with no cure.
2. Materials and Methods
Details of resources used in the research, including antibodies, cell culture media, reagents, and software, are listed in Table 1.

Table 1.
Key resources used in this work.
2.1. Cell Lines and 3D Model
For the majority of our experiments, we used C2C12 myoblasts, which are myoblasts originally established from satellite cells derived from the thigh muscle of a female C3H murine donor following a crush injury [21]. C2C12 cells and selected clones with mutations in exon 4 of Lmna have been grown at 37 °C of temperature and 5% CO2 conditions. The culture medium of myoblasts was based on Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Three human myoblast cell lines, previously described [18], were used in this work. Two were controls isolated from 25 and 38 year old healthy individuals (C25CL48 and AB1079, respectively). A third myoblast cell line was isolated from a 3 year old L-CMD patient who carried a LMNA p.R249W mutation. Human myoblasts were grown at a 37 °C and 5% CO2 conditions, in 1 volume of medium 199 with four volumes of DMEM with 20% FBS and 1% penicillin/streptomycin, and supplemented with 25 µg/mL fetuin, 5 ng/mL human epidermal growth factor (hEGF), 0.5 ng/mL basic fibroblast growth factor (bFGF), 5 µg/mL insulin, and 0.2 µg/mL dexamethasone.
3D muscle models were fabricated by photo-molding technique, as we have described previously [22] (Figure S2). Gelatin-based polymer solution (see Appendix A) was mixed with a C2C12 cell suspension to reach the final cell density of 25 × 106 cell/mL. Briefly, a 6 µL drop of cell-laden polymer was placed on a functionalized coverslip (Appendix A), and a microstructured silicone (PDMS) stamp was pressed lightly on top, filling the microchannels with the solution. The sizes of PDMS stamp consisted of grooves of 100 µm, which were 100 µm in height with and ridges of 100 µm. The hydrogel was photo-crosslinked using a Ultraviolet Product (UVP) Crosslinker (Model CL-1000L, 365 nm, 40 W, from Analytik Jena U.S., Upland, CA, USA) by the exposure of 30 s under UV light. After carefully removing the stamp, the micro-structured cell-laden hydrogels were incubated for 1 day with culture medium. Due to high cell density in the hydrogel, growth medium was changed to differentiation medium, based in DMEM high glucose, supplemented with 2% horse serum and 1% penicillin/streptomycin, after 1 day to induce differentiation into myotubes.
2.2. Generation of Clones with Mutations in Lmna Exon 4 Using CRISPR/Cas9
All single guide RNAs (sgRNAs) were designed to target exon 4 of the LMNA gene using the Breaking Cas Design tool (https://bioinfogp.cnb.csic.es/tools/breakingcas/) [23]. Three sgRNAs for the Lmna exon 4 (sg10, sg11, and sg12; Table 2) were cloned in the pX459 vector. The C2C12 cells were transfected, using Lipofectamine 3000, with the pX459-sgRNA vector with the corresponding template (ssODN) in low (30 pmol) or high (300 pmol) doses. Templates included forms mutated only in the Protospacer Adjacent Motif (PAM) (mut) or mutated in the PAM and in the Protospacer (mut2) (Table 2). After 48 hours of transfection, puromycin (2 µg/mL) was added for 5 days. The selected cells were grown until a pool was obtained per condition. The clones were picked up after seeding at low density of the pools. The pools and clones were expanded to allow the complete analysis. For the control samples, we followed the same process but using an empty px459 vector and no templates.

Table 2.
DNA sequences used in this work.
2.3. DNA Sequencing and Bioinformatic Analysis
To identify mutations at Lmna exon 4, MiSeq DNA sequencing was carried out in the Genomic Unit of Instituto de Salud Carlos III. DNA was isolated from the different pools and clones. Next, Illumina adapters were added to the target region of the Cas9 nuclease by PCR using the DeepSeq-Fw and DeepSeq-Rv primers (Table 2).
The resulting product was used for a second PCR, where the specific indexes were added to every sample. Genomic reads in FASTQ format were analyzed using the CRISPResso platform. This software isolates the part of each read spanning the chosen region, highlights small insertions/deletions, and outputs a count of each regional sequence. We then analyzed the percentage of the sequences showing regional differences in control and with the different sgRNA-exon 4 of LMNA-transduced samples. All the pools and clones were also analyzed by Sanger sequencing of PCR products amplified using Lmna exon 4-specific primers Sanger-mLmna_Ex4_Fw and Sanger-mLmna_Ex4_Rv (Table 2). Sanger sequences were analyzed using the TIDE platform.
2.4. Western Blot Analysis
SDS-PAGE was performed using standard procedures. Briefly, cells were disrupted with RadioImmunoPrecipitation Assay (RIPA) buffer (150 mM NaCl, 1% Nonyl Phenoxypolyethoxylethanol (NP-40), 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl (pH 7.5), and protease and phosphatase inhibitors). The lysates were sonicated with the ultrasonic processor UPH100H (Hielscher, Teltow, Germany) (three pulses of 5 s at 100% amplitude) to allow dissociation of proteins. Samples were centrifuged at 14,000 rpm at 4 °C for 15 min. Protein concentration was quantified with the BCA system. Table 1 lists the references and dilutions of the primary and Horseradish Peroxidase (HRP)-labelled secondary antibodies used for Western blot analyses. Detection was carried out using an ECL western blotting system.
2.5. Immunofluorescence Microscopy, Nuclear Morphology Analyses, and Myogenic Differentiation Studies
For 2D cultures, cells plated onto coverslips were fixed with methanol for 5 min at −20 °C. Cells were incubated in blocking solution (0.1% bovine serum albumin (BSA), 1% FBS, and 1% horse serum in phosphate-buffered saline (PBS), in the case of SUN2; 1% BSA, 2.5% goat serum, 2.5% donkey serum, and 0.3% Triton X-100 in PBS, in the cases of lamin A/C, lamin B1, and emerin) for 30 min. Primary and secondary antibodies used for immunofluorescence detection are detailed in Table 1. Coverslips were mounted using Prolong Gold (ThermoFisher Scientific; Waltham, MA, USA) with DAPI, and images were captured using a Leica TCS SP5 confocal microscope (Leica, Wetzlar, Germany) and a 63× (HCX PL APO 63× 1.4 OIL).
For the 3D models, cells were fixed in a 10% formalin solution (Sigma-Aldrich, St. Louis, MI, USA). Then, hydrogels were washed with Tris-buffered saline (TBS), permeabilized with 0.1% Triton X-100 in TBS for 15 min, and blocked with a blocking buffer consisting of 0.3% Triton X-100 and 3% donkey serum in TBS for 2 h. Afterwards, samples were washed with TBS and incubated with anti-MYH7 in blocking buffer (O/N, 4 °C). Then, samples were washed with TBS and incubated with rhodamine phalloidin and goat anti-rabbit A488 in blocking buffer (O/N, 4 °C). After washing with TBS, nuclei were counterstained with 1 µM DAPI for 15 min.
Cells (5000 cells per well) were plated into a 96-well cell culture black microplate with a glass bottom (Greiner Bio-one, Kremsmünster, Austria). After 24 h, the cells were fixed with methanol for 5 min at −20 °C. The fixed cells were incubated with 2 µL/mL Hoechst 33324 in PBS for 10 min at 37 °C. Finally, nuclear morphology was evaluated by determining the circularity index using the Cytell Cell Imaging System (GE Healthcare Life Sciences, Marlborough, MA, USA). The circularity values were between 0 and 1, with 1 being the value of a perfect circle. For the nuclei morphology in the 3D muscle model, confocal microscopy images were analyzed using ImageJ software (National Institutes of Health and the Laboratory for Optical and Computational Instrumentation, Madison, WI, USA).
