Inflammatory Responses during Tumour Initiation: From Zebrafish Transgenic Models of Cancer to Evidence from Mouse and Man
Abstract
1. Introduction
2. Zebrafish Transgenic Models of Cancer for In Vivo Live Imaging Studies of Tumour Initiation
3. Macrophages and Neutrophils are Co-Opted by Cancer to Perform Tumour-Supporting Activities
3.1. Tumour-Associated Macrophages
3.2. Tumour-Associated Neutrophils
4. Evidence of Tumour-Promoting Inflammation in Early Tumourigenesis of Mouse and Man
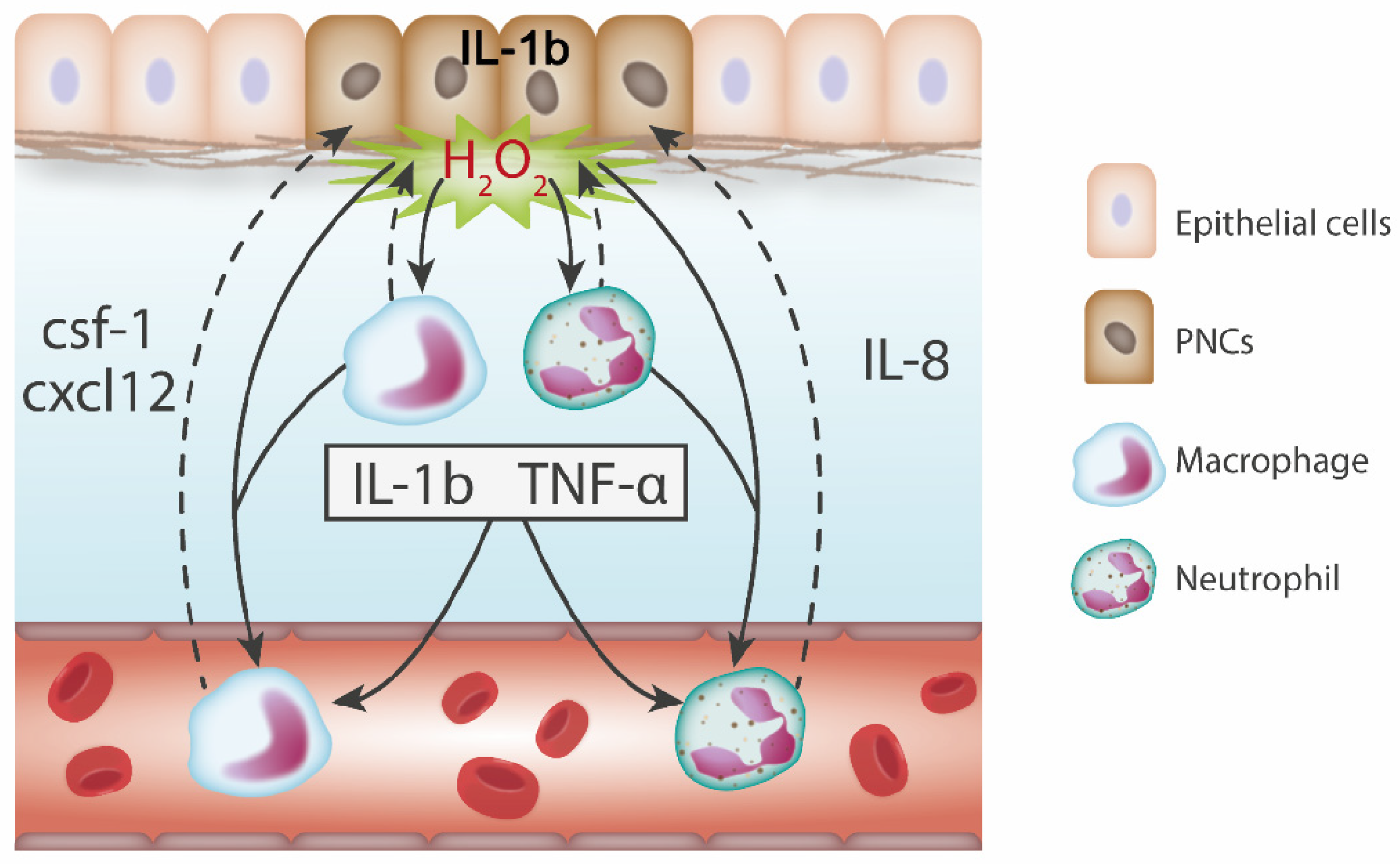
5. Zebrafish Studies Reveal Leukocyte Recruitment and Trophic Function at the Pre-Neoplastic Stage
5.1. Skin
5.2. Liver
5.3. Brain
6. Mechanisms Governing Intrinsic Pro-Tumour Inflammation: Parallels between Zebrafish, Mouse and Man
6.1. Pro-Inflammatory Cytokines are Expressed in Response to Tumour Initiation
6.2. Chemokines Recruit Leukocytes to PNCs
6.3. Plasticity and Heterogeneity of Leukocytes
6.4. PGE2 as Trophic Factor and Immunomodulator
6.5. TGF-β Governs Pro-Tumour Neutrophils
6.6. Extracellular ATP Attracts Pro-Tumour Microglia/Macrophages
7. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flier, J.S.; Underhill, L.H.; Dvorak, H.F. Tumors: Wounds That Do Not Heal. N. Engl. J. Med. 1986, 315, 1650–1659. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Ruffell, B.; Coussens, L.M. Macrophages and Therapeutic Resistance in Cancer. Cancer Cell 2015, 27, 462–472. [Google Scholar] [CrossRef]
- Wu, L.; Saxena, S.; Awaji, M.; Singh, R.K. Tumor-associated neutrophils in cancer: Going pro. Cancers (Basel) 2019, 11, 564. [Google Scholar] [CrossRef]
- Allavena, P.; Garlanda, C.; Borrello, M.G.; Sica, A.; Mantovani, A. Pathways connecting inflammation and cancer. Curr. Opin. Genet. Dev. 2008, 18, 3–10. [Google Scholar] [CrossRef]
- Feng, Y.; Santoriello, C.; Mione, M.; Hurlstone, A.; Martin, P. Live Imaging of Innate Immune Cell Sensing of Transformed Cells in Zebrafish Larvae: Parallels between Tumor Initiation and Wound Inflammation. PLoS Biol. 2010, 8, e1000562. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Renshaw, S.; Martin, P. Live Imaging of Tumor Initiation in Zebrafish Larvae Reveals a Trophic Role for Leukocyte-Derived PGE2. Curr. Biol. 2012, 22, 1253–1259. [Google Scholar] [CrossRef]
- Patton, E.E.; Zon, L.I. The art and design of genetic screens: Zebrafish. Nat. Rev. Genet. 2001, 2, 956–966. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Spitsbergen, J.M.; Buhler, D.R.; Peterson, T.S. Neoplasia and neoplasm-associated lesions in laboratory colonies of zebrafish emphasizing key influences of diet and aquaculture system design. ILAR J. 2012, 53, 114–125. [Google Scholar] [CrossRef]
- Storer, N.Y.; Zon, L.I. Zebrafish models of p53 functions. Cold Spring Harb. Perspect. Biol. 2010, 2, a001123. [Google Scholar] [CrossRef] [PubMed]
- Krens, S.F.G.; He, S.; Spaink, H.P.; Snaar-Jagalska, B.E. Characterization and expression patterns of the MAPK family in zebrafish. Gene Expr. Patterns 2006, 6, 1019–1026. [Google Scholar] [CrossRef] [PubMed]
- Barnes, T.A.; Amir, E. HYPE or HOPE: The prognostic value of infiltrating immune cells in cancer. Br. J. Cancer 2017, 117, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Iwahori, K. Cytotoxic CD8+ Lymphocytes in the Tumor Microenvironment. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2020; Volume 1224, pp. 53–62. [Google Scholar]
- Guisier, F.; Barros-Filho, M.C.; Rock, L.D.; Strachan-Whaley, M.; Marshall, E.A.; Dellaire, G.; Lam, W.L. Janus or Hydra: The Many Faces of T Helper Cells in the Human Tumour Microenvironment. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2020; Volume 1224, pp. 35–51. [Google Scholar]
- White, R.M.; Sessa, A.; Burke, C.; Bowman, T.; LeBlanc, J.; Ceol, C.; Bourque, C.; Dovey, M.; Goessling, W.; Burns, C.E.; et al. Transparent Adult Zebrafish as a Tool for In Vivo Transplantation Analysis. Cell Stem Cell 2008, 2, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Langenau, D.M.; Traver, D.; Ferrando, A.A.; Kutok, J.L.; Aster, J.C.; Kanki, J.P.; Lin, S.; Prochownik, E.; Trede, N.S.; Zon, L.I.; et al. Myc-induced T cell leukemia in transgenic zebrafish. Science (80-. ) 2003, 299, 887–890. [Google Scholar] [CrossRef] [PubMed]
- Langenau, D.M.; Feng, H.; Berghmans, S.; Kanki, J.P.; Kutok, J.L.; Look, A.T. Cre/lox-regulated transgenic zebrafish model with conditional myc-induced T cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2005, 102, 6068–6073. [Google Scholar] [CrossRef]
- Kawakami, K.; Shima, A.; Kawakami, N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc. Natl. Acad. Sci. USA 2000, 97, 11403–11408. [Google Scholar] [CrossRef]
- Balciunas, D.; Wangensteen, K.J.; Wilber, A.; Bell, J.; Geurts, A.; Sivasubbu, S.; Wang, X.; Hackett, P.B.; Largaespada, D.A.; McIvor, R.S.; et al. Harnessing a High Cargo-Capacity Transposon for Genetic Applications in Vertebrates. PLoS Genet. 2006, 2, e169. [Google Scholar] [CrossRef]
- Patton, E.E.; Widlund, H.R.; Kutok, J.L.; Kopani, K.R.; Amatruda, J.F.; Murphey, R.D.; Berghmans, S.; Mayhall, E.A.; Traver, D.; Fletcher, C.D.M.; et al. BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol. 2005, 15, 249–254. [Google Scholar] [CrossRef]
- Dovey, M.; White, R.M.; Zon, L.I. Oncogenic NRAS cooperates with p53 loss to generate melanoma in zebrafish. Zebrafish 2009, 6, 397–404. [Google Scholar] [CrossRef]
- Michailidou, C.; Jones, M.; Walker, P.; Kamarashev, J.; Kelly, A.; Hurlstone, A.F.L. Dissecting the roles of Raf- and PI3K-signalling pathways in melanoma formation and progression in a zebrafish model. DMM Dis. Model. Mech. 2009, 2, 399–411. [Google Scholar] [CrossRef]
- Santoriello, C.; Gennaro, E.; Anelli, V.; Distel, M.; Kelly, A.; Köster, R.W.; Hurlstone, A.; Mione, M. Kita driven expression of oncogenic HRAS leads to early onset and highly penetrant melanoma in zebrafish. PLoS ONE 2010, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.W.; Raghuram, D.; Fong, P.S.A.; Gong, Z. Inducible Intestine-Specific Expression of kras V12 Triggers Intestinal Tumorigenesis In Transgenic Zebrafish. Neoplasia (United States) 2018, 20, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Davison, J.M.; Rhee, J.; Hruban, R.H.; Maitra, A.; Leach, S.D. Oncogenic KRAS Induces Progenitor Cell Expansion and Malignant Transformation in Zebrafish Exocrine Pancreas. Gastroenterology 2008, 134, 2080–2090. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.; Chen, W.; Orr, B.A.; Spitsbergen, J.M.; Jia, S.; Eden, C.J.; Henson, H.E.; Taylor, M.R. Oncogenic KRAS promotes malignant brain tumors in zebrafish. Mol. Cancer 2015, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Osmani, N.; Goetz, J.G. Multiscale Imaging of Metastasis in Zebrafish. Trends Cancer 2019, 5, 766–778. [Google Scholar] [CrossRef]
- Schiavone, M.; Rampazzo, E.; Casari, A.; Battilana, G.; Persano, L.; Moro, E.; Liu, S.; Leach, S.D.; Tiso, N.; Argenton, F. Zebrafish reporter lines reveal in vivo signaling pathway activities involved in pancreatic cancer. DMM Dis. Model. Mech. 2014, 7, 883–894. [Google Scholar] [CrossRef]
- Zhu, S.; Lee, J.S.; Guo, F.; Shin, J.; Perez-Atayde, A.R.; Kutok, J.L.; Rodig, S.J.; Neuberg, D.S.; Helman, D.; Feng, H.; et al. Activated ALK Collaborates with MYCN in Neuroblastoma Pathogenesis. Cancer Cell 2012, 21, 362–373. [Google Scholar] [CrossRef]
- Kaufman, C.K.; Mosimann, C.; Fan, Z.P.; Yang, S.; Thomas, A.J.; Ablain, J.; Tan, J.L.; Fogley, R.D.; Van Rooijen, E.; Hagedorn, E.J.; et al. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science. 2016, 351, aad2197. [Google Scholar] [CrossRef]
- Tanimura, N.; Fujita, Y. Epithelial defense against cancer (EDAC). Semin. Cancer Biol. 2019. [CrossRef]
- Anton, K.A.; Kajita, M.; Narumi, R.; Fujita, Y.; Tada, M. Src-transformed cells hijack mitosis to extrude from the epithelium. Nat. Commun. 2018, 9, 4695. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Shea, J.; Slattum, G.; Firpo, M.A.; Alexander, M.; Golubovskaya, V.M.; Rosenblatt, J. Defective apical extrusion signaling contributes to aggressive tumor hallmarks. Elife 2015, 2015, e04069. [Google Scholar] [CrossRef] [PubMed]
- Knopf, F.; Schnabel, K.; Haase, C.; Pfeifer, K.; Anastassiadis, K.; Weidinger, G. Dually inducible TetON systems for tissue-specific conditional gene expression in zebrafish. Proc. Natl. Acad. Sci. USA 2010, 107, 19933–19938. [Google Scholar] [CrossRef] [PubMed]
