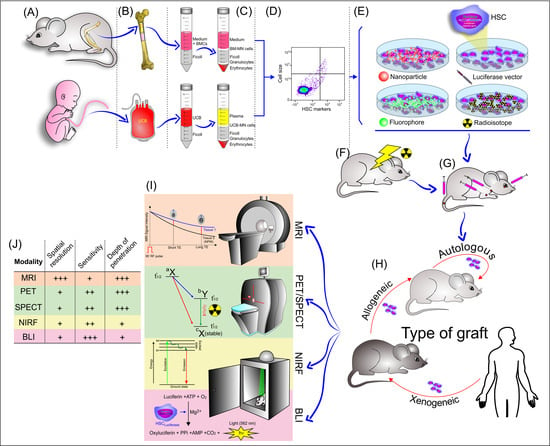
Noninvasive Tracking of Hematopoietic Stem Cells in a Bone Marrow Transplant Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Inclusion Criteria
2.3. Exclusion Criteria
2.4. Data Extraction, Data Collection, and Risk of Bias Assessment
2.5. Data Analysis
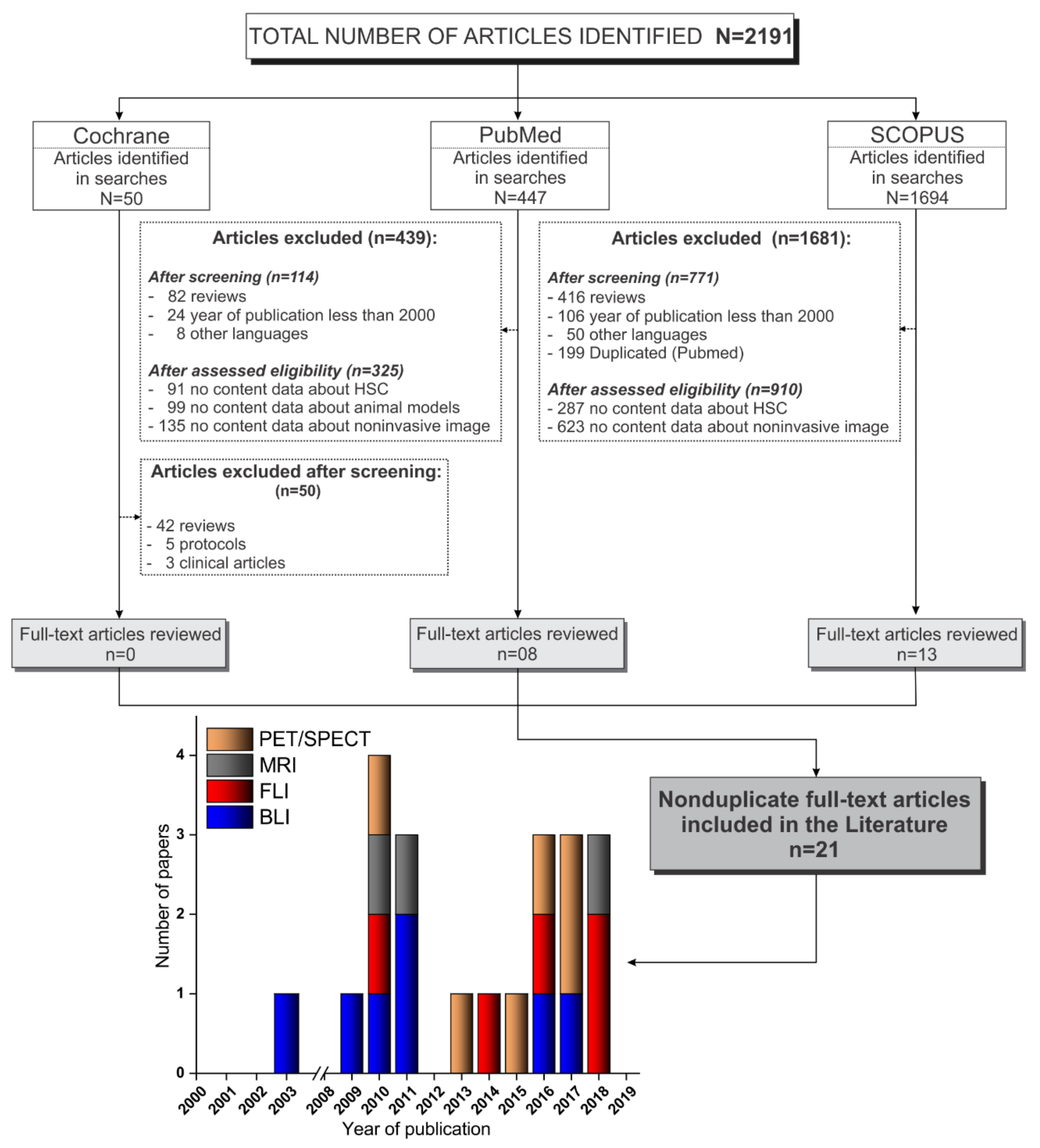
3. Results
3.1. Selection Process of the Articles Identified According to the PRISMA Guidelines
3.2. Extraction and Isolation of Hematopoietic Stem Cells
3.3. Lentiviral Transduction of Hematopoietic Stem Cells
3.4. Labeling Strategies and Techniques of Hematopoietic Stem Cells with Tracers
3.4.1. Labeling of HSCs with Radioisotopes/Radiopharmaceuticals
3.4.2. Labeling of HSCs with Fluorophore
3.4.3. Labeling of HSCs with Nanoparticles
3.5. Administration of HSCs Labeled/Transfected in the Bone Marrow Transplant Model
3.6. Imaging Techniques Used in HSC Migration Homing and Tracking Analysis
3.6.1. Bioluminescence
3.6.2. PET/SPECT
3.6.3. Fluorescence Imaging
3.6.4. Magnetic Resonance Imaging
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Lorenz, E.; Uphoff, D.; Reid, T.R.; Shelton, E. Modification of irradiation injury in mice and guinea pigs by bone marrow injections. J. Natl. Cancer Inst. 1951, 12, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.D.; Lochte, H.L., Jr.; Cannon, J.H.; Sahler, O.D.; Ferrebee, J.W. Supralethal whole body irradiation and isologous marrow transplantation in man. J. Clin. Investig. 1959, 38, 1709–1716. [Google Scholar] [CrossRef]
- Gratwohl, A.; Pasquini, M.C.; Aljurf, M.; Atsuta, Y.; Baldomero, H.; Foeken, L.; Gratwohl, M.; Bouzas, L.F.; Confer, D.; Frauendorfer, K.; et al. One million haemopoietic stem-cell transplants: A retrospective observational study. Lancet. Haematol. 2015, 2, e91–e100. [Google Scholar] [CrossRef]
- Copelan, E.A. Hematopoietic stem-cell transplantation. N. Engl. J. Med. 2006, 354, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, J.L.; Levine, J.E.; Reddy, P.; Holler, E. Graft-versus-host disease. Lancet 2009, 373, 1550–1561. [Google Scholar] [CrossRef]
- McDonald-Hyman, C.; Turka, L.A.; Blazar, B.R. Advances and challenges in immunotherapy for solid organ and hematopoietic stem cell transplantation. Sci. Transl. Med. 2015, 7, 280rv282. [Google Scholar] [CrossRef]
- Brown, V.I. Graft Failure. In Hematopoietic Stem Cell Transplantation for the Pediatric Hematologist/Oncologist; Brown, V.I., Ed.; Springer International Publishing: Cham, Switzerland, 2018; pp. 187–194. [Google Scholar] [CrossRef]
- Askenasy, N.; Farkas, D.L. Optical imaging of PKH-labeled hematopoietic cells in recipient bone marrow in vivo. Stem Cells 2002, 20, 501–513. [Google Scholar] [CrossRef]
- Wang, X.; Rosol, M.; Ge, S.; Peterson, D.; McNamara, G.; Pollack, H.; Kohn, D.B.; Nelson, M.D.; Crooks, G.M. Dynamic tracking of human hematopoietic stem cell engraftment using in vivo bioluminescence imaging. Blood 2003, 102, 3478–3482. [Google Scholar] [CrossRef]
- Kalchenko, V.; Shivtiel, S.; Malina, V.; Lapid, K.; Haramati, S.; Lapidot, T.; Brill, A.; Harmelin, A. Use of lipophilic near-infrared dye in whole-body optical imaging of hematopoietic cell homing. J. Biomed. Opt. 2006, 11, 050507. [Google Scholar] [CrossRef]
- Steiner, D.; Gelovani, J.; Savoldo, B.; Robinson, S.N.; Decker, W.K.; Brouard, N.; Najjar, A.; Xing, D.; Yang, H.; Li, S.; et al. Noninvasive bioluminescent imaging demonstrates long-term multilineage engraftment of ex vivo-expanded CD34-selected umbilical cord blood cells. Stem Cells 2009, 27, 1932–1940. [Google Scholar] [CrossRef]
- Cen, P.; Chen, J.; Hu, C.; Fan, L.; Wang, J.; Li, L. Noninvasive in-vivo tracing and imaging of transplanted stem cells for liver regeneration. Stem Cell Res. Ther. 2016, 7, 143. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Marciscano, A.E.; Thorek, D.L.J. Role of noninvasive molecular imaging in determining response. Adv. Radiat. Oncol. 2018, 3, 534–547. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Shu, J. Multimodal Molecular Imaging: Current Status and Future Directions. Contrast Media Mol. Imaging 2018, 2018, 1382183. [Google Scholar] [CrossRef] [PubMed]
- Youn, H.; Hong, K.J. In vivo Noninvasive Small Animal Molecular Imaging. Osong Public Health Res. Perspect. 2012, 3, 48–59. [Google Scholar] [CrossRef]
