A Functional Binding Domain in the Rbpr2 Receptor Is Required for Vitamin A Transport, Ocular Retinoid Homeostasis, and Photoreceptor Cell Survival in Zebrafish
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Homology Modeling and Molecular Docking
2.3. Cloning of Zebrafish Rbpr2 cDNA
2.4. Site-Directed Mutagenesis and Generation of Stable Cell Lines Expressing Mutant Rbpr2 binding Residue Recombinant Proteins
2.5. Indirect Immunofluorescence and Confocal Microscopy
2.6. RBP4 binding and Retinol Uptake Studies
2.7. RBP4 binding to Rbpr2 in Co-immunoprecipitation Assays and UV Crosslinking
2.8. Animal Study Approval
2.9. Zebrafish Strains and Maintenance
2.10. Zebrafish Immunohistochemistry and Fluorescence Imaging
2.11. Generation of Zebrafish rbpr2 mutants using CRISPR/Cas9
2.12. rbpr2fs-muz99 Mutant Rescue Experiments Using WT or Mutant-rbpr2 mRNA
2.13. All-Trans Retinoic Acid Rescue Experiments in rbpr2fs-muz99 Mutant Zebrafish
2.14. Apoptosis/TUNEL Assay
2.15. Transmission Electron Microscopy (TEM)
2.16. Western Blot Analysis and Densitometry
2.17. Quantitative Real-Time PCR
2.18. HPLC Analysis for Retinoids
2.19. Statistical Analysis
3. Results
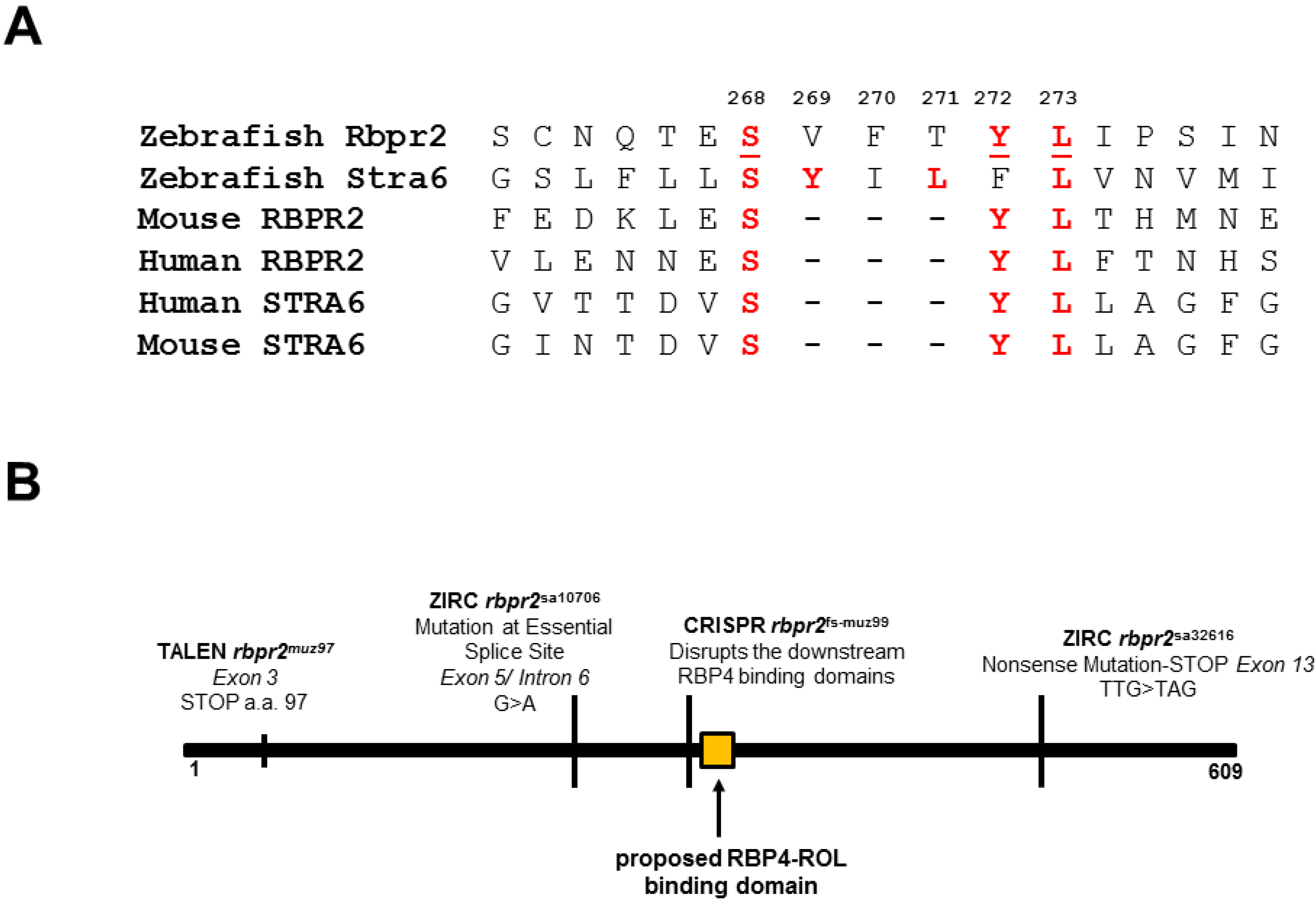
3.1. Zebrafish Rbpr2 Contains Consensus RBP4 Binding Residues
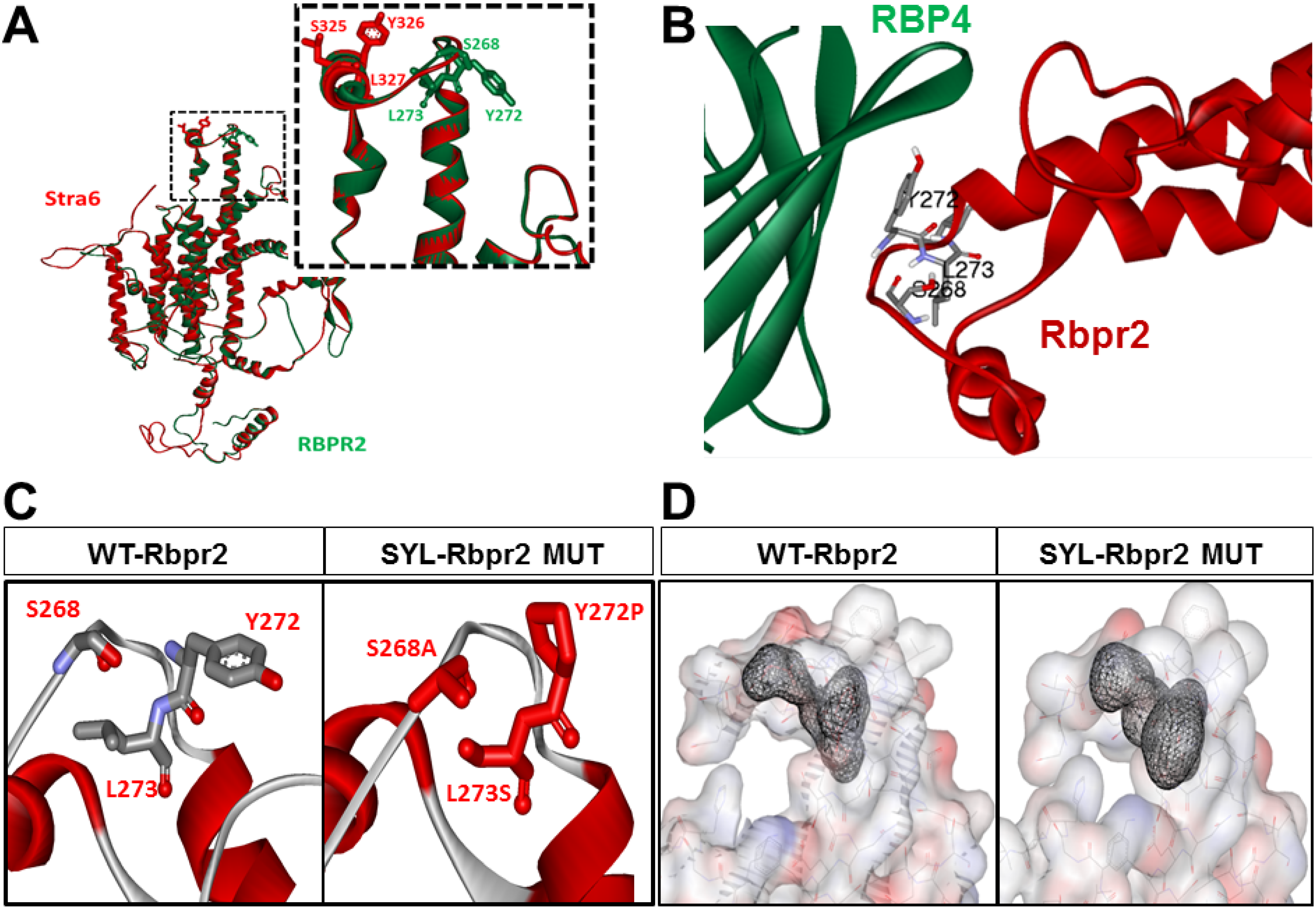
3.2. Homology Modeling and Molecular Docking Analysis Confirm Importance of a Proposed RBP4 Binding Domain in Rbpr2 for Vitamin A Transport
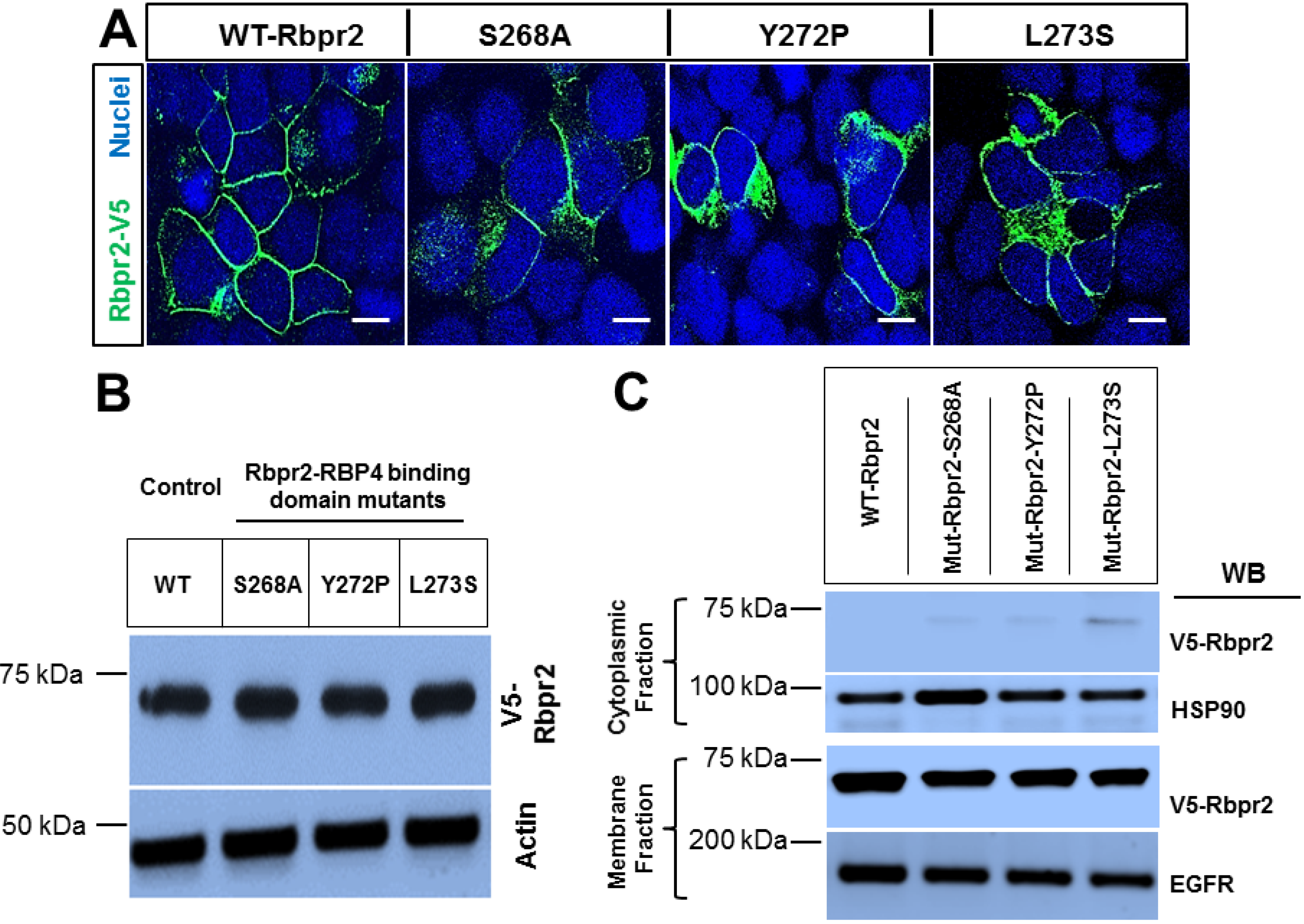
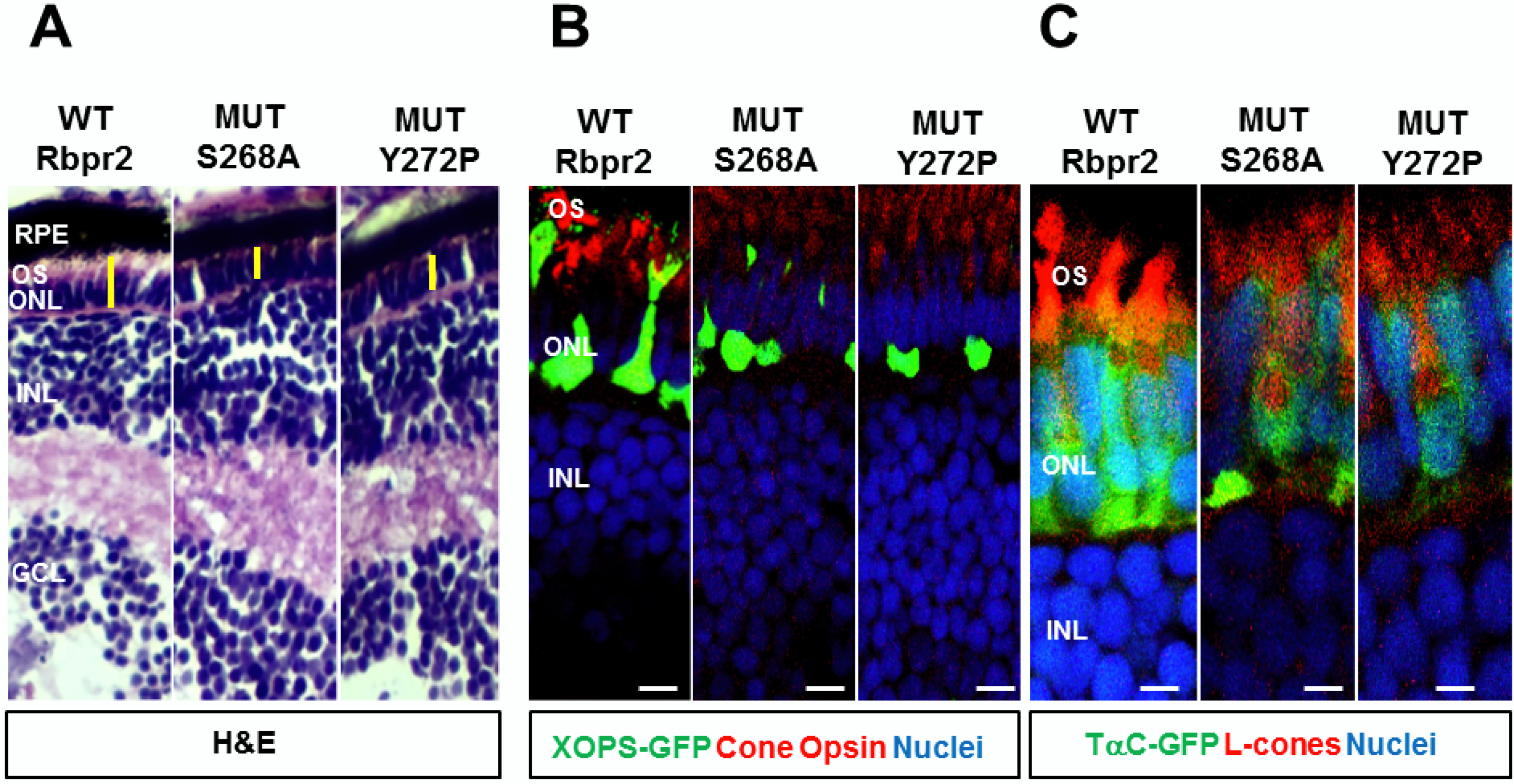
3.3. Rbpr2 Mutants Encompassing the Proposed RBP4 Binding Domain in Rbpr2 Show Normal Membrane Trafficking
3.4. Rbpr2 Mutants Encompassing the Proposed RBP4 Binding Domain are Defective in [3H]ROL-RBP4 Uptake
3.5. Co-Immunoprecipitation Assays Show that Rbpr2 mutants Encompassing the Proposed RBP4 Binding Domain have Defective RBP4-ROL Binding Capabilities
3.6. Generation of rbpr2 Mutant Zebrafish Lines targeting the RBP4 Binding Domain
3.7. rbpr2fs-muz99 Mutant Zebrafish Show Eye Phenotypes at Late Larval Stages Typically, Associated with Low Ocular Vitamin A Content and Defective Retinoid Signaling
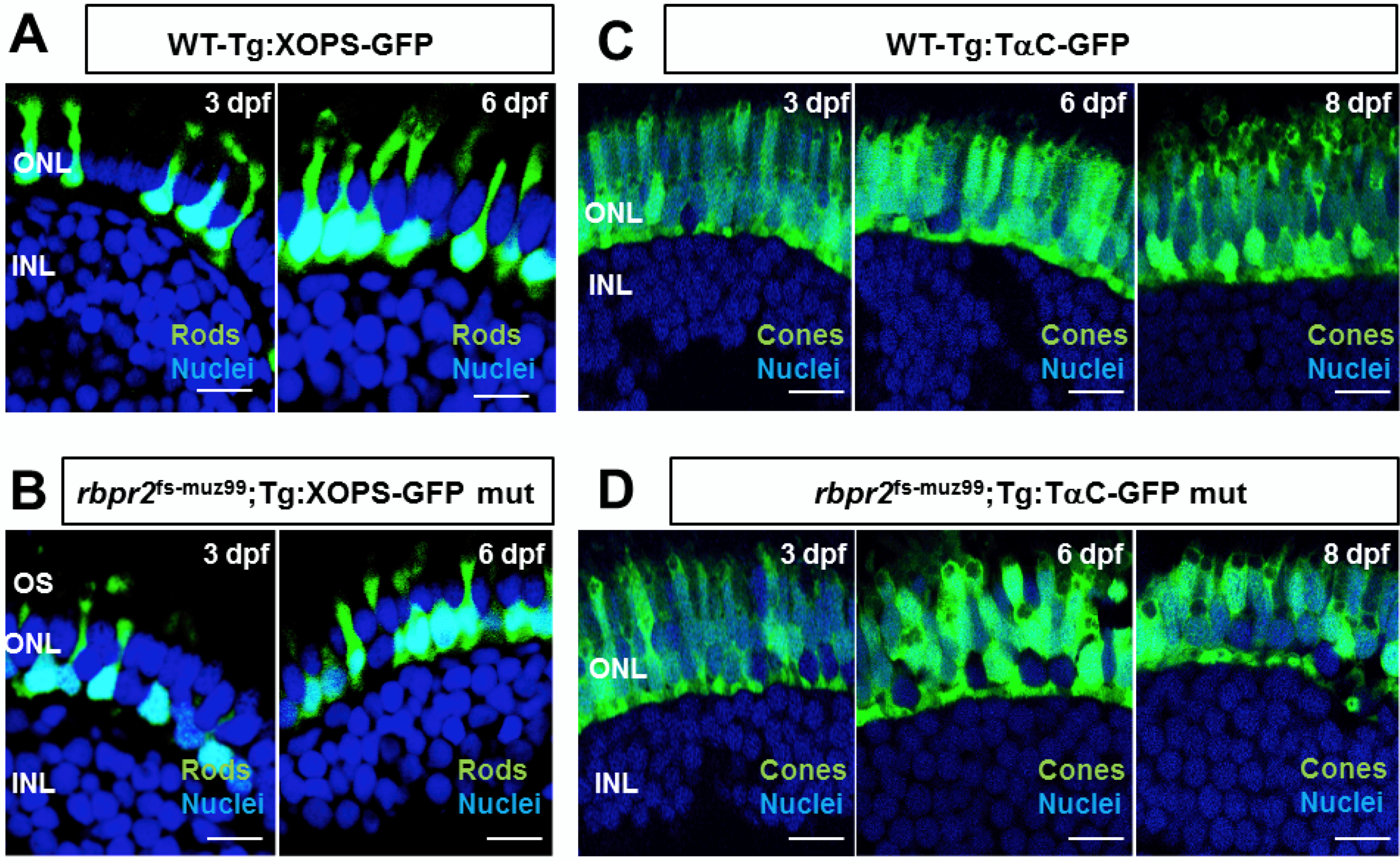
3.8. rbpr2fs-muz99 Mutant Zebrafish Show Retinal Phenotypes at Larval Stages
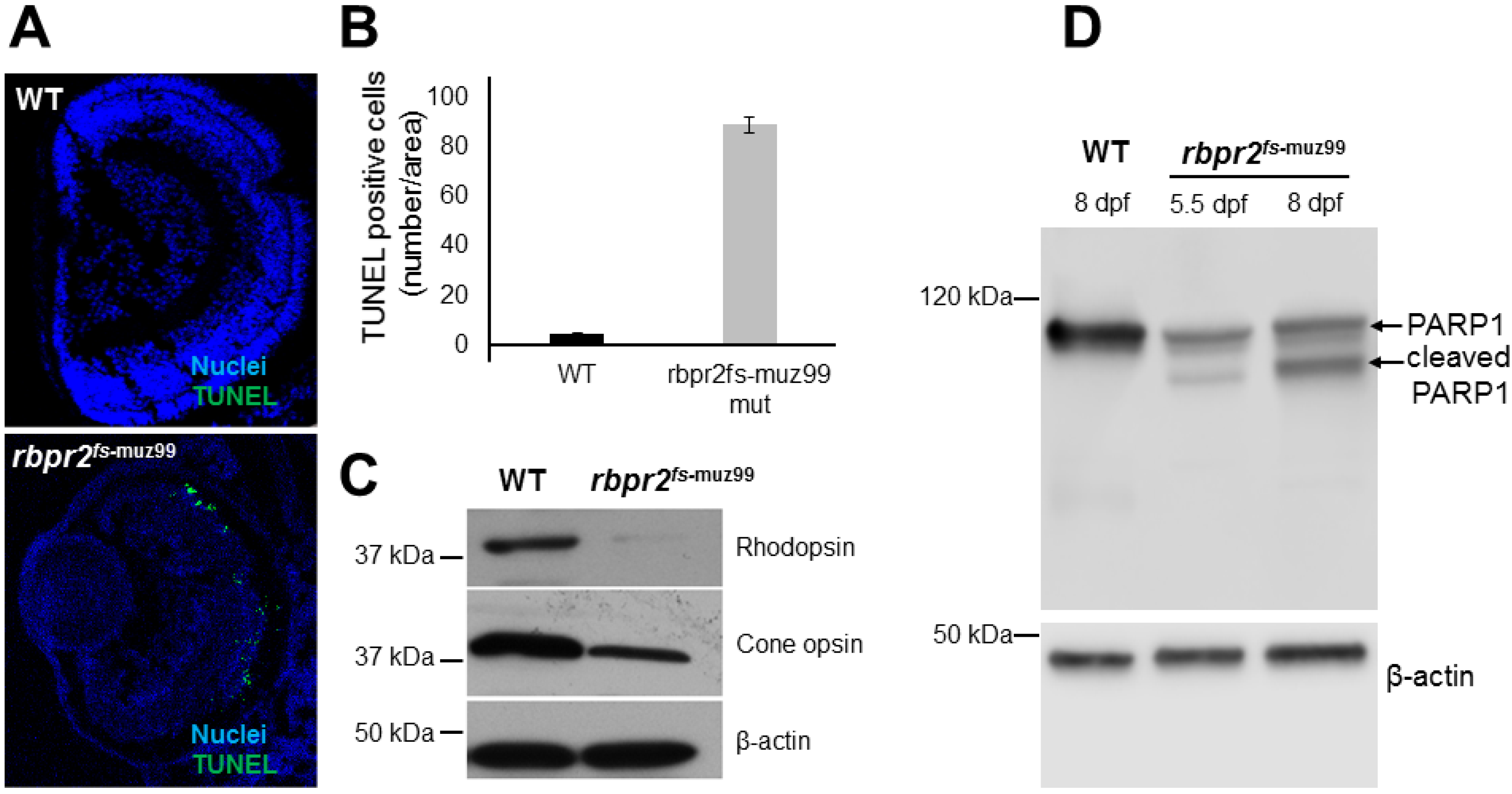
3.9. rbpr2fs-muz99 Mutant Zebrafish Show Rod and Cone Dystrophy
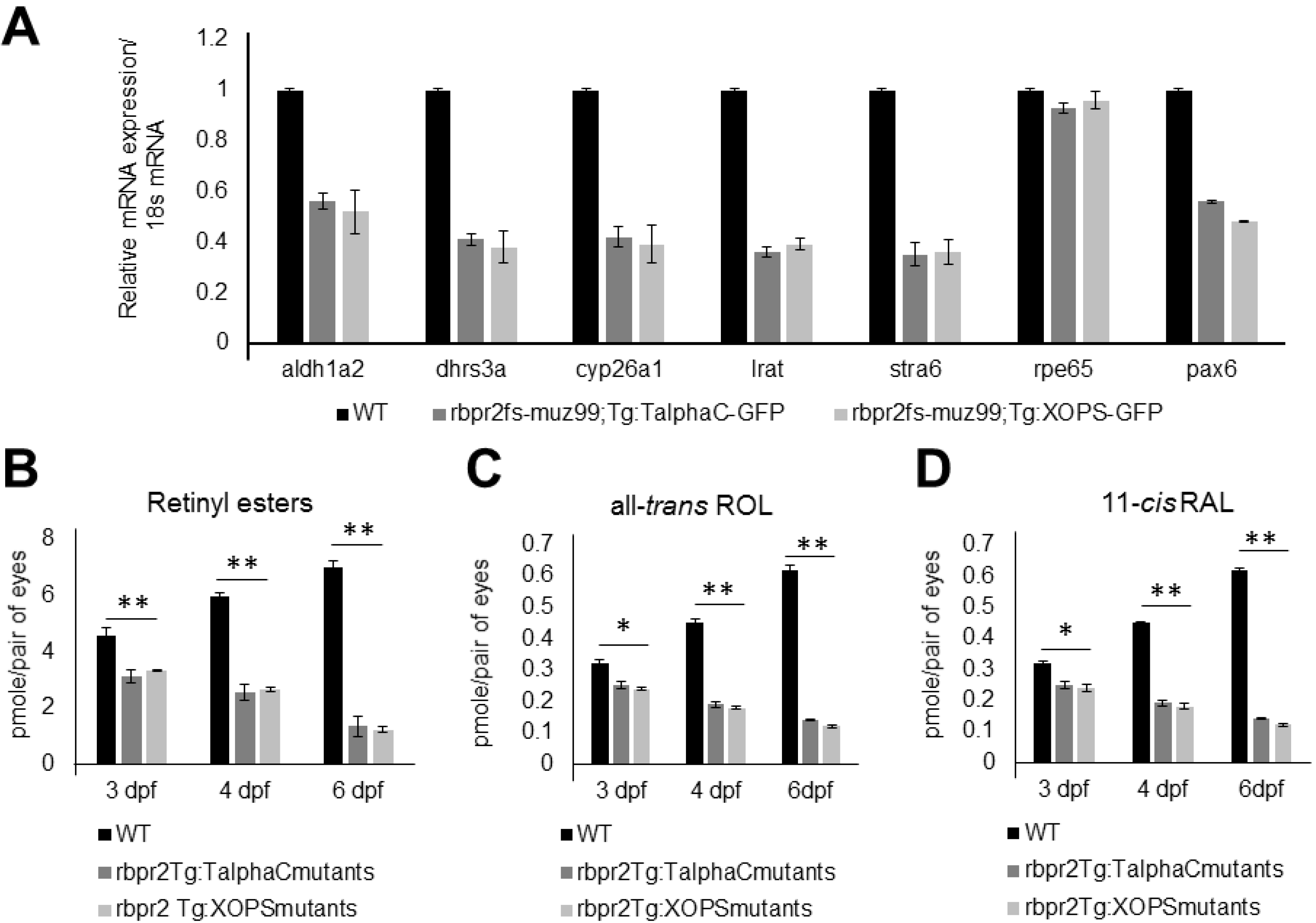
3.10. rbpr2fs-muz99 Mutant Zebrafish show Decreased Ocular Retinoid Content and Signaling
3.11. Apoptosis Pattern of Retinas in rbpr2fs-muz99 Mutant Zebrafish
3.12. WT-Rbpr2 mRNA and All-trans Retinoic Acid (atRA) rescue the rbpr2fs-muz99 Mutant Phenotype
4. Discussion
4.1. Importance of Dietary Vitamin A Transporters for Vision
4.2. Importance of RBP4-ROL Binding Residues in Vitamin A Receptors
4.3. Rbpr2 Contains a Putative RBP4-ROL Binding Domain Important for Yolk Vitamin A Transport to the Eye
4.4. Stra6 and Rbpr2
4.5. RBP4 Binding Residues on Rbpr2 are Necessary for Systemic Vitamin A Transport
4.6. Limitations of the rbpr2 mutant in Identifying “SYL” Residues as the Putative Rbp4 Binding Domain