- Emelyanov, A.; Parinov, S. Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev. Biol. 2008, 320, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Lepage, S.E.; Tada, M.; Bruce, A.E.E. Zebrafish Dynamin is required for maintenance of enveloping layer integrity and the progression of epiboly. Dev. Biol. 2014, 385, 52–66. [Google Scholar] [CrossRef]
- Hans, S.; Freudenreich, D.; Geffarth, M.; Kaslin, J.; Machate, A.; Brand, M. Generation of a non-leaky heat shock-inducible Cre line for conditional Cre/lox strategies in zebrafish. Dev. Dyn. 2011, 240, 108–115. [Google Scholar] [CrossRef]
- Laux, D.W.; Kelly, L.; Bravo, I.R.; Ramezani, T.; Feng, Y. Live imaging the earliest host innate immune response to preneoplastic cells using a zebrafish inducible KalTA4-ERT2/UAS system. Methods Cell Biol. 2017, 138, 137–150. [Google Scholar]
- Ramezani, T.; Laux, D.W.; Bravo, I.R.; Tada, M.; Feng, Y. Live Imaging of Innate Immune and Preneoplastic Cell Interactions Using an Inducible Gal4/UAS Expression System in Larval Zebrafish Skin. J. Vis. Exp. 2015, e52107. [Google Scholar] [CrossRef]
- Yan, C.; Huo, X.; Wang, S.; Feng, Y.; Gong, Z. Stimulation of hepatocarcinogenesis by neutrophils upon induction of oncogenic kras expression in transgenic zebrafish. J. Hepatol. 2015, 63, 420–428. [Google Scholar] [CrossRef]
- Gore, A.V.; Pillay, L.M.; Venero Galanternik, M.; Weinstein, B.M. The zebrafish: A fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, 1–17. [Google Scholar] [CrossRef]
- Novoa, B.; Figueras, A. Zebrafish: Model for the study of inflammation and the innate immune response to infectious diseases. Adv. Exp. Med. Biol. 2012, 946, 253–275. [Google Scholar] [PubMed]
- Rosowski, E.E. Determining macrophage versus neutrophil contributions to innate immunity using larval zebrafish. Dis. Model. Mech. 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Henry, K.M.; Loynes, C.A.; Whyte, M.K.B.; Renshaw, S.A. Zebrafish as a model for the study of neutrophil biology. J. Leukoc. Biol. 2013, 94, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Herbomel, P.; Thisse, B.; Thisse, C. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 1999, 126, 3735–3745. [Google Scholar] [PubMed]
- Crowhurst, M.O.; Layton, J.E.; Lieschke, G.J. Developmental biology of zebrafish myeloid cells. Int. J. Dev. Biol. 2002, 46, 483–492. [Google Scholar] [PubMed]
- Lieschke, G.J.; Oates, A.C.; Crowhurst, M.O.; Ward, A.C.; Layton, J.E. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood 2001, 98, 3087–3096. [Google Scholar] [CrossRef]
- Robertson, A.L.; Avagyan, S.; Gansner, J.M.; Zon, L.I. Understanding the regulation of vertebrate hematopoiesis and blood disorders—big lessons from a small fish. FEBS Lett. 2016, 590, 4016–4033. [Google Scholar] [CrossRef]
- Torraca, V.; Mostowy, S. Zebrafish Infection: From Pathogenesis to Cell Biology. Trends Cell Biol. 2018, 28, 143–156. [Google Scholar] [CrossRef]
- Martin, P.; Nunan, R. Cellular and molecular mechanisms of repair in acute and chronic wound healing. Br. J. Dermatol. 2015, 173, 370–378. [Google Scholar] [CrossRef]
- Richardson, R.J. Parallels between vertebrate cardiac and cutaneous wound healing and regeneration. NPJ Regen. Med. 2018, 3, 1–9. [Google Scholar] [CrossRef]
- Ellett, F.; Pase, L.; Hayman, J.W.; Andrianopoulos, A.; Lieschke, G.J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 2011, 117, e49–e56. [Google Scholar] [CrossRef] [PubMed]
- Walton, E.M.; Cronan, M.R.; Beerman, R.W.; Tobin, D.M. The Macrophage-Specific Promoter mfap4 Allows Live, Long-Term Analysis of Macrophage Behavior during Mycobacterial Infection in Zebrafish. PLoS ONE 2015, 10, e0138949. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, S.A.; Loynes, C.A.; Trushell, D.M.I.; Elworthy, S.; Ingham, P.W.; Whyte, M.K.B. Atransgenic zebrafish model of neutrophilic inflammation. Blood 2006, 108, 3976–3978. [Google Scholar] [CrossRef]
- Kitaguchi, T.; Kawakami, K.; Kawahara, A. Transcriptional regulation of a myeloid-lineage specific gene lysozyme C during zebrafish myelopoiesis. Mech. Dev. 2009, 126, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Sabaawy, H.E.; Azuma, M.; Embree, L.J.; Tsai, H.J.; Starost, M.F.; Hickstein, D.D. TEL-AML1 transgenic zebrafish model of precursor B cell lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2006, 103, 15166–15171. [Google Scholar] [CrossRef] [PubMed]
- Ceol, C.J.; Houvras, Y.; Jane-Valbuena, J.; Bilodeau, S.; Orlando, D.A.; Battisti, V.; Fritsch, L.; Lin, W.M.; Hollmann, T.J.; Ferré, F.; et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 2011, 471, 513–518. [Google Scholar] [CrossRef]
- White, R.M.; Cech, J.; Ratanasirintrawoot, S.; Lin, C.Y.; Rahl, P.B.; Burke, C.J.; Langdon, E.; Tomlinson, M.L.; Mosher, J.; Kaufman, C.; et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 2011, 471, 518–522. [Google Scholar] [CrossRef]
- Kenyon, A.; Gavriouchkina, D.; Zorman, J.; Chong-Morrison, V.; Napolitani, G.; Cerundolo, V.; Sauka-Spengler, T. Generation of a double binary transgenic zebrafish model to study myeloid gene regulation in response to oncogene activation in melanocytes. DMM Dis. Model. Mech. 2018, 11, dmm030056. [Google Scholar] [CrossRef]
- Evason, K.J.; Francisco, M.T.; Juric, V.; Balakrishnan, S.; Pazmino, M.D.P.L.; Gordan, J.D.; Kakar, S.; Spitsbergen, J.; Goga, A.; Stainier, D.Y.R. Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish. PLOS Genet. 2015, 11, e1005305. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Emelyanov, A.; Koh, C.H.V.; Spitsbergen, J.M.; Lam, S.H.; Mathavan, S.; Parinov, S.; Gong, Z. A high level of liver-specific expression of oncogenic Kras(V12) drives robust liver tumorigenesis in transgenic zebrafish. Dis. Model. Mech. 2011, 4, 801–813. [Google Scholar] [CrossRef]
- Nguyen, A. An inducible kras(V12) transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis. Model. Mech. 2012, 5, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Chew, T.W.; Liu, X.J.; Liu, L.; Spitsbergen, J.M.; Gong, Z.; Low, B.C. Crosstalk of Ras and Rho: Activation of RhoA abates Kras-induced liver tumorigenesis in transgenic zebrafish models. Oncogene 2014, 33, 2717–2727. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Huang, X.; Zhan, H.; Zeng, Z.; Li, C.; Spitsbergen, J.M.; Meierjohann, S.; Schartl, M.; Gong, Z. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. J. Hepatol. 2012, 56, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Li, Z.; Nguyen, A.T.; Li, C.; Emelyanov, A.; Gong, Z. Xmrk, Kras and Myc Transgenic Zebrafish Liver Cancer Models Share Molecular Signatures with Subsets of Human Hepatocellular Carcinoma. PLoS ONE 2014, 9, e91179. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zheng, W.; Wang, Z.; Zeng, Z.; Zhan, H.; Li, C.; Zhou, L.; Yan, C.; Spitsbergen, J.M.; Gong, Z. A transgenic zebrafish liver tumor model with inducible Myc expression reveals conserved Myc signatures with mammalian liver tumors. DMM Dis. Model. Mech. 2013, 6, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Powell, D.; Lou, M.; Barros Becker, F.; Huttenlocher, A. Cxcr1 mediates recruitment of neutrophils and supports proliferation of tumor-initiating astrocytes in vivo. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Liu, N.A.; Jiang, H.; Ben-Shlomo, A.; Wawrowsky, K.; Fan, X.M.; Lin, S.; Melmed, S. Targeting zebrafish and murine pituitary corticotroph tumors with a cyclin-dependent kinase (CDK) inhibitor. Proc. Natl. Acad. Sci. USA 2011, 108, 8414–8419. [Google Scholar] [CrossRef]
- Langenau, D.M.; Keefe, M.D.; Storer, N.Y.; Guyon, J.R.; Kutok, J.L.; Le, X.; Goessling, W.; Neuberg, D.S.; Kunkel, L.M.; Zon, L.I. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev. 2007, 21, 1382–1395. [Google Scholar] [CrossRef]
- Ignatius, M.S.; Chen, E.; Elpek, N.M.; Fuller, A.Z.; Tenente, I.M.; Clagg, R.; Liu, S.; Blackburn, J.S.; Linardic, C.M.; Rosenberg, A.E.; et al. In Vivo Imaging of Tumor-Propagating Cells, Regional Tumor Heterogeneity, and Dynamic Cell Movements in Embryonal Rhabdomyosarcoma. Cancer Cell 2012, 21, 680–693. [Google Scholar] [CrossRef]
- Storer, N.Y.; White, R.M.; Uong, A.; Price, E.; Petur Nielsen, G.; Langenau, D.M.; Zon, L.I. Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogene expression during myogenesis. Development 2013, 140, 3040–3050. [Google Scholar] [CrossRef]
