MicrobioLink: An Integrated Computational Pipeline to Infer Functional Effects of Microbiome–Host Interactions
Abstract
1. Introduction
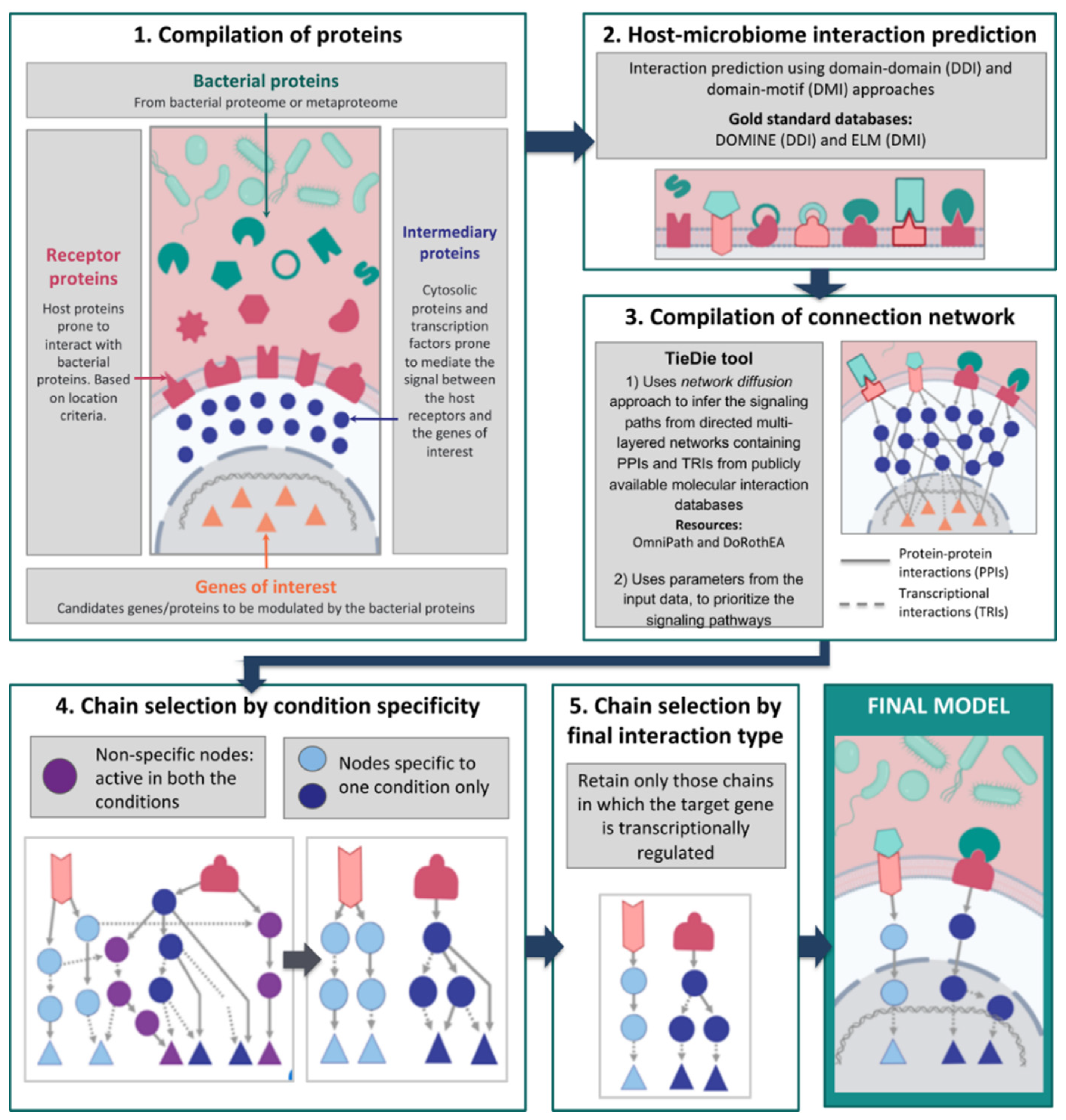
2. Materials and Methods
2.1. Compilation of Input Proteins and Genes
2.2. Bacterial–Host Interaction Prediction
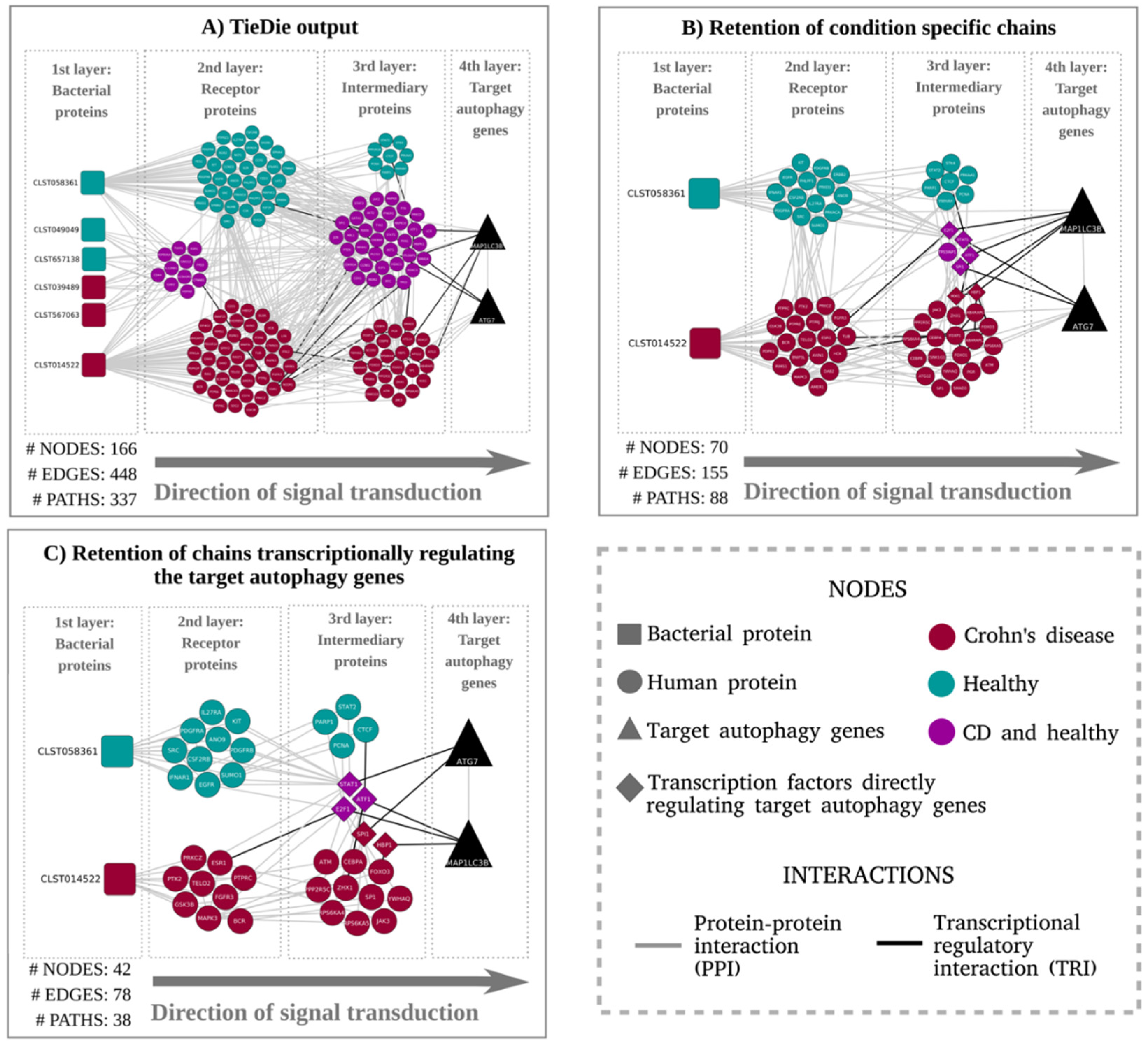
2.3. Network Compilation and Path Tracing Using Diffusion
3. Results
4. Discussion
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data and Software Availability
References
- Weimer, B.C.; Chen, P.; Desai, P.T.; Chen, N.; Shah, J. Whole Cell Cross-Linking to Discover Host–Microbe Protein Cognate Receptor/Ligand Pairs. Front. Microbiol. 2018, 9, 1585. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Marchesi, J.R.; Mougel, C.; Selosse, M.-A. Host-microbiota interactions: from holobiont theory to analysis. Microbiome 2019, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Wooley, J.C.; Godzik, A.; Friedberg, I. A Primer on Metagenomics. PLoS Comput. Boil. 2010, 6, e1000667. [Google Scholar] [CrossRef] [PubMed]
- Krajmalnik-Brown, R.; Ilhan, Z.-E.; Kang, D.-W.; DiBaise, J. Effects of gut microbes on nutrient absorption and energy regulation. Nutr. Clin. Pr. 2012, 27, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, S.; Hemarajata, P.; Versalovic, J. The Human Gut Microbiome and Body Metabolism: Implications for Obesity and Diabetes. Clin. Chem. 2013, 59, 617–628. [Google Scholar] [CrossRef]
- Loh, G.; Blaut, M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes 2012, 3, 544–555. [Google Scholar] [CrossRef]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Nunez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Heinken, A.; Sahoo, S.; Fleming, R.M.T.; Thiele, I. Systems-level characterization of a host-microbe metabolic symbiosis in the mammalian gut. Gut Microbes 2013, 4, 28–40. [Google Scholar] [CrossRef]
- Wojcik, J.; Schächter, V. Protein-protein interaction map inference using interacting domain profile pairs. Bioinformatics 2001, 17, S296–S305. [Google Scholar] [CrossRef]
- Evans, P.; Dampier, W.; Ungar, L.; Tozeren, A. Prediction of HIV-1 virus-host protein interactions using virus and host sequence motifs. BMC Med. Genom. 2009, 2, 27. [Google Scholar] [CrossRef]