4.7. Mechanism(s) of Photoreceptor Cell Death in rbpr2 fs-muz99 Mutants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ablonczy, Z.; Crouch, R.K.; Goletz, P.W.; Redmond, T.M.; Knapp, D.R.; Ma, J.X.; Rohrer, B. 11-cis-retinal reduces constitutive opsin phosphorylation and improves quantum catch in retinoid-deficient mouse rod photoreceptors. J. Biol. Chem. 2002, 277, 40491–40498. [Google Scholar] [CrossRef] [PubMed Central]
- Alapatt, P.; Guo, F.; Komanetsky, S.M.; Wang, S.; Cai, J.; Sargsyan, A.; Díaz, E.R.; Bacon, B.T.; Aryal, P.; Graham, T.E. Liver retinol transporter and receptor for serum retinol-binding protein (RBP4). J. Biol. Chem. 2013, 288, 1250–1265. [Google Scholar] [CrossRef] [PubMed Central]
- Amengual, J.; Golczak, M.; Palczewski, K.; von Lintig, J. Lecithin:retinol acyltransferase is critical for cellular uptake of vitamin A from serum retinol-binding protein. J. Biol. Chem. 2012, 287, 24216–24227. [Google Scholar] [CrossRef] [PubMed Central]
- Blomhoff, R.; Green, M.H.; Berg, T.; Norum, K.R. Transport and storage of vitamin A. Science 1990, 250, 399–404. [Google Scholar] [CrossRef] [PubMed Central]
- Borel, P.; Desmarchelier, C. Genetic Variations Associated with Vitamin A Status and Vitamin A Bioavailability. Nutrients 2017, 9, 246. [Google Scholar] [CrossRef] [PubMed Central]
- D’Ambrosio, D.N.; Clugston, R.D.; Blaner, W.S. Vitamin A metabolism. An update. Nutrients 2011, 3, 63–103. [Google Scholar] [PubMed Central]
- During, A.; Harrison, E.H. Mechanisms of provitamin A (carotenoid) and vitamin A (retinol) transport into and out of intestinal Caco-2 cells. J. Lipid Res. 2007, 48, 2283–2294. [Google Scholar] [CrossRef] [PubMed Central]
- Harrison, E.H. Mechanisms involved in the intestinal absorption of dietary vitamin A and provitamin A carotenoids. Biochim. Biophys. Acta 2012, 1821, 70–77. [Google Scholar] [CrossRef] [PubMed Central]
- Kawaguchi, R.; Yu, J.; Wiita, P.; Honda, J.; Sun, H. An essential ligand-binding domain in the membrane receptor for retinol-binding protein revealed by large-scale mutagenesis and a human polymorphism. J. Biol. Chem. 2008, 30, 15160–15168. [Google Scholar] [CrossRef] [PubMed Central]
- Kawaguchi, R.; Yu, J.; Wiita, P.; Ter-Stepanian, M.; Sun, H. Mapping the membrane topology and extracellular ligand binding domains of the retinol binding protein receptor. Biochemistry 2008, 47, 5387–5395. [Google Scholar] [CrossRef] [PubMed Central]
- Quadro, L.; Blaner, W.S.; Salchow, D.J.; Vogel, S.; Piantedosi, R.; Gouras, P.; Freeman, S.; Cosma, M.P.; Colantuoni, V.; Gottesman, M.E. Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. EMBO J. 1999, 18, 4633–4644. [Google Scholar] [CrossRef] [PubMed Central]
- Ross, A.C.; Zolfaghari, R.; Weisz, J. Recent advances in the biotransformation, transport, and metabolism of retinoids. Recent advances in the biotransformation, transport, and metabolism of retinoids. Curr. Opin. Gastroenterol. 2001, 17, 184–192. [Google Scholar] [CrossRef] [PubMed Central]
- Zhong, M.; Kawaguchi, R.; Kassai, M.; Sun, H. Retina, retinol, retinal and the natural history of vitamin A as a light sensor. Nutrients 2012, 19, 2069–2096. [Google Scholar] [CrossRef] [PubMed Central]