- Mathias, J.R.; Perrin, B.J.; Liu, T.-X.; Kanki, J.; Look, A.T.; Huttenlocher, A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J. Leukoc. Biol. 2006, 80, 1281–1288. [Google Scholar] [CrossRef]
- Yoo, S.K.; Deng, Q.; Cavnar, P.J.; Wu, Y.I.; Hahn, K.M.; Huttenlocher, A. Differential Regulation of Protrusion and Polarity by PI(3)K during Neutrophil Motility in Live Zebrafish. Dev. Cell 2010, 18, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.Y.; Harvie, E.A.; Huttenlocher, A. Heat shock modulates neutrophil motility in zebrafish. PLoS ONE 2013, 8, e84436. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, A.; Gavriouchkina, D.; Zorman, J.; Napolitani, G.; Cerundolo, V.; Sauka-Spengler, T. Active nuclear transcriptome analysis reveals inflammasome-dependent mechanism for early neutrophil response to Mycobacterium marinum. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.; Flores, M.; Storm, T.; Crosier, K.; Crosier, P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol. 2007, 7, 42. [Google Scholar] [CrossRef]
- de Oliveira, S.; Houseright, R.A.; Graves, A.L.; Golenberg, N.; Korte, B.G.; Miskolci, V.; Huttenlocher, A. Metformin modulates innate immune-mediated inflammation and early progression of NAFLD-associated hepatocellular carcinoma in zebrafish. J. Hepatol. 2019, 70, 710–721. [Google Scholar] [CrossRef]
- Bojarczuk, A.; Miller, K.A.; Hotham, R.; Lewis, A.; Ogryzko, N.V.; Kamuyango, A.A.; Frost, H.; Gibson, R.H.; Stillman, E.; May, R.C.; et al. Cryptococcus neoformans Intracellular Proliferation and Capsule Size Determines Early Macrophage Control of Infection. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef]
- Harvie, E.A.; Green, J.M.; Neely, M.N.; Huttenlocher, A. Innate immune response to Streptococcus iniae infection in zebrafish larvae. Infect. Immun. 2013, 81, 110–121. [Google Scholar] [CrossRef]
- Antonio, N.; Bønnelykke-Behrndtz, M.L.; Ward, L.C.; Collin, J.; Christensen, I.J.; Steiniche, T.; Schmidt, H.; Feng, Y.; Martin, P. The wound inflammatory response exacerbates growth of pre-neoplastic cells and progression to cancer. EMBO J. 2015, 34, 2219–2236. [Google Scholar] [CrossRef]
- Kelly, P.M.A.; Davison, R.S.; Bliss, E.; McGee, J.O.D. Macrophages in human breast disease: A quantitative immunohistochemical study. Br. J. Cancer 1988, 57, 174–177. [Google Scholar] [CrossRef]
- Van Overmeire, E.; Laoui, D.; Keirsse, J.; Van Ginderachter, J.A.; Sarukhan, A. Mechanisms driving macrophage diversity and specialization in distinct tumor microenvironments and parallelisms with other tissues. Front. Immunol. 2014, 5, 127. [Google Scholar] [CrossRef] [PubMed]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Ojalvo, L.S.; King, W.; Cox, D.; Pollard, J.W. High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. Am. J. Pathol. 2009, 174, 1048–1064. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Pollard, J.W. Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Invest. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Yang, M.; McKay, D.; Pollard, J.W.; Lewis, C.E. Diverse functions of macrophages in different tumor microenvironments. Cancer Res. 2018, 78, 5492–5503. [Google Scholar] [CrossRef]
- Coffelt, S.B.; Hughes, R.; Lewis, C.E. Tumor-associated macrophages: Effectors of angiogenesis and tumor progression. Biochim. Biophys. Acta.–Rev. Cancer 2009, 1796, 11–18. [Google Scholar] [CrossRef]
- Lin, E.Y.; Pollard, J.W. Tumor-associated macrophages press the angiogenic switch in breast cancer. Cancer Res. 2007, 67, 5064–5066. [Google Scholar] [CrossRef]
- Kitamura, T.; Qian, B.Z.; Pollard, J.W. Immune cell promotion of metastasis. Nat. Rev. Immunol. 2015, 15, 73–86. [Google Scholar] [CrossRef]
- Britto, D.D.; Wyroba, B.; Chen, W.; Lockwood, R.A.; Tran, K.B.; Shepherd, P.R.; Hall, C.J.; Crosier, K.E.; Crosier, P.S.; Astin, J.W. Macrophages enhance Vegfa-driven angiogenesis in an embryonic zebrafish tumour xenograft model. DMM Dis. Model. Mech. 2018, 11, dmm035998. [Google Scholar] [CrossRef] [PubMed]
- Roh-Johnson, M.; Shah, A.N.; Stonick, J.A.; Poudel, K.R.; Kargl, J.; Yang, G.H.; di Martino, J.; Hernandez, R.E.; Gast, C.E.; Zarour, L.R.; et al. Macrophage-Dependent Cytoplasmic Transfer during Melanoma Invasion In Vivo. Dev. Cell 2017, 43, 549–562.e6. [Google Scholar] [CrossRef] [PubMed]
- Masucci, M.T.; Minopoli, M.; Carriero, M.V. Tumor Associated Neutrophils. Their Role in Tumorigenesis, Metastasis, Prognosis and Therapy. Front. Oncol. 2019, 9, 1146. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Hu, P.; Donskov, F.; Wang, G.; Liu, Q.; Du, J. Tumor-Associated Neutrophils as a New Prognostic Factor in Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2014, 9, e98259. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef]
- Galdiero, M.R.; Varricchi, G.; Loffredo, S.; Mantovani, A.; Marone, G. Roles of neutrophils in cancer growth and progression. J. Leukoc. Biol. 2018, 103, 457–464. [Google Scholar] [CrossRef]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef]
- Finisguerra, V.; Di Conza, G.; Di Matteo, M.; Serneels, J.; Costa, S.; Thompson, A.A.R.; Wauters, E.; Walmsley, S.; Prenen, H.; Granot, Z.; et al. MET is required for the recruitment of anti-tumoural neutrophils. Nature 2015, 522, 349–353. [Google Scholar] [CrossRef]
- Weinberg, F.; Ramnath, N.; Nagrath, D. Reactive oxygen species in the tumor microenvironment: An overview. Cancers (Basel) 2019, 11, 1191. [Google Scholar] [CrossRef]
- Koga, Y.; Matsuzaki, A.; Suminoe, A.; Hattori, H.; Hara, T. Neutrophil-Derived TNF-Related Apoptosis-Inducing Ligand (TRAIL): A Novel Mechanism of Antitumor Effect by Neutrophils. Cancer Res. 2004, 64, 1037–1043. [Google Scholar] [CrossRef]
- Governa, V.; Trella, E.; Mele, V.; Tornillo, L.; Amicarella, F.; Cremonesi, E.; Muraro, M.G.; Xu, H.; Droeser, R.; Däster, S.R.; et al. The interplay between neutrophils and CD8+ T cells improves survival in human colorectal cancer. Clin. Cancer Res. 2017, 23, 3847–3858. [Google Scholar] [CrossRef] [PubMed]
- Freisinger, C.M.; Huttenlocher, A. Live Imaging and Gene Expression Analysis in Zebrafish Identifies a Link between Neutrophils and Epithelial to Mesenchymal Transition. PLoS ONE 2014, 9, e112183. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Lamers, G.E.M.; Beenakker, J.W.M.; Cui, C.; Ghotra, V.P.S.; Danen, E.H.J.; Meijer, A.H.; Spaink, H.P.; Snaar-Jagalska, B.E. Neutrophil-mediated experimental metastasis is enhanced by VEGFR inhibition in a zebrafish xenograft model. J. Pathol. 2012, 227, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Tulotta, C.; Snaar-Jagalska, B.E. CXCR4 signalling, metastasis and immunotherapy: Zebrafish xenograft model as translational tool for anti-cancer discovery. J. Cancer Metastasis Treat. 2019, 2019. [Google Scholar] [CrossRef]
- Huo, X.; Li, H.; Li, Z.; Yan, C.; Agrawal, I.; Mathavan, S.; Liu, J.; Gong, Z. Transcriptomic profiles of tumor-associated neutrophils reveal prominent roles in enhancing angiogenesis in liver tumorigenesis in zebrafish. Sci. Rep. 2019, 9, 1509. [Google Scholar] [CrossRef] [PubMed]
- Peñaloza, H.F.; Alvarez, D.; Muñoz-Durango, N.; Schultz, B.M.; González, P.A.; Kalergis, A.M.; Bueno, S.M. The role of myeloid-derived suppressor cells in chronic infectious diseases and the current methodology available for their study. J. Leukoc. Biol. 2019, 105, 857–872. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Patel, S.; Tcyganov, E.; Gabrilovich, D.I. The Nature of Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Trends Immunol. 2016, 37, 208–220. [Google Scholar] [CrossRef]
- Giese, M.A.; Hind, L.E.; Huttenlocher, A. Neutrophil plasticity in the tumor microenvironment. Blood 2019, 133, 2159–2167. [Google Scholar] [CrossRef]
- Cassetta, L.; Fragkogianni, S.; Sims, A.H.; Swierczak, A.; Forrester, L.M.; Zhang, H.; Soong, D.Y.H.; Cotechini, T.; Anur, P.; Lin, E.Y.; et al. Human Tumor-Associated Macrophage and Monocyte Transcriptional Landscapes Reveal Cancer-Specific Reprogramming, Biomarkers, and Therapeutic Targets. Cancer Cell 2019, 35, 588–602.e10. [Google Scholar] [CrossRef]
- Brandau, S.; Moses, K.; Lang, S. The kinship of neutrophils and granulocytic myeloid-derived suppressor cells in cancer: Cousins, siblings or twins? Semin. Cancer Biol. 2013, 23, 171–182. [Google Scholar] [CrossRef]
- Kinzler, K.W.; Vogelstein, B. Lessons from hereditary colorectal cancer. Cell 1996, 87, 159–170. [Google Scholar] [CrossRef]