Myoblast differentiation was induced by growth medium exchange for the differentiation medium (DMEM with 2% horse serum and 1% of penicillin/streptomycin). The cells were differentiated for 5 days. The cells (1 × 105 cells per well) were fixed with methanol for 5 min at −20°C, permeabilized with 0.05% Triton X-100 in PBS for 5 min at room temperature, and washed with PBS for 5 min. Then, coverslips were blocked in blocking solution (15% FBS in PBS) for 30 min at room temperature. The cells were incubated with primary antibody against myosin heavy chain (MF20) for 1 h at room temperature. The cells were washed with PBS three times for 5 min, incubated with secondary antibody anti-mouse Ig for 45 min at room temperature and in the dark, and washed with PBS three times for 5 min. Coverslips were mounted using Prolong Gold (Thermo Fisher Scientific) with DAPI. Stained myotubes were visualized in a Leica DM4B microscope (Leica-Microsystems, Wetzlar, Germany) using the 10× (N PLAN 10×/0.25 DRY) and 20× objectives (HC PL FLUOTAR 20×/0.55 DRY).
2.6. Quantification and Statistical Analysis
The levels of Lamin A/C, Lamin B1, Emerin, SUN1, and SUN2 at the nucleus were analyzed using ImageJ software (National Institutes of Health). In brief, individual lines of constant length were plotted perpendicular to the nucleus and used to evaluate peak fluorescence intensity using the PlotProfile feature from ImageJ. Random individual plot profiles of conditions were generated and quantified at the nuclei using the PlotProfile feature from ImageJ. In every set of experiments, all immunofluorescence images were taken from single confocal acquisitions under the same exposure conditions.
All the graphs and statistical analyses were carried out with GraphPad 8.0 software (GraphPad, San Diego, CA, USA). Error bars represent standard error of the mean (SEM). For statistical analyses, the level of significance was determined by two-tailed, unpaired Mann–Whitney U test or Student’s t-test.
3. Results
3.1. Generation of Lmna Exon 4 Mutations in C2C12 Cells
In order to produce Lmna exon 4 mutants by CRISPR/Cas9 technology, we designed three sgRNAs to target the Cas9 endonuclease to this Lmna region (Figure 1A). Following the scheme depicted in Figure 1B and detailed in Section 2, we produced a total of 18 pools and 210 clones in which CRISPR/Cas9 activity (insertions and deletions, Indels) was measured by TIDE and deep sequence analyses (Table S1). It is interesting to note that each sgRNA induced a different Indel pattern (Figure S3). One single nucleotide deletion was the most frequent event when using sg12 (present in 77% of the clones), whereas a wider spectrum of deletions and insertions was observed for sg10 and sg11 guides. Interestingly, deletions of 3 and 6 nucleotides, which produce in-frame deletions mutants, were mainly observed in sg11-transfected cells (they were detected in 20% and 35% of the clones, respectively). Sequences with no Indels were detected in 3%, 5%, and 23% of the clones transfected with s10, sg11, and sg12 guides, respectively.
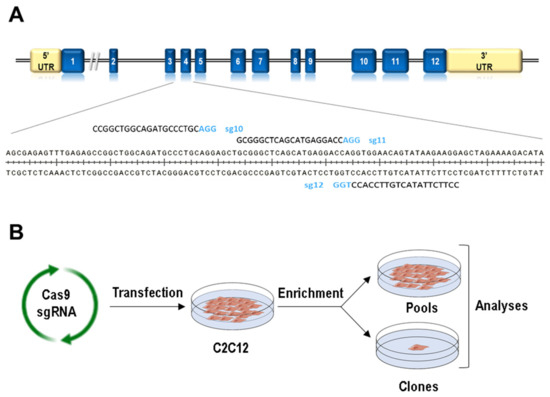
Figure 1.
Generation of Lamin A/C gene (Lmna) exon 4 mutations using CRISPR/Cas technology. (A) Scheme for exon 4 Lmna sgRNA localization. (B) Work-flow followed for the generation of C2C12 cells carrying mutations in Lmna exon 4.
We also aimed at introducing the R249W mutation, one of the human variants associated with a severe type of laminopathy, LMNA-associated congenital muscular dystrophy. In all the transfections, we incorporated a donor template including the mutant nucleotide. Although five clones were positive for the R249W mutation, only one (g11H-D6; Table 3) was free of additional nucleotide changes and, therefore, allowed the expression of this mutant form.

Table 3.
Molecular properties of the selected clones.
A total of 113 clones, named as “null”, only carried frameshift mutations that produced truncated proteins due to premature stop codons. Sixteen clones, classified as “delta”, were characterized by the only presence of triplet deletions compatible with the formation of Lamin A/C proteins with deletions of 1, 7, 14, 15, or 57 amino acids (Figure S1). Ten clones that were transfected with Cas9 expressing vectors with no sgRNAs showed no mutations in Lmna exon 4 and were used and named as “controls”. Table 3 lists the control, null, delta, and R249W clones that were selected for a detailed study on the effects caused by the mutations in Lmna exon 4.
3.2. Components of the Nuclear Lamina Were Abnormally Distributed in Lmna Exon 4 Mutant Myoblasts, Lamin A/C Abnormal Expression, and Subcellular Localization in Lmna Exon 4 Mutants
We first analyzed the total protein levels and nuclear sub-cellular localization of Lamin A/C. As shown in Figure 2A, all the Lmna exon 4 mutants were characterized by a significant reduction in the amount of both Lamin A and C proteins. This feature was accompanied by an abnormal subcellular localization of Lamin A/C in all delta and R249W mutants (Figure 2B). Whereas control and R249W myoblasts showed a predominant Lamin A/C signal in the nuclear periphery, delta clones showed no differences in Lamin A/C intensity between perinuclear and nucleoplasmic regions. Moreover, in the different delta clones, we were able to detect a wide spectrum of abnormalities regarding Lamin A/C staining including honeycomb-like patterns, asymmetric distribution, capping, and bright foci (Figure S4). Although Lamin A/C subcellular localization in R249W cells followed the same pattern found in control myoblasts, a detailed analysis showed lower fluorescence intensity of Lamin A/C at the nuclear periphery when compared with the control clones (Figure 2B).
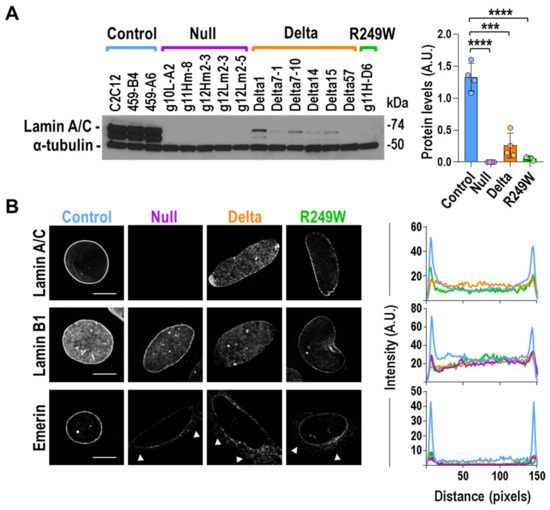
Figure 2.