- Zile, M.H. Function of vitamin A in vertebrate embryonic development. J. Nutr. 2001, 131, 705–708. [Google Scholar] [CrossRef] [PubMed Central]
- Rhinn, M.; Dollé, P. Retinoic acid signalling during development. Development 2012, 139, 843–845. [Google Scholar] [CrossRef] [PubMed Central]
- Kiser, P.D.; Palczewski, K. Retinoids and Retinal Diseases. Annu. Rev. Vis. Sci. 2016, 2, 197–234. [Google Scholar] [CrossRef] [PubMed Central]
- Kumar, S.; Duester, G. SnapShot: Retinoic acid signaling. Cell 2011, 147, 1422–1438. [Google Scholar] [CrossRef] [PubMed Central]
- See, A.W.; Clagett-Dame, M. The temporal requirement for vitamin A in the developing eye: Mechanism of action in optic fissure closure and new roles for the vitamin in regulating cell proliferation and adhesion in the embryonic retina. Dev. Biol. 2009, 325, 94–105. [Google Scholar] [CrossRef] [PubMed Central]
- Travis, G.H.; Golczak, M.; Moise, A.R.; Palczewski, K. Diseases Caused by Defects in the Visual Cycle: Retinoids as Potential Therapeutic Agents. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 469–512. [Google Scholar] [CrossRef] [PubMed Central]
- Warkany, J.; Schraffenberger, E. Congenital malformations induced in rats by maternal vitamin A deficiency; defects of the eye. Arch. Ophthal. 1946, 35, 150–169. [Google Scholar] [CrossRef] [PubMed Central]
- Wilson, J.G.; Warkany, J. Cardiac and aortic arch anomalies in the offspring of vitamin A deficient rats correlated with similar human anomalies. Pediatrics 1950, 5, 708–725. [Google Scholar] [PubMed Central]
- Dowling, J.E. Night blindness. Sci. Am 1966, 215, 78–84. [Google Scholar] [CrossRef] [PubMed Central]
- Dowling, J.E. Nutritional and Inherited Blindness in the Rat. Exp. Eye Res. 1964, 3, 348–356. [Google Scholar] [CrossRef] [PubMed Central]
- Chowdhury, S.; Kumar, R.; Ganguly, N.K.; Kumar, L.; Walia, B.N. Effect of vitamin A supplementation on childhood morbidity and mortality. Indian J. Med. Sci. 2002, 56, 259–264. [Google Scholar] [PubMed Central]
- Christian, P. Recommendations for indicators: Night blindness during pregnancy--a simple tool to assess vitamin A deficiency in a population. J. Nutr. 2002, 132, 2884S–2888S. [Google Scholar] [CrossRef] [PubMed Central]
- Le, H.G.; Dowling, J.E.; Cameron, D.J. Early retinoic acid deprivation in developing zebrafish results in microphthalmia. Vis. Neurosci. 2012, 29, 219–228. [Google Scholar] [CrossRef] [PubMed Central]
- Yahyavi, M.; Abouzeid, H.; Gawdat, G.; De Preux, A.S.; Xiao, T.; Bardakjian, T.; Schneider, A.; Choi, A.; Jorgenson, E.; Baier, H.; et al. ALDH1A3 loss of function causes bilateral anophthalmia/microphthalmia and hypoplasia of the optic nerve and optic chiasm. Hum. Mol. Genet. 2013, 22, 3250–3258. [Google Scholar] [CrossRef] [PubMed Central]
- Kawaguchi, R.; Yu, J.; Honda, J.; Hu, J.; Whitelegge, J.; Ping, P.; Wiita, P.; Bok, D.; Sun, H. Membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science 2007, 315, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M.; von Lintig, J. STRA6: Role in cellular retinol uptake and efflux. Hepatobiliary Surg. Nutr. 2015, 4, 229–242. [Google Scholar] [PubMed Central]