- Solomon, E.; Voss, R.; Hall, V.; Bodmer, W.F.; Jass, J.R.; Jeffreys, A.J.; Lucibello, F.C.; Patel, I.; Rider, S.H. Chromosome 5 allele loss in human colorectal carcinomas. Nature 1988, 328, 616–619. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, W.F.; Bailey, C.J.; Bodmer, J.; Bussey, H.J.R.; Ellis, A.; Gorman, P.; Lucibello, F.C.; Murday, V.A.; Rider, S.H.; Scambler, P.; et al. Localization of the gene for familial adenomatous polyposis on chromosome 5. Nature 1988, 328, 614–616. [Google Scholar] [CrossRef] [PubMed]
- Moser, A.R.; Pitot, H.C.; Dove, W.F. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 1990, 247, 322–324. [Google Scholar] [CrossRef] [PubMed]
- Rowan, A.J.; Lamlum, H.; Ilyas, M.; Wheeler, J.; Straub, J.; Papadopoulou, A.; Bicknell, D.; Bodmer, W.F.; Tomlinson, I.P.M. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc. Natl. Acad. Sci. USA 2000, 97, 3352–3357. [Google Scholar] [CrossRef]
- Takahashi, M.; Mutoh, M.; Kawamori, T.; Sugimura, T.; Wakabayashi, K. Altered expression of β-catenin, inducible nitric oxide synthase and cyclooxygenase-2 in azoxymethane-induced rat colon carcinogenesis. Carcinogenesis 2000, 21, 1319–1327. [Google Scholar] [CrossRef]
- Pugh, S.; Thomas, G.A.O. Patients with adenomatous polyps and carcinomas have increased colonic mucosal prostaglandin E2. Gut 1994, 35, 675–678. [Google Scholar] [CrossRef]
- Rao, C.V.; Indranie, C.; Simi, B.; Reddy, B.S.; Manning, P.T.; Connor, J.R. Chemopreventive properties of a selective inducible nitric oxide synthase inhibitor in colon carcinogenesis, administered alone or in combination with celecoxib, a selective cyclooxygenase-2 inhibitor. Cancer Res. 2002, 62, 165–170. [Google Scholar]
- Mutoh, M.; Watanabe, K.; Kitamura, T.; Shoji, Y.; Takahashi, M.; Kawamori, T.; Sugimura, T.; Wakabayashi, K.; Tani, K.; Kobayashi, M.; et al. Involvement of prostaglandin E receptor subtype EP4 in colon carcinogenesis. Cancer Res. 2002, 62, 28–32. [Google Scholar]
- Oshima, M.; Dinchuk, J.E.; Kargman, S.L.; Oshima, H.; Hancock, B.; Kwong, E.; Trzaskos, J.M.; Evans, J.F.; Taketo, M.M. Suppression of intestinal polyposis in Apc delta716 knockout mice by inhibition of cyclooxygenase 2 (COX-2). Cell 1996, 87, 803–809. [Google Scholar] [CrossRef]
- Sonoshita, M.; Takaku, K.; Sasaki, N.; Sugimoto, Y.; Ushikubi, F.; Narumiya, S.; Oshima, M.; Taketo, M.M. Acceleration of intestinal polyposis through prostaglandin receptor EP2 in ApcΔ716 knockout mice. Nat. Med. 2001, 7, 1048–1051. [Google Scholar] [CrossRef] [PubMed]
- Chulada, P.C.; Thompson, M.B.; Mahler, J.F.; Doyle, C.M.; Gaul, B.W.; Lee, C.; Tiano, H.F.; Morham, S.G.; Smithies, O.; Langenbach, R. Genetic disruption of Ptgs-1, as well as Ptgs-2, reduces intestinal tumorigenesis in Min mice. Cancer Res. 2000, 60, 4705–4708. [Google Scholar] [PubMed]
- Steinbach, G.; Lynch, P.M.; Phillips, R.K.S.; Wallace, M.H.; Hawk, E.; Gordon, G.B.; Wakabayashi, N.; Saunders, B.; Shen, Y.; Fujimura, T.; et al. The effect of celecoxib, a cyclooxygenase-2 inhibitor, in familial adenomatous polyposis. N. Engl. J. Med. 2000, 342, 1946–1952. [Google Scholar] [CrossRef]
- Meyskens, F.L.; McLaren, C.E.; Pelot, D.; Fujikawa-Brooks, S.; Carpenter, P.M.; Hawk, E.; Kelloff, G.; Lawson, M.J.; Kidao, J.; McCracken, J.; et al. Difluoromethylornithine plus sulindac for the prevention of sporadic colorectal adenomas: A randomized placebo-controlled, double-blind trial. Cancer Prev. Res. 2008, 1, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Bertagnolli, M.M.; Eagle, C.J.; Zauber, A.G.; Redston, M.; Breazna, A.; Kim, K.M.; Tang, J.; Rosenstein, R.B.; Umar, A.; Bagheri, D.; et al. Five-year efficacy and safety analysis of the adenoma prevention with celecoxib trial. Cancer Prev. Res. 2009, 2, 310–321. [Google Scholar] [CrossRef]
- Gala, M.K.; Chan, A.T. Molecular Pathways Molecular Pathways: Aspirin and Wnt Signaling-A Molecularly Targeted Approach to Cancer Prevention and Treatment. Clin Cancer Res 2015, 21, 1543–1548. [Google Scholar] [CrossRef]
- Senior, K. COX-2 inhibitors: Cancer prevention or cardiovascular risk? Lancet Oncol. 2005, 6, 68. [Google Scholar] [CrossRef]
- Stasinopoulos, I.; Shah, T.; Penet, M.-F.; Krishnamachary, B.; Bhujwalla, Z.M. COX-2 in cancer: Gordian knot or Achilles heel? Front. Pharmacol. 2013, 4, 34. [Google Scholar] [CrossRef]
- Martín-Sanz, P.; Casado, M.; Boscá, L. Cyclooxygenase 2 in liver dysfunction and carcinogenesis: Facts and perspectives. World J. Gastroenterol. 2017, 23, 3572–3580. [Google Scholar] [CrossRef]
- Krysan, K.; Reckamp, K.; Sharma, S.; Dubinett, S. The Potential and Rationale for COX-2 Inhibitors in Lung Cancer. Anticancer. Agents Med. Chem. 2008, 6, 209–220. [Google Scholar] [CrossRef]
- Jiang, J.; Qiu, J.; Li, Q.; Shi, Z. Prostaglandin E2 Signaling: Alternative Target for Glioblastoma? Trends Cancer 2017, 3, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Omura, N.; Griffith, M.; Vincent, A.; Li, A.; Hong, S.M.; Walter, K.; Borges, M.; Goggins, M. Cyclooxygenase-deficient pancreatic cancer cells use exogenous sources of prostaglandins. Mol. Cancer Res. 2010, 8, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.; Li, Y.; Tran, L.M.; Dry, S.; Calvopina, J.H.; Garcia, A.; Kim, C.; Wang, Y.; Donahue, T.R.; Herschman, H.R.; et al. Cell intrinsic role of COX-2 in pancreatic cancer development. Mol. Cancer Ther. 2012, 11, 2127–2137. [Google Scholar] [CrossRef] [PubMed]
- McClellan, J.L.; Davis, J.M.; Steiner, J.L.; Enos, R.T.; Jung, S.H.; Carson, J.A.; Pena, M.M.; Carnevale, K.A.; Berger, F.G.; Murphy, E.A. Linking tumor-associated macrophages, inflammation, and intestinal tumorigenesis: Role of MCP-1. Am. J. Physiol. Liver Physiol. 2012, 303, G1087–G1095. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, T.; Clarke, M.; Steele, C.W.; Samuel, M.S.; Neumann, J.; Jung, A.; Huels, D.; Olson, M.F.; Das, S.; Nibbs, R.J.B.; et al. Inhibition of CXCR2 profoundly suppresses inflammation-driven and spontaneous tumorigenesis. J. Clin. Invest. 2012, 122, 3127–3144. [Google Scholar] [CrossRef] [PubMed]
- Jayakumar, A.; Bothwell, A.L.M. Stat6 Promotes Intestinal Tumorigenesis in a Mouse Model of Adenomatous Polyposis by Expansion of MDSCs and Inhibition of Cytotoxic CD8 Response. Neoplasia (United States) 2017, 19, 595–605. [Google Scholar] [CrossRef]
- Katoh, H.; Wang, D.; Daikoku, T.; Sun, H.; Dey, S.K.; DuBois, R.N. CXCR2-Expressing Myeloid-Derived Suppressor Cells Are Essential to Promote Colitis-Associated Tumorigenesis. Cancer Cell 2013, 24, 631–644. [Google Scholar] [CrossRef]
- Yan, C.; Yang, Q.; Gong, Z. Tumor-associated neutrophils and macrophages promote gender disparity in hepatocellular carcinoma in zebrafish. Cancer Res. 2017, 77, 1395–1407. [Google Scholar] [CrossRef]
- Yang, Q.; Yan, C.; Gong, Z. Activation of liver stromal cells is associated with male-biased liver tumor initiation in xmrk and Myc transgenic zebrafish. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef]
- Chia, K.; Mazzolini, J.; Mione, M.; Sieger, D. Tumor initiating cells induce cxcr4- mediated infiltration of pro-tumoral macrophages into the brain. Elife 2018, 7, e31918. [Google Scholar] [CrossRef]
- Chia, K.; Keatinge, M.; Mazzolini, J.; Sieger, D. Brain tumours repurpose endogenous neuron to microglia signalling mechanisms to promote their own proliferation. Elife 2019, 8, e46912. [Google Scholar] [CrossRef] [PubMed]
- Van Den Berg, M.C.W.; Maccarthy-Morrogh, L.; Carter, D.; Morris, J.; Bravo, I.R.; Feng, Y.; Correspondence, P.M. Proteolytic and Opportunistic Breaching of the Basement Membrane Zone by Immune Cells during Tumor Initiation. CellReports 2019, 27, 2837–2846. [Google Scholar] [CrossRef] [PubMed]
- Fridlender, Z.G.; Sun, J.; Mishalian, I.; Singhal, S.; Cheng, G.; Kapoor, V.; Horng, W.; Fridlender, G.; Bayuh, R.; Worthen, G.S.; et al. Transcriptomic analysis comparing tumor-associated neutrophils with granulocytic myeloid-derived suppressor cells and normal neutrophils. PLoS ONE 2012, 7, e31524. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, S.; Houseright, R.A.; Korte, B.G.; Huttenlocher, A. DnaJ-PKAc fusion induces liver inflammation in a zebrafish model of Fibrolamellar Introduction. Dis. Model. Mech. 2020. [Google Scholar] [CrossRef]
- Li, Y.; Du, X.F.; Liu, C.S.; Wen, Z.L.; Du, J.L. Reciprocal Regulation between Resting Microglial Dynamics and Neuronal Activity In Vivo. Dev. Cell 2012, 23, 1189–1202. [Google Scholar] [CrossRef]