- Jones, E.J.; Matthews, Z.; Gul, L.; Sudhakar, P.; Treveil, A.; Divekar, D.; Buck, J.; Wrzesinski, T.; Jefferson, M.; Armstrong, S.D.; et al. Integrative analysis of Paneth cell proteomic and transcriptomic data from intestinal organoids reveals functional processes dependent on autophagy. Dis. Model. Mech. 2019, 12, dmm037069. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar, P.; Jacomin, A.-C.; Hautefort, I.; Samavedam, S.; Fatemian, K.; Ari, E.; Gul, L.; Demeter, A.; Jones, E.; Korcsmaros, T.; et al. Targeted interplay between bacterial pathogens and host autophagy. Autophagy 2019, 15, 1620–1633. [Google Scholar] [CrossRef] [PubMed]
- Katiyar-Agarwal, S.; Jin, H. Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 2010, 48, 225–246. [Google Scholar] [CrossRef] [PubMed]
- Ruch, T.R.; Engel, J.N. Targeting the Mucosal Barrier: How Pathogens Modulate the Cellular Polarity Network. Cold Spring Harb. Perspect. Boil. 2017, 9, a027953. [Google Scholar] [CrossRef][Green Version]
- Killick, K.E.; Magee, D.A.; Park, S.D.E.; Taraktsoglou, M.; Browne, J.; Conlon, K.M.; Nalpas, N.C.; Gormley, E.; Gordon, S.V.; MacHugh, D.E.; et al. Key Hub and Bottleneck Genes Differentiate the Macrophage Response to Virulent and Attenuated Mycobacterium bovis. Front. Immunol. 2014, 5, 422. [Google Scholar] [CrossRef]
- Tekir, S.D.; Cakir, T.; Ülgen, K.Ö. Infection Strategies of Bacterial and Viral Pathogens through Pathogen–Human Protein–Protein Interactions. Front. Microbiol. 2012, 3, 46. [Google Scholar] [CrossRef]
- Foley, S.L.; Johnson, T.; Ricke, S.C.; Nayak, R.; Danzeisen, J. Salmonella Pathogenicity and Host Adaptation in Chicken-Associated Serovars. Microbiol. Mol. Boil. Rev. 2013, 77, 582–607. [Google Scholar] [CrossRef]
- Nourani, E.; Khunjush, F.; Durmuş, S. Computational approaches for prediction of pathogen-host protein-protein interactions. Front. Microbiol. 2015, 6, 94. [Google Scholar] [CrossRef]
- Gould, A.; Zhang, V.; Lamberti, L.; Jones, E.; Obadia, B.; Korasidis, N.; Gavryushkin, A.; Carlson, J.M.; Beerenwinkel, N.; Ludington, W.B. Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. USA 2018, 115, E11951–E11960. [Google Scholar]
- Dyer, M.D.; Neff, C.; Dufford, M.; Rivera, C.G.; Shattuck, N.; Bassaganya-Riera, J.; Murali, T.M.; Sobral, B. The Human-Bacterial Pathogen Protein Interaction Networks of Bacillus anthracis, Francisella tularensis, and Yersinia pestis. PLoS ONE 2010, 5, e12089. [Google Scholar] [CrossRef]
- Dyer, M.D.; Murali, T.M.; Sobral, B. The Landscape of Human Proteins Interacting with Viruses and Other Pathogens. PLOS Pathog. 2008, 4, e32. [Google Scholar] [CrossRef]