- Amengual, J.; Zhang, N.; Kemerer, M.; Maeda, T.; Palczewski, K.; von Lintig, J. STRA6 is critical for cellular vitamin A uptake and homeostasis. Hum. Mol. Genet. 2014, 23, 5402–5417. [Google Scholar] [CrossRef] [PubMed Central]
- Isken, A.; Golczak, M.; Oberhauser, V.; Hunzelmann, S.; Driever, W.; Imanishi, Y.; Palczewski, K.; von Lintig, J. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell. Metab. 2008, 7, 258–268. [Google Scholar] [CrossRef] [PubMed Central]
- Lobo, G.P.; Pauer, G.; Lipschutz, J.H.; Hagstrom, S.A. The Retinol-Binding Protein Receptor 2 (Rbpr2) Is Required for Photoreceptor Survival and Visual Function in the Zebrafish. Adv. Exp. Med. Biol. 2018, 1074, 569–576. [Google Scholar] [PubMed]
- Shi, Y.; Obert, E.; Rahman, B.; Rohrer, B.; Lobo, G.P. The Retinol Binding Protein Receptor 2 (Rbpr2) is required for Photoreceptor Outer Segment Morphogenesis and Visual Function in Zebrafish. Sci. Rep. 2017, 7, 16207–16217. [Google Scholar] [CrossRef] [PubMed Central]
- Kiefer, C.; Sumser, E.; Wernet, M.F.; von Lintig, J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. USA 2002, 99, 10581–10586. [Google Scholar] [CrossRef] [PubMed Central]
- Lobo, G.P.; Amengual, J.; Baus, D.; Shivdasani, R.A.; Taylor, D.; Von Lintig, J. Genetics and diet regulate vitamin A production via the homeobox transcription factor ISX. J. Biol. Chem. 2013, 288, 9017–9027. [Google Scholar] [CrossRef] [PubMed Central]
- Lobo, G.P.; Hessel, S.; Eichinger, A.; Noy, N.; Moise, A.R.; Wyss, A.; Palczewski, K.; von Lintig, J. ISX is a retinoic acid-sensitive gatekeeper that controls intestinal beta,beta-carotene absorption and vitamin A production. FASEB J. 2010, 24, 1656–1666. [Google Scholar] [CrossRef] [PubMed Central]
- Widiaja-Adhi, M.; Lobo, G.P.; Golczak, M.; von Lintig, J. A diet responsive regulatory network controls intestinal fat-soluble vitamin and carotenoid absorption. Hum. Mol. Genet. 2015, 24, 3206–3219. [Google Scholar] [CrossRef] [PubMed Central]
- Van Zundert, G.C.; Rodrigues, J.P.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.; van Dijk, M.; De Vries, S.J.; Bonvin, A.M. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed Central]
- Lobo, G.P.; Au, A.; Kiser, P.D.; Hagstrom, S.A. Involvement of Endoplasmic Reticulum Stress in TULP1 Induced Retinal Degeneration. PLoS ONE 2016, 17, e0151806. [Google Scholar] [CrossRef] [PubMed Central]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Lunt, S.C.; Haynes, T.; Perkins, B.D. Zebrafish ift57, ift88, and ift172 intraflagellar transport mutants disrupt cilia but do not affect hedgehog signaling. Dev. Dyn. 2009, 238, 1744–1759. [Google Scholar] [CrossRef] [PubMed Central]
- Chen, Y.; Clarke, O.B.; Kim, J.; Stowe, S.; Kim, Y.K.; Assur, Z.; Cavalier, M.; Godoy-Ruiz, R.; Desiree, C.; Manzini, C.; et al. Structure of the STRA6 receptor for retinol uptake. Science 2016, 353, 19–33. [Google Scholar] [CrossRef] [PubMed Central]
- Hwang, W.Y. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef] [PubMed Central]
- Talbot, J.C.; Amacher, S.L. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish 2014, 11, 583–585. [Google Scholar] [CrossRef] [PubMed Central]