Components of the nuclear lamin were abnormally distributed in Lmna exon 4 mutant myoblasts. (A) Lamin A and C levels were reduced in Lmna exon 4 mutants. Quantification of Lamin A and C protein levels. Left pictures show one of the three independent biological replicates analyzed. The other replicates are included in Figure S5. Quantification of Lamin A/C signal in the three independent analyses, by mutant type, is shown on the right. Bars are mean ± SEM. Two-tailed, unpaired Student’s t-test: **** p < 0.0001, *** p = 0.003, n = 4. (B) Mouse myoblasts were immunostained for Lamin A/C, Lamin B1, and Emerin. Representative single confocal images from nuclei are shown for each Lmna group. Extra-nuclear signal was observed for Emerin in null, delta, and R249W mutants (arrows). Scale bar = 8 μm. Right plots show average fluorescence intensity across nuclei by mutant type (control (blue): three clones; null (purple): five clones; delta (orange): six clones; R249W (green): one clone; n = 5 nuclei per clone). Peaks at both ends of the plot represent peripheral staining. Representative images from other clones are shown in Figure S4 (Lamin A/C), Figure S6 (Lamin B1), and Figure S7 (Emerin).
To investigate the consequences of the Lamin A/C mislocalization and low expression in Lmna exon 4 mutants, we studied, using single confocal imaging, the sub-cellular localization of Lamin B1 and Emerin, two main inner nuclear membrane proteins. In control clones, Lamin B1 accumulated at the nuclear periphery and was also detected, to a lesser extent, at the nucleoplasm. Lamin B1 distribution was slightly altered in Lmna exon 4 mutants, which did not show a sharp contrast between the Lamin B1 signal detected in Lamin area and nucleoplasm (Figure 2B). A significant effect was also expected for Emerin because the localization of this protein is dependent on Lamin A/C [24]. Indeed, Emerin sub-cellular distribution was affected in null, delta, and R249W clones (Figure 2B). In all the cases, Emerin extranuclear signal was compatible with the endoplasmic reticulum pattern previously reported for Lmna null and mutant cells [25,26,27].
Our results indicated that a reduction and abnormal sub-cellular localization of the main lamina components—Lamin A/C, Lamin B1, and Emerin—was common to all the Lmna exon 4 mutants analyzed.
3.3. Nuclear Morphology Abnormalities Were Common for All the Clones with Mutations in Lmna Exon 4
The abnormalities described for all the Lmna exon 4 mutants were compatible with a defective lamin that might affect the nuclear membrane integrity. To test this hypothesis, we investigated the nuclear morphology of all the clones analyzed in this work. Whereas nuclei of controls are circular or ovoid, mutant clones are characterized by irregular nuclei with multiple herniations and, more frequently, by elongated nuclei. To quantify these differences in terms of nuclear morphology, we calculated the circularity index of the nuclei of myoblasts asynchronously growing in 2D cultures. As expected, all the mutants showed a significant reduction in the circularity index of their nuclei (Figure 3A).
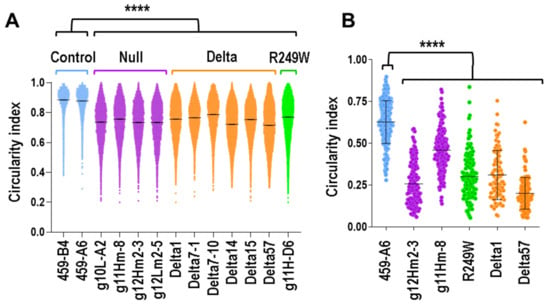
Figure 3.
Nuclear morphology was altered in mouse myoblasts carrying Lmna exon 4 mutations. (A) Circularity index was calculated in nuclei from clones asynchronously growing in 2D cultures (**** p < 0.0001; n > 5000 nuclei per clone). (B) Plot shows the circularity index from clones encapsulated in a 3D pattern. Data were compared using an unpaired Mann–Whitney U test (**** p < 0.0001). Blue, purple, green, and orange colors indicate control, null, R249W, and delta clones, respectively.
To study nuclear morphology in a more physiologic condition, we encapsulated the myoblasts in a mimetic 3D architecture that recreated a native skeletal muscle (Figure S2A). Under this 3D cellular organization, all type of clones formed highly elongated and aligned fibers (Figure S2B). For both control and mutant clones, the circularity was reduced when compared to the values obtained in 2D experiments (Figure 3B). This effect was probably caused by the geometric constriction of the pattern and the compliant nature of the hydrogel used to create the 3D structure. Importantly, circularity was significantly reduced in mutant clones when they were compared to controls (Figure 3B).
These results demonstrated that mutations in Lmna exon 4 negatively affect nuclear morphology in myoblasts growing in both 3D and 2D conditions.
3.4. Abnormal Myogenic Differentiation Was Common to All Myoblasts Carrying Lmna Exon 4 Mutations
LMNA mutations have been associated not only with structural abnormalities, such as the nuclear integrity defects described above, but also with functional deficiencies, including differentiation capacity to myogenic fibers [28,29]. To test if this was also the case for the mutants generated in this work, we induced myogenic differentiation in all the myoblasts under study. In a first set of experiments, we studied the differentiation capacity of control and mutant clones growing in 2D conditions. As shown in Figure 4A, 5 days post-induction of differentiation, control myoblasts formed myogenic fibers that were positive for myosin heavy chain (MHC). However, in null, R249W and the majority of the delta clones, we were not able to detect either myogenic fibers or MHC expression. Interestingly, delta57 clone, which is missing the whole Lmna exon 4, showed a moderate formation of fibers and MHC expression.
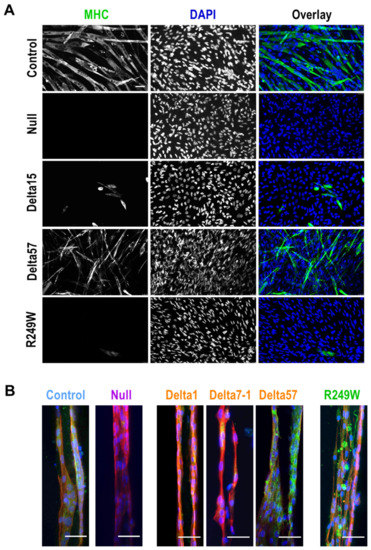
Figure 4.
Myogenic differentiation was impaired in mouse myoblasts with mutations in Lmna exon 4. (A) Myoblasts growing in 2D conditions were induced to differentiate to myogenic fiber as described in Section 2. Representative pictures show myosin heavy chain (MHC, green) and nuclei (DAPI, blue). Scale bar = 60 µm. (B) Myogenic fiber formation was also affected in 3D models. Representative pictures show, at 7 days post-differentiation, myosin heavy chain 7 (green), nuclei (blue), and phalloidin staining (red). Control and null samples correspond to 459-A6 and g12Lm2-3 clones. Scale bar = 50 µm. Single color channels for this panel are shown in Figure S8.
Similar results were obtained from myoblasts growing in 3D structures (Figure 4B). In this case, MHC was only detected in control clones as well as in delta57 and R249W mutants. Importantly, although R249W cells were positive for MHC, they showed an abnormal distribution of this protein that could indicate the formation of abnormal myogenic fibers.
Our results indicate that Lmna exon 4 mutations significantly affect the capacity of C2C12 cells to differentiate to myogenic fibers.
3.5. Expression of Lmna Exon 4 Mutants Was Associated with Abnormal Subcellular Localization of SUN1 and SUN2
In an effort to understand the mechanisms underlying the nuclear morphology and myogenic differentiation defects that characterize the Lmna exon 4 mutants, we studied the status of SUN1 and SUN2. These proteins, together with Lamin A/C, Emerin, and Nesprins, among others, constitute the LINC complex, a molecular link that connects the nucleus to the cyto-skeleton, and have been associated with muscular dystrophies [30]. The total protein levels of SUN1 and SUN2 were not altered in any of the mutant types in null, delta, or R249W (Figure 5A). However, their subcellular distribution was significantly affected in all the mutants when compared with controls (Figure 5B). Whereas SUN1 and SUN2 accumulated in the nuclear envelope of control myoblasts, they were barely detected in the same region of Lmna exon 4 mutants.