- Sieger, D.; Moritz, C.; Ziegenhals, T.; Prykhozhij, S.; Peri, F. Long-Range Ca2+ Waves Transmit Brain-Damage Signals to Microglia. Dev. Cell 2012, 22, 1138–1148. [Google Scholar] [CrossRef]
- Eyo, U.B.; Peng, J.; Swiatkowski, P.; Mukherjee, A.; Bispo, A.; Wu, L.J. Neuronal hyperactivity recruits microglial processes via neuronal NMDA receptors and microglial P2Y12 receptors after status epilepticus. J. Neurosci. 2014, 34, 10528–10540. [Google Scholar] [CrossRef]
- Eyo, U.B.; Gu, N.; De, S.; Dong, H.; Richardson, J.R.; Wu, L.J. Modulation of microglial process convergence toward neuronal dendrites by extracellular calcium. J. Neurosci. 2015, 35, 2417–2422. [Google Scholar] [CrossRef]
- Massara, M.; Persico, P.; Bonavita, O.; Poeta, V.M.; Locati, M.; Simonelli, M.; Bonecchi, R. Neutrophils in gliomas. Front. Immunol. 2017, 8, 1349. [Google Scholar] [CrossRef]
- Krelin, Y.; Voronov, E.; Dotan, S.; Elkabets, M.; Reich, E.; Fogel, M.; Huszar, M.; Iwakura, Y.; Segal, S.; Dinarello, C.A.; et al. Interleukin-1β-driven inflammation promotes the development and invasiveness of chemical carcinogen-induced tumors. Cancer Res. 2007, 67, 1062–1071. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Medzhitov, R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007, 317, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Cataisson, C.; Salcedo, R.; Hakim, S.; Moffitt, B.; Andrea, A.; Wright, L.; Yi, M.; Stephens, R.; Dai, R.M.; Lyakh, L.; et al. IL-1R-MyD88 signaling in keratinocyte transformation and carcinogenesis. J. Exp. Med. 2012, 209, 1689–1702. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.; Wasiliew, P.; Kracht, M. Interleukin-1 (IL-1) pathway. Sci. Signal. 2010, 3, cm1–cm1. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef] [PubMed]
- Demetri, G.D.; Ernst, T.J.; Pratt, E.S.; Zenzie, B.W.; Rheinwald, J.G.; Griffin, J.D. Expression of ras oncogenes in cultured human cells alters the transcriptional and posttranscriptional regulation of cytokine genes. J. Clin. Invest. 1990, 86, 1261–1269. [Google Scholar] [CrossRef]
- Castelli, C.; Sensi, M.; Lupetti, R.; Mortarini, R.; Panceri, P.; Anichini, A.; Parmiani, G. Expression of interleukin 1 alpha, interleukin 6, and tumor necrosis factor alpha genes in human melanoma clones is associated with that of mutated N-RAS oncogene. Cancer Res. 1994, 54, 4785–4790. [Google Scholar]
- Liu, J.; Yang, G.; Thompson-Lanza, J.A.; Glassman, A.; Hayes, K.; Patterson, A.; Marquez, R.T.; Auersperg, N.; Yu, Y.; Hahn, W.C.; et al. A genetically defined model for human ovarian cancer. Cancer Res. 2004, 64, 1655–1663. [Google Scholar] [CrossRef]
- Ancrile, B.B.; O’Hayer, K.M.; Counter, C.M. Oncogenic ras-induced expression of cytokines: A new target of anti-cancer therapeutics. Mol. Interv. 2008, 8, 22–27. [Google Scholar] [CrossRef]
- Ancrile, B.; Lim, K.H.; Counter, C.M. Oncogenic Ras-induced secretion of IL6 is required for tumorigenesis. Genes Dev. 2007, 21, 1714–1719. [Google Scholar] [CrossRef]
- Moore, R.J.; Owens, D.M.; Stamp, G.; Arnott, C.; Burke, F.; East, N.; Holdsworth, H.; Turner, L.; Rollins, B.; Pasparakis, M.; et al. Mice deficient in tumor necrosis factor-α are resistant to skin carcinogenesis. Nat. Med. 1999, 5, 828–831. [Google Scholar] [CrossRef]
- Arnott, C.H.; Scott, K.A.; Moore, R.J.; Robinson, S.C.; Thompson, R.G.; Balkwill, F.R. Expression of both TNF-α receptor subtypes is essential for optimal skin tumour development. Oncogene 2004, 23, 1902–1910. [Google Scholar] [CrossRef] [PubMed]
- Knight, B.; Yeoh, G.C.T.; Husk, K.L.; Ly, T.; Abraham, L.J.; Yu, C.; Rhim, J.A.; Fausto, N. Impaired preneoplastic changes and liver tumor formation in tumor necrosis factor receptor type 1 knockout mice. J. Exp. Med. 2000, 192, 1809–1818. [Google Scholar] [CrossRef] [PubMed]
- Sedger, L.M.; McDermott, M.F. TNF and TNF-receptors: From mediators of cell death and inflammation to therapeutic giants—past, present and future. Cytokine Growth Factor Rev. 2014, 25, 453–472. [Google Scholar] [CrossRef] [PubMed]
- Manthey, C.L.; Johnson, D.L.; Illig, C.R.; Tuman, R.W.; Zhou, Z.; Baker, J.F.; Chaikin, M.A.; Donatelli, R.R.; Franks, C.F.; Zeng, L.; et al. JNJ-28312141, a novel orally active colony-stimulating factor-1 receptor/FMS-related receptor tyrosine kinase-3 receptor tyrosine kinase inhibitor with potential utility in solid tumors, bone metastases, and acute myeloid leukemia. Mol. Cancer Ther. 2009, 8, 3151–3161. [Google Scholar] [CrossRef]
- Nowicki, A.; Szenajch, J.; Ostrowska, G.; Wojtowicz, A.; Wojtowicz, K.; Kruszewski, A.A.; Maruszynski, M.; Aukerman, S.L.; Wiktor-Jedrzejczak, W. Impaired tumor growth in colony-stimulating factor 1 (CSF-1)-deficient, macrophage-deficient op/op mouse: Evidence for a role of CSF-1-dependent macrophages in formation of tumor stroma. Int. J. Cancer 1996, 65, 112–119. [Google Scholar] [CrossRef]
- Aharinejad, S.; Abraham, D.; Paulus, P.; Abri, H.; Hofmann, M.; Grossschmidt, K.; Schäfer, R.; Stanley, E.R.; Hofbauer, R. Colony-stimulating Factor-1 Antisense Treatment Suppresses Growth of Human Tumor Xenografts in Mice 1. Cancer Res. 2002, 62, 5317–5324. [Google Scholar]
- Aharinejad, S.; Paulus, P.; Sioud, M.; Hofmann, M.; Zins, K.; Schäfer, R.; Stanley, E.R.; Abraham, D. Colony-stimulating factor-1 blockade by antisense oligonucleotides and small interfering RNAs suppresses growth of human mammary tumor xenografts in mice. Cancer Res. 2004, 64, 5378–5384. [Google Scholar] [CrossRef]
- Cannarile, M.A.; Weisser, M.; Jacob, W.; Jegg, A.M.; Ries, C.H.; Rüttinger, D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother. Cancer 2017, 5, 53. [Google Scholar] [CrossRef]
- Pyonteck, S.M.; Gadea, B.B.; Wang, H.W.; Gocheva, V.; Hunter, K.E.; Tang, L.H.; Joyce, J.A. Deficiency of the macrophage growth factor CSF-1 disrupts pancreatic neuroendocrine tumor development. Oncogene 2012, 31, 1459–1467. [Google Scholar] [CrossRef]
- Ryder, M.; Gild, M.; Hohl, T.M.; Pamer, E.; Knauf, J.; Ghossein, R.; Joyce, J.A.; Fagin, J.A. Genetic and Pharmacological Targeting of CSF-1/CSF-1R Inhibits Tumor-Associated Macrophages and Impairs BRAF-Induced Thyroid Cancer Progression. PLoS ONE 2013, 8, e54302. [Google Scholar] [CrossRef]
- Lin, E.Y.; Nguyen, A.V.; Russell, R.G.; Pollard, J.W. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 2001, 193, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Oguma, K.; Oshima, H.; Aoki, M.; Uchio, R.; Naka, K.; Nakamura, S.; Hirao, A.; Saya, H.; Taketo, M.M.; Oshima, M. Activated macrophages promote Wnt signalling through tumour necrosis factor-α in gastric tumour cells. EMBO J. 2008, 27, 1671–1681. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Bowman, R.L.; Akkari, L.; Quick, M.L.; Schuhmacher, A.J.; Huse, J.T.; Holland, E.C.; Sutton, J.C.; Joyce, J.A. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science 2016, 352, aad3018. [Google Scholar] [CrossRef] [PubMed]
- Boimel, P.J.; Smirnova, T.; Zhou, Z.N.; Wyckoff, J.; Park, H.; Coniglio, S.J.; Qian, B.-Z.; Stanley, E.R.; Cox, D.; Pollard, J.W.; et al. Contribution of CXCL12 secretion to invasion of breast cancer cells. Breast Cancer Res. 2012, 14, R23. [Google Scholar] [CrossRef]
- Arwert, E.N.; Harney, A.S.; Entenberg, D.; Wang, Y.; Sahai, E.; Pollard, J.W.; Condeelis, J.S. A Unidirectional Transition from Migratory to Perivascular Macrophage Is Required for Tumor Cell Intravasation. Cell Rep. 2018, 23, 1239–1248. [Google Scholar] [CrossRef]
- Ping, Y.F.; Yao, X.H.; Jiang, J.Y.; Zhao, L.T.; Yu, S.C.; Jiang, T.; Lin, M.C.M.; Chen, J.H.; Wang, B.; Zhang, R.; et al. The chemokine CXCL12 and its receptor CXCR4 promote glioma stem cell-mediated VEGF production and tumour angiogenesis via PI3K/AKT signalling. J. Pathol. 2011, 224, 344–354. [Google Scholar] [CrossRef]
- Gravina, G.L.; Mancini, A.; Colapietro, A.; Vitale, F.; Vetuschi, A.; Pompili, S.; Rossi, G.; Marampon, F.; Richardson, P.J.; Patient, L.; et al. The novel CXCR4 antagonist, PRX177561, reduces tumor cell proliferation and accelerates cancer stem cell differentiation in glioblastoma preclinical models. Tumor Biol. 2017, 39, 101042831769552. [Google Scholar] [CrossRef]
- Balkwill, F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin. Cancer Biol. 2004, 14, 171–179. [Google Scholar] [CrossRef]
- Walenkamp, A.M.E.; Lapa, C.; Herrmann, K.; Wester, H.J. CXCR4 ligands: The next big hit? J. Nucl. Med. 2017, 58, 77S–82S. [Google Scholar] [CrossRef]