- Kennedy, B.N.; Alvarez, Y.; Brockerhoff, S.E.; Stearns, G.W.; Sapetto-Rebow, B.; Taylor, M.R.; Hurley, J.B. Identification of a zebrafish cone photoreceptor-specific promoter and genetic rescue of achromatopsia in the nof mutant. Investig. Ophthalmol. Vis. Sci. 2007, 48, 522–529. [Google Scholar] [CrossRef] [PubMed Central]
- Perkins, B.D.; Fadool, J.M. Photoreceptor structure and development analyses using GFP transgenes. Methods Cell Biol. 2010, 100, 205–218. [Google Scholar] [PubMed Central]
- Lobo, G.P.; Fulmer, D.; Guo, L.; Zuo, X.; Dang, Y.; Kim, S.H.; Su, Y.; George, K.; Obert, E.; Fogelgren, B.; et al. The exocyst is required for photoreceptor ciliogenesis and retinal development. J. Biol. Chem. 2017, 292, 14814–14826. [Google Scholar] [CrossRef] [PubMed Central]
- Lobo, G.P.; Isken, A.; Hoff, S.; Babino, D.; von Lintig, J. BCDO2 acts as a carotenoid scavenger and gatekeeper for the mitochondrial apoptotic pathway. Development 2012, 139, 2966–2977. [Google Scholar] [CrossRef] [PubMed Central]
- Kim, Y.K.; Quadro, L. Reverse-phase high-performance liquid chromatography (HPLC) analysis of retinol and retinyl esters in mouse serum and tissues. Methods Mol. Biol. 2010, 652, 263–275. [Google Scholar] [PubMed Central]
- Berry, D.C.; Jacobs, H.; Marwarha, G.; Gely-Pernot, A.; O’Byrne, S.M.; DeSantis, D.; Klopfenstein, M.; Feret, B.; Dennefeld, C.; Blaner, W.S.; et al. The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye. J. Biol. Chem. 2013, 23, 24528–24539. [Google Scholar] [CrossRef] [PubMed Central]
- Bibliowicz, J.; Tittle, R.K.; Gross, J.M. Towards a better understanding of human eye disease: Insights from the zebrafish, Danio rerio. Prog. Mol. Biol. Transl. Sci. 2011, 100, 287–330. [Google Scholar] [PubMed Central]
- Biehlmaier, O.; Lampert, J.M.; von Lintig, J.; Kohler, K. Photoreceptor morphology is severely affected in the beta,beta-carotene-15,15’-oxygenase (bcox) zebrafish morphant. Eur. J. Neurosci. 2005, 21, 59–68. [Google Scholar] [CrossRef] [PubMed Central]
- Duester, G. Keeping an eye on retinoic acid signaling during eye development. Chem. Biol. Interact. 2009, 178, 178–181. [Google Scholar] [CrossRef] [PubMed Central]
- Drager, U.C.; Wagner, E.; Andreadis, A.; McCaffery, P. The Role and Evolutionary Development of Retinoic-Acid Signalling in the Eye; Birkhauser Verlag: Basel, Switzerland, 2000; pp. 73–82. [Google Scholar]
- Fain, G.L. Why photoreceptors die (and why they don’t). Bioessays 2006, 28, 344–354. [Google Scholar] [CrossRef] [PubMed Central]
- Isken, A.; Holzschuh, J.; Lampert, J.M.; Fischer, L.; Oberhauser, V.; Palczewski, K.; von Lintig, J. Sequestration of retinyl esters is essential for retinod signaling in the zebrafish embryos. J. Biol. Chem. 2007, 282, 1144–1151. [Google Scholar] [CrossRef] [PubMed Central]
- Lampert, J.M.; Holzschuh, J.; Hessel, S.; Driever, W.; Vogt, K.; von Lintig, J. Provitamin A conversion to retinal via the beta,beta-carotene-15,15′-oxygenase (bcox) is essential for pattern formation and differentiation during zebrafish embryogenesis. Development 2003, 130, 2173–2186. [Google Scholar] [CrossRef] [PubMed Central]