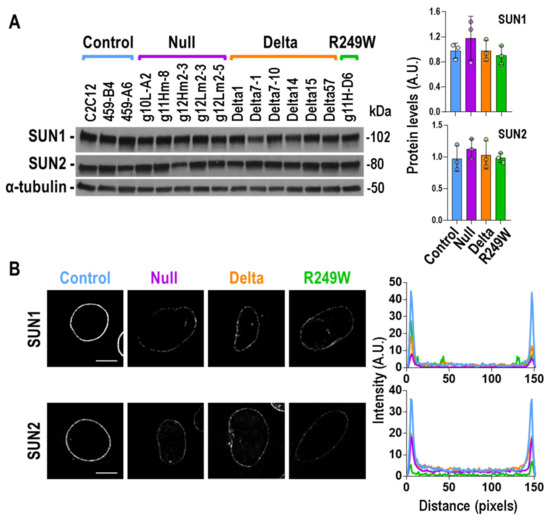
Figure 5.
SUN1 and SUN2 protein levels and sub-cellular localization in Lmna exon 4 mutant myoblasts. (A) Total amount of SUN1 and SUN2 proteins was determined by Western blot. One of the three biological replicates is shown. The rest of the Western blots are shown in Figure S5. Quantification, by mutant type, of SUN1 and SUN2 signals for the three independent analyses is shown in the graphs on the right. Bars are mean ± SEM, n = 3. No significant differences were detected when mutants were compared with controls using a two-tailed, unpaired Student’s t-test. Representative, single, confocal, images of SUN1 (B) and SUN2 (C) detected by immunostaining are shown. A decreased nuclear envelope signal was observed in null, delta, and R249W mutants. Scale bar = 8 μm. Right plots show average fluorescence intensity across nuclei by mutant type (control (blue): three clones; null (purple): five clones; delta (orange): six clones; R249W (green): one clone; n = 5 nuclei per clone). Peaks at both ends of the plot represent peripheral staining. Representative images for SUN1 and SUN2 in all the clones are shown in Figures S9 and S10, respectively.
These results, together with the ones obtained for the localization of Emerin and Lamin proteins, indicated that the defects induced at the nuclear envelope by the Lmna exon 4 mutations significantly impact the subcellular localization of LINC complex components.
3.6. MAPK Signaling Was Not Altered in Lmna Exon 4 Mutant Myoblasts
Thanks to the analysis of the LmnaH222P mouse model, a connection between LMNA mutations and signaling pathways has been discovered [31]. Specifically, enhanced phosphorylation of ERK1/2 and AKT has been reported in the heart of LmnaH222P mice [10,32]. Therefore, we decided to study the status of these key signaling molecules in Lmna exon 4 mutants. Although the activation of ERK1/2, estimated by the quantification of phosphorylated-ERK1/2, was very heterogeneous for inter and intra clones (Figure S5), no significant differences were observed when control, null, delta, and R249W groups were compared (Figure 6A). Likewise, there were no significant differences in AKT activation (Figure 6B).
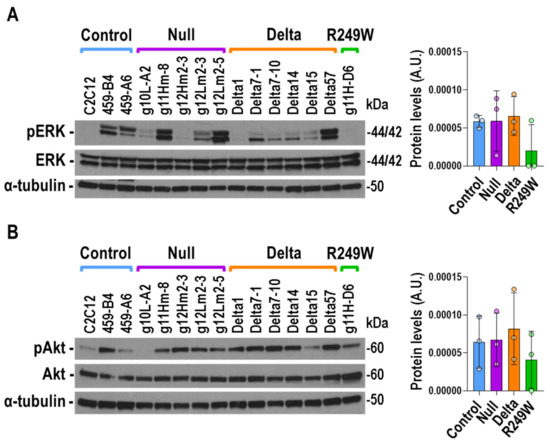
Figure 6.
No differences in ERK1/2 and AKT activation were detected in Lmna exon 4 mutant myoblasts. (A) Representative immunoblot for the detection of phosphorylated (pERK) and total (ERK) ERK1/2 in Lmna exon 4 mutants. (B) Representative immunoblot for the detection of phosphorylated (pAKT) and total (AKT) AKT. Detection of α-tubulin was used in all the cases as loading control. Data in bar graphs are mean ± SEM of all the clones of each type, n = 3. No significant differences were detected when mutants were compared with controls using a two-tailed, unpaired Student’s t-test. The remainder of the Western blots analyzed in (A) and (B) are shown in Figure S5.
Therefore, the Lmna exon 4 mutations studied here were not characterized by homogenous and significant changes in the activation of the signaling proteins ERK1/2 and AKT.
3.7. High Levels of DNA Damage Were Associated with R249W Mutation
We also wanted to study the effect of Lmna exon 4 mutations at the DNA level, as progeroid and striated muscle laminopathies have been associated with high levels of DNA damage [19,33,34,35]. Quantification of DNA damage by detection of γH2AX (phosphor-H2A.X Variant Histone) in asynchronously growing myoblasts revealed elevated levels of DNA damage in the R249W clone, whereas no signal was detected in the rest of mutants (null and delta) or controls (Figure 7A). To further analyze the connection between the R249W mutation and DNA damage, we also explored it in human myoblasts carrying a LMNA p.R249W mutation causing L-CMD. Interestingly, the human R249W mutant showed a clear increase in γH2AX levels when compared with the controls (Figure 7B).
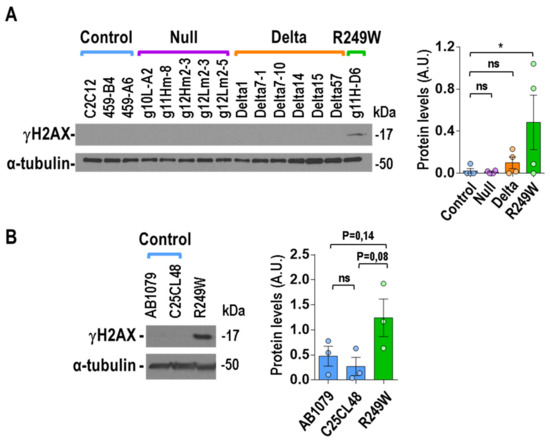
Figure 7.
High levels of DNA damage were specifically associated with LMNA R249W mutation. (A) Representative immunoblot showing γH2AX levels in Lmna exon 4 mutant myoblasts. (B) Immunoblot showing γH2AX levels in human myoblasts from two controls and one patient carrying the p.R249W mutation. Detection of α-tubulin was used in all the cases as loading control. Data in bar graphs are mean ± SEM, n = 3. In (A) and (B), data were compared using a two-tailed, unpaired Student’s t-test. * p < 0.05. The remainder of the Western blots analyzed in (A) and (B) are shown in Figure S5.
These results indicate that high levels of DNA damage are a specific feature of LMNA p.R249W myoblasts.
4. Discussion
4.1. Comparison of Lmna Exon 4 Mutants Generated in This Study with Previously Reported LMNA Mutants
In this study, using CRISPR/Cas9 technology, we analyzed a collection of Lmna exon 4 mutants in C2C12 mouse myoblasts. Three different types of mutants were generated: null mutants, due to frameshift mutations; delta mutants, caused by in-frame deletions; and an already known R249W mutant. Interestingly, none of the delta and null mutants generated in this work have been previously reported in patients. Upon studying a number of molecular, cellular, and functional features, we can conclude that the subcellular localization of key members of the LINC complex and nuclear envelope, including Lamin A/C, Lamin B1, Emerin, SUN1, and SUN2, as well as the nuclear membrane integrity and myogenic differentiation capacity, were negatively affected.