- Guven-Maiorov, E.; Tsai, C.-J.; Nussinov, R. Structural host-microbiota interaction networks. PLoS Comput. Boil. 2017, 13, e1005579. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Hou, Y.; Shen, J.; Huang, Y.; Martin, W.; Cheng, F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020, 6, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Tekir, S.D.; Cakir, T.; Ardıç, E.; Sayılırbaş, A.S.; Konuk, G.; Sarıyer, H.; Uğurlu, A.; Karadeniz, I.; Özgür, A.; Sevilgen, F.E.; et al. PHISTO: pathogen–host interaction search tool. Bioinformatics 2013, 29, 1357–1358. [Google Scholar] [CrossRef]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.P.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2013, 42, D581–D591. [Google Scholar] [CrossRef] [PubMed]
- Vialás, V.; Gil, C. Proteopathogen2, a database and web tool to store and display proteomics identification results in the mzIdentML standard. EuPA Open Proteom. 2015, 8, 22–27. [Google Scholar] [CrossRef]
- Li, Y.; Wang, C.; Miao, Z.; Bi, X.; Wu, D.; Jin, N.; Wang, L.; Wu, H.; Qian, K.; Li, C.; et al. ViRBase: a resource for virus–host ncRNA-associated interactions. Nucleic Acids Res. 2014, 43, D578–D582. [Google Scholar] [CrossRef]
- Kumar, R.; Nanduri, B. HPIDB - a unified resource for host-pathogen interactions. BMC Bioinform. 2010, 11, S16. [Google Scholar] [CrossRef]
- Heirendt, L.; Arreckx, S.; Pfau, T.; Mendoza, S.N.; Richelle, A.; Heinken, A.; Haraldsdóttir, H.S.; Wachowiak, J.; Keating, S.M.; Vlasov, V.; et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 2019, 14, 639–702. [Google Scholar] [CrossRef]
- Ågren, R.; Liu, L.; Shoaie, S.; Vongsangnak, W.; Nookaew, I.; Nielsen, J. The RAVEN Toolbox and Its Use for Generating a Genome-scale Metabolic Model for Penicillium chrysogenum. PLoS Comput. Boil. 2013, 9, e1002980. [Google Scholar] [CrossRef]
- Levy, R.; Carr, R.; Kreimer, A.; Freilich, S.; Borenstein, E. NetCooperate: a network-based tool for inferring host-microbe and microbe-microbe cooperation. BMC Bioinform. 2015, 16, 164. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.H.; Pérez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Sen, P.; Orešič, M. Metabolic Modeling of Human Gut Microbiota on a Genome Scale: An Overview. Metabolites 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Samaras, P.; Frejno, M.; Gessulat, S.; Barnert, M.; Kienegger, H.; Krcmar, H.; Schlegl, J.; Ehrlich, H.-C.; Aiche, S.; et al. ProteomicsDB. Nucleic Acids Res. 2018, 46, D1271–D1281. [Google Scholar] [CrossRef] [PubMed]