- Andrade, J.; Ge, S.; Symbatyan, G.; Rosol, M.S.; Olch, A.J.; Crooks, G.M. Effects of sublethal irradiation on patterns of engraftment after murine bone marrow transplantation. Biol. Blood Marrow Transplant. J. Am. Soc. Blood Marrow Transplant. 2011, 17, 608–619. [Google Scholar] [CrossRef]
- Astuti, Y.; Kramer, A.C.; Blake, A.L.; Blazar, B.R.; Tolar, J.; Taisto, M.E.; Lund, T.C. A Functional Bioluminescent Zebrafish Screen for Enhancing Hematopoietic Cell Homing. Stem Cell Rep. 2017, 8, 177–190. [Google Scholar] [CrossRef]
- Lin, Y.; Cheung, P.; Roth, J.C.; Wilson, D.L.; Gerson, S.L. Imaging stem cell-derived persistent foci after in vivo selection of lentiviral MGMT-P140K transduced murine bone marrow cells. Mol. Ther. J. Am. Soc. Gene Ther. 2011, 19, 1342–1352. [Google Scholar] [CrossRef]
- Ohmori, T.; Kashiwakura, Y.; Ishiwata, A.; Madoiwa, S.; Mimuro, J.; Furukawa, Y.; Sakata, Y. Vinculin is indispensable for repopulation by hematopoietic stem cells, independent of integrin function. J. Biol. Chem. 2010, 285, 31763–31773. [Google Scholar] [CrossRef]
- Saia, M.; Termanini, A.; Rizzi, N.; Mazza, M.; Barbieri, E.; Valli, D.; Ciana, P.; Gruszka, A.M.; Alcalay, M. AML1/ETO accelerates cell migration and impairs cell-to-cell adhesion and homing of hematopoietic stem/progenitor cells. Sci. Rep. 2016, 6, 34957. [Google Scholar] [CrossRef]
- Hamilton, N.; Sabroe, I.; Renshaw, S.A. A method for transplantation of human HSCs into zebrafish, to replace humanised murine transplantation models [version 1; referees: 2 approved]. F1000Research 2018, 7. [Google Scholar] [CrossRef]
- Lopez, D.; Lin, L.; Monaghan, J.R.; Cogle, C.R.; Bova, F.J.; Maden, M.; Scott, E.W. Mapping hematopoiesis in a fully regenerative vertebrate: The axolotl. Blood 2014, 124, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Parada-Kusz, M.; Penaranda, C.; Hagedorn, E.J.; Clatworthy, A.; Nair, A.V.; Henninger, J.E.; Ernst, C.; Li, B.; Riquelme, R.; Jijon, H.; et al. Generation of mouse-zebrafish hematopoietic tissue chimeric embryos for hematopoiesis and host-pathogen interaction studies. Dis. Models Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Staal, F.J.T.; Spaink, H.P.; Fibbe, W.E. Visualizing Human Hematopoietic Stem Cell Trafficking in Vivo Using a Zebrafish Xenograft Model. Stem Cells Dev. 2016, 25, 360–365. [Google Scholar] [CrossRef] [PubMed]
- Ushiki, T.; Kizaka-Kondoh, S.; Ashihara, E.; Tanaka, S.; Masuko, M.; Hirai, H.; Kimura, S.; Aizawa, Y.; Maekawa, T.; Hiraoka, M. Noninvasive tracking of donor cell homing by near-infrared fluorescence imaging shortly after bone marrow transplantation. PLoS ONE 2010, 5, e11114. [Google Scholar] [CrossRef] [PubMed]
- Pantin, J.M.; Hoyt, R.F., Jr.; Aras, O.; Sato, N.; Chen, M.Y.; Hunt, T.; Clevenger, R.; Eclarinal, P.; Adler, S.; Choyke, P.; et al. Optimization of intrabone delivery of hematopoietic progenitor cells in a swine model using cell radiolabeling with zirconium. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2015, 15, 606–617. [Google Scholar] [CrossRef]
- Faivre, L.; Chaussard, M.; Vercellino, L.; Vanneaux, V.; Hosten, B.; Teixera, K.; Parietti, V.; Merlet, P.; Sarda-Mantel, L.; Rizzo-Padoin, N.; et al. 18F-FDG labelling of hematopoietic stem cells: Dynamic study of bone marrow homing by PET–CT imaging and impact on cell functionality. Curr. Res. Transl. Med. 2016, 64, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Asiedu, K.O.; Koyasu, S.; Szajek, L.P.; Choyke, P.L.; Sato, N. Bone Marrow Cell Trafficking Analyzed by (89)Zr-oxine Positron Emission Tomography in a Murine Transplantation Model. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 2759–2768. [Google Scholar] [CrossRef]
- Massollo, M.; Podestà, M.; Marini, C.; Morbelli, S.; Cassanelli, C.; Pinto, V.; Ubezio, G.; Curti, G.; Uccelli, A.; Frassoni, F.; et al. Contact with the bone marrow microenvironment readdresses the fate of transplanted hematopoietic stem cells. Exp. Hematol. 2010, 38, 968–977. [Google Scholar] [CrossRef]
- Sambuceti, G.; Massollo, M.; Marini, C.; Podesta, M.; Cassanelli, C.; Morbelli, S.; Fiz, F.; Buschiazzo, A.; Capitanio, S.; Augeri, C.; et al. Trafficking and homing of systemically administered stem cells: The need for appropriate analysis tools of radionuclide images. Q. J. Nucl. Med. Mol. Imaging Off. Publ. Ital. Assoc. Nucl. Med. (AIMN) Int. Assoc. Radiopharmacol. (IAR) Sect. Soc. 2013, 57, 207–215. [Google Scholar]