The comparison of the features of the Lmna mutants described here with those previously reported by others resulted in coincidences and differences. For instance, the Emerin extranuclear localization, compatible with endoplasmic reticulum, detected in null, delta, and R249W clones, is similar to the previously reported for Lmna null, LmnaL530P/L530P, LmnadK32, and LmnaN195K mouse myoblasts, as well as LMNA p.Y259X patient cells [15,25,26,27,36]. Contradictory results have been described for Lamin B1 localization. It has been reported that the loss of A-type lamins allows the assembly of B-type lamins [25]. In addition, more recently, LMNB1 localization defects have been shown in induced pluripotent stem cells carrying LMNA mutations [17], which is in line with the results we obtained in the present work. On the other hand, the Lmna null, delta, and R249W analyzed in this work showed a nuclear, although low intensity, signal for SUN1 and SUN2 at the nuclear envelope. A regulation by Lamin A of SUN2 localization has been previously reported in mouse fibroblasts [37]. In addition, SUN2 nuclear distribution has been dramatically altered in synaptic nuclei of Lmna−/− and LmnaH222P/H222P muscle [38]. On the other hand, the weak nuclear envelope localization characteristic for Lmna exon 4 mutant C2C12 cells differs from initial reports showing that localization of SUN1 does not depend on A-type lamins in mouse fibroblasts and HeLa cells [38,39,40]. However, other works in Lmna null or LmnaL530P/L530P mouse fibroblasts showed an extranuclear signal or loss of nuclear envelope signal [27,41,42]. The differences observed between these reports and the present work might be due to the different cell system used—fibroblasts versus myoblasts. Importantly, it has been reported that LMNA mutations causing Emery–Dreifuss muscular dystrophy are associated with low SUN1 levels in human myoblasts [43], an outcome similar to the one obtained in the Lmna exon 4 mutants generated and characterized in this work. In summary, different conclusions might be obtained for the SUN1–Lamin A/C connection that could be a consequence of different cellular or molecular scenarios. Further studies are needed to better clarify the role that Lamin A/C proteins play on SUN1 localization and function.
LMNA p.H222P, one of the most studied LMNA mutations, is also located in exon 4. However, some of its features differ from the data obtained with the exon 4 mutants analyzed here. For instance, the alterations of MAPK and AKT signaling pathways described in the LmnaH222P–H222P model [10,32] were not detected in the Lmna exon 4 mutants analyzed in this work. Moreover, Lmna knockdown in C2C12 cells has been previously associated with a significant increase in ERK1/2 activation [44], an abnormality that is not found in the null C2C12 clones studied here (Figure 6A). To note, the siRNA (small interfering Ribonucleic Acid) used in the former did not fully inhibit Lamin A/C expression, whereas the use CRISPR/Cas9 induced the complete elimination of Lamin A/C expression (Figure 2A). In our study, we observed a high heterogeneity both inter and intra clones for p-ERK and p-AKT levels. Because this was not the case for other proteins analyzed here (Figure S5), we conclude that none of these two pathways are homogeneously altered in the Lmna exon 4 mutants studied in this work, whereas Lmna p.H222P mutation is specifically associated with an increased activation of both pathways. Another possible explanation, would be the fact that the majority of the studies for Lmna p.H222P have been carried out using cardiomyocytes from a LmnaH222P knockin mouse model, whereas the cells used in the present study were mouse myoblasts. The detailed characterization of the same cell type from a mouse LmnaR249W model (currently under study) will help us to discern the correct explanation of these differences.
4.2. Mechanistic and Functional Defects Associated with Lmna Exon 4 Mutations
Myogenic differentiation is the most important function of myoblasts. It has been previously shown that the expression of Lamin A mutants associated with different laminopathies inhibits the in vitro differentiation of C2C12 myoblasts [28,29]. Abnormal myoblast differentiation has also been reported in LmnadK32 cells [15]. Moreover, Lmna null satellite cells are characterized by a defective myogenic differentiation [45]. The same defects were observed for the majority of the null, delta, and R249W mutants analyzed in this work, which confirmed the fact that mutations and/or downregulation of Lamin A/C are associated with an impairment in myogenic differentiation that might explain a significant part of the pathological phenotype of laminopathies. However, other scenarios cannot be ruled out. For instance, a premature differentiation associated to SMAD6 (SMAD Family Member 6) overexpression and BMP4 (Bone Morphogenetic Protein 4) downregulation has been reported in LmnaΔ8–11 myoblasts [46]. Although this seems not to be the case for Lmna exon 4 mutant myoblasts (data not shown), further studies will help to fully understand the differentiation problems induced by these Lmna mutations.
Other potential mechanisms underlying the abnormal differentiation of the Lmna exon 4 mutants is an abnormal function of the LINC complex. The deficient subcellular localization observed for many of the LINC complex components (Emerin, Lamin A/C, Lamin B1, and SUN1/SUN2) in Lmna exon 4 mutant myoblasts could be the cause of the defects detected in nuclear membrane integrity and myogenic differentiation. Interestingly, the LINC complex, a molecular link between the nucleo- and cyto-skeleton, has been associated with muscular dystrophies [30]. Furthermore, the alteration of the nuclear transport due to a malfunction of the nuclear pores has already been associated with LINC complexes defects [47,48], and might be the functional cause of the pathological phenotype associated with Lmna exon 4 mutants. However, additional studies are needed to confirm the causal relationship between Lmna exon 4 mutations and LINC complex in laminopathies.
Other mechanisms and pathways previously associated with muscular dystrophies and cardiomyopathies might be associated with the Lmna exon 4 mutations. For instance, an altered WNT (Wingless and Int-1)/β-catenin signaling has been found in hearts of LmnaH222P mice [10,11], and pharmacological activation of this pathway improves heart dysfunction in this mouse model [49]. On the other hand, disruption of the nucleocytoplasmic Ran gradient and consequent nuclear protein import defects have been reported in cells carrying LMNA mutations [5,50]. Moreover, alterations of the nuclear morphology, such as the ones associated with Lmna exon 4 mutants, are associated with an abnormal distribution of the chromatin and, subsequently, changes in the epigenetics marks [51,52]. Whether all these pathways are also causally connected with the Lmna exon 4 mutations we have described herein remains to be determined and will be part of further studies.
4.3. DNA Damage: A Feature Specific for the R249W Mutation?
The nuclear lamina is a critical structure for the stability and proper nuclear distribution of DNA [53]. A weak nuclear envelope affects DNA integrity, inducing the accumulation of DNA damage, a feature linked to LMNA-associated diseases [19,54,55,56]. The majority of the mouse myoblasts studied in this work did not show elevated levels of DNA damage, which is consistent with the fact that we have explored this property only in undifferentiated myoblasts, and increased levels DNA damage are mainly observed in differentiated cells and tissues of HGPS and LMNA-related, muscular dystrophies [19,55]. Surprisingly, even in this undifferentiated myoblast state, the Lmna p.R249W mutant under study showed significantly elevated levels of DNA damage. The fact that this feature was also observed in human LMNA p.R249W myoblasts (Figure 7) and in muscle biopsies from a 1 year old patient carrying a LMNA p.R249W mutation [19I] points to a specific association with DNA integrity and/or repair mechanisms. Further studies will help to validate this connection and explore therapeutic opportunities related to DNA damage and repair.
4.4. CRISPR/Cas9 Activity and Gene Therapy Implications
The delta and R249W clones analyzed in this work showed a significant reduction in the levels of Lamin A/C (Figure 2). This was probably due to the CRISPR-mediated deletion of one or more Lmna alleles (C2C12 cells used in this work were hypertriploid), as mRNA expression was significantly reduced in all of them (data not shown). Therefore, the phenotypes observed in the Lmna exon 4 mutants could have been due not only to the mutations generated in Lmna, but also to a defective expression of lamin proteins. It is important to note that Lmna haploinsufficiency is associated with dilated cardiomyopathy [57]. On the other hand, all the clones analyzed in this work lost their wild type alleles and showed a single mutant allele. This genomic scenario strongly differs from the predominantly autosomal dominant scenario of LMNA-associated diseases in which missense mutations could lead to Lamin or Prelamin A accumulation. To note, a completely different situation is found in the null mutations generated by CRISPR/Cas9. All this evidence has important implications for the therapeutic potential that CRISPR technology could have for the treatment of laminopathies and the potential use as disease models of the clones we generated.