- Veres, D.; Gyurko, D.M.; Thaler, B.; Szalay, K.; Fazekas, D.; Korcsmaros, T.; Csermely, P. ComPPI: a cellular compartment-specific database for protein–protein interaction network analysis. Nucleic Acids Res. 2014, 43, D485–D493. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Launay, G.; Salza, R.; Multedo, D.; Thierry-Mieg, N.; Ricard-Blum, S. MatrixDB, the extracellular matrix interaction database: updated content, a new navigator and expanded functionalities. Nucleic Acids Res. 2014, 43, D321–D327. [Google Scholar] [CrossRef]
- Peabody, M.; Laird, M.; Vlasschaert, C.; Lo, R.; Brinkman, F. PSORTdb: expanding the bacteria and archaea protein subcellular localization database to better reflect diversity in cell envelope structures. Nucleic Acids Res. 2015, 44, D663–D668. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Bendtsen, J.D.; Kiemer, L.; Fausbøll, A.; Brunak, S. Non-classical protein secretion in bacteria. BMC Microbiol. 2005, 5, 58. [Google Scholar] [CrossRef]
- Bader, S.; Kühner, S.; Gavin, A.-C. Interaction networks for systems biology. FEBS Lett. 2008, 582, 1220–1224. [Google Scholar] [CrossRef]
- Korcsmáros, T.; Dunai, Z.A.; Vellai, T.; Csermely, P. Teaching the bioinformatics of signaling networks: an integrated approach to facilitate multi-disciplinary learning. Briefings Bioinform. 2013, 14, 618–632. [Google Scholar] [CrossRef] [PubMed]
- Raghavachari, B.; Tasneem, A.; Przytycka, T.M.; Jothi, R. DOMINE: a database of protein domain interactions. Nucleic Acids Res. 2007, 36, D656–D661. [Google Scholar] [CrossRef] [PubMed]
- Gouw, M.; Michael, S.; Sámano-Sánchez, H.; Kumar, M.; Zeke, A.; Lang, B.; Bely, B.; Chemes, L.B.; E Davey, N.; Deng, Z.; et al. The eukaryotic linear motif resource - 2018 update. Nucleic Acids Res. 2018, 46, D428–D434. [Google Scholar] [CrossRef] [PubMed]
- Dinkel, H.; Van Roey, K.; Michael, S.; E Davey, N.; Weatheritt, R.; Born, D.; Speck, T.; Krueger, D.; Grebnev, G.; Kubań, M.; et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2013, 42, D259–D266. [Google Scholar] [CrossRef] [PubMed]
- Weatheritt, R.; Jehl, P.; Dinkel, H.; Gibson, T.J. iELM—a web server to explore short linear motif-mediated interactions. Nucleic Acids Res. 2012, 40, W364–W369. [Google Scholar] [CrossRef] [PubMed]
- Mészáros, B.; Erdos, G.; Dosztányi, Z. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018, 46, W329–W337. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.; Qureshi, M.; Richardson, L.J.; A Salazar, G.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; I Fraser, M.; et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2018, 47, D351–D360. [Google Scholar] [CrossRef]
- Paull, E.O.; Carlin, D.E.; Niepel, M.; Sorger, P.; Haussler, D.; Stuart, J.M. Discovering causal pathways linking genomic events to transcriptional states using Tied Diffusion Through Interacting Events (TieDIE). Bioinformatics 2013, 29, 2757–2764. [Google Scholar] [CrossRef]