- Li, Z.; Korzh, V.; Gong, Z. Localized rbp4 expression in the yolk syncytial layer plays a role in yolk cell extension and early liver development. BMC Dev. Biol. 2007, 7, 117–130. [Google Scholar] [CrossRef] [PubMed Central]
- Ward, R.; Sundaramurthi, H.; Di Giacomo, V.; Kennedy, B.N. Enhancing Understanding of the Visual Cycle by Applying CRISPR/Cas9 Gene Editing in Zebrafish. Front. Cell Dev. Biol. 2018, 6, 37–50. [Google Scholar] [CrossRef] [PubMed Central]
- Schonthaler, H.B.; Lampert, J.M.; Isken, A.; Rinner, O.; Mader, A.; Gesemann, M.; Oberhauser, V.; Golczak, M.; Biehlmaier, O.; Palczewski, K.; et al. Evidence for RPE65-independent vision in the cone-dominated zebrafish retina. Eur. J. Neurosci. 2007, 26, 1940–1949. [Google Scholar] [CrossRef] [PubMed Central]
- Brockerhoff, S.E.; Fadool, J.M. Genetics of photoreceptor degeneration and regeneration in zebrafish. Cell Mol. Life Sci. 2011, 68, 651–659. [Google Scholar] [CrossRef] [PubMed Central]
- Campo-Paysaa, F.; Marletaz, F.; Laudet, V.; Schubert, M. Retinoic acid signaling in development: Tissue-specific functions and evolutionary origins. Genesis 2008, 46, 640–656. [Google Scholar] [CrossRef] [PubMed Central]
- Biesalski, H.K.; Frank, J.; Beck, S.C.; Heinrich, F.; Illek, B.; Reifen, R.; Gollnick, H.; Seeliger, M.W.; Wissinger, B.; Zrenner, E. Biochemical but not clinical vitamin A deficiency results from mutations in the gene for retinol binding protein. Am. J. Clin. Nutr. 1999, 69, 931–939. [Google Scholar] [CrossRef] [PubMed Central]
- Dobbs-McAuliffe, B.; Zhao, Q.; Linney, E. Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech. Dev. 2004, 121, 339–350. [Google Scholar] [CrossRef] [PubMed Central]
- Armstrong, G.A.; Liao, M.; You, Z.; Lissouba, A.; Chen, B.E.; Drapeau, P. Homology Directed Knockin of Point Mutations in the Zebrafish tardbp and fus Genes in ALS Using the CRISPR/Cas9 System. PLoS ONE 2016, 11, e0150188. [Google Scholar] [CrossRef] [PubMed Central]
- Fadool, J.M. Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Dev. Biol. 2003, 258, 277–290. [Google Scholar] [CrossRef] [PubMed Central]
- Hale, F. Pigs born without eyeballs. J. Hered. 1933, 24, 105–106. [Google Scholar] [CrossRef]
- Hyatt, G.A.; Schmitt, E.A.; Fadool, J.M.; Dowling, J.E. Retinoic acid alters photoreceptor development in vivo. Proc. Natl. Acad. Sci. USA 1996, 93, 13298–13303. [Google Scholar] [CrossRef] [PubMed Central]
- Kam, R.K.; Shi, W.; Chan, S.O.; Chen, Y.; Xu, G.; Bik-San Lau, C.; Fung, K.P.; Chan, W.Y.; Zhao, H. Dhrs3 protein attenuates retinoic acid signaling and is required for early embryonic patterning. J. Biol. Chem. 2013, 288, 31477–31487. [Google Scholar] [CrossRef] [PubMed Central]
- Hu, P.; Tian, M.; Bao, J.; Xing, G.; Gu, X.; Gao, X.; Linney, E.; Zhao, Q. Retinoid Regulation of the Zebrafish cyp26a1 Promoter. Dev. Dyn. 2008, 237, 3798–3808. [Google Scholar] [CrossRef] [PubMed Central]
- Humphries, M.M.; Rancourt, D.; Farrar, G.J.; Kenna, P.; Hazel, M.; Bush, R.A.; Sieving, P.A.; Sheils, D.M.; Creighton, P.; Erven, A.; et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat. Genet. 1997, 15, 216–219. [Google Scholar] [CrossRef] [PubMed Central]