- Lange, S.; Steder, A.; Killian, D.; Knuebel, G.; Sekora, A.; Vogel, H.; Lindner, I.; Dunkelmann, S.; Prall, F.; Murua Escobar, H.; et al. Engraftment Efficiency after Intra-Bone Marrow versus Intravenous Transplantation of Bone Marrow Cells in a Canine Nonmyeloablative Dog Leukocyte Antigen-Identical Transplantation Model. Biol. Blood Marrow Transplant. J. Am. Soc. Blood Marrow Transplant. 2017, 23, 247–254. [Google Scholar] [CrossRef]
- Bengtsson, N.E.; Kim, S.; Lin, L.; Walter, G.A.; Scott, E.W. Ultra-high-field MRI real-time imaging of HSC engraftment of the bone marrow niche. Leukemia 2011, 25, 1223–1231. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Niemeyer, M.; Oostendorp, R.A.J.; Kremer, M.; Hippauf, S.; Jacobs, V.R.; Baurecht, H.; Ludwig, G.; Piontek, G.; Bekker-Ruz, V.; Timmer, S.; et al. Non-invasive tracking of human haemopoietic CD34+ stem cells in vivo in immunodeficient mice by using magnetic resonance imaging. Eur. Radiol. 2010, 20, 2184–2193. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, S.K.; Manzar, G.S.; Zavazava, N.; Assouline, J.G. Tracking embryonic hematopoietic stem cells to the bone marrow: Nanoparticle options to evaluate transplantation efficiency. Stem Cell Res. Ther. 2018, 9, 204. [Google Scholar] [CrossRef] [PubMed]
- Choy, G.; Choyke, P.; Libutti, S.K. Current advances in molecular imaging: Noninvasive in vivo bioluminescent and fluorescent optical imaging in cancer research. Mol. Imaging 2003, 2, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Mezzanotte, L.; van ‘t Root, M.; Karatas, H.; Goun, E.A.; Lowik, C. In Vivo Molecular Bioluminescence Imaging: New Tools and Applications. Trends Biotechnol. 2017, 35, 640–652. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Van Zant, G.; Szilvassy, S.J. Effects of aging on the homing and engraftment of murine hematopoietic stem and progenitor cells. Blood 2005, 106, 1479–1487. [Google Scholar] [CrossRef] [PubMed]
- Hillman, E.M.; Amoozegar, C.B.; Wang, T.; McCaslin, A.F.; Bouchard, M.B.; Mansfield, J.; Levenson, R.M. In vivo optical imaging and dynamic contrast methods for biomedical research. Philos. Trans. A Math. Phys. Eng. Sci. 2011, 369, 4620–4643. [Google Scholar] [CrossRef]
- Jung, J.S.; Jo, D.; Jo, G.; Hyun, H. Near-Infrared Contrast Agents for Bone-Targeted Imaging. Tissue Eng. Regen. Med. 2019, 16, 443–450. [Google Scholar] [CrossRef]
- Shah, K.; Weissleder, R. Molecular optical imaging: Applications leading to the development of present day therapeutics. NeuroRx 2005, 2, 215–225. [Google Scholar] [CrossRef]
- Balas, C. Review of biomedical optical imaging—A powerful, non-invasive, non-ionizing technology for improvingin vivodiagnosis. Meas. Sci. Technol. 2009, 20, 104020. [Google Scholar] [CrossRef]
- James, M.L.; Gambhir, S.S. A molecular imaging primer: Modalities, imaging agents, and applications. Physiol. Rev. 2012, 92, 897–965. [Google Scholar] [CrossRef] [PubMed]
- Yahyapour, R.; Farhood, B.; Graily, G.; Rezaeyan, A.; Rezapoor, S.; Abdollahi, H.; Cheki, M.; Amini, P.; Fallah, H.; Najafi, M.; et al. Stem Cell Tracing Through MR Molecular Imaging. Tissue Eng. Regen. Med. 2018, 15, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Hengerer, A.; Grimm, J. Molecular magnetic resonance imaging. Biomed. Imaging Interv. J. 2006, 2, e8. [Google Scholar] [CrossRef] [PubMed]
- Ntziachristos, V.; Bremer, C.; Weissleder, R. Fluorescence imaging with near-infrared light: New technological advances that enable in vivo molecular imaging. Eur. Radiol. 2003, 13, 195–208. [Google Scholar] [CrossRef]
- Lu, F.-M.; Yuan, Z. PET/SPECT molecular imaging in clinical neuroscience: Recent advances in the investigation of CNS diseases. Quant. Imaging Med. Surg. 2015, 5, 433–447. [Google Scholar]
- Varani, M.; Auletta, S.; Signore, A.; Galli, F. State of the Art of Natural Killer Cell Imaging: A Systematic Review. Cancers 2019, 11, 967. [Google Scholar] [CrossRef]
- Artemov, D. Molecular magnetic resonance imaging with targeted contrast agents. J. Cell. Biochem. 2003, 90, 518–524. [Google Scholar] [CrossRef]
- Zhang, X.-A.; Lovejoy, K.S.; Jasanoff, A.; Lippard, S.J. Water-soluble porphyrins as a dual-function molecular imaging platform for MRI and fluorescence zinc sensing. Proc. Natl. Acad. Sci. USA 2007, 104, 10780. [Google Scholar] [CrossRef]
- Louie, A. Multimodality imaging probes: Design and challenges. Chem. Rev. 2010, 110, 3146–3195. [Google Scholar] [CrossRef]