One of the most interesting results we obtained in this work was the high percentage of delta clones isolated upon CRISPR/Cas9 activity on Lmna exon 4 (Figure S1). This indicated that these types of in-frame mutations might have some selective advantage over the rest of the Indels generated by CRISPR/Cas9 activity. Moreover, one of these clones, delta57, is characterized by the expression of a Lmna exon 4 null gene. Interestingly, although delta57 shows nuclear morphology and LINC complex abnormalities, it does retain some myogenic differentiation capacity. This could indicate that an exon 4 skipping strategy for the elimination of mutations located in this exon could be compatible with a partially functional myoblast. As far as we know, in LMNA-associated disease, successful exon skipping has only been shown for exon 5 in human cells [20]. The potential of this therapeutic strategy needs further experimentation in appropriate in vivo models.
In conclusion, in this work, we characterized in detail the consequences associated with mutations in Lmna exon 4 in myoblasts. This new knowledge has opened the possibility of exploring new therapeutic approaches based on the DNA damage specifically induced by the Lmna R249W mutation and the use of exon skipping strategies for the elimination of Lmna exon 4 mutations.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/9/5/1286/s1. Table S1: Sequencing information of Lmna exon 4 mutant clones and pools. Figure S1: Scheme of the Lmna gene and protein structure and the mutants generated in this work. Figure S2: Generation of 3D skeletal muscle models. Figure S3: Deletions/insertions detected in C2C12 pools and clones. Figure S4: Lamin A-C subcellular localization and expression in delta mutants. Figure S5: All the Western blots analyzed in this work for Lamin A/C, ERK1/2, AKT, SUN1, SUN2, and γH2AX. Figure S6: Lamin B1 subcellular localization in Lmna exon 4 mutants. Figure S7: Emerin subcellular localization in Lmna exon 4 mutants. Figure S8: Single color channels for Figure 5b panel. Figure S9: SUN1 sub-cellular localization in Lmna exon 4 mutants. Figure S10: SUN2 sub-cellular localization in Lmna exon 4 mutants.
Author Contributions
Conceptualization, I.P.d.C.; Methodology, D.G.-D., C.E., F.d.M., A.G.C, J.R.-A., I.P.d.C.; Investigation, D.G.-D., C.E., F.d.M., A.G.C., B.V.-M., A.M., S.A.-Q., M.A.R.-M.; Resources, A.T.B., G.B.; Writing—Original Draft Preparation, D.G.-D., C.E., I.P.d.C.; Writing—Review and Editing, D.G.-D., C.E., F.d.M., A.G.C, B.V.-M., A.M., S.A.-Q., A.B., G.B., J.R.-A, M.A.R.-M, I.P.d.C.; Supervision, I.P.d.C.; Project Administration, I.P.d.C.; Funding Acquisition, F.d.M., I.P.d.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Ministerio de Ciencia e Innovación (Acción estratégica en Salud intramural PI16III/00017-TPY1348/16) and by Fundación Andrés Marcio, niños contra la laminopatía (TPY-259/19). F.M. was funded by a Miguel Servet II contract from Instituto de Salud Carlos III (ISCIII).
Acknowledgments
We thank all the members of the Human Genetics Area of the Instituto de Investigación de Enfermedades Raras (IIER) for their continuous support and help. We are deeply grateful to Beatriz Martínez (IIER) for the logistic and scientific support at the beginning of the project. The authors thank Óscar Zaragoza for confocal microscopy assistance. We are very grateful to Eric Schirmer and. Colin Stewart for kindly providing SUN2 and SUN1 antibodies.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Appendix A
Synthesis of Prepolymer Precursors
Gelatin methacryloyl (GelMA) was synthesized with a 40% degree of methacrylation, as previously described (García-Lizarribar, Fernández-Garibay et al. 2018). The material was then dialyzed in Milli Q water with 6–8 kDa molecular weight cut-off (MWCO) membranes (Spectrumlabs, Pireas, Greece, 08-700-142) for 4 days. Sodium carboxymethylcellulose (CMC) (Sigma-Aldrich, 419273) was methacrylated at a maximum degree of methacrylation, as previously described [58]. The methacrylation reaction was performed by mixing a solution of 1% (w/v) of the polymer in 50 mM MES (2-(N-morpholino)ethanesulfonic acid) buffer at pH 6.5 with 20 mM EDC (1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride), 10 mM N-hydroxysuccinimide (Sigma-Aldrich, 130672), and 10 mM 2 aminoethylmethacrylate (Sigma-Aldrich, 479659). The reaction was stopped after 24 h with the addition of acetone (Panreac, 161007, Barcelona, Spain) and filtered using a vacuum flask. The precipitate was dissolved in 10 mM PBS and dialyzed in Milli Q water with 3.5 kDa MWCO membranes (Thermofisher, 68035). Finally, the solutions of methacrylated polymers (GelMA and CMCMA) were lyophilized and stored at −20°C.
Preparation of Prepolymer Solutions
A mixture of the prepolymer precursors (GelMA and CMCMA) was diluted in growth medium, containing the photoinitiator lithium phenyl(2,4,6-trimethylbenzoyl) phosphinate (LAP; TCI EUROPE N.V, L0290), at 65 °C for 3 h to obtain a homogeneous solution. The concentrations of GelMA, CMCMA, and LAP were fixed to obtain final concentrations of 5%, 1%, and 0.1% (w/v), respectively.