- Kemp, C.M.; Jacobson, S.G.; Faulkner, D.J.; Walt, R.W. Visual function and rhodopsin levels in humans with vitamin A deficiency. Exp. Eye Res. 1998, 46, 185–197. [Google Scholar] [CrossRef] [PubMed Central]
- Maeda, A.; Palczewski, K. Retinal degeneration in animal models with a defective visual cycle. Retinal degeneration in animal models with a defective visual cycle. Drug Discov. Today Dis. Model. 2013, 10, e163–e172. [Google Scholar] [CrossRef] [PubMed Central]
- Maeda, T.; Cideciyan, A.V.; Maeda, A.; Golczak, M.; Aleman, T.S.; Jacobson, S.G.; Palczewski, K. Loss of cone photoreceptors caused by chromophore depletion is partially prevented by the artificial chromophore pro-drug, 9-cis-retinyl acetate. Hum. Mol. Genet. 2009, 18, 2277–2287. [Google Scholar] [CrossRef] [PubMed Central]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B.; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: Phi, psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed Central]
- McAbee, G.N.; Prieto, D.M.; Kirby, J.; Santilli, A.M.; Setty, R. Permanent visual loss due to dietary vitamin A deficiency in an autistic adolescent. J. Child Neurol. 2009, 24, 1288–1289. [Google Scholar] [CrossRef] [PubMed Central]
- Mitchell DMStevens, C.B.; Frey, R.A.; Hunter, S.S.; Ashino, R.; Kawamura, S.; Stenkamp, D.L. Retinoic Acid Signaling Regulates Differential Expression of the Tandemly-Duplicated Long Wavelength-Sensitive Cone Opsin Genes in Zebrafish. PLoS Genet. 2015, 21, e1005483. [Google Scholar] [CrossRef] [PubMed Central]
- Moise, A.R.; Golczak, M.; Imanishi, Y.; Palczewski, K. Topology and membrane association of lecithin: Retinol acyltransferase. J. Biol. Chem. 2007, 19, 2081–2090. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, P.J.; Cowley, G.S.; McGee, T.L.; Sandberg, M.A.; Berson, E.L.; Dryja, T.P. A null mutation in the rhodopsin gene causes rod photoreceptor dysfunction and autosomal recessive retinitis pigmentosa. Nat. Genet. 1992, 3, 209–213. [Google Scholar] [CrossRef] [PubMed Central]
- Stenkamp, D.L. Development of the Vertebrate Eye and Retina. Prog. Mol. Biol. Transl. Sci. 2015, 134, 397–414. [Google Scholar] [PubMed Central]
- Sun, H.; Sun, C.; Galicia, C.; Stenkamp, D.L. Transcripts within rod photoreceptors of the Zebrafish retina. Biochim. Biophys. Acta 2012, 1821, 99–112. [Google Scholar] [CrossRef] [PubMed Central]
- Tang, P.H.; Kono, M.; Koutalos, Y.; Ablonczy, Z.; Crouch, R.K. New insights into retinoid metabolism and cycling within the retina. Prog. Retin. Eye Res. 2013, 32, 48–63. [Google Scholar] [CrossRef] [PubMed Central]
- von Lintig, J. Metabolism of carotenoids and retinoids related to vision. J. Biol. Chem. 2012, 287, 1627–1634. [Google Scholar] [CrossRef] [PubMed Central]
- Wang, J.S.; Kefalov, V.J. The cone-specific visual cycle. Prog. Retin. Eye Res. 2011, 30, 115–128. [Google Scholar] [CrossRef] [PubMed Central]
- Wassef, L.; Quadro, L. Uptake of dietary retinoids at the maternal-fetal barrier: In vivo evidence for the role of lipoprotein lipase and alternative pathways. J. Biol. Chem. 2011, 286, 32198–32207. [Google Scholar] [CrossRef] [PubMed Central]