- Mezzanotte, L.; Que, I.; Kaijzel, E.; Branchini, B.; Roda, A.; Lowik, C. Sensitive dual color in vivo bioluminescence imaging using a new red codon optimized firefly luciferase and a green click beetle luciferase. PLoS ONE 2011, 6, e19277. [Google Scholar] [CrossRef]
- Saito, K.; Chang, Y.F.; Horikawa, K.; Hatsugai, N.; Higuchi, Y.; Hashida, M.; Yoshida, Y.; Matsuda, T.; Arai, Y.; Nagai, T. Luminescent proteins for high-speed single-cell and whole-body imaging. Nat. Commun. 2012, 3, 1262. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gotzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. PLoS Med. 2009, 6, e1000100. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Ann. Intern. Med. 2009, 151, 264–269. [Google Scholar] [CrossRef]
- Morikawa, T.; Takubo, K. Use of Imaging Techniques to Illuminate Dynamics of Hematopoietic Stem Cells and Their Niches. Front. Cell Dev. Biol. 2017, 5, 62. [Google Scholar] [CrossRef] [PubMed]
- Morrison, S.J.; Scadden, D.T. The bone marrow niche for haematopoietic stem cells. Nature 2014, 505, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Arber, C.; Brenner, M.K.; Reddy, P. Mouse models in bone marrow transplantation and adoptive cellular therapy. Semin. Hematol. 2013, 50, 131–144. [Google Scholar] [CrossRef][Green Version]
- Nilsson, S.K.; Johnston, H.M.; Coverdale, J.A. Spatial localization of transplanted hemopoietic stem cells: Inferences for the localization of stem cell niches. Blood 2001, 97, 2293–2299. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Yin, T.; Wiegraebe, W.; He, X.C.; Miller, D.; Stark, D.; Perko, K.; Alexander, R.; Schwartz, J.; Grindley, J.C.; et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature 2009, 457, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Beeres, S.L.; Bengel, F.M.; Bartunek, J.; Atsma, D.E.; Hill, J.M.; Vanderheyden, M.; Penicka, M.; Schalij, M.J.; Wijns, W.; Bax, J.J. Role of imaging in cardiac stem cell therapy. J. Am. Coll. Cardiol. 2007, 49, 1137–1148. [Google Scholar] [CrossRef]
- Schroeder, T. Imaging stem-cell-driven regeneration in mammals. Nature 2008, 453, 345–351. [Google Scholar] [CrossRef]
- Rizzo, S.; Petrella, F.; Politi, L.S.; Wang, P. Molecular Imaging of Stems Cells: In Vivo Tracking and Clinical Translation. Stem Cells Int. 2017, 2017, 1783841. [Google Scholar] [CrossRef] [PubMed]
- Bose, R.J.C.; Mattrey, R.F. Accomplishments and challenges in stem cell imaging in vivo. Drug Discov. Today 2019, 24, 492–504. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Petrella, F.; Nicosia, L.; Bellomi, M.; Rizzo, S. Molecular Imaging of Stem Cell Transplantation for Liver Diseases: Monitoring, Clinical Translation, and Theranostics. Stem Cells Int. 2016, 2016, 4058656. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, H.R.; Mamani, J.B.; Nucci, M.P.; Nucci, L.P.; Kondo, A.T.; Fantacini, D.M.C.; de Souza, L.E.B.; Picanco-Castro, V.; Covas, D.T.; Kutner, J.M.; et al. Triple-modal imaging of stem-cells labeled with multimodal nanoparticles, applied in a stroke model. World J. Stem Cells 2019, 11, 100–123. [Google Scholar] [CrossRef]
- Mamani, J.B.; Malheiros, J.M.; Cardoso, E.F.; Tannús, A.; Silveira, P.H.; Gamarra, L.F. In vivo magnetic resonance imaging tracking of C6 glioma cells labeled with superparamagnetic iron oxide nanoparticles. Einstein 2012, 10, 164–170. [Google Scholar] [CrossRef]
- Sibov, T.T.; Pavon, L.F.; Miyaki, L.A.; Mamani, J.B.; Nucci, L.P.; Alvarim, L.T.; Silveira, P.H.; Marti, L.C.; Gamarra, L. Umbilical cord mesenchymal stem cells labeled with multimodal iron oxide nanoparticles with fluorescent and magnetic properties: Application for in vivo cell tracking. Int. J. Nanomed. 2014, 9, 337–350. [Google Scholar] [CrossRef]
- Chen, K.; Chen, X. Design and development of molecular imaging probes. Curr. Top. Med. Chem. 2010, 10, 1227–1236. [Google Scholar] [CrossRef]
- Perrone-Filardi, P.; Dellegrottaglie, S.; Rudd, J.; Costanzo, P.; Marciano, C.; Vassallo, E.; Marsico, F.; Ruggiero, D.; Petretta, M.; Chiariello, M.; et al. Molecular imaging of atherosclerosis in translational medicine. Eur. J. Nucl. Med. Mol. Imaging 2010, 38, 969–975. [Google Scholar] [CrossRef][Green Version]
- Welling, M.M.; Hensbergen, A.W.; Bunschoten, A.; Velders, A.H.; Roestenberg, M.; van Leeuwen, F.W.B. An update on radiotracer development for molecular imaging of bacterial infections. Clin. Transl. Imaging 2019, 7, 105–124. [Google Scholar] [CrossRef]
- Deshpande, N.; Pysz, M.A.; Willmann, J.K. Molecular ultrasound assessment of tumor angiogenesis. Angiogenesis 2010, 13, 175–188. [Google Scholar] [CrossRef]