- Türei, D.; Korcsmáros, T.; Saez-Rodriguez, J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods 2016, 13, 966–967. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Türei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef] [PubMed]
- Tschurtschenthaler, M.; Adolph, T.E. The Selective Autophagy Receptor Optineurin in Crohn’s Disease. Front. Immunol. 2018, 9, 766. [Google Scholar] [CrossRef]
- Kuballa, P.; Huett, A.; Rioux, J.D.; Daly, M.J.; Xavier, R.J. Impaired Autophagy of an Intracellular Pathogen Induced by a Crohn’s Disease Associated ATG16L1 Variant. PLoS ONE 2008, 3, e3391. [Google Scholar] [CrossRef]
- Li, Q.; Wang, C.; Tang, C.; He, Q.; Li, N.; Li, J. Dysbiosis of Gut Fungal Microbiota is Associated With Mucosal Inflammation in Crohn’s Disease. J. Clin. Gastroenterol. 2014, 48, 513–523. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Shreiner, A.B.; Gillilland, M.G.; Kitamoto, S.; Ishii, C.; Hirayama, A.; Kuffa, P.; El-Zaatari, M.; Grasberger, H.; Seekatz, A.M.; et al. Functional Characterization of Inflammatory Bowel Disease-Associated Gut Dysbiosis in Gnotobiotic Mice. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 468–481. [Google Scholar] [CrossRef] [PubMed]
- Erickson, A.R.; Cantarel, B.L.; Lamendella, R.; Darzi, Y.; Mongodin, E.F.; Pan, C.; Shah, M.; Halfvarson, J.; Tysk, C.; Henrissat, B.; et al. Integrated Metagenomics/Metaproteomics Reveals Human Host-Microbiota Signatures of Crohn’s Disease. PLoS ONE 2012, 7, e49138. [Google Scholar] [CrossRef] [PubMed]
- Durães, C.; Machado, J.C.; Portela, F.; Rodrigues, S.; Lago, P.; Cravo, M.; Ministro, P.; Marques, M.; Cremers, I.; Freitas, J.; et al. Phenotype–Genotype Profiles in Crohn’s Disease Predicted by Genetic Markers in Autophagy-Related Genes (GOIA Study II). Inflamm. Bowel Dis. 2013, 19, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Rioux, J.D.; Xavier, R.J.; Taylor, K.D.; Silverberg, M.S.; Goyette, P.; Huett, A.; Green, T.; Kuballa, P.; Barmada, M.M.; Datta, L.W.; et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 2007, 39, 596–604. [Google Scholar] [CrossRef]
- Kubisch, J.; Türei, D.; Földvári-Nagy, L.; Dunai, Z.A.; Zsákai, L.; Varga, M.; Vellai, T.; Csermely, P.; Korcsmáros, T. Complex regulation of autophagy in cancer – Integrated approaches to discover the networks that hold a double-edged sword. Semin. Cancer Boil. 2013, 23, 252–261. [Google Scholar] [CrossRef]