References
- De Leeuw, R.; Gruenbaum, Y.; Medalia, O. Nuclear Lamins: Thin Filaments with Major Functions. Trends Cell Biol. 2018, 28, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.L.; Zullo, J.M.; Bertolino, E.; Singh, H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 2008, 452, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Dechat, T.; Pfleghaar, K.; Sengupta, K.; Shimi, T.; Shumaker, D.K.; Solimando, L.; Goldman, R.D. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008, 22, 832–853. [Google Scholar] [CrossRef] [PubMed]
- Swift, J.; Discher, D.E. The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J. Cell Sci. 2014, 127, 3005–3015. [Google Scholar] [CrossRef]
- Busch, A.; Kiel, T.; Heupel, W.-M.; Wehnert, M.; Hübner, S. Nuclear protein import is reduced in cells expressing nuclear envelopathy-causing lamin A mutants. Exp. Cell Res. 2009, 315, 2373–2385. [Google Scholar] [CrossRef]
- Worman, H.J. Nuclear lamins and laminopathies. J. Pathol. 2012, 226, 316–325. [Google Scholar] [CrossRef]
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear Intermediate Filament Proteins with Fundamental Functions in Nuclear Mechanics and Genome Regulation. Annu. Rev. Biochem. 2015, 84, 131–164. [Google Scholar] [CrossRef]
- Cenni, V.; Capanni, C.; Mattioli, E.; Columbaro, M.; Wehnert, M.; Ortolani, M.; Fini, M.; Novelli, G.; Bertacchini, J.; Maraldi, N.M.; et al. Rapamycin treatment of Mandibuloacral dysplasia cells rescues localization of chromatin-associated proteins and cell cycle dynamics. Aging 2014, 6, 755–770. [Google Scholar] [CrossRef]
- Pellegrini, C.; Columbaro, M.; Capanni, C.; D’Apice, M.R.; Cavallo, C.; Murdocca, M.; Lattanzi, G.; Squarzoni, S. All-trans retinoic acid and rapamycin normalize Hutchinson Gilford progeria fibroblast phenotype. Oncotarget 2015, 6, 29914–29928. [Google Scholar] [CrossRef]
- Muchir, A.; Pavlidis, P.; Decostre, V.; Herron, A.J.; Arimura, T.; Bonne, G.; Worman, H.J. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J. Clin. Invest. 2007, 117, 1282–1293. [Google Scholar] [CrossRef]
- Muchir, A.; Wu, W.; Choi, J.C.; Iwata, S.; Morrow, J.; Homma, S.; Worman, H.J. Abnormal p38 mitogen-activated protein kinase signaling in dilated cardiomyopathy caused by lamin A/C gene mutation. Hum. Mol. Genet. 2012, 21, 4325–4333. [Google Scholar] [CrossRef] [PubMed]
- Larrieu, D.; Britton, S.; Demir, M.; Rodriguez, R.; Jackson, S.P. Chemical Inhibition of NAT10 Corrects Defects of Laminopathic Cells. Science 2014, 344, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Beyret, E.; Liao, H.-K.; Yamamoto, M.; Hernandez-Benitez, R.; Fu, Y.; Erikson, G.; Reddy, P.; Izpisua Belmonte, J.C. Single-dose CRISPR–Cas9 therapy extends lifespan of mice with Hutchinson–Gilford progeria syndrome. Nat. Med. 2019, 25, 419–422. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Fernández, O.; Osorio, F.G.; Quesada, V.; Rodríguez, F.; Basso, S.; Maeso, D.; Rolas, L.; Barkaway, A.; Nourshargh, S.; Folgueras, A.R.; et al. Development of a CRISPR/Cas9-based therapy for Hutchinson–Gilford progeria syndrome. Nat. Med. 2019, 25, 423–426. [Google Scholar] [CrossRef]
- Bertrand, A.T.; Brull, A.; Azibani, F.; Benarroch, L.; Chikhaoui, K.; Stewart, C.L.; Medalia, O.; Ben Yaou, R.; Bonne, G. Lamin A/C Assembly Defects in LMNA-Congenital Muscular Dystrophy Is Responsible for the Increased Severity of the Disease Compared with Emery-Dreifuss Muscular Dystrophy. Cells 2020, 9, 844. [Google Scholar] [CrossRef]
- Scharner, J.; Brown, C.A.; Bower, M.; Iannaccone, S.T.; Khatri, I.A.; Escolar, D.; Gordon, E.; Felice, K.; Crowe, C.A.; Grosmann, C.; et al. Novel LMNA mutations in patients with Emery-Dreifuss muscular dystrophy and functional characterization of four LMNA mutations. Hum. Mutat. 2011, 32, 152–167. [Google Scholar] [CrossRef]
- Steele-Stallard, H.B.; Pinton, L.; Sarcar, S.; Ozdemir, T.; Maffioletti, S.M.; Zammit, P.S.; Tedesco, F.S. Modeling Skeletal Muscle Laminopathies Using Human Induced Pluripotent Stem Cells Carrying Pathogenic LMNA Mutations. Front. Physiol. 2018, 9, 1332. [Google Scholar] [CrossRef]
- Bertrand, A.T.; Ziaei, S.; Ehret, C.; Duchemin, H.; Mamchaoui, K.; Bigot, A.; Mayer, M.; Quijano-Roy, S.; Desguerre, I.; Lainé, J.; et al. Cellular microenvironments reveal defective mechanosensing responses and elevated YAP signaling in LMNA-mutated muscle precursors. J. Cell Sci. 2014, 127, 2873–2884. [Google Scholar] [CrossRef]
- Earle, A.J.; Kirby, T.J.; Fedorchak, G.R.; Isermann, P.; Patel, J.; Iruvanti, S.; Moore, S.A.; Bonne, G.; Wallrath, L.L.; Lammerding, J. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat. Mater. 2019, 19, 464–473. [Google Scholar] [CrossRef]
- Scharner, J.; Figeac, N.; Ellis, J.A.; Zammit, P.S. Ameliorating pathogenesis by removing an exon containing a missense mutation: A potential exon-skipping therapy for laminopathies. Gene Ther. 2015, 22, 503–515. [Google Scholar] [CrossRef]
- Yaffe, D.; Saxel, O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 1977, 270, 725–727. [Google Scholar] [CrossRef] [PubMed]
- Ortega, M.A.; Fernández-Garibay, X.; Castaño, A.G.; De Chiara, F.; Hernández-Albors, A.; Balaguer-Trias, J.; Ramón-Azcón, J. Muscle-on-a-chip with an on-site multiplexed biosensing system for in situ monitoring of secreted IL-6 and TNF-α. Lab. Chip 2019, 19, 2568–2580. [Google Scholar] [CrossRef] [PubMed]
- Oliveros, J.C.; Franch, M.; Tabas-Madrid, D.; San-León, D.; Montoliu, L.; Cubas, P.; Pazos, F. Breaking-Cas—interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 2016, 44, W267–W271. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, A.; Alvarez-Reyes, M.; Bridger, J.M.; Broers, J.L.; Ramaekers, F.C.; Wehnert, M.; Morris, G.E.; Whitfield, W.G.F.; Hutchison, C.J. Both emerin and lamin C depend on lamin A for localization at the nuclear envelope. J. Cell Sci. 2001, 114, 2577–2590. [Google Scholar]
- Sullivan, T.; Escalante-Alcalde, D.; Bhatt, H.; Anver, M.; Bhat, N.; Nagashima, K.; Stewart, C.L.; Burke, B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999, 147, 913–920. [Google Scholar] [CrossRef]
- Muchir, A.; van Engelen, B.G.; Lammens, M.; Mislow, J.M.; McNally, E.; Schwartz, K.; Bonne, G. Nuclear envelope alterations in fibroblasts from LGMD1B patients carrying nonsense Y259X heterozygous or homozygous mutation in lamin A/C gene. Exp. Cell Res. 2003, 291, 352–362. [Google Scholar] [CrossRef]
- Hale, C.M.; Shrestha, A.L.; Khatau, S.B.; Stewart-Hutchinson, P.J.; Hernandez, L.; Stewart, C.L.; Hodzic, D.; Wirtz, D. Dysfunctional Connections Between the Nucleus and the Actin and Microtubule Networks in Laminopathic Models. Biophys. J. 2008, 95, 5462–5475. [Google Scholar] [CrossRef]
- Favreau, C.; Higuet, D.; Courvalin, J.-C.; Buendia, B. Expression of a Mutant Lamin A That Causes Emery-Dreifuss Muscular Dystrophy Inhibits In Vitro Differentiation of C2C12 Myoblasts. Mol. Cell. Biol. 2004, 24, 1481–1492. [Google Scholar] [CrossRef]