- Fernandez-Friera, L.; Garcia-Alvarez, A.; Ibanez, B. Imagining the future of diagnostic imaging. Rev. Esp. De Cardiol. 2013, 66, 134–143. [Google Scholar] [CrossRef]
- Gangadaran, P.; Ahn, B.-C. Molecular Imaging: A Useful Tool for the Development of Natural Killer Cell-Based Immunotherapies. Front. Immunol. 2017, 8, 1090. [Google Scholar] [CrossRef] [PubMed]
- Sakhtianchi, R.; Minchin, R.F.; Lee, K.B.; Alkilany, A.M.; Serpooshan, V.; Mahmoudi, M. Exocytosis of nanoparticles from cells: Role in cellular retention and toxicity. Adv. Colloid Interface Sci. 2013, 201, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Gurevitch, O.; Prigozhina, T.B.; Pugatsch, T.; Slavin, S. Transplantation of allogeneic or xenogeneic bone marrow within the donor stromal microenvironment. Transplantation 1999, 68, 1362–1368. [Google Scholar] [CrossRef]
- Ozdemir, Z.N.; Civriz Bozdag, S. Graft failure after allogeneic hematopoietic stem cell transplantation. Transfus. Apher. Sci. 2018, 57, 163–167. [Google Scholar] [CrossRef]
- Huang, X.; Guo, B.; Capitano, M.; Broxmeyer, H.E. Past, present, and future efforts to enhance the efficacy of cord blood hematopoietic cell transplantation. F1000Research 2019, 8. [Google Scholar] [CrossRef]


| Ref. | Year | Extraction of HSCs | Isolation of HSCs | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor | Source of Cells | Donor Age | Donor Gender | Harvest | Medium and Supplementation | Separation | Immunophenotypic Characterization | Technique for Sorting | Purity (%) | ||
| Parada-Kusz et al. [23] | 2018 | Mice (C57BL/6 or CByJ.B6- UBI-GFP) | BM | 6-8 w | M-F | Maceration | PBS (0.5% BSA/BFS, 2 mM EDTA) | NR | CD45+; c-kit+; CD11b+ | MACS | NR |
| Sweeney et al. [34] | 2018 | Mice (129/SvJ) | ESC | NR | NR | Differentiation | StemPro34 with cytokines | NR | CD41+; CD45+; Sca-1+ c-Kit+ | NR | NR |
| Asiedu et al. [28] | 2017 | Mice (C57BL/6 CD45.2) | BM | NR | M-F | NR | NR | NR | NR | NR | NR |
| Lange et al. [31] | 2017 | Dog (Beagle) | BM | 1–3 y | M | Aspirate | NR | Ficoll | CD34+ | NR | NR |
| Astuti et al. [17] | 2017 | Fish (zebrafish ubi:luc) | WKM | NR | NR | Maceration | PBS | NR | NR | NR | NR |
| Saia et al. [20] | 2016 | Mice (Ubi-Luc2KI) | BM | 8–10 w | NR | NR | NR | NR | Lin- | MACS | NR |
| Lopez et al. [22] | 2014 | Axolotl (GFP+; nucCherryRed+) | SP; LV | NR | NR | Maceration | APBS (5% FBS) | Ficoll | NR | FACS | NR |
| Sambuceti et al. [30] | 2013 | Rat (Lewis) | BM | 7 w | M | Flushing | DMEN (10% FBS, UFH) | Ficoll | CD90+ | FACS | NR |
| Lin et al. [18] | 2011 | Mice (Balb/c) | BM | 6–8 w | F | Flushing | α-MEM (10% UFH) | NR | NR | NR | NR |
| Andrade et al. [16] | 2011 | Mice (FVB H-2Kq) | BM | NR | NR | NR | NR | NR | NR | NR | NR |
| Bengtsson et al. [32] | 2011 | Mice (TgCAGDsRed) | BM | NR | M | Flushing | NR | NR | lin- c-kit+ Sca-1+ | MACS; FACS | NR |
| Ohmori et al. [19] | 2010 | Mice (C57BL/6 Ly5.1) | BM | NR | NR | NR | NR | NR | lin- c-kit+ Sca-1+ | MACS; FACS | NR |
| Ushiki et al. [25] | 2010 | Mice (Balb/c nu/nu) | BM; SP | 7–9 w | NR | NR | NR | Lympholyte | NR | MACS | NR |
| Massolo et al. [29] | 2010 | Rat (Lewis) | BM | 7 w | M | Flushing | DMEN (10% FBS, UFH) | Ficoll | CD90+ | FACS | NR |
| Hamilton et al. [21] | 2018 | Human | PB | 18-40 y | M-F | NR | NR | Percoll | CD34+ | MACS; FACS | NR |
| Faivre et al. [27] | 2016 | Human | CB | NR | NR | NR | NR | NR | CD34+ | MACS | 94.6 |
| Staal et al. [24] | 2016 | Human | CB | NR | NR | NR | NR | Ficoll | CD34+ | MACS; FACS | 95 |
| Pantin et al. [26] | 2015 | Human | PB | NR | NR | Apheresis | NR | NR | CD133+ | MACS | NR |
| Niemeyer et al. [33] | 2010 | Human | CB | NR | NR | NA | Citrate buffer | Ficoll | CD34+ | MACS | >95 |
| Steiner et al. [11] | 2008 | Human | CB | NR | NR | NR | NR | Ficoll | CD34+ | MACS; FACS | >95 |
| Wang et al. [9] | 2003 | Human | CB | NR | NR | NR | NR | NR | CD34+ CD38-; CD34+ | MACS; FACS | 80-90 |
| Ref. | Tracking Agent | Vector | Cell Dose | Transfection Agent | MOI | Medium Supplemented | Cytokines SDT (ng/mL) | Incubation (Hours) | Evaluation |
|---|---|---|---|---|---|---|---|---|---|
| Lin et al. [18] | FLUC | Lentiviral plasmid, pCSO-rre-cppt-MCU3-LUC | NR | Polybrene 8 μg/mL | 0.5–1 | DMEN; Iscove’s MEM (20% FBS) | mIL-3: 20; mIL-6: 50; rSCF: 50 | 48 | FCT; CFU; PCR |