- Carey, R.; Jurickova, I.; Ballard, E.; Bonkowski, E.; Han, X.; Xu, H.; A Denson, L. Activation of an IL-6:STAT3-dependent transcriptome in pediatric-onset inflammatory bowel disease. Inflamm. Bowel Dis. 2008, 14, 446–457. [Google Scholar] [CrossRef]
- Montero-Melendez, T.; Llor, X.; Garcia-Planella, E.; Perretti, M.; Suarez, A. Identification of Novel Predictor Classifiers for Inflammatory Bowel Disease by Gene Expression Profiling. PLoS ONE 2013, 8, e76235. [Google Scholar] [CrossRef] [PubMed]
- Vancamelbeke, M.; Vanuytsel, T.; Farré, R.; Verstockt, S.; Ferrante, M.; Van Assche, G.; Rutgeerts, P.; Schuit, F.; Vermeire, S.; Arijs, I.; et al. Genetic and Transcriptomic Bases of Intestinal Epithelial Barrier Dysfunction in Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2017, 23, 1718–1729. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Breast Cancer 2016, 1418, 93–110. [Google Scholar] [CrossRef]
- Blander, J.M. Death in the intestinal epithelium-basic biology and implications for inflammatory bowel disease. FEBS J. 2016, 283, 2720–2730. [Google Scholar] [CrossRef]
- Fitzwalter, B.; Towers, C.G.; Sullivan, K.D.; Andrysik, Z.; Hoh, M.; Ludwig, M.; O’Prey, J.; Ryan, K.M.; Espinosa, J.M.; Morgan, M.J.; et al. Autophagy Inhibition Mediates Apoptosis Sensitization in Cancer Therapy by Relieving FOXO3a Turnover. Dev. Cell 2018, 44, 555–565.e3. [Google Scholar] [CrossRef]
- Cho, S.-J.; Yun, S.-M.; Jo, C.; Lee, D.-H.; Choi, K.J.; Song, J.C.; Park, S.I.; Kim, Y.-J.; Koh, Y.H. SUMO1 promotes Aβ production via the modulation of autophagy. Autophagy 2014, 11, 100–112. [Google Scholar] [CrossRef]
- Polager, S.; Ofir, M.; Ginsberg, D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 2008, 27, 4860–4864. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lee, A.; Kim, K.L.; Murray, J.; Shrinidhi, A.; Sung, G.; Park, K.M.; Kim, K. Autophagy Caught in the Act: A Supramolecular FRET Pair Based on an Ultrastable Synthetic Host-Guest Complex Visualizes Autophagosome-Lysosome Fusion. Angew. Chem. Int. Ed. 2018, 57, 2120–2125. [Google Scholar] [CrossRef]
- Carroll, I.M.; Maharshak, N. Enteric bacterial proteases in inflammatory bowel disease- pathophysiology and clinical implications. World J. Gastroenterol. 2013, 19, 7531–7543. [Google Scholar] [CrossRef]
- Jablaoui, A.; Kriaa, A.; Mkaouar, H.; Akermi, N.; Soussou, S.; Wysocka, M.; Wołoszyn, D.; Amouri, A.; Gargouri, A.; Maguin, E.; et al. Fecal Serine Protease Profiling in Inflammatory Bowel Diseases. Front. Microbiol. 2020, 10, 21. [Google Scholar] [CrossRef]