- Yu, X.; Wang, D.; Wang, X.; Sun, S.; Zhang, Y.; Wang, S.; Miao, R.; Xu, X.; Qu, X. CXCL12/CXCR4 promotes inflammation-driven colorectal cancer progression through activation of RhoA signaling by sponging miR-133a-3p. J. Exp. Clin. Cancer Res. 2019, 38, 32. [Google Scholar] [CrossRef]
- Schmid, M.C.; Avraamides, C.J.; Foubert, P.; Shaked, Y.; Kang, S.W.; Kerbel, R.S.; Varner, J.A. Combined blockade of integrin-α4β1 plus cytokines SDF-1α or IL-1β potently inhibits tumor inflammation and growth. Cancer Res. 2011, 71, 6965–6975. [Google Scholar] [CrossRef] [PubMed]
- Tulotta, C.; Stefanescu, C.; Chen, Q.; Torraca, V.; Meijer, A.H.; Snaar-Jagalska, B.E. CXCR4 signaling regulates metastatic onset by controlling neutrophil motility and response to malignant cells. Sci. Rep. 2019, 9, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Jablonska, J.; Leschner, S.; Westphal, K.; Lienenklaus, S.; Weiss, S. Neutrophils responsive to endogenous IFN-β regulate tumor angiogenesis and growth in a mouse tumor model. J. Clin. Invest. 2010, 120, 1151–1164. [Google Scholar] [CrossRef] [PubMed]
- Benedicto, A.; Romayor, I.; Arteta, B. CXCR4 receptor blockage reduces the contribution of tumor and stromal cells to the metastatic growth in the liver. Oncol. Rep. 2018, 39, 2022–2030. [Google Scholar] [CrossRef]
- Shive, H.R.; West, R.R.; Embree, L.J.; Sexton, J.M.; Hickstein, D.D. Expression of KRASG12V in zebrafish gills induces hyperplasia and CXCL8-associated inflammation. Zebrafish 2015, 12, 221–229. [Google Scholar] [CrossRef]
- Sparmann, A.; Bar-Sagi, D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell 2004, 6, 447–458. [Google Scholar] [CrossRef]
- Gong, L.; Cumpian, A.M.; Caetano, M.S.; Ochoa, C.E.; De la Garza, M.M.; Lapid, D.J.; Mirabolfathinejad, S.G.; Dickey, B.F.; Zhou, Q.; Moghaddam, S.J. Promoting effect of neutrophils on lung tumorigenesis is mediated by CXCR2 and neutrophil elastase. Mol. Cancer 2013, 12, 154. [Google Scholar] [CrossRef]
- Lee, L.-F.; Hellendall, R.P.; Wang, Y.; Haskill, J.S.; Mukaida, N.; Matsushima, K.; Ting, J.P.-Y. IL-8 Reduced Tumorigenicity of Human Ovarian Cancer In Vivo Due to Neutrophil Infiltration. J. Immunol. 2000, 164, 2769–2775. [Google Scholar] [CrossRef]
- Ichikawa, M.; Williams, R.; Wang, L.; Vogl, T.; Srikrishna, G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol. Cancer Res. 2011, 9, 133–148. [Google Scholar] [CrossRef]
- Huh, S.J.; Liang, S.; Sharma, A.; Dong, C.; Robertson, G.P. Transiently entrapped circulating tumor cells interact with neutrophils to facilitate lung metastasis development. Cancer Res. 2010, 70, 6071–6082. [Google Scholar] [CrossRef]
- Kowanetz, M.; Wu, X.; Lee, J.; Tan, M.; Hagenbeek, T.; Qu, X.; Yu, L.; Ross, J.; Korsisaari, N.; Cao, T.; et al. Granulocyte-colony stimulating factor promotes lung metastasis through mobilization of Ly6G+Ly6C+ granulocytes. Proc. Natl. Acad. Sci. USA 2010, 107, 21248–21255. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; McCampbell, K.K.; David, J.M.; Palena, C. Neutralization of IL-8 decreases tumor PMN-MDSCs and reduces mesenchymalization of claudin-low triple-negative breast cancer. JCI Insight 2017, 2, e94296. [Google Scholar] [CrossRef] [PubMed]
- Tazzyman, S.; Barry, S.T.; Ashton, S.; Wood, P.; Blakey, D.; Lewis, C.E.; Murdoch, C. Inhibition of neutrophil infiltration into A549 lung tumors in vitro and in vivo using a CXCR2-specific antagonist is associated with reduced tumor growth. Int. J. Cancer 2011, 129, 847–858. [Google Scholar] [CrossRef] [PubMed]
- Shang, K.; Bai, Y.-P.; Wang, C.; Wang, Z.; Gu, H.-Y.; Du, X.; Zhou, X.-Y.; Zheng, C.-L.; Chi, Y.-Y.; Mukaida, N.; et al. Crucial Involvement of Tumor-Associated Neutrophils in the Regulation of Chronic Colitis-Associated Carcinogenesis in Mice. PLoS ONE 2012, 7, e51848. [Google Scholar] [CrossRef] [PubMed]
- Asfaha, S.; Dubeykovskiy, A.N.; Tomita, H.; Yang, X.; Stokes, S.; Shibata, W.; Friedman, R.A.; Ariyama, H.; Dubeykovskaya, Z.A.; Muthupalani, S.; et al. Mice that express human interleukin-8 have increased mobilization of immature myeloid cells, which exacerbates inflammation and accelerates colon carcinogenesis. Gastroenterology 2013, 144, 155–166. [Google Scholar] [CrossRef]
- Mariani, F.; Sena, P.; Roncucci, L. Inflammatory pathways in the early steps of colorectal cancer development. World J. Gastroenterol. 2014, 20, 9716–9731. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Wang, Z.; Fu, L.; Xu, T. Macrophage polarization in the development and progression of ovarian cancers: An overview. Front. Oncol. 2019, 9, 421. [Google Scholar] [CrossRef]
- Mishalian, I.; Bayuh, R.; Levy, L.; Zolotarov, L.; Michaeli, J.; Fridlender, Z.G. Tumor-associated neutrophils (TAN) develop pro-tumorigenic properties during tumor progression. Cancer Immunol. Immunother. 2013, 62, 1745–1756. [Google Scholar] [CrossRef]
- Hagemann, T.; Wilson, J.; Burke, F.; Kulbe, H.; Li, N.F.; Plüddemann, A.; Charles, K.; Gordon, S.; Balkwill, F.R. Ovarian Cancer Cells Polarize Macrophages Toward A Tumor-Associated Phenotype. J. Immunol. 2006, 176, 5023–5032. [Google Scholar] [CrossRef]
- Nguyen-Chi, M.; Laplace-Builhe, B.; Travnickova, J.; Luz-Crawford, P.; Tejedor, G.; Phan, Q.T.; Duroux-Richard, I.; Levraud, J.P.; Kissa, K.; Lutfalla, G.; et al. Identification of polarized macrophage subsets in zebrafish. Elife 2015, 4, e07288. [Google Scholar] [CrossRef]
- Krzyszczyk, P.; Schloss, R.; Palmer, A.; Berthiaume, F. The role of macrophages in acute and chronic wound healing and interventions to promote pro-wound healing phenotypes. Front. Physiol. 2018, 9, 419. [Google Scholar] [CrossRef] [PubMed]
- Wilgus, T.A.; Roy, S.; McDaniel, J.C. Neutrophils and Wound Repair: Positive Actions and Negative Reactions. Adv. Wound Care 2013, 2, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Hull, M.A.; Booth, J.K.; Tisbury, A.; Scott, N.; Bonifer, C.; Markham, A.F.; Coletta, P.L. Cyclooxygenase 2 is up-regulated and localized to macrophages in the intestine of Min mice. Br. J. Cancer 1999, 79, 1399–1405. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bamba, H.; Ota, S.; Kato, A.; Adachi, A.; Itoyama, S.; Matsuzaki, F. High expression of cyclooxygenase-2 in macrophages of human colonic adenoma. Int. J. Cancer 1999, 83, 470–475. [Google Scholar] [CrossRef]
- Tanaka, S.; Tatsuguchi, A.; Futagami, S.; Gudis, K.; Wada, K.; Seo, T.; Mitsui, K.; Yonezawa, M.; Nagata, K.; Fujimori, S.; et al. Monocyte chemoattractant protein 1 and macrophage cyclooxygenase 2 expression in colonic adenoma. Gut 2006, 55, 54–61. [Google Scholar] [CrossRef]
- Hull, M.A.; Cuthbert, R.J.; Ko, C.W.S.; Scott, D.J.; Cartwright, E.J.; Hawcroft, G.; Perry, S.L.; Ingram, N.; Carr, I.M.; Markham, A.F.; et al. Paracrine cyclooxygenase-2 activity by macrophages drives colorectal adenoma progression in the Apc Min/+ mouse model of intestinal tumorigenesis. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Smakman, N.; Kranenburg, O.; Vogten, J.M.; Bloemendaal, A.L.A.; van Diest, P.; Borel Rinkes, I.H. Cyclooxygenase-2 Is a Target of KRAS D12, Which Facilitates the Outgrowth of Murine C26 Colorectal Liver Metastases. Clin. Cancer Res. 2005, 11, 41–48. [Google Scholar]
- Liu, W.; Reinmuth, N.; Stoeltzing, O.; Parikh, A.A.; Tellez, C.; Williams, S.; Jung, Y.D.; Fan, F.; Takeda, A.; Akagi, M.; et al. Cyclooxygenase-2 is up-regulated by interleukin-1 beta in human colorectal cancer cells via multiple signaling pathways. Cancer Res. 2003, 63, 3632–3636. [Google Scholar]
- Attiq, A.; Jalil, J.; Husain, K.; Ahmad, W. Raging the war against inflammation with natural products. Front. Pharmacol. 2018, 9, 976. [Google Scholar] [CrossRef]
- Lee, K.W.; Kim, M.S.; Kang, N.J.; Kim, D.H.; Surh, Y.J.; Lee, H.J.; Moon, A. H-Ras selectively up-regulates MMP-9 and COX-2 through activation of EKK1/2 and NF-κB: An implication for invasive phenotype in rat liver epithelial cells. Int. J. Cancer 2006, 119, 1767–1775. [Google Scholar] [CrossRef]
- Medeiros, A.; Peres-Buzalaf, C.; Fortino Verdan, F.; Serezani, C.H. Prostaglandin E2 and the suppression of phagocyte innate immune responses in different organs. Mediators Inflamm. 2012, 2012, 327568. [Google Scholar] [CrossRef] [PubMed]