| Ohmori et al. [19] | LUC | LentiLox vectors | NR | Polybrene 8 μg/mL | 20 | StemPro 34 SFM | SFC: 100; TPO: 100; IL-6: 100; Flt-3L: 100; sIL-6R: 200 | 24 | FCT |
| Steiner et al. [11] | FLUC | Plasmid pHIV-GFPFFLuc | 3-8 × 105 per well | Retronectin 100 μg/mL | NR | α-MEM (10% FCS; 2mM L-Gln) | NR | Overnight | FCT |
| Wang et al. [9] | FLUC | Lentiviral vector SMPU-R-MNCU3-LUC | 1–10 × 104 per plate; 3–30 × 102 per well | NR | NR | NR | IL-3: 5; IL-6: 16.5; SCF: 25 | 24 (2 cycles) | IHC |
| Ref. | Radioisotope | Radiopharmaceuticals | Half-Life (h) | Manufacture | Labeling Activity (MBq) | Activity after Labeling (MBq) | Cell Dose | Incubation (min) | Yield (%) |
|---|---|---|---|---|---|---|---|---|---|
| Asiedu et al. [28] | 89Zr | Oxime | 78.4 | Cyclotron | 0.01–5.55 | 0.0036- 1.7 | 1 × 106 | 20 | 26-30 |
| Lange et al. [31] | 99mTc | HMPAO | 6.03 | Ceretec, GE Healthcare | 550–583; 669–1350 | NR | NR | NR | NR |
| Faivre et al. [27] | 18F | FDG | 1.83 | Cyclotron | 301.8–945.9 | 5–10 | NR | 30 | 94.6 ± 6 |
| Pantin et al. [26] | 89Zr | NA | 78.4 | Cyclotron | 0.37 | 0.28 | 2x108 | 30 | NR |
| Sambuceti et al. [30] | 99mTc | HMPAO | 6.03 | Ceretec, GE Healthcare | 37 | 5.55 | 2x106 | 30 | 15 ± 3 |
| Massolo et al. [29] | 99mTc | HMPAO | 6.03 | Ceretec, GE Healthcare | 37 | 5.55 | 2x106 | 30 | 15 ± 3 |
| Ref. | Agent | Manufacture | Excitation/ Emission (nm) | Concentration | Incubation (min) | Evaluation |
|---|---|---|---|---|---|---|
| Parada-Kusz et al. [23] | CellTrace™ Violet Cell Proliferation Kit | Thermo Fisher Scientific Inc. | 405/450 | 5 µM | 20–30 | NR |
| CellTrace™ CSFE | Thermo Fisher Scientific Inc. | 495/519 | 5 µM | 20–30 | NR | |
| Hamilton et al. [21] | Fluorocein | NR | 495/519 | NR | 10 | NR |
| Staal et al. [24] | PKH26 (#PKH26GL) | Sigma Aldrich | 551/567 | 2 µM | 2 | NR |
| Lopez et al. [22] | GFP* | NA | 475/509 | NA | NA | FCT; PCR |
| Ushiki et al. [25] | Cy5.5 | GE Healthcare UK Ltd., Buckinghamshire, UK | 675/694 | 0.4 mg/mL | 15 | FCT |
| AF750 | Invitrogen, Eugene, OR, USA | 749/775 | 0.1 mg/mL | 15 | FCT |
| Ref. | Particle | Size Core (nm) | Coating | DH (nm) | Manufacture | PDI | Transfection Agent | Concentration (µg/mL) | Incubation (h) | Evaluation |
|---|---|---|---|---|---|---|---|---|---|---|
| Sweeney et al. [34] | Gd2O3-MSN with pores 24Å | NR | Pore functionalized with: (a) TRITC; (b) FITC; (c) TRITC and PEG | 177 | Synthetized | 0.535 | Polybrene or protamine sulfate | 125 | 2–4 | Fluorescent microscopy |
| Bengtsson et al. [32] | Magnetite | 4.8 | Dextran | 80–150 | Feridex (Berlex Laboratories, Montville, NJ, USA) | 0.29 | Protamine sulfate | NR | Overnight | Prussian blue |
| Niemeyer et al. [33] | Magnetite/maghemite | 3-5 | Carboxydextran | 60 | Resovist (Bayer Schering Pharma AG, Berlin, Germany) | 0.207 | Lipofectamine | 25 | 4 | Prussian blue |
| Magnetite | 4.8 | Dextran | 120–180 | Endorem (Guerbet S.A., Roissy, France) | 0.266 | Lipofectamine | 25 | 4 | Prussian blue |
| Ref. | Animal Receptor | Irradiation | Cells Transplantation | Graft Assessment | Graft Efficiency (% or Number of Cells) At Time | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Specie | Age | Gender | Conditioning | Source | Dose (Gy) | Dose-Rate (Gy/min) | Type of Graft | Delay for Cells Infusion (Hours) | Cell Dose | Route | Vehicle | |||
| Parada-Kusz et al. [23] | Zebrafish (nacre−/-) | 3–5 h | NR | NA | NA | NA | NA | Xenogeneic | NA | 1-6 × 104 | Blastoderm | PBS | FCT (CD45); TUNEL | 0 at 4 d |
| Sweeney et al. [34] | Mice (129/SvJ) | NR | NR | TBI | 137-Cs | 2 × (7–8) | NR | Allogeneic | 24 | 7.2 × 106 | IV (RO) | NR | FCT (GFP) | 0 at 20 d |
| Hamilton et al. [21] | Zebrafish (Tg-kdrl:EGFP) | 52 h | NR | NA | NA | NA | NA | Xenogeneic | NA | NR | DC yolk sac | NR | NA | NA |
| Asiedu et al. [28] | Mice (C57BL/6 CD45.1) | 8–12 w | F-M | TBI | NR | 9.5 | NR | Allogeneic | 24 | 2 × 107 | IV | NR | FCT (CD45.1, CD45.2, linage specify) | 85 at 70 d |
| Lange et al. [31] | Dog (Beagle) | 1–3 y | M | NA | NA | NA | NA | Autologous | NA | 1.5 × 106/kg | IB (humerus) | NR | NA | NR |
| Astuti et al. [17] | Zebrafish | NR | NR | TBI | X-rays | 20 | 2.7 | Allogeneic | 48 | 5–50 × 104 | IV (IC) | NR | IF, FCT (GFP) | 306.5 ± 136.6 cells at 14 d |
| Saia et al. [20] | Mice (B6 albino) | NR | NR | TBI | NR | 6.5 | NR | Allogeneic | NA | 5 × 105 | IV (tail) | NR | qPCR (LUC2) | 53 at 120 d |
| Faivre et al. [27] | Mice (NSG) | 8 w | F | TBI | 137-Cs | 2.25 | NR | Xenogeneic | 24 | 2 × 106 | IV (tail) | Saline | FCT (CD34, CD45) | CD34: 1.5 ± 0.6; CD45: 20 ± 6 at 30 d |