- Tapias, N.S.; Vergnolle, N.; Denadai-Souza, A.; Barreau, F. The Interplay Between Genetic Risk Factors and Proteolytic Dysregulation in the Pathophysiology of Inflammatory Bowel Disease. J. Crohn’s Colitis 2020. [Google Scholar] [CrossRef]
- Achuthan, A.; Aslam, A.S.M.; Nguyen, Q.; Lam, P.-Y.; Fleetwood, A.J.; Frye, A.; Louis, C.; Lee, K.M.-C.; Smith, J.E.; Cook, A.D.; et al. Glucocorticoids promote apoptosis of proinflammatory monocytes by inhibiting ERK activity. Cell Death Dis. 2018, 9, 267. [Google Scholar] [CrossRef] [PubMed]
- Zanzoni, A.; Spinelli, L.; Braham, S.; Brun, C. Perturbed human sub-networks by Fusobacterium nucleatum candidate virulence proteins. Microbiome 2017, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, P.; Palumbo, E.; Chatzou, M.; Prieto, P.; Heuer, M.L.; Notredame, C. The impact of Docker containers on the performance of genomic pipelines. PeerJ 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Lynch, S.V. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 2018, 216, 20–40. [Google Scholar] [CrossRef] [PubMed]
- Celi, P.; Cowieson, A.; Fru-Nji, F.; Steinert, R.; Kluenter, A.-M.; Verlhac, V. Gastrointestinal functionality in animal nutrition and health: New opportunities for sustainable animal production. Anim. Feed. Sci. Technol. 2017, 234, 88–100. [Google Scholar] [CrossRef]
- Turner, T.R.; James, E.; Poole, P.S. The plant microbiome. Genome Boil. 2013, 14, 209. [Google Scholar] [CrossRef]
- Ni, Y.; Yu, G.; Chen, H.; Deng, Y.; Wells, P.M.; Steves, C.J.; Ju, F.; Fu, J. M2IA: a Web Server for Microbiome and Metabolome Integrative Analysis. Bioinformatics 2020. [Google Scholar] [CrossRef]
- Harcombe, W.R.; Riehl, W.; Dukovski, I.; Granger, B.R.; Betts, A.; Lang, A.H.; Bonilla, G.; Kar, A.; Leiby, N.; Mehta, P.; et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014, 7, 1104–1115. [Google Scholar] [CrossRef]

| Resource/Tool | Standalone Version? | Description | Can User-Provided Datasets Be Handled? | Nonpathogenic Species Included/Handling? | Protein–Protein Interactions? | Inferring Downstream Effects? | Microorganisms Supported | Host Organisms Supported |
|---|---|---|---|---|---|---|---|---|
| PHISTO [24] | online | Web-tool for mining and retrieving host–pathogen interactions | no | no | yes | no | Viral, bacterial, fungal, and protozoan pathogens | Human |
| PATRIC [25] | online | Genome-focussed infectious disease research database | yes | no | yes | no | Bacterial pathogens | Actinoptergii, Arachnida, Chromadorea, Insecta, and Mammalia |