- Kalinski, P. Regulation of Immune Responses by Prostaglandin E 2. J. Immunol. 2012, 188, 21–28. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, K.F.; Clark, K.; Naqvi, S.; McGuire, V.A.; Nöehren, G.; Kristariyanto, Y.; van den Bosch, M.; Mudaliar, M.; McCarthy, P.C.; Pattison, M.J.; et al. PGE 2 Induces Macrophage IL-10 Production and a Regulatory-like Phenotype via a Protein Kinase A–SIK–CRTC3 Pathway. J. Immunol. 2013, 190, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Luan, B.; Yoon, Y.S.; Le Lay, J.; Kaestner, K.H.; Hedrick, S.; Montminy, M. CREB pathway links PGE2 signaling with macrophage polarization. Proc. Natl. Acad. Sci. USA 2015, 112, 15642–15647. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Scambler, T.E.; Smallie, T.; Cunliffe, H.E.; Ross, E.A.; Rosner, D.R.; O’Neil, J.D.; Clark, A.R. Macrophage responses to lipopolysaccharide are modulated by a feedback loop involving prostaglandin E2, dual specificity phosphatase 1 and tristetraprolin. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Agard, M.; Asakrah, S.; Morici, L.A. PGE2 suppression of innate immunity during mucosal bacterial infection. Front. Cell. Infect. Microbiol. 2013, 4, 45. [Google Scholar] [CrossRef]
- Loynes, C.A.; Lee, J.A.; Robertson, A.L.; Steel, M.J.G.; Ellett, F.; Feng, Y.; Levy, B.D.; Whyte, M.K.B.; Renshaw, S.A. PGE2 production at sites of tissue injury promotes an anti-inflammatory neutrophil phenotype and determines the outcome of inflammation resolution in vivo. Sci. Adv. 2018, 4, eaar8320. [Google Scholar] [CrossRef]
- Van Dalen, F.J.; Van Stevendaal, M.H.M.E.; Fennemann, F.L.; Verdoes, M.; Ilina, O. Molecular repolarisation of tumour-associated macrophages. Molecules 2019, 24, 9. [Google Scholar] [CrossRef]
- Nakanishi, Y.; Nakatsuji, M.; Seno, H.; Ishizu, S.; Akitake-Kawano, R.; Kanda, K.; Ueo, T.; Komekado, H.; Kawada, M.; Minami, M.; et al. COX-2 inhibition alters the phenotype of tumor-associated macrophages from M2 to M1 in Apc Min/1 mouse polyps. Carcinogenesis 2011, 32, 1333–1339. [Google Scholar] [CrossRef]
- Na, Y.R.; Yoon, Y.N.; Son, D.I.; Seok, S.H. Cyclooxygenase-2 Inhibition Blocks M2 Macrophage Differentiation and Suppresses Metastasis in Murine Breast Cancer Model. PLoS ONE 2013, 8, e63451. [Google Scholar] [CrossRef]
- Sinha, P.; Clements, V.K.; Fulton, A.M.; Ostrand-Rosenberg, S. Prostaglandin E2 promotes tumor progression by inducing myeloid-derived suppressor cells. Cancer Res. 2007, 67, 4507–4513. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Ubreva, J.; Garcia-Gomez, A.; Ballestar, E. Epigenetic mechanisms of myeloid differentiation in the tumor microenvironment. Curr. Opin. Pharmacol. 2017, 35, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Obermajer, N.; Muthuswamy, R.; Odunsi, K.; Edwards, R.P.; Kalinski, P. PGE 2-induced CXCL 12 production and CXCR4 expression controls the accumulation of human MDSCs in ovarian cancer environment. Cancer Res. 2011, 71, 7463–7470. [Google Scholar] [CrossRef] [PubMed]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of Tumor-Associated Neutrophil Phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef]
- Li, Z.; Pang, Y.; Gara, S.K.; Achyut, B.R.; Heger, C.; Goldsmith, P.K.; Lonning, S.; Yang, L. Gr-1+CD11b+ cells are responsible for tumor promoting effect of TGF-β in breast cancer progression. Int. J. Cancer 2012, 131, 2584–2595. [Google Scholar] [CrossRef]
- Pang, Y.; Gara, S.K.; Achyut, B.R.; Li, Z.; Yan, H.H.; Day, C.P.; Weiss, J.M.; Trinchieri, G.; Morris, J.C.; Yang, L. TGF-β Signaling in myeloid cells is required for tumor metastasis. Cancer Discov. 2013, 3, 936–951. [Google Scholar] [CrossRef]
- Haider, C.; Hnat, J.; Wagner, R.; Huber, H.; Timelthaler, G.; Grubinger, M.; Coulouarn, C.; Schreiner, W.; Schlangen, K.; Sieghart, W.; et al. Transforming Growth Factor-β and Axl Induce CXCL5 and Neutrophil Recruitment in Hepatocellular Carcinoma. Hepatology 2019, 69, 222–236. [Google Scholar] [CrossRef]
- Fang, K.M.; Wang, Y.L.; Huang, M.C.; Sun, S.H.; Cheng, H.; Tzeng, S.F. Expression of macrophage inflammatory protein-1α and monocyte chemoattractant protein-1 in glioma-infiltrating microglia: Involvement of ATP and P2X7 receptor. J. Neurosci. Res. 2011, 89, 199–211. [Google Scholar] [CrossRef]
- Haage, V.; Semtner, M.; Vidal, R.O.; Hernandez, D.P.; Pong, W.W.; Chen, Z.; Hambardzumyan, D.; Magrini, V.; Ly, A.; Walker, J.; et al. Comprehensive gene expression meta-analysis identifies signature genes that distinguish microglia from peripheral monocytes/macrophages in health and glioma. Acta Neuropathol. Commun. 2019, 7, 20. [Google Scholar] [CrossRef]
- Desai, B.N.; Leitinger, N. Purinergic and calcium signaling in macrophage function and plasticity. Front. Immunol. 2014, 5, 580. [Google Scholar] [CrossRef]
- Di Virgilio, F.; Sarti, A.C.; Falzoni, S.; De Marchi, E.; Adinolfi, E. Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat. Rev. Cancer 2018, 18, 601–618. [Google Scholar] [CrossRef] [PubMed]
- De Marchi, E.; Orioli, E.; Pegoraro, A.; Sangaletti, S.; Portararo, P.; Curti, A.; Colombo, M.P.; Di Virgilio, F.; Adinolfi, E. The P2X7 receptor modulates immune cells infiltration, ectonucleotidases expression and extracellular ATP levels in the tumor microenvironment. Oncogene 2019, 38, 3636–3650. [Google Scholar] [CrossRef] [PubMed]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Kornete, M.; Joyce, J.A. Re-education of macrophages as a therapeutic strategy in cancer. Immunotherapy 2019, 11, 677–689. [Google Scholar] [CrossRef]
- Ablain, J.; Durand, E.M.; Yang, S.; Zhou, Y.; Zon, L.I. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell 2015, 32, 756–764. [Google Scholar] [CrossRef]
- Cornet, C.; Di Donato, V.; Terriente, J. Combining Zebrafish and CRISPR/Cas9: Toward a more efficient drug discovery pipeline. Front. Pharmacol. 2018, 9, 703. [Google Scholar] [CrossRef]
- Khalil, D.N.; Smith, E.L.; Brentjens, R.J.; Wolchok, J.D. The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 2016, 13, 273–290. [Google Scholar] [CrossRef]
- Tan, S.; Li, D.; Zhu, X. Cancer immunotherapy: Pros, cons and beyond. Biomed. Pharmacother. 2020, 124, 109821. [Google Scholar] [CrossRef]
- Bajoghli, B.; Aghaallaei, N.; Hess, I.; Rode, I.; Netuschil, N.; Tay, B.H.; Venkatesh, B.; Yu, J.K.; Kaltenbach, S.L.; Holland, N.D.; et al. Evolution of Genetic Networks Underlying the Emergence of Thymopoiesis in Vertebrates. Cell 2009, 138, 186–197. [Google Scholar] [CrossRef]
- Aghaallaei, N.; Bajoghli, B. Making thymus visible: Understanding T-cell development from a new perspective. Front. Immunol. 2018, 9, 375. [Google Scholar] [CrossRef]
- Lam, S.H.; Chua, H.L.; Gong, Z.; Lam, T.J.; Sin, Y.M. Development and maturation of the immune system in zebrafish, Danio rerio: A gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 2004, 28, 9–28. [Google Scholar] [CrossRef]
- Langenau, D.M.; Ferrando, A.A.; Traver, D.; Kutok, J.L.; Hezel, J.P.D.; Kanki, J.P.; Zon, L.I.; Thomas Look, A.; Trede, N.S. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc. Natl. Acad. Sci. USA 2004, 101, 7369–7374. [Google Scholar] [CrossRef]
- Meeker, N.D.; Trede, N.S. Immunology and zebrafish: Spawning new models of human disease. Dev. Comp. Immunol. 2008, 32, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Meeker, N.D.; Smith, A.C.H.; Frazer, J.K.; Bradley, D.F.; Rudner, L.A.; Love, C.; Trede, N.S. Characterization of the zebrafish T cell receptor β locus. Immunogenetics 2010, 62, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Dee, C.T.; Nagaraju, R.T.; Athanasiadis, E.I.; Gray, C.; Fernandez del Ama, L.; Johnston, S.A.; Secombes, C.J.; Cvejic, A.; Hurlstone, A.F.L. CD4-Transgenic Zebrafish Reveal Tissue-Resident Th2- and Regulatory T Cell–like Populations and Diverse Mononuclear Phagocytes. J. Immunol. 2016, 197, 3520–3530. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Mitra, S.; Wyse, C.; Alnabulsi, A.; Zou, J.; Weerdenburg, E.M.; van der Sar, A.; Wang, D.; Secombes, C.J.; Bird, S. First Demonstration of Antigen Induced Cytokine Expression by CD4-1+ Lymphocytes in a Poikilotherm: Studies in Zebrafish (Danio rerio). PLoS ONE 2015, 10, e0126378. [Google Scholar] [CrossRef]
- Hui, S.P.; Sheng, D.Z.; Sugimoto, K.; Gonzalez-Rajal, A.; Nakagawa, S.; Hesselson, D.; Kikuchi, K. Zebrafish Regulatory T Cells Mediate Organ-Specific Regenerative Programs. Dev. Cell 2017, 43, 659–672.e5. [Google Scholar] [CrossRef]
- Kasheta, M.; Painter, C.A.; Moore, F.E.; Lobbardi, R.; Bryll, A.; Freiman, E.; Stachura, D.; Rogers, A.B.; Houvras, Y.; Langenau, D.M.; et al. Identification and characterization of T reg–like cells in zebrafish. J. Exp. Med. 2017, 214, 3519–3530. [Google Scholar] [CrossRef]