| Staal et al. [24] | Zebrafish (casper Fli-GFP) | 48 h | NR | NA | NA | NA | NA | Xenogeneic | NA | 0.5–5 × 102 | DC yolk sac IV (RO) | NR | FCT (CD3, CD14, CD34, CD38, CD45) | NR |
| Pantin et al. [26] | Porcine (domestic swine) | NR | F-M | NA | NA | NA | NA | Xenogeneic | NA | 2 × 108 | IV (jugular) IB (iliac crest)IA (iliac) | Saline | NA | NA |
| Lopez et al. [22] | Salamander (white mutant (d/d)) | >12 w | NR | TBI | NR | 9.5 | NR | Allogeneic | NR | 1-50 × 104 | IV (IC) | NR | FCT (GFP); TUNEL; HC | NR |
| Sambuceti et al. [30] | Rat (Lewis) | 7 w | M | TBI | X-rays | 9.5 | 0.9 | Allogeneic | 24 | 2 × 106 | IV (NA) | NR | CA | LV: 0.28 ± 0.18; SP: 0.19 ± 0.12; LG: 0.03 ± 0.42 |
| Lin et al. [18] | Mice (Balb/c) | NR | F | BG + BCNU; TBI | 137-Cs | 7.5 | NR | Allogeneic | 48; 1–5 | 1–10 × 105 | NA | NR | FCT; CFU | NR |
| Andrade et al. [16] | Mice (NSG) | 8–10 w | NR | TBI; RHLIR | 137-Cs; X-rays | 2.7 | NR | Allogeneic | NR | 1 × 104; 1 × 105; 1 × 106 | IV (tail) IB (femur) | NR | FCT (CD11b, Ly-6c/g, CD3ε, B220, NK1.1) | 69 ± 5 at 98 d |
| Bengtsson et al. [32] | Mice (C57BL/6j) | 6–8 w | F | TBI | NR | 9.5 | NR | Allogeneic | NR | 1 × 103; 1 × 106 | IV (RO) | NR | HA | NA |
| Ohmori et al. [19] | Mice (C57BL/6 Ly5.2) | 8–12 w | NR | TBI | NR | 9.5 | NR | Allogeneic | NR | 1 × 105 | IV (carotid) | NR | FCT; IF | 58 at 30 d |
| Ushiki et al. [25] | Mice (Balb/c) | 7–9 w | NR | TBI | 137-Cs | 8 | NR | Allogeneic | 6-8 | 1 × 107 | IV (tail) IB (tibia) | NR | FCT (eGFP); HA | 43.5 at 3 d |
| Massolo et al. [29] | Rat (Lewis) | 7 w | M | TBI | X-rays | 9.5 | 0.9 | Allogeneic | 24 | 2 × 106 | IV (tail) IB (tibia) | Saline | Blood count | NR |
| Niemeyer et al. [33] | Mice (Balb/c) | NR | NR | NA | NA | NA | NA | Xenogeneic | NA | 1-5 × 106; 5 × 106; 1 × 107 | IV (tail) | NR | HA; IHC; FCT (CD34, CD45, CD71) | 0.08 at 1d |
| Steiner et al. [11] | Mice (NSG) | 6–12 w | NR | TBI | 137-Cs | 2.7 | NR | Xenogeneic | NR | 8-35 × 104 | IV (tail) | NR | FCT (CD45, linage specify); IHC | 39.6 at 365 d |
| Wang et al. [9] | Mice (NSG) | 8–10 w | NR | TBI | NR | 3 | NR | Xenogeneic | 2 | 4 × 104; 1 × 105 | IV (tail) | NR | FCT (CD 45) | 1.3 at 105 d |
| Ref. | Substrate | Dose (mg/kg) | Substrate Administration Route | Time Before Image (min) | Equipment | Software | Animal Position | Exposure Time (min) | Binning | Homing Evaluation Time | Outcome (Cells Migration) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Astuti et al. [17] | Luciferin | 75# | IP | 10–15 | Xenogen IVIS50 system (Caliper Life Sciences) | Living Image | Dorsal and lateral | 1 | 4; 8 | 98 d | + |
| Saia et al. [20] | Luciferin | 80 | IP | 15 | Xenogen IVIS Lumina system (Caliper, PerkinElmer) | Living Image | Ventral and dorsal | 1 | NR | 1-10 d (20, 30, 40-120 d) | + |
| Lin et al. [18] | Luciferin | 125 | IP | 7 | Xenogen IVIS 200 Imaging system (Caliper Life Sciences) | Living Image | Ventral and dorsal | 5 | NR | 146 d | + |
| Andrade et al. [16] | Luciferin | 125 | IP | 8 | Xenogen IVIS 100 imaging system (Caliber Life Sciences) | Living Image | Ventral and dorsal | 3-1 | NR | 1-65 d | + |
| Ohmori et al. [19] | Luciferin | 1.5§ | IP | NR | IVIS Imaging system (Xenogen, Alameda, CA) | Living Image | Ventral | NR | NR | 3, 7, 14, 21, …, 256 d | + |
| Steiner et al. [11] | Luciferin | 150 | IP | NR | Xenogen-IVIS Imaging system (Caliper Life Sciences Hopkinton, MA) | Living Image | Ventral, dorsal and lateral | 3 -1 s | NR | 7, 14 d (3, 6, 12 months) | + |
| Wang et al. [9] | Luciferin | 125 | IV | 2 | IVIS 3-D optical imaging system (Xenogen, Alameda, CA) | Living Image | Ventral and dorsal | 3-1 | NR | 1, 8 d (7-15 weeks) | + |
| Ref. | Image Modality | RP | Equipament | Reconstruction Parameters | Type of Image | Energy Window(KeV) | Image Acquisition Time | Homing Evaluation Time | Uptake Distribution | Outcome (Cells Migration) |
|---|---|---|---|---|---|---|---|---|---|---|
| Asiedu et al. [28] | PET-CT | 89Zr-oxime | MicroPET-CT (BioPET, Bioscan) | 3D; Iterative reconstruction | Wholebody | 400–700 | 5 min per bed for 4 h; 6.5, 7.5, 12.5, 15.5 min per bed for 1, 2, 5,7 d | 0, 2, 4, 24, 48 h (5, 7 d) | Bone marrow, spleen and liver | + |
| Lange et al. [31] | SPECT | 99mTc-HMPAO | PRISM 2000 XP gamma camera (Phillips, Hamburg, Germany) | NR | Wholebody | 140 ± 5% | NR | 1, 6, 24 h | Bone marrow, lungs, ribs and spines | + |