- Markiewicz, E.; Ledran, M.; Hutchison, C.J. Remodelling of the nuclear lamina and nucleoskeleton is required for skeletal muscle differentiation in vitro. J. Cell Sci. 2005, 118, 409–420. [Google Scholar] [CrossRef]
- Meinke, P.; Thuy, D.N.; Wehnert, M.S. The LINC complex and human disease. Biochem. Soc. Trans. 2011, 39, 1693–1697. [Google Scholar] [CrossRef]
- Gerbino, A.; Procino, G.; Svelto, M.; Carmosino, M. Role of Lamin A/C Gene Mutations in the signaling defects leading to cardiomyopathies. Front. Physiol. 2018, 9, 1356. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.C.; Wu, W.; Muchir, A.; Iwata, S.; Homma, S.; Worman, H.J. Dual specificity phosphatase 4 mediates cardiomyopathy caused by lamin A/C (LMNA) gene mutation. J. Biol. Chem. 2012, 287, 40513–40524. [Google Scholar] [CrossRef] [PubMed]
- Burla, R.; La Torre, M.; Merigliano, C.; Vernì, F.; Saggio, I. Genomic instability and DNA replication defects in progeroid syndromes. Nucleus 2018, 9, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Graziano, S.; Kreienkamp, R.; Coll-Bonfill, N.; Gonzalo, S. Causes and consequences of genomic instability in laminopathies: Replication stress and interferon response. Nucleus 2018, 9, 289–306. [Google Scholar] [CrossRef]
- Chen, S.N.; Lombardi, R.; Karmouch, J.; Tsai, J.Y.; Czernuszewicz, G.; Taylor, M.R.G.; Mestroni, L.; Coarfa, C.; Gurha, P.; Marian, A.J. DNA Damage Response/TP53 Pathway Is Activated and Contributes to the Pathogenesis of Dilated Cardiomyopathy Associated with LMNA (Lamin A/C) Mutations. Circ. Res. 2019, 124, 856–873. [Google Scholar] [CrossRef]
- Ho, C.Y.; Jaalouk, D.E.; Vartiainen, M.K.; Lammerding, J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature 2013, 497, 507–511. [Google Scholar] [CrossRef]
- Liang, Y.; Chiu, P.H.; Yip, K.Y.; Chan, S.Y. Subcellular Localization of SUN2 Is Regulated by Lamin A and Rab5. PLoS ONE 2011, 6, e20507. [Google Scholar] [CrossRef]
- Méjat, A.; Decostre, V.; Li, J.; Renou, L.; Kesari, A.; Hantaï, D.; Stewart, C.L.; Xiao, X.; Hoffman, E.; Bonne, G.; et al. Lamin A/C–mediated neuromuscular junction defects in Emery-Dreifuss muscular dystrophy. J. Cell Biol. 2009, 184, 31–44. [Google Scholar] [CrossRef]
- Haque, F.; Lloyd, D.J.; Smallwood, D.T.; Dent, C.L.; Shanahan, C.M.; Fry, A.M.; Trembath, R.C.; Shackleton, S. SUN1 Interacts with Nuclear Lamin A and Cytoplasmic Nesprins To Provide a Physical Connection between the Nuclear Lamina and the Cytoskeleton. Mol. Cell. Biol. 2006, 26, 3738. [Google Scholar] [CrossRef]
- Hasan, S.; Güttinger, S.; Mühlhäusser, P.; Anderegg, F.; Bürgler, S.; Kutay, U. Nuclear envelope localization of human UNC84A does not require nuclear lamins. FEBS Lett. 2006, 580, 1263–1268. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Chi, Y.-H.; Mutalif, R.A.; Starost, M.F.; Myers, T.G.; Anderson, S.A.; Stewart, C.L.; Jeang, K.-T. Accumulation of the inner nuclear envelope protein Sun1 is pathogenic in progeric and dystrophic laminopathies. Cell 2012, 149, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Ostlund, C.; Folker, E.S.; Choi, J.C.; Gomes, E.R.; Gundersen, G.G.; Worman, H.J. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J. Cell Sci. 2009, 122, 4099–4108. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, E.; Columbaro, M.; Capanni, C.; Maraldi, N.M.; Cenni, V.; Scotlandi, K.; Marino, M.T.; Merlini, L.; Squarzoni, S.; Lattanzi, G. Prelamin A-mediated recruitment of SUN1 to the nuclear envelope directs nuclear positioning in human muscle. Cell Death Differ. 2011, 18, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Muchir, A.; Wu, W.; Worman, H.J. Reduced expression of A-type lamins and emerin activates extracellular signal-regulated kinase in cultured cells. Biochim. Biophys. Acta 2009, 1792, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Frock, R.L.; Kudlow, B.A.; Evans, A.M.; Jameson, S.A.; Hauschka, S.D.; Kennedy, B.K. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev. 2006, 20, 486–500. [Google Scholar] [CrossRef] [PubMed]
- Janin, A.; Bauer, D.; Ratti, F.; Valla, C.; Bertrand, A.; Christin, E.; Chopin, E.; Streichenberger, N.; Bonne, G.; Gache, V.; et al. SMAD6 overexpression leads to accelerated myogenic differentiation of LMNA mutated cells. Sci. Rep. 2018, 8, 5618. [Google Scholar] [CrossRef]
- Liu, Q.; Pante, N.; Misteli, T.; Elsagga, M.; Crisp, M.; Hodzic, D.; Burke, B.; Roux, K.J. Functional association of Sun1 with nuclear pore complexes. J. Cell Biol. 2007, 178, 785–798. [Google Scholar] [CrossRef]
- Jahed, Z.; Soheilypour, M.; Peyro, M.; Mofrad, M.R.K. The LINC and NPC relationship—It’s complicated! J. Cell Sci. 2016, 129, 3219–3229. [Google Scholar] [CrossRef]
- Le Dour, C.; Macquart, C.; Sera, F.; Homma, S.; Bonne, G.; Morrow, J.P.; Worman, H.J.; Muchir, A. Decreased WNT/β-catenin signalling contributes to the pathogenesis of dilated cardiomyopathy caused by mutations in the lamin a/C gene. Hum. Mol. Genet. 2017, 26, 333–343. [Google Scholar] [CrossRef]
- Chen, H.; Li, C.; Peng, X.; Zhou, Z.; Weinstein, J.N.; Liang, H.; Caesar-Johnson, S.J.; Demchok, J.A.; Felau, I.; Kasapi, M.; et al. A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples. Cell 2018, 173, 386–399. [Google Scholar] [CrossRef]
- Goldman, R.D.; Shumaker, D.K.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Gordon, L.B.; Gruenbaum, Y.; Khuon, S.; Mendez, M.; Varga, R.; et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson–Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 8963–8968. [Google Scholar] [CrossRef] [PubMed]
- Camozzi, D.; Capanni, C.; Cenni, V.; Mattioli, E.; Columbaro, M.; Squarzoni, S.; Lattanzi, G. Diverse lamin-dependent mechanisms interact to control chromatin dynamics. Nucleus 2014, 5, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, J.; Chan, K.M.; Tjia, W.M.; Deng, W.; Guan, X.; Huang, J.; Li, K.M.; Chau, P.Y.; Chen, D.J.; et al. Genomic instability in laminopathy-based premature aging. Nat. Med. 2005, 11, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Musich, P.R.; Zou, Y. DNA-damage accumulation and replicative arrest in Hutchinson–Gilford progeria syndrome. Biochem. Soc. Trans. 2011, 39, 1764–1769. [Google Scholar] [CrossRef] [PubMed]
- Saha, B.; Zitnik, G.; Johnson, S.; Nguyen, Q.; Risques, R.A.; Martin, G.M.; Oshima, J. DNA damage accumulation and TRF2 degradation in atypical Werner syndrome fibroblasts with LMNA mutations. Front. Genet. 2013, 4, 129. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.M.; Wang, L.; Alcalai, R.; Pizard, A.; Burgon, P.G.; Ahmad, F.; Sherwood, M.; Branco, D.M.; Wakimoto, H.; Fishman, G.I.; et al. Lamin A/C haploinsufficiency causes dilated cardiomyopathy and apoptosis-triggered cardiac conduction system disease. J. Mol. Cell. Cardiol. 2008, 44, 293–303. [Google Scholar] [CrossRef]
- García-Lizarribar, A.; Fernández-Garibay, X.; Velasco-Mallorquí, F.; Castaño, A.G.; Samitier, J.; Ramon-Azcon, J. Composite Biomaterials as Long-Lasting Scaffolds for 3D Bioprinting of Highly Aligned Muscle Tissue. Macromol. Biosci. 2018, 18, 1–13. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).