- Wan, F.; Hu, C.B.; Ma, J.X.; Gao, K.; Xiang, L.X.; Shao, J.Z. Characterization of γδ T cells from zebrafish provides insights into their important role in adaptive humoral immunity. Front. Immunol. 2017, 7, 675. [Google Scholar] [CrossRef]
- de Sousa Andrade, L.N.; Otake, A.H.; Cardim, S.G.B.; da Silva, F.I.; Sakamoto, M.M.I.; Furuya, T.K.; Uno, M.; Pasini, F.S.; Chammas, R. Extracellular Vesicles Shedding Promotes Melanoma Growth in Response to Chemotherapy. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Shinohara, H.; Kuranaga, Y.; Kumazaki, M.; Sugito, N.; Yoshikawa, Y.; Takai, T.; Taniguchi, K.; Ito, Y.; Akao, Y. Regulated Polarization of Tumor-Associated Macrophages by miR-145 via Colorectal Cancer–Derived Extracellular Vesicles. J. Immunol. 2017, 199, 1505–1515. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Lam, E.W.F.; Sun, Y. Extracellular vesicles in the tumor microenvironment: Old stories, but new tales. Mol. Cancer 2019, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hyenne, V.; Ghoroghi, S.; Collot, M.; Bons, J.; Follain, G.; Harlepp, S.; Mary, B.; Bauer, J.; Mercier, L.; Busnelli, I.; et al. Studying the Fate of Tumor Extracellular Vesicles at High Spatiotemporal Resolution Using the Zebrafish Embryo. Dev. Cell 2019, 48, 554–572. [Google Scholar] [CrossRef] [PubMed]
- Collot, M.; Ashokkumar, P.; Anton, H.; Boutant, E.; Faklaris, O.; Galli, T.; Mély, Y.; Danglot, L.; Klymchenko, A.S. MemBright: A Family of Fluorescent Membrane Probes for Advanced Cellular Imaging and Neuroscience. Cell Chem. Biol. 2019, 26, 600–614. [Google Scholar] [CrossRef]
- Frederik Verweij, A.J.; Revenu, C.; Arras, G.; Del Bene, F.; Raposo, G.; van Niel Correspondence, G. Live Tracking of Inter-organ Communication by Endogenous Exosomes In Vivo. Dev. Cell 2019, 48, 573–589. [Google Scholar] [CrossRef]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Yang, Q.; Yan, C.; Gong, Z. Interaction of hepatic stellate cells with neutrophils and macrophages in the liver following oncogenic kras activation in transgenic zebrafish. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Precazzini, F.; Pancher, M.; Gatto, P.; Tushe, A.; Adami, V.; Anelli, V.; Mione, M.C. Automated in vivo screen in zebrafish identifies Clotrimazole as targeting a metabolic vulnerability in a melanoma model. Dev. Biol. 2020, 457, 215–225. [Google Scholar] [CrossRef]
| Organ | Cell Type | Promoter | Oncogene | Marker | Regulation | Ref. |
|---|---|---|---|---|---|---|
| Blood | Lymphoblasts | Xef1a | (Hs) ETV6-Runx1 | eGFP | Promoter-driven | [57] |
| Actb2 | (Hs) ETV6-Runx1 | eGFP | Promoter-driven | [57] | ||
| T-Lymphoblasts | Rag2 | (Mm) c-Myc | eGFP | Promoter Driven | [17] | |
| Skin | Melanocytes | Mitfa | (Hs) BRAFV600E, p53−/− | None * | Promoter-driven | [21] |
| (Hs) BRAFV600E, p53−/− | eGFP | Promoter-driven | [58,59] | |||
| (Hs) HRASG12V | GFP | Promoter-driven | [23] | |||
| (Hs) HRASG12V | mCherry | Promoter-driven | [23] | |||
| (Hs) NRASQ61K | mCherry | Inducible LexPR | [60] | |||
| Melanocytes and Goblet Cells | KITa | (Hs) HRASG12V | eGFP | GALTA4/UAS | [24] | |
| (Hs) NRASQ61K | mCherry | Inducible LexPR | [60] | |||
| Liver | Hepatocytes | Fabp10 | (Xl) pt-β-cat | None * | Promoter-driven | [61] |
| (Dr) KRASG12V | eGFP | Promoter-driven | [62] | |||
| (Dr) KRASG12V | eGFP | Inducible LexPR | [63] | |||
| (Dr) KRASG12V | eGFP | Inducible Tet-On | [64] | |||
| (X) Xmrk | None * | Inducible Tet-On | [65,66] | |||
| (Mm) c-Myc | None * | Inducible Tet-On | [66,67] | |||
| Intestine | - | Fabp2 | (Dr) KRASG12V | eGFP | Inducible LexPR | [25] |
| Brain | Glial cells | Gfap | (Hs) KRASG12V | mCherry | GAL4/UAS | [27] |
| (Hs) KRASG12V | GFP | Promoter-driven | [68] | |||
| Brain & PNS | Glial cells | Krt5 | (Hs) KRASG12V | mCherry | GAL4/UAS | [27] |
| Adrenal Gland | Neuroblasts | Dβh | (Hs) n-Myc | eGFP | Promoter-driven | [30] |
| Pancreas | Progenitor cells | Ptf1a | (Hs) KRASG12V | eGFP | Promoter-driven | [26] |
| (Hs) KRASG12D | eGFP | GAL4/UAS | [29] | |||
| Pituitary Gland | Corticotrophs | Pomc | (Dr) PTTG | None * | Promoter-driven | [69] |
| Muscle | Progenitor cells | Rag2 | (Hs) KRASG12D | None * | Promoter-driven | [70,71,72] |
| Cdh15 | (Hs) KRASG12D | None * | Promoter-driven | [72] | ||
| Mylz2 | (Hs) KRASG12D | None * | Promoter-driven | [72] |
| Promoter | Marker | Notes | Ref. | |
|---|---|---|---|---|
| Neutrophil | Mpx/Mpo | GFP | - | [73] |
| eGFP | - | [55] | ||
| mCherry | - | [74] | ||
| eGFP-L10a | Ribosomes and polysomes | [75] | ||
| BirA-Citrine | Biotin-tagging | [76] | ||
| LysC/Lyz | dsRed | - | [77] | |
| eGFP | - | [56,77] | ||
| BFP | - | [78] | ||
| Macrophage | Mpeg1.1 | eGFP | - | [53] |
| mCherry | - | [53] | ||
| mCherry-F | Membrane Bound | [79] | ||
| Dendra | Photoconvertible | [80] | ||
| CFP-DEVD-YFP | FRET, caspase cleavable | [81] | ||
| BirA-Citrine | Biotin-tagging | [60] | ||
| Mfap4 | tdTomato-CAAX | Membrane Bound | [54] | |
| Turquoise2 | - | [54] | ||
| dLanYFP-CAAX | Membrane Bound | [54] |
| Zebrafish (Pre-neoplastic Stage) | Mouse (Early Neoplastic Stage) | |
|---|---|---|
| H2O2 | Released by PNCs and neighbouring cells in the skin [6]. | Unknown |
| Promotes PNC proliferation and local leukocyte recruitment [6]. | ||
| IL-1β | Expressed in response to tumour initiation in the skin, liver and brain [6,41,68,140,146]. | Promotes neoplasm formation and leukocyte recruitment in the skin and colon [152,153,154]. |
| Promotes PNC proliferation and leukocyte recruitment in the liver [146]. | Upregulates pro-inflammatory cytokines and growth factors [155]. | |
| TNF-α | Expressed in response to tumour initiation in the skin and liver [6,78,140,146]. | Promotes neoplasm formation and leukocyte recruitment in the skin and liver [162,163,164]. |
| Promotes PNC proliferation and leukocyte recruitment in the liver [146]. | ||
| CSF-1 | Expressed in response to tumour initiation in the liver [140]. | Promotes neoplasm formation and macrophage recruitment in thyroid and pancreas [171,172]. |
| Promotes PNC proliferation and leukocyte recruitment in the brain [142]. | Only promotes later stages of breast and colon cancers [173,174]. | |
| Cxcl12-Cxcr4 | Cxcl12 is expressed in response to tumour initiation in the brain [142]. | Promotes neoplasm formation and macrophage recruitment in colon and lung [182,183]. |
| Promotes PNC proliferation by macrophage recruitment [142]. | ||
| IL-8- Cxcl1/Cxcl2 | IL-8 is expressed in response to tumour initiation in the skin, liver and brain [6,41,68,104,140]. | Promotes neoplasm formation and the recruitment of neutrophils and G-MDSCs in the colon [137,196,197]. |
| Cxcr1/2 signalling promotes PNC proliferation and neutrophil recruitment in the brain and skin respectively [68,104]. | ||
| PGE2 | Produced by leukocytes in response to tumour initiation in the skin [7]. | Secreted by neoplastic cells and macrophages in the colon [205,206,207,208]. |
| Directly promotes PNC proliferation [7]. | Directly promotes proliferation [128]. | |
| TGF-β | Expressed in response to tumour initiation in the liver [41,104,140,141]. | Unknown |
| Governs neutrophil phenotype and promotes PNC proliferation [41]. | ||
| ATP | Released by PNCs in response to tumour initiation in the brain [143]. | Unknown |
| Promotes PNC proliferation and microglia contact via purinergic signalling [143]. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elliot, A.; Myllymäki, H.; Feng, Y. Inflammatory Responses during Tumour Initiation: From Zebrafish Transgenic Models of Cancer to Evidence from Mouse and Man. Cells 2020, 9, 1018. https://doi.org/10.3390/cells9041018
Elliot A, Myllymäki H, Feng Y. Inflammatory Responses during Tumour Initiation: From Zebrafish Transgenic Models of Cancer to Evidence from Mouse and Man. Cells. 2020; 9(4):1018. https://doi.org/10.3390/cells9041018
Chicago/Turabian StyleElliot, Abigail, Henna Myllymäki, and Yi Feng. 2020. "Inflammatory Responses during Tumour Initiation: From Zebrafish Transgenic Models of Cancer to Evidence from Mouse and Man" Cells 9, no. 4: 1018. https://doi.org/10.3390/cells9041018
APA StyleElliot, A., Myllymäki, H., & Feng, Y. (2020). Inflammatory Responses during Tumour Initiation: From Zebrafish Transgenic Models of Cancer to Evidence from Mouse and Man. Cells, 9(4), 1018. https://doi.org/10.3390/cells9041018