| Faivre et al. [27] | PET-CT | 18F-FDG | Micro-PET-CT (Inveon, Siemens) | OSEM 3D | Dynamic | 511 ± 5% | Dynamic study for 2.5 h | 0, 1, 2, 3 h | Lung, kidney, spleen, liver, femur and vertebrae | + |
| Pantin et al. [26] | PET-CT | 89Zr | Gemini TF clinical PET/CT (Philips Healthcare, Andover, MA) | 3D; Iterative reconstruction; Scatter and attenuation correction; Time of Flight; Spatial resolution: 4.8mm | Wholebody | 400–700 | NR | 5, 10, 15 h | Lungs, bone marrow | + |
| Sambuceti et al. [30] | SPECT | 99mTc-HMPAO | Gamma-camera (GE Millennium, Milwaukee, USA) | Parallel hole collimator; Image size 128 × 128 × 16 | Dynamic | 140 ± 5% | 240 img of 0.5 sec, 60 img of 2 sec, 36 img of 10 sec, 5 img of 120 sec, 2 img of 300 sec | 30 min | Heart, lung, liver, spleen | + |
| Massolo et al. [29] | SPECT | 99mTc-HMPAO | Gamma-camera (GE Millennium, Milwaukee, USA) | Parallel hole collimator; Image size: 128 × 128 × 16 | Dynamic | 140 ± 5% | 240 img of 0.5 sec, 60 img of 2 sec, 36 img of 10 sec, 5 img of 120 sec, 2 img of 300 sec | 30 min | Heart, liver, spleen and forelimb, maxillary lymph node | + |
| Ref. | Tracking Agent | Equipment | Parameters | Homing Evaluation Time | Uptake Distribution | Outcome (Cells Migration) |
|---|---|---|---|---|---|---|
| Parada-Kusz et al. [23] | CellTrace™ Violet Cell; CellTrace™ CSFE | Epifluorescence microscope (Zeiss Axio Observer), an A1R; C2 (Nikon) confocal microscope; Eclipse Ti (Nikon) spinning disk confocal microscope | NR | [1, 2, 3 d]*, 4, 5, 6 d | Yolk sac, tail, ICM, PBI and AGM | + |
| Hamilton et al. [21] | FITC | Spinning Disk confocal microscope | NR | 1, 4, 7, 10, 13 h | Tail | + |
| Staal et al. [24] | PKH26 | Leica fluorescent microscope | NR | NR | Yolk sac, tail | + |
| Lopez et al. [22] | GFP | Leica MZ16FA microscope, using a Hamamatsu digital camera model C7780 and Volocity Imaging software (Perkin Elmer) | NR | NR | NR | + |
| Ushiki et al. [25] | Cy5.5; AF750 | IVIS Spectrum system (Xenogen, Alameda, CA, USA) | Excitation/emission (nn): 640/700 for Cy5.5, 710/780 for AF750. Exposure time: 5 s, lamp level: high, binning: medium, FOV: 12.9612.9 cm, and f/stop: 1. | 0.5, 1, 3, 6, 12, 18, 24 h | Bone marrow, lung, spleen, liver, kidney | + |
| Ref. | Equipment | Software | MF (T) | Sequence | Weighted Images (TR/TE; ms) | FOV;MT;ST | Homing Evaluation Time | Uptake Distribution | Outcome (Cells Migration) |
|---|---|---|---|---|---|---|---|---|---|
| Sweeney et al. [34] | Varian® Unity/INOVA 4.7 T small animal scanner | MIPAV | 4.7 | Fast Spin Echo; Gradient Echo | T2: 2100/15 T2*: NR | NR; 256x512; NR | 3, 24 h (6, 9 d) | Bone marrow, spleen, and liver | + |
| Bengtsson et al. [32] | Bruker BioSpin, Madison, WI, USA | Paravision 4.0 (Bruker, Madison, WI, USA) and OsiriX v.3.5 | 17.6 | 3D Gradient Echo | T2*: 80/2.5 | 1.1x0.6x0.5cm3; 393x214x83; NR | 7, 14 d | Bone marrow | + |
| Niemeyer et al. [33] | 1.5-T imaging MR scanner (ACS NT; Philips, Best, the Netherlands) | NR | 1.5 | 3D Fast Field Echo | T2*: 32/14 | 100x80mm; 512x512; 0.4mm | 2, 24 h | Bone marrow and liver | + |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira, F.A.; Nucci, M.P.; Filgueiras, I.S.; Ferreira, J.M.; Nucci, L.P.; Mamani, J.B.; Alvieri, F.; Souza, L.E.B.; Rego, G.N.A.; Kondo, A.T.; et al. Noninvasive Tracking of Hematopoietic Stem Cells in a Bone Marrow Transplant Model. Cells 2020, 9, 939. https://doi.org/10.3390/cells9040939
Oliveira FA, Nucci MP, Filgueiras IS, Ferreira JM, Nucci LP, Mamani JB, Alvieri F, Souza LEB, Rego GNA, Kondo AT, et al. Noninvasive Tracking of Hematopoietic Stem Cells in a Bone Marrow Transplant Model. Cells. 2020; 9(4):939. https://doi.org/10.3390/cells9040939
Chicago/Turabian StyleOliveira, Fernando A., Mariana P. Nucci, Igor S. Filgueiras, João M. Ferreira, Leopoldo P. Nucci, Javier B. Mamani, Fernando Alvieri, Lucas E. B. Souza, Gabriel N. A. Rego, Andrea T. Kondo, and et al. 2020. "Noninvasive Tracking of Hematopoietic Stem Cells in a Bone Marrow Transplant Model" Cells 9, no. 4: 939. https://doi.org/10.3390/cells9040939
APA StyleOliveira, F. A., Nucci, M. P., Filgueiras, I. S., Ferreira, J. M., Nucci, L. P., Mamani, J. B., Alvieri, F., Souza, L. E. B., Rego, G. N. A., Kondo, A. T., Hamerschlak, N., & Gamarra, L. F. (2020). Noninvasive Tracking of Hematopoietic Stem Cells in a Bone Marrow Transplant Model. Cells, 9(4), 939. https://doi.org/10.3390/cells9040939