- Yin, J.; Brocher, J.; Linder, B.; Hirmer, A.; Sundaramurthi, H.; Fischer, U.; Winkler, C. The 1D4 antibody labels outer segments of long double cone but not rod photoreceptors in zebrafish. Investig. Ophthalmol. Vis. Sci. 2012, 26, 4943–4951. [Google Scholar] [CrossRef] [PubMed Central]
- Zhao, Q.; Dobbs-McAuliffe, B.; Linney, E. Expression of cyp26b1 during zebrafish early development. Gene Expr. Patterns 2005, 5, 363–369. [Google Scholar] [CrossRef] [PubMed Central]
- Znoiko, S.L.; Rohrer, B.; Lu, K.; Lohr, H.R.; Crouch, R.K.; Ma, J.X. Downregulation of cone-specific gene expression and degeneration of cone photoreceptors in the Rpe65-/- mouse at early ages. Investig. Ophthalmol. Vis. Sci. 2005, 46, 1473–1479. [Google Scholar] [CrossRef] [PubMed Central]
- Cukras, C.; Gaasterland, T.; Lee, P.; Gudiseva, H.V.; Chavali, V.R.; Pullakhandam, R.; Maranhao, B.; Edsall, L.; Soares, S.; Reddy, G.B.; et al. Exome analysis identified a novel mutation in the RBP4 gene in a consanguineous pedigree with retinal dystrophy and developmental abnormalities. PLoS ONE 2012, 7, e50205. [Google Scholar] [CrossRef] [PubMed Central]




| RBP4-Rbpr2 Docking | RBP4-STRA6 Docking | |
|---|---|---|
| HADDOCK score | −85.6 +/− 2.0 | −81.4 +/− 3.1 |
| RMSD from the overall lowest-energy structure | 0.6 +/− 0.3 | 16.0 +/− 0.3 |
| Van der Waals energy | −33.3 +/− 1.5 | −39.8 +/− 1.8 |
| Electrostatic energy | −67.2 +/− 14.1 | −16.6 +/− 2.0 |
| Desolvation energy | −41.6 +/− 1.9 | −38.7 +/− 3.5 |
| Restraints violation energy | 27.3 +/− 13.23 | 3.8 +/− 2.54 |
| Buried Surface Area | 1127.8 +/− 74.6 | 902.8 +/− 35.0 |
| Z-Score | –1.6 | –1.4 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solanki, A.K.; Kondkar, A.A.; Fogerty, J.; Su, Y.; Kim, S.-H.; Lipschutz, J.H.; Nihalani, D.; Perkins, B.D.; Lobo, G.P. A Functional Binding Domain in the Rbpr2 Receptor Is Required for Vitamin A Transport, Ocular Retinoid Homeostasis, and Photoreceptor Cell Survival in Zebrafish. Cells 2020, 9, 1099. https://doi.org/10.3390/cells9051099
Solanki AK, Kondkar AA, Fogerty J, Su Y, Kim S-H, Lipschutz JH, Nihalani D, Perkins BD, Lobo GP. A Functional Binding Domain in the Rbpr2 Receptor Is Required for Vitamin A Transport, Ocular Retinoid Homeostasis, and Photoreceptor Cell Survival in Zebrafish. Cells. 2020; 9(5):1099. https://doi.org/10.3390/cells9051099
Chicago/Turabian StyleSolanki, Ashish K., Altaf A. Kondkar, Joseph Fogerty, Yanhui Su, Seok-Hyung Kim, Joshua H. Lipschutz, Deepak Nihalani, Brian D. Perkins, and Glenn P. Lobo. 2020. "A Functional Binding Domain in the Rbpr2 Receptor Is Required for Vitamin A Transport, Ocular Retinoid Homeostasis, and Photoreceptor Cell Survival in Zebrafish" Cells 9, no. 5: 1099. https://doi.org/10.3390/cells9051099
APA StyleSolanki, A. K., Kondkar, A. A., Fogerty, J., Su, Y., Kim, S.-H., Lipschutz, J. H., Nihalani, D., Perkins, B. D., & Lobo, G. P. (2020). A Functional Binding Domain in the Rbpr2 Receptor Is Required for Vitamin A Transport, Ocular Retinoid Homeostasis, and Photoreceptor Cell Survival in Zebrafish. Cells, 9(5), 1099. https://doi.org/10.3390/cells9051099