| Proteopathogen2 [26] | online | Database and web application to store and display fungal pathogen proteomics data | no | no | no | no | Fungal pathogens | Mammalian species |
| VirBase [27] | online | Database of virus–host ncRNA-associated interactions and interaction networks during viral infections | no | yes | no | no | Virus | Vertebrates, plants, and arthropods |
| NetCoperate [31] | python module | Web-based tool and software package for determining host–microbe and microbe–microbe cooperative potential from metabolic networks | yes | yes | no | yes | Any microorganism | Any host species |
| Kbase [32] | Online, python, and java | Software and data platform that enables data sharing, integration, and analysis of microbes, plants, and their communities by creating workflows consisting of a series of analysis tool runs and code blocks | yes | yes | no | yes | Any microorganism | Any host |
| M²IA [79] | web-based server | Statistical analysis methods for microbiome and metabolome data integration, including correlation analysis and functional network analysis | yes | yes | no | yes | Any microorganism | Any host species |
| COMETS [80] | Matlab and a python toolbox | Modelling framework that integrates dynamic flux balance analysis with diffusion to communities | yes | yes | no | yes | Any microorganism | Any host species |
| MicrobioLink (this paper) | Python and Docker | Integrated evaluation of microbe–host interaction networks | yes | yes | yes | yes | Any microorganism | Any host species |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrighetti, T.; Bohar, B.; Lemke, N.; Sudhakar, P.; Korcsmaros, T. MicrobioLink: An Integrated Computational Pipeline to Infer Functional Effects of Microbiome–Host Interactions. Cells 2020, 9, 1278. https://doi.org/10.3390/cells9051278
Andrighetti T, Bohar B, Lemke N, Sudhakar P, Korcsmaros T. MicrobioLink: An Integrated Computational Pipeline to Infer Functional Effects of Microbiome–Host Interactions. Cells. 2020; 9(5):1278. https://doi.org/10.3390/cells9051278
Chicago/Turabian StyleAndrighetti, Tahila, Balazs Bohar, Ney Lemke, Padhmanand Sudhakar, and Tamas Korcsmaros. 2020. "MicrobioLink: An Integrated Computational Pipeline to Infer Functional Effects of Microbiome–Host Interactions" Cells 9, no. 5: 1278. https://doi.org/10.3390/cells9051278
APA StyleAndrighetti, T., Bohar, B., Lemke, N., Sudhakar, P., & Korcsmaros, T. (2020). MicrobioLink: An Integrated Computational Pipeline to Infer Functional Effects of Microbiome–Host Interactions. Cells, 9(5), 1278. https://doi.org/10.3390/cells9051278





